Exo70B2, a subunit of the exocyst complex, which is involved in exocytosis, is targeted for degradation by the ubiquitin ligase PUB22 upon activation of the immune system. This work also reveals that Exo70B2 is required for the activation of PAMP-triggered responses.
Abstract
Plant pathogens are perceived by pattern recognition receptors, which are activated upon binding to pathogen-associated molecular patterns (PAMPs). Ubiquitination and vesicle trafficking have been linked to the regulation of immune signaling. However, little information exists about components of vesicle trafficking involved in immune signaling and the mechanisms that regulate them. In this study, we identified Arabidopsis thaliana Exo70B2, a subunit of the exocyst complex that mediates vesicle tethering during exocytosis, as a target of the plant U-box–type ubiquitin ligase 22 (PUB22), which acts in concert with PUB23 and PUB24 as a negative regulator of PAMP-triggered responses. We show that Exo70B2 is required for both immediate and later responses triggered by all tested PAMPs, suggestive of a role in signaling. Exo70B2 is also necessary for the immune response against different pathogens. Our data demonstrate that PUB22 mediates the ubiquitination and degradation of Exo70B2 via the 26S Proteasome. Furthermore, degradation is regulated by the autocatalytic turnover of PUB22, which is stabilized upon PAMP perception. We therefore propose a mechanism by which PUB22-mediated degradation of Exo70B2 contributes to the attenuation of PAMP-induced signaling.
INTRODUCTION
Immunity in plants is initially activated by pathogen-associated molecular patterns (PAMPs), which are perceived by pattern recognition receptors (PRRs). The best described PRR in plants is the flagellin receptor the flagellin sensing2, FLS2 (Zipfel et al., 2004). Flagellin binding triggers the rapid heterodimerization of the FLS2 receptor with the regulatory protein brassinosteroid insensitive1-associated kinase1 (BAK1/SERK3) and BAK1-like1 (BKK1/SERK4), which is required for downstream signaling (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011). Early responses (seconds to minutes) triggered by PAMPs include the oxidative burst, ion fluxes across the plasma membrane, and the activation of mitogen-activated protein kinases (MAPKs). These are followed by late responses (hours to days), such as gene induction and callose deposition (Nicaise et al., 2009). Ubiquitination has been shown to be involved in many aspects of the regulation of immune responses, including PAMP-triggered immunity (PTI); Trujillo and Shirasu, 2010. One example is the related plant U-box–type E3 ubiquitin ligases PUB22, PUB23, and PUB24. These contribute additively to the downregulation of PAMP responses, as reflected by the increased and prolonged signaling in loss-of-function mutant lines (Trujillo et al., 2008). In addition, an enhanced oxidative burst, after elicitation with various PAMPs, suggests that the PUB triplet targets components of a process that is required for signaling mediated by diverse PRRs (Trujillo et al., 2008). Further examples of PUBs acting as negative regulators of PTI were subsequently reported, suggesting that PUBs regulate PRR function at multiple levels (Lu et al., 2011).
Amplitude and duration of signaling events triggered by PAMP perception must be tightly regulated to ensure an appropriate response. Internalization of receptors by endocytosis, followed by their degradation, has emerged as a general mechanism for attenuating signaling provoked by extracellular stimuli (MacGurn et al., 2012). Recent studies suggested that FLS2 is subject to this type of regulation during a refractory phase that follows initial activation by the PAMP epitope flg22 (Robatzek et al., 2006). In metazoans, ubiquitination is known to play key roles in the various steps of the vesicle trafficking of receptors (Clague and Urbé, 2010; MacGurn et al., 2012). The first indication that ubiquitination is also involved in the regulation of vesicle trafficking in plants came from the bacterial Type III effector AvrPtoB, which can ubiquitinate the PRRs FLS2 and CERK1, resulting in their degradation (Göhre et al., 2008; Gimenez-Ibanez et al., 2009). More support was provided by a report that Arabidopsis thaliana PUB12 and PUB13 are the endogenous E3 ligases that ubiquitinate FLS2 and thereby mediate signal attenuation (Lu et al., 2011).
After vesicle formation (or budding), vesicle trafficking entails transport, tethering, and fusion to the target membrane (exocytosis). Exocytosis has received most attention in plant immunity for its key role in the secretion of antimicrobial metabolites and polypeptides during late immune responses (>4 h; Bednarek et al., 2010). One prominent example is provided by studies describing the PEN1/SYP121 soluble N-ethylmaleimide–sensitive factor attachment protein receptor protein (SNARE), which is required for penetration resistance against powdery mildew fungi (Collins et al., 2003). PEN1 is believed to function by forming a ternary SNARE complex with the synaptosome-associated membrane protein 33 and vesicle-associated membrane protein 721 (VAMP721) and VAMP722 (Kwon et al., 2008).
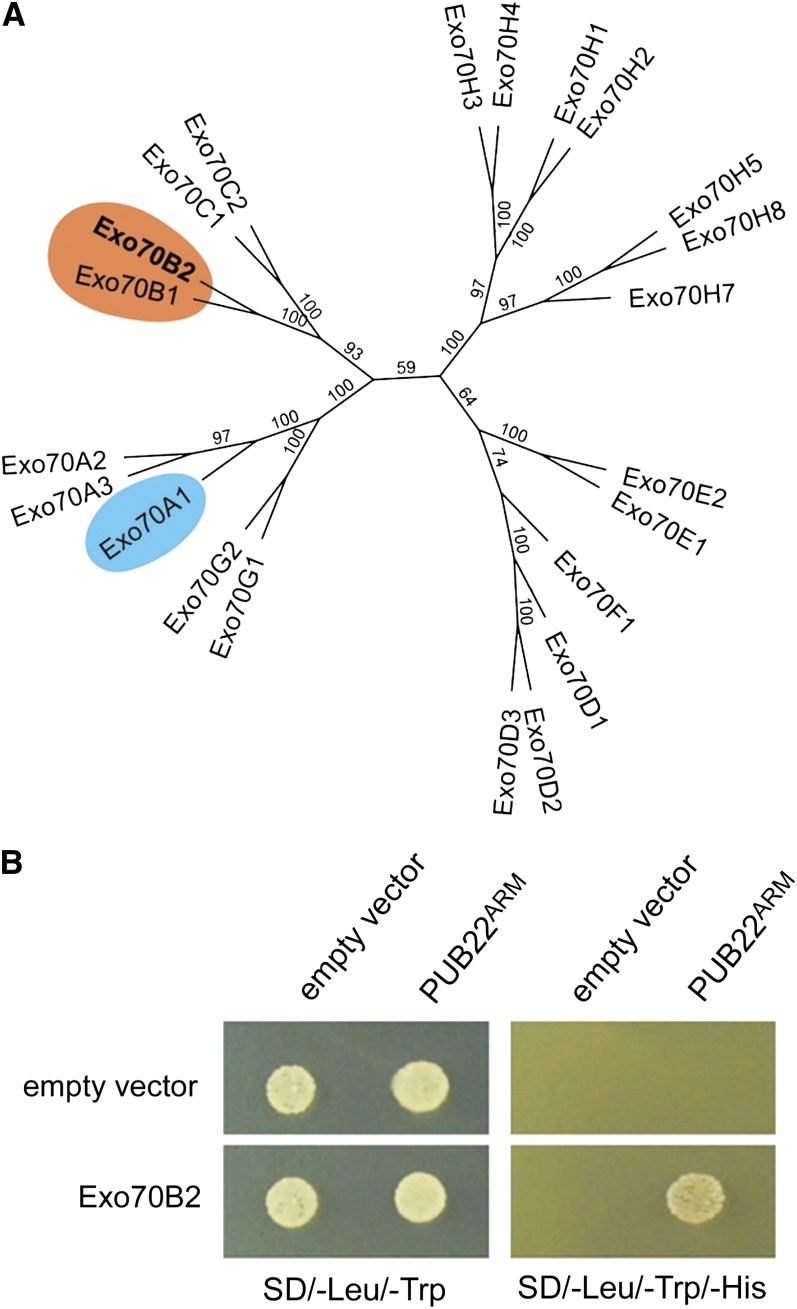
The exocyst is an octameric complex that mediates early steps of exocytosis before SNARE-mediated membrane fusion (He and Guo, 2009). It was initially defined in yeast and later linked to several mammalian cellular processes, including innate immunity (TerBush et al., 1996). In plants, exocyst function has mainly been related to polar secretion required for growth and pollen incompatibility (Samuel et al., 2009; Zárský et al., 2009). Exocyst subunits have been proposed to exist as subcomplexes that are brought together to mediate vesicle tethering at specific sites of exocytosis. Exo70 and Sec3 are thought to associate with the acceptor membrane, where they function as a spatial landmark to guide tethering (He and Guo, 2009). Most metazoans possess only one Exo70 gene. By contrast, the Exo70 gene family underwent a substantial expansion unique to plants (Elias et al., 2003; Synek et al., 2006; Chong et al., 2010). In Arabidopsis, 23 homologs can be found (Figure 1A), and rice (Oryza sativa) contains at least 32 (Cvrčková et al., 2012). This expansion suggests that plants have evolved functionally specialized Exo70 genes (Zárský et al., 2009). Accordingly, a recent report demonstrated that Arabidopsis Exo70B2 and Exo70H1 genes are induced in response to pathogens and PAMPS and are genetically necessary for a functional immune response (Pecenková et al., 2011).
Figure 1.
Identification of the Exocyst Subunit Exo70B2.
(A) Phylogenetic relations between Arabidopsis Exo70 proteins. The tree was generated using the neighbor-joining method, and bootstrap values are shown as percentages (see Supplemental Data Set 1 online). Highlighted are Exo70B2 and its nearest homolog Exo70B1. Exo70A1, which has been shown to be required for cell growth, is also highlighted.
(B) Y2H assay showing the interaction of PUB22ARM with Exo70B2. Yeast growth on SD-Leu/-Trp confirms the presence of both vectors for protein expression. Growth on SD-Leu/-Trp/-His indicates protein–protein interaction.
[See online article for color version of this figure.]
Components of vesicle trafficking required for PAMP responses have not been identified in plants. Here, we report that Exo70B2 is ubiquitinated by PUB22, and we provide evidence that PUB22 targets Exo70B2 for proteasomal degradation. We show that PUB22 is stabilized in response to flg22 treatment and that Exo70B2 levels consequently decrease. Moreover, Exo70B2 is required for full activation of immune signaling, and mutant plants fail to mount a full immune response, resulting in enhanced susceptibility to pathogens. Hence, data presented here support a mechanism in which PUB22 contributes to the negative regulation of PAMP-triggered responses by targeting Exo70B2, a component of the exocytic machinery.
RESULTS
PUB22 Interacts with the Exo70B2 Subunit of the Exocyst
In order to gain insight into the function of PUB22, we performed a yeast two-hybrid screen (Y2H) to identify proteins targeted for ubiquitination. We chose PUB22 as bait because our previous genetic data indicated that PUB22, together with PUB24, played the main role in the downregulation of PAMP-triggered signaling (Trujillo et al., 2008). PUB22 is a modular protein that consists of a U-box domain that mediates the interaction with a ubiquitin-conjugating enzyme (E2) and four armadillo (ARM) repeats responsible for the interaction with the target protein (Azevedo et al., 2001; Andersen et al., 2004; Mudgil et al., 2004). We used as bait only the ARM repeats domain without the U-box (PUB22ARM, residues 76 to 435) in order to prevent ubiquitination and the potential degradation of the target in yeast. We obtained several potential targets, including two clones encoding the N terminus of Exo70B2, a homolog of the yeast Exo70p subunit of the exocyst complex (Figure 1B; see Supplemental Table 1 online). These clones define the interaction site to the first 144 amino acids of Exo70B2 (see Supplemental Figure 1A online). We also confirmed via Y2H that the full-length PUB22 was able to interact with the full-length Exo70B2 (see Supplemental Figures 1B and 1C online). Exo70B2 also interacts with other subunits of the exocyst complex, supporting its role as a functional component of the exocyst complex (Pecenková et al., 2011).
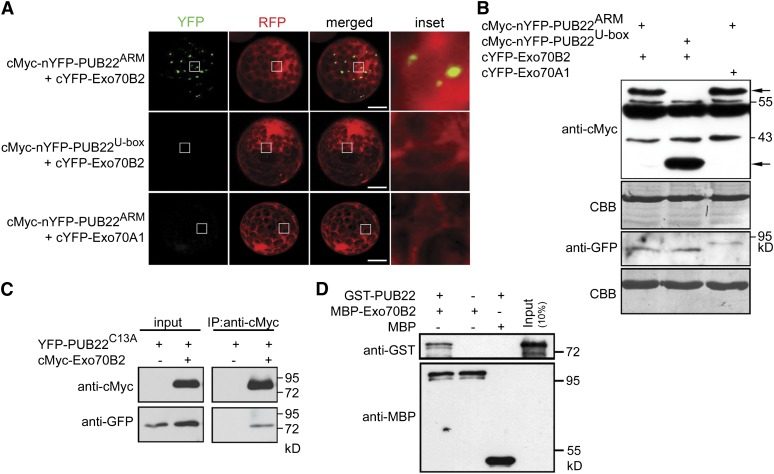
To confirm the interaction between PUB22 and Exo70B2 in vivo, we employed bimolecular fluorescence complementation (BiFC) with split yellow fluorescent protein (YFP). Coexpression of cMyc-nYFP-PUB22ARM, containing only the ARM repeats domain, with cYFP-Exo70B2 in Arabidopsis protoplasts resulted in the complementation of fluorescence localized to punctae, supporting an interaction in vivo (Figure 2A). This result corroborates that the ARM repeats are responsible and sufficient for the interaction with Exo70B2. By contrast, no signal could be observed by coexpression of PUB22U-box, containing only the U-box domain, with cYFP-Exo70B2 (Figure 2A). nYFP-PUB22ARM was also not able to interact with the homologous exocyst subunit Exo70A1 (Figures 2A and 1A). Protein accumulation was confirmed by immunoblot (Figure 2B). We further confirmed the interaction by coimmunoprecipitation (Co-IP). cMyc epitope-tagged Exo70B2 coimmunoprecipitated YFP epitope-tagged PUB22 (Figure 2C).
Figure 2.
PUB22 Interacts with the Exocyst Subunit Exo70B2.
(A) BiFC of split YFP following coexpression of nYFP-PUB22ARM or nYFP-PUB22U-box with cYFP-Exo70B2 or cYFP-Exo70A1 as indicated. Interactions were assayed using transient expression in Arabidopsis protoplasts. Free RFP was coexpressed to label the cytoplasm and nucleus. Pictures are representative of three independent experiments with similar results. Bars = 50 µm.
(B) Protein accumulation of the indicated recombinant proteins from experiments in (A), with equal loading shown by Coomassie blue (CBB). Arrows indicate nYFP- PUB22ARM (61.1 kD; top) and nYFP- PUB22U-box (33.5 kD; bottom) bands as detected by anti-cMyc antibody. cYFP-Exo70B2 (80.7 kD) and cYFP-Exo70A1 (88.7 kD) proteins were detected by anti-GFP antibody.
(C) PUB22 interacts with Exo70B2 in a Co-IP assay. The ligase-inactive mutant YFP-PUB22C13A (75.8 kD) was coexpressed with cMyc-Exo70B2 (84.7 kD) in protoplasts. Co-IP was performed using anti-cMyc antibodies. Proteins were analyzed in an immunoblot using antibodies as indicated.
(D) In vitro pull-down assay showing physical interaction of PUB22 with Exo70B2. GST-PUB22 (76.0 kD) and MBP-Exo70B2 (110.7 kD) were expressed in bacteria. Pull down was performed using amylose resin. Proteins were detected in an immunoblot using antibodies as indicated.
To test whether the interaction between Exo70B2 and PUB22 is direct, recombinant Exo70B2 tagged with maltose binding protein (MBP) was incubated in vitro with recombinant PUB22 tagged with glutathione S-transferase (GST). GST-PUB22 was readily pulled down, showing that it physically bound MBP-Exo70B2, confirming that the interaction is direct (Figure 2D).
Using Y2H, no interaction was detected with any other subunits of the exocyst complex with the exception of the C terminus of Exo84 (see Supplemental Figures 1B and 1C online). Interestingly, structural modules with a high degree of homology in the N terminus of Exo70p and the C terminus of Exo84p have been determined in yeast (Dong et al., 2005).
PUB22 was shown to act in concert with its close homologs PUB23 and PUB24 in the regulation of PAMP-triggered responses (Trujillo et al., 2008). We therefore tested PUB23C18A and PUB24C30A, inactive ligase mutant variants. In both cases, they were able to complement fluorescence in the presence of cYFP-Exo70B2, suggesting that they also interact in vivo (see Supplemental Figure 2 online).
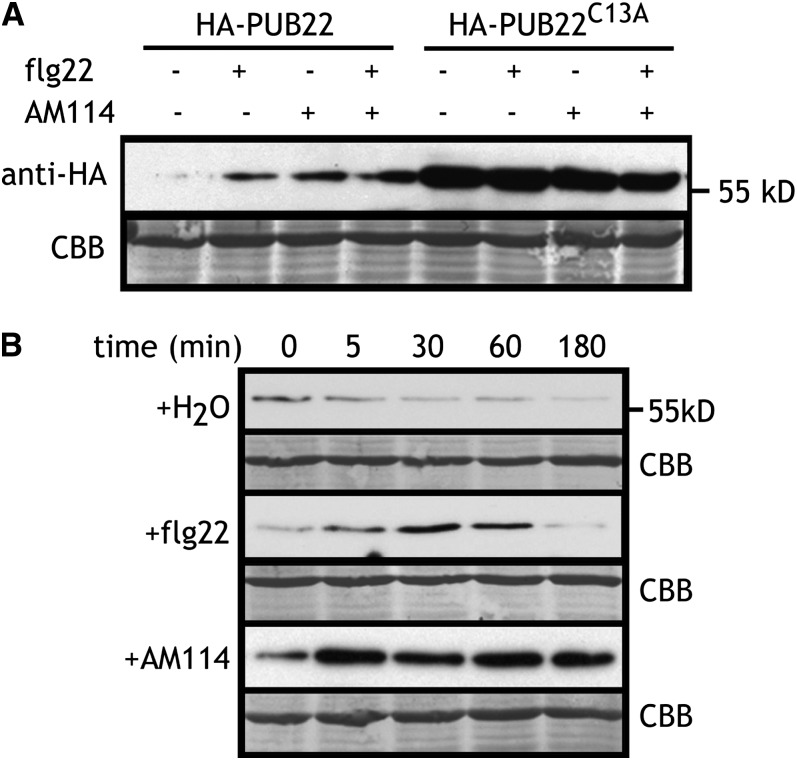
PUB22 Is Stabilized by flg22
PUBs act as scaffolds by bringing the ubiquitin conjugating enzyme and the target protein into close proximity, thus mediating ubiquitination and conveying specificity to the process (Vierstra, 2009). Our data suggested that the interaction between PUB22 and Exo70B2 does not require an external stimulus, such as PAMPs, and is therefore to some extent constitutive. We previously showed that PUB22 is an active ubiquitin ligase with the ability to autoubiquitinate (Trujillo et al., 2008). Autoubiquitination activity can lead to protein degradation by the 26S proteasome (Galan and Peter, 1999). To test whether autocatalytic ubiquitination mediates PUB22 stability, we expressed HA-PUB22 and an inactive mutant variant HA-PUB22C13A, which is unable to bind ubiquitin conjugating enzymes. This mutant accumulated to much higher levels than wild-type PUB22, indicating that autocatalytic ubiquitination may indeed occur and result in degradation (Figure 3A). A logical consequence of this observation would be that PUB22 must be stabilized in order to be able to ubiquitinate its target with high efficiency. We therefore tested whether treatment with flg22, the conserved epitope of the bacterial PAMP flagellin, led to the stabilization of PUB22. Indeed, the abundance of PUB22 increased markedly 1 h after treatment with flg22 (Figure 3A). Inhibition of the 26S proteasome also increased PUB22 abundance, indicating that it mediates PUB22 turnover (Figure 3A). Since PUB22 is involved in the regulation of early PAMP-triggered responses, we analyzed the kinetics of its stabilization. Treatment with flg22 resulted in the transient accumulation of PUB22, which became evident as early as 5 min after treatment and decreased back to basal levels after 180 min (Figure 3B). Proteasome inhibition also resulted in a rapid increase of PUB22 levels, which in contrast with the results of flg22 treatment, remained high at 3 h after treatment (Figure 3B). Therefore, PAMP-mediated PUB22 stabilization potentially represents a regulatory mechanism of target ubiquitination.
Figure 3.
PUB22 Is Stabilized upon flg22 Treatment.
(A) Expression of recombinant HA-PUB22 or HA-PUB22C13A in Arabidopsis protoplasts 1 h after treatment with flg22 (1 µM), AM114 (20 µM), or equivalent volume of DMSO (−) as indicated. CBB, Coomassie blue.
(B) Expression kinetic of recombinant HA-PUB22 in Arabidopsis protoplasts after treatment with flg22 (1 µM), the proteasomal inhibitor AM114 (20 µM), or equivalent volume of DMSO (−) as indicated. Experiments were repeated at least three times with similar results. Recombinant protein was detected with anti-HA antibody. Equal loading is shown by Coomassie blue staining.
PUB22 Mediates the Ubiquitination and Degradation of Exo70B2
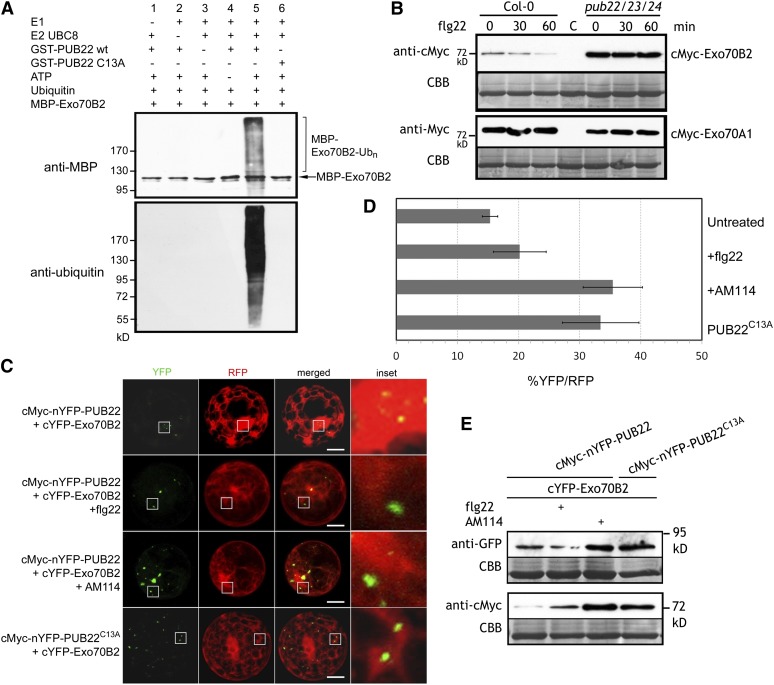
We next assayed the ability of PUB22 to use Exo70B2 as a substrate in an in vitro ubiquitination assay. In the presence of all required components, recombinant GST-PUB22 ubiquitinated MBP-Exo70B2, as evidenced by the increase in the molecular size of the target (Figure 4A). No ubiquitination was detectable in absence of any of the required components. Furthermore, a point mutation in a conserved amino acid in the PUB22 U-box domain abolished ubiquitination of MBP-Exo70B2 (Figure 4A). Thus, MBP-Exo70B2 can be used as a substrate by GST-PUB22. Moreover, the presence of Exo70B2 reduced PUB22 autoubiquitination in vitro (see Supplemental Figure 3 online).
Figure 4.
PUB22 Ubiquitinates Exo70B2 and Mediates Its Degradation via the 26S Proteasome.
(A) In vitro ubiquitination of Exo70B2 by PUB22. GST-PUB22, the mutant variant GST-PUB22C13A, and MBP-Exo70B2 were tested using the Arabidopsis His-UBA1 and His-UBC8. Lanes 1 to 4 and 6 are negative controls. Proteins were separated by SDS-PAGE and detected by immunoblotting using the indicated antibodies.
(B) Expression of recombinant cMyc-Exo70B2 and cMyc-Exo70A1 in wild-type Col-0 and pub22 pub23 pub24 Arabidopsis protoplasts 0, 30, and 60 min after treatment with flg22 (1 µM). CBB, Coomassie blue.
(C) BiFC of split YFP following coexpression of cMyc-nYFP-PUB22 and nYFP-PUB22C13A with cYFP-Exo70B2 or cYFP-Exo70A1 after treatment with flg22 (1 µM) or AM114 (20 µM) as indicated. Proteins were expressed transiently in Arabidopsis protoplasts. Coexpressed free RFP labels cytoplasm and nucleus. Pictures are representative of three independent experiments with similar results. Bars = 50 µm.
(D) Percentage of total transformed protoplasts measured by cells with RFP fluorescence and YFP reconstitution. Data shown as mean percentage ± sd of three independent biological replications each with at least 70 cells/treatment.
(E) Immunoblot showing abundance of indicated recombinant proteins from experiments in (C). Results are representative of two independent experiments.
We then assayed Exo70B2 levels after PAMP perception in vivo. Treatment with flg22 induced the gradual reduction of cMyc-Exo70B2 within the first 60 min (Figure 4B). By contrast, protein levels remained unchanged in the pub22 pub23 pub24 triple mutant background. Moreover, overall Exo70B2 protein accumulation was higher in the pub triple mutant background compared with the wild type (Figure 4B). To rule out that this was a general effect, we examined the protein accumulation of cMyc-Exo70A1, an Exo70B2 homolog that does not interact with PUB22 (Figure 1A). Exo70A1 accumulation was independent of the genetic background and was not altered by flg22 treatment (Figure 4B).
We next analyzed the effect of PUB22 specifically on Exo70B2 levels. Coexpression of the full-length cMyc-nYFP-PUB22 and cYFP-Exo70B2 resulted in a weak complementation of fluorescence (Figures 4C and 4D). Surprisingly, flg22 treatment did not lead to major changes in the number of cells displaying fluorescence (Figure 4D). However, Exo70B2 protein levels moderately decreased, in line with previous results (Figure 4E). PUB22 levels, on the other hand, displayed a clear increase, as it was stabilized by flg22 (Figure 4E). Hence, the absence of clear changes in cells displaying fluorescence reflects the increase of PUB22 levels. Accordingly, inhibition of the 26S proteasome or coexpression with an inactive PUB22C13A mutant resulted in higher levels of Exo70B2 (Figure 4E) and a higher number of cells with fluorescence (Figure 4D).
Together, these data indicate that Exo70B2 is a ubiquitination substrate of PUB22 and that Exo70B2 ubiquitination by PUB22 leads to its degradation in vivo. This process is potentially regulated by the stability of PUB22 through autocatalytic ubiquitination leading to its degradation, which is inhibited by treatment with flg22.
EXO70B2 Mutants Are Impaired in PAMP-Triggered Responses
To determine whether Exo70B2 plays a role in PTI, we analyzed independent T-DNA insertion mutant lines (exo70B2-1, SALK_091877; and exo70B2-3, GK-726G07; see Supplemental Figure 4A online). Both alleles do not display any obvious developmental defects; flowering and seed yield under normal growth conditions are comparable to wild-type Columbia-0 (Col-0), similar to the previously described pub22 pub23 pub24 mutant (see Supplemental Figure 4B online).
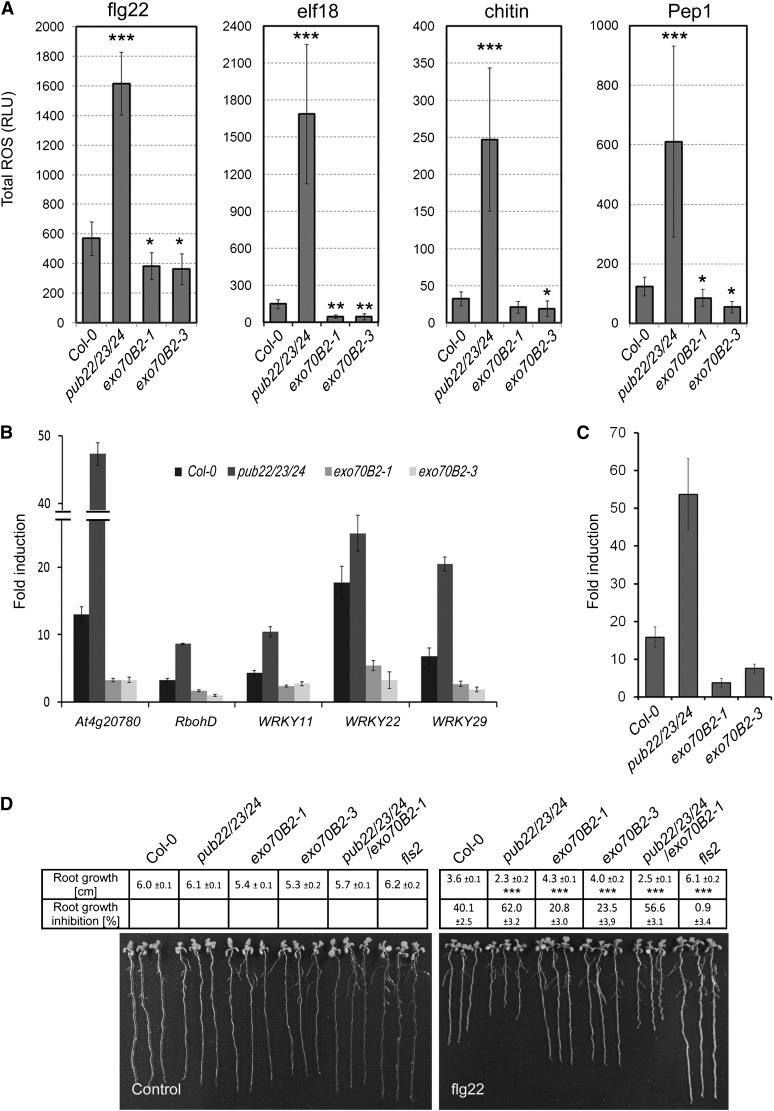
Because PUB22 was defined genetically as a negative regulator of PTI and it targets Exo70B2 for degradation, we predicted that Exo70B2 acts as a positive regulator of PTI. Thus, exo70B2 null mutants should display attenuated PTI. To test this, we first assayed the production of reactive oxygen species (ROS). Perception of flg22 is mediated by the PRR FLS2 (Gómez-Gómez and Boller, 2000). Activation of FLS2 leads to an oxidative burst, one of the first measurable responses of plants in response to PAMP perception (Nicaise et al., 2009). Mutation of pub22, pub23, and pub24 additively derepresses this response, which is strongest in the pub22 pub23 pub24 triple mutant (Trujillo et al., 2008). As predicted, both exo70B2 alleles were less sensitive than wild-type Col-0 to flg22, as evidenced by a reduced production of ROS (Figure 5A). This indicates that Exo70B2 functions as a positive regulator of signaling in immunity and supports the idea that it contributes to the enhanced responsiveness of the pub triple mutant. Derepressed responses in pub22 pub23 pub24 extended to all tested PAMPs and also oligogalacturonides (Trujillo et al., 2008). Correspondingly, the oxidative burst was compromised in exo70B2 in response to all tested elicitors, including elf18, the conserved epitope of bacterial transcription factor EF-Tu (Kunze et al., 2004); chitin, a cell wall component of fungi (Kaku et al., 2006; Miya et al., 2007); and Pep1, an endogenous DAMP (Figure 5A; Yamaguchi et al., 2006). This also indicates that Exo70B2 is involved in the regulation of signaling mediated by different PRRs, independently of BAK1 because BAK1-indepenent chitin responses were also affected (Shan et al., 2008).
Figure 5.
Exo70B2 Is Required for PAMP- and DAMP-Triggered Responses.
(A) Total production of ROS in relative light units (RLU) during 60 min after treatment with 100 nM PAMP peptides flg22 or elf18, 100 µg/mL chitin, or 100 nM DAMP peptide Pep1 as indicated. Statistical significance compared with Col-0 plants is indicated by asterisks (Student’s t test; *P < 0.05, **P < 0.01, and **P < 0.001). Data shown as mean ± sd (n = 8). ROS production for each elicitor was evaluated in at least three independent experiments with similar results.
(B) Quantitative RT-PCR of ROS and immune response marker genes 60 min after flg22 treatment. Fourteen-day-old seedlings were treated with 1 µM flg22 or water (control). ACT2 was used as a reference gene. Similar results were obtained in three independent experiments. Data shown as mean ± sd (n = 3). Changes in fold expression are significant for all genes in comparison to the wild type (Student’s t test at least P < 0.01).
(C) PR1 expression is decreased in exo70B2 mutant lines in response to Pst. Plants were syringe infiltrated with a bacterial solution containing 1 × 107 cfu/mL or water (control) and harvested 24 h after inoculation. Data shown as mean ± sd (n = 3).
(D) Root growth inhibition by 1 μM flg22 in exo70B2 plants. Length of the main root was measured 7 d after transplanting. Data shown as mean of three independent experiments ± se (n ≥ 60). Statistical significance compared with Col-0 plants is indicated by asterisks (Student’s t test; ***P < 0.001).
We further confirmed a function of Exo70B2 in early PAMP responses by testing MAPK activity after flg22 elicitation. exo70B2-3 plants exhibited reduced MAPK3 activity compared with Col-0 (see Supplemental Figure 5 online). Thus, exo70B2 mutants displayed reduced PAMP-triggered signaling in all early signaling markers, which is the opposite of the pub22 pub23 pub24 mutant (Trujillo et al., 2008).
Later responses to PAMP perception include the transcriptional activation of genes involved in immunity (Nicaise et al., 2009). Similar to the decreased oxidative burst and MAPK activity, the transcriptional response in exo70B2 to flg22 was reduced for five PTI marker genes: At4g20780 (encoding a Ca2+ binding protein), RbohD (encoding an NADPH oxidase), and the transcription factor genes WRKY11, WRKY22, and WRKY29 (Figure 5B). Furthermore, the induction of PR1, a marker gene for the salicylic acid pathway, was also compromised after infection with Pseudomonas syringae pv tomato DC3000 (Pst; Figure 5C).
To assay the impact on sustained flg22-triggered FLS2 signaling, we tested the root growth inhibition by flg22. Wild-type Col plants displayed a 40.1% decrease in root growth compared with untreated controls, consistent with previous reports (Bethke et al., 2009). Enhanced PTI signaling in pub22 pub23 pub24 resulted in a stronger growth inhibition of 62%. In line with the previous data, root growth inhibition in exo70B2 mutants was distinctly reduced to almost half compared with the wild type, with 20.8 in exo70B2-1 and 23.5 in exo70B2-3 (Figure 5D), further demonstrating the importance of Exo70B2 in PAMP-triggered signaling.
Nevertheless, exo70B2 mutants displayed mild phenotypes for some of the assayed responses. We hypothesized that this is due to overlapping function of homologs, such as Exo70B1, which shows the highest sequence similarity (53% amino acid identity; Figure 1A). To test this, we generated exo70B1 alleles and the exo70B1-1 exo70B2-1 double mutant (see Supplemental Figure 6A online). exo70B1 mutants showed a reduction of the flg22-triggered ROS production (see Supplemental Figure 6B online). However, this phenotype was not significantly enhanced in exo70B1-1 exo70B2-1 double mutants when compared with exo70B1 alleles. Similarly, root growth inhibition by flg22 in exo70B1-1 was reduced in comparison to the wild type (see Supplemental Figure 6C online). Nevertheless, the double mutant phenotype was also comparable to exo70B2-1, indicating that Exo70B1 and Exo70B2 apparently do not functionally overlap, although both are required for PTI signaling.
Exo70B2 Mutants Are More Susceptible to Pathogens
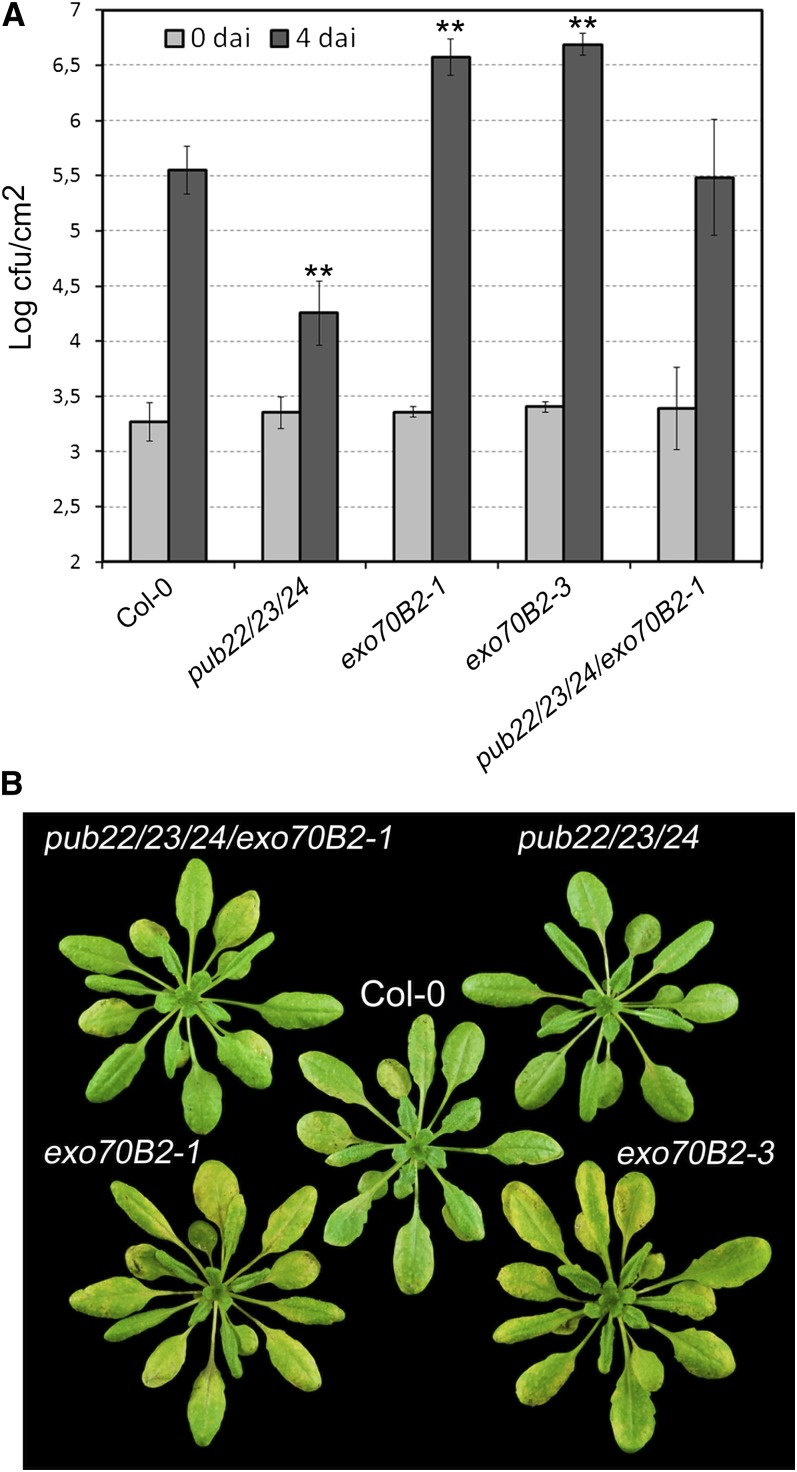
To assay further the role of Exo70B2 in immunity, we analyzed phenotypes of exo70B2 in response to infection by pathogens of Arabidopsis. To mimic a natural Pst infection, we spray-inoculated plants, which allows the detection of resistance phenotypes related to PAMP perception, as previously shown for FLS2 receptor-like kinase mutants (Zipfel et al., 2004). As reported previously, pub22 pub23 pub24 plants were more resistant to the virulent Pst as a consequence of the hyperactivation of immune responses, which result from derepressed PAMP-triggered signaling (Figure 6A). By contrast, both exo70B2 alleles were immunocompromised and showed a clear increase in the bacterial growth (Figure 6A). This was also reflected by the stronger development of disease symptoms, such as leaf yellowing (Figure 6B).
Figure 6.
Exo70B2 Is Required for Resistance against Pst.
(A) Infection assays with the virulent bacterial pathogen Pst DC3000. Col-0, pub22 pub23 pub24, exo70B2-1, exo70B2-3, and pub22 pub23 pub24 exo70B2-1 plants were spray inoculated with a bacterial suspension of 5 × 108 cfu/mL. Infection and bacterial growth were assessed at 0 and 4 d after inoculation (dai). Data shown as mean ± sd (n = 5). Similar results were obtained in four independent experiments. Statistical significance between wild-type Col-0 and mutant lines was assessed with the Student’s t test (**P < 0.01).
(B) Disease phenotypes from spray-inoculated plants in (A) at 4 d after inoculation.
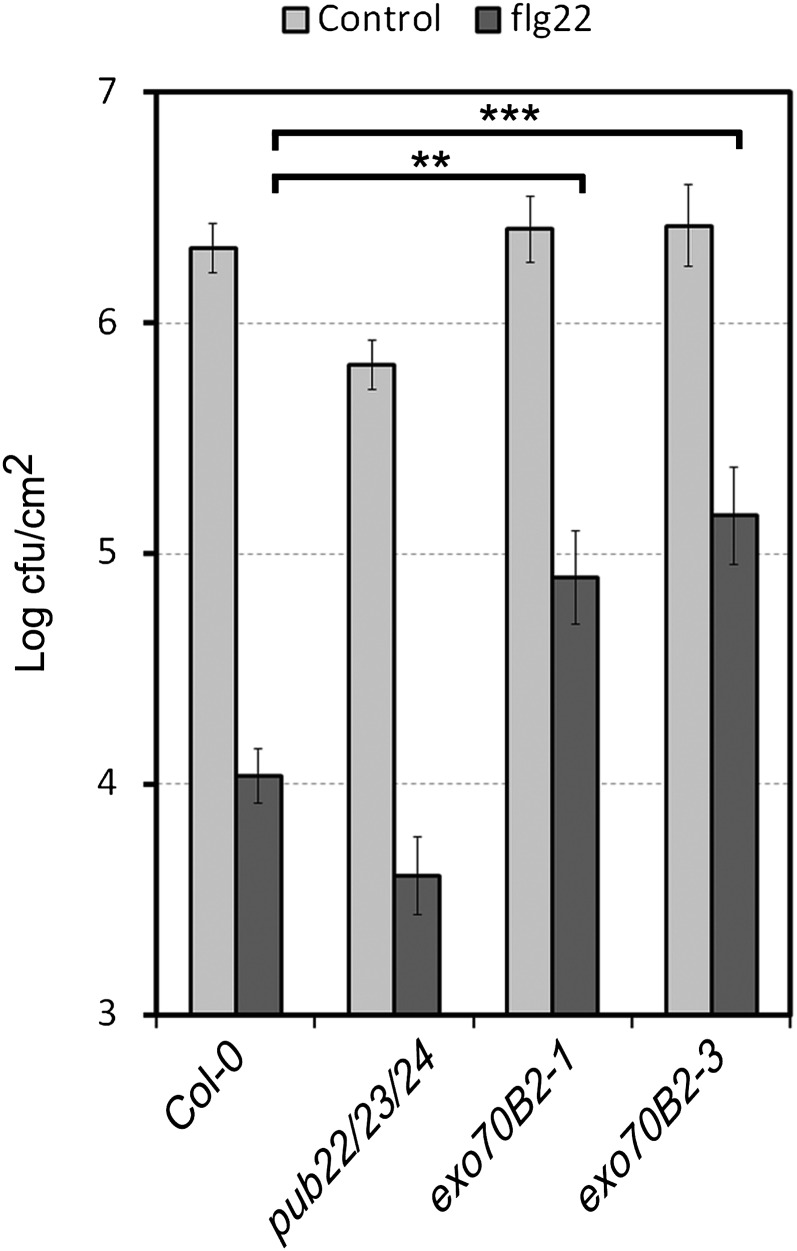
Activation of immunity by PAMPs prior to infection has a protective effect by priming immune responses for posterior infection (Zipfel et al., 2004). This is manifested by reduced growth of virulent Pst in plants that have been pretreated with flg22. We performed this experiment with exo70B2 plants and observed that the mutants displayed an enhanced bacterial growth in comparison to the wild-type parent (Figure 7). Thus, the protective effect of flg22 pretreatment prior to infection with Pst was less effective in exo70B2 plants.
Figure 7.
Protective Effect of flg22 Is Compromised in exo70B2 Mutants.
Growth of Pst on Col-0, pub22 pub23 pub24, exo70B2-1, and exo70B2-3 plants pretreated with water or 100 nM flg22 for 24 h and then syringe infiltrated with 1 × 106 cfu/mL of bacteria. Bacterial growth was determined 2 d after inoculation. Data are shown as the mean of three independent experiments ± se (n = 18). Statistical significance between samples is indicated by brackets and asterisks (Student’s t test; **P < 0.01 and ***P < 0.001).
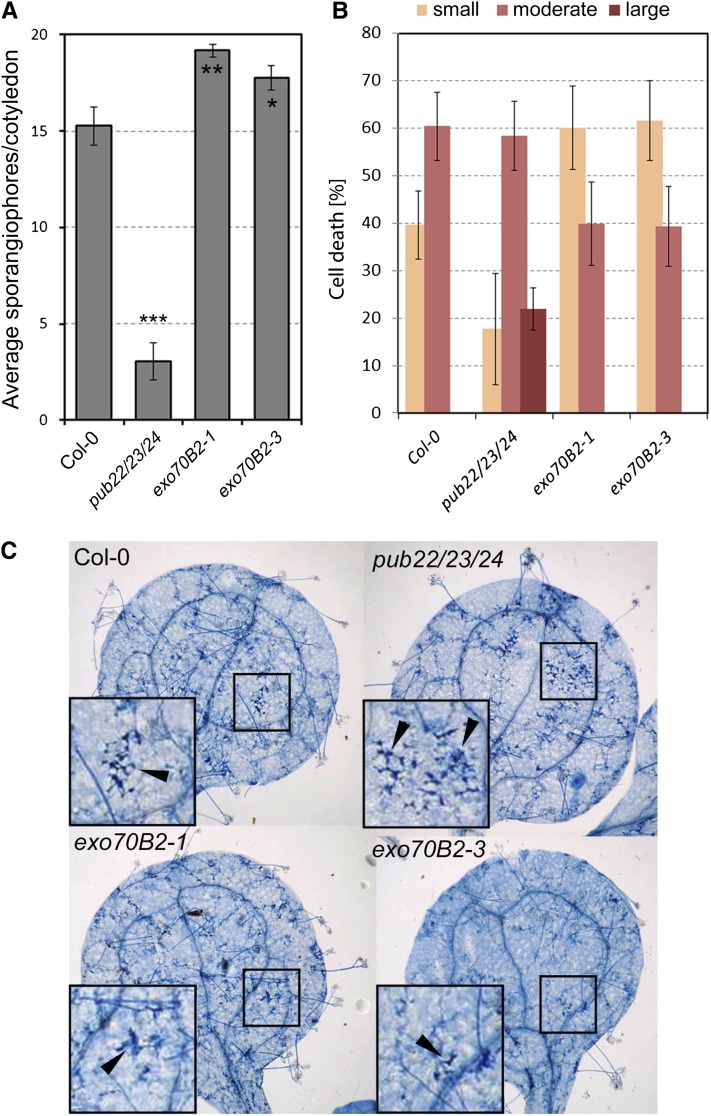
To complement our analysis, we challenged mutant plants with the obligate biotrophic oomycete Hyaloperonospora arabidopsidis (Hpa) isolate Emco5, which is virulent on wild-type Col-0 (McDowell et al., 2005). exo70B2 plants exhibited an enhanced disease susceptibility phenotype compared with Col-0 (Figure 8A). This result indicates that Exo70B2 plays an important role in immunity against bacterial and oomycete pathogens (Figures 6A and 8A).
Figure 8.
Exo70B2 Is Required for Resistance against Hpa and Cell Death Response.
(A) Infection assay with the oomycete pathogen Hpa isolate Emco5. Two-week-old seedlings were inoculated with a solution of 5 × 104 spores/mL. Hpa growth was assessed by the number of sporangiophores 7 d after inoculation ± se, on at least 50 cotyledons. Experiments were repeated five times with similar results.
(B) Cotyledons were stained with trypan blue and cell death lesions were scored as none/small, moderate, or large. Shown are the percentages for average of each category of cell death from three independent experiments ± se (n ≥ 60).
(C) Representative pictures of trypan blue–stained cotyledons with cell death reactions (arrowheads) in wild-type Col-0 and mutant lines.
[See online article for color version of this figure.]
The pub22 pub23 pub24 plants display enhanced cell death in response to Hpa infection (Trujillo et al., 2008). By contrast, exo70B2 plants were not able to mount the same degree of cell death as the wild type (Figures 8B and 8C). Cell death foci, as detected by Trypan blue staining, were divided into three categories according to size. The size of cell death foci in exo70B2 plants was reduced, as seen by a 20% reduction of cotyledons with moderate size lesions and a corresponding increase in those with small lesions (Figures 8B and 8C). Because cell death is normally associated with effector-triggered immunity, our observations support a model in which PTI is mechanistically similar to effector-triggered immunity and, therefore, that PAMP signaling can induce cell death, as previously proposed (Shen et al., 2007). Together, Exo70B2 contributes to the activation of PTI during pathogen invasion and to the activation of cell death during Hpa infection.
If Exo70B2 is a target of PUB22, then the exo70B2 mutant should partially suppress the phenotypes of the pub22 pub23 pub24 triple mutant. To address this hypothesis, we generated a pub22 pub23 pub24 exo70B2-1 quadruple mutant that we used to test epistasis. Bacterial growth was higher in the quadruple mutant than in the pub22 pub23 pub24 parent, indicating that exo70B2 was able to suppress partially the enhanced resistance phenotype of the triple mutant (Figure 6A). Moreover, the oxidative burst triggered by flg22 was also reduced in pub22 pub23 pub24 exo70B2-1 compared with pub22 pub23 pub24 (see Supplemental Figure 7 online). In addition, a similar, although mild, epistatic effect of exo70B2 on pub22 pub23 pub24 was present in the root growth inhibition (Figure 5D). Suppression of tested responses was only partial, suggesting additional important targets that participate in PAMP-triggered responses. The genetic interaction of Exo70B2 with pub22 pub23 pub24 underscores its importance in PTI, while at the same time it reveals a higher order complexity highlighted by the manifold ligase and target homologs and potentially additional targets yet to be identified.
DISCUSSION
The function of exocytosis in plant immunity has been previously linked to the deployment of defense responses by the characterization of SNARE complexes. Responses include the secretion of toxic compounds and cell wall reinforcement to impede cell wall penetration (Bednarek et al., 2010). However, exocytosis and its role in vesicle trafficking plays additional fundamental functions by maintaining membrane integrity and contributing to membrane remodeling in response to altered environmental conditions. Plants quickly react to changes in light or gravitropism as illustrated by the relocalization of the phytohormone auxin efflux carrier PIN3, thus regulating auxin concentrations, which result in a different growth direction (Friml et al., 2002; Ding et al., 2011). They can also quickly adapt to changes in nutrient availability by regulating the number of nutrient transporters (Takano et al., 2005).
In this study, we show that PUB22 targets Exo70B2 for degradation upon flg22 perception, suggesting a mechanism for the attenuation of immune signaling and, thus, adaption/response to pathogen invasion. We show that Exo70B2 is required for full activation of early immune signaling events, such as the ROS burst and MPK3 activity, as well as later responses, such as the transcriptional activation of marker genes and reduced root growth inhibition in combination with an immunocompromised phenotype of mutants. The involvement of Exo70B2 in PAMP-signaling events supports the view that both intracellular vesicle trafficking and signaling are intertwining molecular networks, as shown for animal cells (Scita and Di Fiore, 2010). Clues as to the potential function of Exo70B2 in plants are given by studies in other organisms, which show the involvement of the exocyst in the delivery of the N-methyl-D-aspartate (NMDA) receptor, the glucose transporter type 4 (Glut4), and the α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) receptor to the cell surface (Inoue et al., 2003; Sans et al., 2003; Gerges et al., 2006). Further studies have reported that the complex can also localize to recycling endosomes and have implicated it in the recycling of the α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate receptor (Fölsch et al., 2003; Langevin et al., 2005; Mao et al., 2010). In such a scenario, Exo70B2 could contribute to recycling of plasma membrane proteins involved in PAMP-triggered signaling. These could include NADPH oxidases, ion channels, and potentially receptor-like kinases, such as FLS2, as well as yet unidentified components. Degradation of Exo70B2 by PUB22 would attenuate the recycling pathway in order to redirect positive signaling components into the vacuolar degradation pathway to downregulate signaling. Recently, it was shown that PUB12 and 13 directly ubiquitinate FLS2 and mediate its degradation and that, in parallel to PUB22, PUB23, and PUB24, they also contribute to the downregulation of immune signaling (Trujillo et al., 2008; Lu et al., 2011). The degradation of integral membrane proteins is mediated by the vacuole; therefore ubiquitination of FLS2 is expected to modulate its intracellular trafficking. Hence, signal attenuation is potentially regulated simultaneously at various levels/stages of vesicle trafficking.
Each stage of vesicle trafficking requires a multitude of proteins to execute these processes, many of which have paralogs. Our genetic data showed that exo70B2 partially suppressed the enhanced responsiveness phenotype of pub22 pub23 pub24. Partial suppression could not be explained by redundancy between Exo70B2 and its nearest homolog Exo70B1 (see Supplemental Figure 6B online). However, a complete functional overlap between PUB22, PUB23, and PUB24 is unlikely. PUB22, PUB23, and PUB24 are anticipated to have other and/or additional targets that contribute to processes required for PAMP responses. Indeed, the yeast two-hybrid screen with PUB22 identified additional candidate targets that are currently under investigation (see Supplemental Table 1 online). Some of these putative targets have functions in vesicle trafficking, suggesting that PUB22 might regulate trafficking protein complexes.
The self-incompatibility response, which promotes outcrossing, is mediated by receptor kinases such as the self-incompatibility S-locus receptor kinase and mechanistically resembles PTI in various instances. One similarity is the ligand-dependent endocytosis of S-locus receptor kinase that is reminiscent of the endocytosis of FLS2 (Robatzek et al., 2006; Ivanov and Gaude, 2009). Several mechanistic parallels can be observed between the proposed role of the Brassica napus U-box–type ligase ARC1, which is required for the pollen self-incompatibility response in Brassica species, and PUB22 (Stone et al., 1999). ARC1 was shown to interact with the Exo70B2 homolog Exo70A1 and was suggested to mediate its degradation (Samuel et al., 2009). Degradation of Exo70A1 by ARC1 was proposed to function in the rejection of self-pollen, by inhibiting the exocytosis of compatibility factors. It is therefore conceivable that Exo70B2 in Arabidopsis and Exo70A1 in Brassica play similar functions although acting on different pathways.
Several lines of evidence document a function of the exocyst complex, or subunits thereof, in immune signaling in mammals. A complex containing the exocyst Sec5 subunit directly participates in the activation of innate immune signaling during Toll-like receptor activation (Chien et al., 2006). Furthermore, signaling via the innate immunity components TANK binding kinase 1 and stimulator of interferon genes is supported by Sec5 (Ishikawa and Barber, 2008; Ishikawa et al., 2009). The importance of the exocyst in immunity is further supported by the fact that it is targeted by pathogens in mammalian systems. The Bacillus anthracis toxins EF and LF cooperatively inhibit the endocytic recycling through the Rab11/Sec15 exocyst, leading to reduction of Notch signaling and levels of cadherin at adheren junctions (Guichard et al., 2010). In plants, there are also indications that the exocyst is targeted by the Avr3a effector protein of Phytophthora infestans (Bos et al., 2010). Interestingly, in the same study, it was demonstrated that Avr3a targets a Nicotiana benthamiana PUB called CMPG1, a homolog of PUB22, and that silencing of CMPG1 led to increased resistance against P. infestans, which is biotrophic during early stages of infection. Similarly to PUB22, CMPG1 displayed high turnover, and interaction with Avr3a resulted in its stabilization.
PUB22 was shown to mediate its own degradation by autocatalytic ubiquitination, which was rapidly reduced by flg22 treatment, leading to its accumulation. This suggests a mechanism by which Exo70B2 levels are regulated by quick changes in the turnover of PUB22 in response to PAMPs and thus supports its function in the regulation of PAMP-triggered immune responses. Autocatalytic ubiquitination has been shown to be responsible for the intrinsic instability of F-box proteins that are subunits of SKP1-cullin-F-box ubiquitin ligases in yeast (Galan and Peter, 1999). Autoubiquitination has also been reported for RING-type ligases, as for example Mdm2, which mediates the degradation of p53 in mice (Fang et al., 2000), or KEG1, which mediates the degradation of the drought response transcription factor ABI5 in plants (Liu and Stone, 2010). Treatment with phytohormone abscisic acid led to the reduction of KEG1 levels via autoubiquitination (Liu and Stone, 2010). By contrast, PUB22 protein levels are stabilized after treatment with flg22, potentially by inhibition of autocatalytic ubiquitination activity.
Identification of ubiquitin ligase target proteins remains a major challenge and will reveal whether PUBs specialize in the regulation of intracellular vesicle trafficking. Furthermore, isolation of the signaling components transported by Exo70B2-positive vesicles and the identification of the destination organelles in future studies will provide insight into the function of the exocyst complex in plant immunity.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana exo70B2-1 (SALK091877), exo70B2-3 (GK726G07), exo70B1-1 (GK114C03), and exo70B1-2 (GK156G02) mutants (Col-0 background) were obtained from the European Arabidopsis Stock Centre. The pub22 pub23 pub24 mutant was previously described (Trujillo et al., 2008). All plants for pathogen inoculation experiments, oxidative burst assays, and transient transformation via microbombardment were grown at 21°C and 70% relative humidity under short-day conditions (8 h light/16 h dark). exo70B1 alleles and exo70B1-1/exo70B2-1 for ROS assays were grown under long-day conditions (16 h light/8 h dark). For sterile conditions, seedlings were grown on Murashige and Skoog (MS) agar media supplemented with vitamins and 1% Suc at 22°C in short-day conditions (8 h light/16 h dark). Protoplasts were isolated from 5-week-old plants as previously described (He et al., 2007).
Y2H Screen
Y2H screening was performed by Hybrigenics. The coding sequence for amino acids 76 to 435 of PUB22 (At3G52450) was cloned into pB27 as a C-terminal fusion to LexA (N-LexA-prolyl 4-hydroxylase-C) and transformed into the yeast strain L40. A random-primed Arabidopsis (wild-type Col-0) 1-week-old seedling cDNA library constructed into pP6 was transformed into yeast strain Y187. After mating, 62.4 million clones (6.2-fold the complexity of the library) were screened. Prey fragments of the positive clones were amplified by PCR and sequenced at 5′ and 3′ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBank database (National Center for Biotechnology Information) using a fully automated procedure.
Transient Protoplast Transformation, BiFC, and PUB22 Stabilization
PUB22, PUB22C13A, PUB22ARM, and PUB22U-box were cloned into pESPYNEgw (nYFP) and Exo70B2 into pESPYCEgw (cYFP; Ehlert et al., 2006) via LR reaction (Invitrogen). Constructs were transformed in mesophyll protoplasts as described previously (Yoo et al., 2007). Plasmid DNA (1 µg/µL) of each construct was mixed with mCherry coding construct (1 µg/µL), and 100 µg total DNA was used to transform 1 mL of mesophyll protoplasts derived from 5- to 6-week-old plants. Reconstitution of split YFP was assessed by confocal imaging 1 d after transformation. For imaging, a LSM710 (Zeiss) was employed with the following settings: YFP excitation, 488 nm; emission band pass, 505 to 550 nm; red fluorescent protein (RFP) excitation, 543 nm; and emission band pass, 560 to 615 nm. Flg22 treatment and inhibition of the 26S proteasome was performed 1 d after transformation. For proteasome inhibition 20 µM AM114 (Calbiochem) was used. Confocal imaging was conducted 1 h after flg22 treatment and 3 h after proteasome inhibition. To analyze protein levels, protoplasts were pelleted (200g, 1min) and resuspended in protein extraction buffer (50 mM Tris-HCl, pH 6.8, 4% SDS, 8 M urea, 30% glycerol, 0.1 M DTT, and 0.005% OrangeG). After denaturing (65°C, 10 min) samples were analyzed by SDS-PAGE and protein gel blots using anti-cMyc (Sigma-Aldrich) and polyclonal anti-green fluorescent protein (GFP; Invitrogen) antibodies. cMyc-tagged Exo70B2 and HA-tagged PUB22 constructs were generated by cloning the coding sequences into pGWB418 and pGWB415 (Nakagawa et al., 2007) via LR reaction (Invitrogen). Constructs were transformed in protoplasts (50 µg DNA/1 mL protoplasts), and protein levels were assessed as described above using anti-cMyc and anti-HA (Sigma-Aldrich) antibodies.
Co-IP Assay
cMyc-tagged Exo70B2 and YFP-tagged PUB22C13A under the control of a 35S promoter were generated by cloning the coding sequence of PUB22C13A into pGWB418 (Nakagawa et al., 2007) and pEARLEYGATE104 (Earley et al., 2006) via LR reaction (Invitrogen). Constructs were transformed in mesophyll protoplasts as described in the Supplemental Methods 1 online. For Immunoprecipitation, 1 mL of transfected protoplasts were pelleted (200g, 1min) and resuspended in extraction buffer (20 mM HEPES, 50 mM KCl, 2.5 mM MgCl2, 10 µM ZnCl2, 2.5 mM EDTA, 5 mM DTT, 0.1% Triton X-100, 10 µM AM114, 100 µM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride, 1 mM NaF, 0.5 mM Na3VO4, 15 mM β-glycerophosphate, and 1% Protease Inhibitor Cocktail). The lysate was cleared by centrifugation (15.000g, 10 min), and the supernatant was incubated with anti-cMyc beads (Clontech) for 3 h at 4°C with gentle shaking. Beads were collected and washed five times with extraction buffer. Proteins were eluted with 2× LDS buffer (12.5 mM Tris HCl, pH 6.8, 2.5% SDS, 20% glycerol, 50 mM DTT, and 0.005% OrangeG) and analyzed by SDS-PAGE and immunoblotting using anti-cMyc (Sigma-Aldrich) and anti-GFP (Invitrogen) antibodies.
ROS Measurement
Leaf discs (0.125 cm2) of 7-week-old plants were incubated overnight in water in a 96-well titer plate using one leaf disc per well. ROS produced by leaf discs were measured by a luminol-based assay (20 µg/mL horseradish peroxidase and 30 µg/mL luminol). Luminescence is shown as relative light units and was measured with a 1450 Micro Beta Jet Luminescence Counter (Perkin-Elmer) and signal integration time of 2 s.
cDNA Synthesis and Real-Time Quantitative PCR
Total RNA was extracted using TriFast peqGOLD (Peqlab) following the manufacturer’s instructions. Two micrograms of DNaseI (Fermentas) treated RNA was reverse transcribed with Maxima Reverse transcriptase following the manufacturer’s instructions (Fermentas).
Real-Time quantitative PCR was performed using the Maxima SYBR Green mix (Fermentas) in a CFX384 qPCR System (Bio-Rad). Gene induction was calculated using the Bio-Rad software. Reaction volume was 20 μL and contained 10 μL of the reaction mix, 0.5 nmol of each primer, and 2 μL of cDNA. PCR conditions were as follows: 2 min incubation at 50°C and 3 min denaturation at 95°C followed by 45 cycles of 95°C for 30 s, 60°C for 10 s, and 72°C for 20 s. Subsequently, a dissociation curve was performed. All reactions were performed in triplicate.
Root Growth Assays
Seeds were sterilized and grown on 1× MS agar media supplemented with vitamins and 1% Suc. Seeds were stratified at 4°C for 2 d and grown for 5 d in short-day conditions vertically (8 h light/16 h dark). Seedlings were then transplanted on square Petri dishes with MS agar media supplemented with flg22 (1 µM) or DMSO (0.01%). Length of the main root was scored 7 d after transfer root growth was measured using Image J software.
Pathogen Infection Assays
For bacterial growth experiments, 6-week-old plants were spray inoculated with a solution of 5 × 108 colony-forming units (cfu)/mL Pst as described by Zipfel et al. (2004). Bacterial growth was measured 4 d after inoculation. To analyze flg22-induced resistance effect, 6-week old plants were infiltrated with 100 nm flg22. After 24 h, the same leaves were challenged by infiltrating 1 × 105 cfu/mL Pst DC3000. Bacterial growth was assessed 2 d after inoculation.
Hpa isolate Emco5 infections were performed as described (McDowell et al., 2011) with suspensions of 5 × 104 spores/mL, sprayed on 2-week-old seedlings under short-day conditions and scored 2 weeks after infection by counting sporangiophores. For the visualization of cell death, plants were briefly boiled in a 50% ethanol trypan blue-lactophenol solution and cleared with chloral hydrate overnight as described.
Pull-Down Assays
For pull-downs, recombinant proteins from Escherichia coli lysates were immobilized on amylose resins (New England Biolabs), incubated for 1 h at 4°C with E. coli lysate containing interacting partner, eluted, and analyzed by immunoblotting.
Recombinant Protein Purification and E3 Ubiquitin Ligase Activity Assay
The generation of the GST-PUB22, GST-PUB23, and GST-PUB24 fusion constructs and mutant variants and purification was performed as previously described (Trujillo et al., 2008). Exo70B2 was cloned into pMAL-c2X using EcoRI and HindIII restriction sites. Recombinant MBP-Exo70B2 proteins were expressed in E. coli BL21(DE3) and purified by affinity chromatography using amylose resin (New England Biolabs). Recombinant His-UBA1 and His-UBC8 were purified using Ni-Ted resin (Macherey-Nagel).
Purified proteins were used for in vitro ubiquitination assays. Each reaction of 30 μL final volume contained 40 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 50 mM KCl, 2 mM ATP, 1 mM DTT, 10% glycerol, 200 ng E1 His-UBA1, 200 ng E2 His-UBC8, 500 ng of E3s, and 1 µg of MBP-Exo70B2. The reactions were incubated at 30°C for 3 h and stopped by adding SDS-PAGE sample buffer and incubating for 5 min at 65°C. Samples were separated by SDS-PAGE electrophoresis using 7% SDS-PAGE followed by detection of ubiquitinated substrate by immunoblotting using anti-MBP (New England Biolabs) and antiubiquitin (Santa Cruz Biotechnology) antibodies.
Phylogenetic Analysis
An alignment between Arabidopsis Exo70 proteins (available as Supplemental Data Set 1 online) was generated using ClustalW with a Blosum62 cost matrix, and the tree was generated using the neighbor-joining method, with bootstrap values (1000 replicates) shown as percentages.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers as specified in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PUB22 Interacts with the N-Terminal Domain of Exo70B2 and the Exo84 Subunit.
Supplemental Figure 2. Exo70B2 Interacts with PUB23 and PUB24.
Supplemental Figure 3. Autoubiquitination of PUB22 Is Decreased in the Presence of Exo70B2.
Supplemental Figure 4. Characterization of exo70B2 Mutant Plants.
Supplemental Figure 5. The exo70B2 Mutants Display Reduced MPK3 Activity.
Supplemental Figure 6. Mutants of the Exo70B1 Homolog Display Reduced Responses to flg22.
Supplemental Figure 7. exo70B2 Is Epistatic to pub22 pub23 pub24 in Terms of the Enhanced ROS Burst Phenotype.
Supplemental Table 1. Putative PUB22-Interacting Proteins Identified by Y2H Screen.
Supplemental Table 2. Oligonucleotides Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Figure 1A.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft SFB 567 (M.S., P.R., and M.T.), Leibniz Gemeinschaft (M.T.), KAKENHI 24228008 (K.S.), National Science Foundation, Integrative and Organismal Biology Award 0744875 (R.G.A. and J.M.M.), and Czech Science Foundation (Grantová agentura České republiky) P501/10/2081 (T.P. and V.Z.). We thank Ben Schwessinger and Cyril Zipfel for critical reading and helpful comments.
AUTHOR CONTRIBUTIONS
M.S., M.T., R.G.A., K.I., T.P., and P.R. performed the experiments. V.Z., J.M.M., K.S., and M.T. analyzed the data. M.T. planned the project and wrote the article.
Glossary
- PAMP
pathogen-associated molecular pattern
- PRR
pattern recognition receptor
- MAPK
mitogen-activated protein kinase
- PTI
to be defined
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptor protein
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- Co-IP
coimmunoprecipitation
- Col-0
Columbia-0
- ROS
reactive oxygen species
- Pst
Pseudomonas syringae pv tomato DC3000
- Hpa
Hyaloperonospora arabidopsidis
- MS
Murashige and Skoog
- GFP
green fluorescent protein
- cfu
colony-forming units
- RFP
red fluorescent protein
References
- Andersen P., Kragelund B.B., Olsen A.N., Larsen F.H., Chua N.H., Poulsen F.M., Skriver K. (2004). Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 279: 40053–40061 [DOI] [PubMed] [Google Scholar]
- Azevedo C., Santos-Rosa M.J., Shirasu K. (2001). The U-box protein family in plants. Trends Plant Sci. 6: 354–358 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Kwon C., Schulze-Lefert P. (2010). Not a peripheral issue: Secretion in plant-microbe interactions. Curr. Opin. Plant Biol. 13: 378–387 [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.I., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 107: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y., et al. (2006). RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127: 157–170 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Chong Y.T., Gidda S.K., Sanford C., Parkinson J., Mullen R.T., Goring D.R. (2010). Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol. 185: 401–419 [DOI] [PubMed] [Google Scholar]
- Clague M.J., Urbé S. (2010). Ubiquitin: Same molecule, different degradation pathways. Cell 143: 682–685 [DOI] [PubMed] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., Schulze-Lefert P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Cvrčková F., Grunt M., Bezvoda R., Hála M., Kulich I., Rawat A., Zárský V. (2012). Evolution of the land plant exocyst complexes. Front. Plant Sci. 3: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Galván-Ampudia C.S., Demarsy E., Łangowski L., Kleine-Vehn J., Fan Y., Morita M.T., Tasaka M., Fankhauser C., Offringa R., Friml J. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Dong G., Hutagalung A.H., Fu C., Novick P., Reinisch K.M. (2005). The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat. Struct. Mol. Biol. 12: 1094–1100 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Ehlert A., Weltmeier F., Wang X., Mayer C.S., Smeekens S., Vicente-Carbajosa J., Dröge-Laser W. (2006). Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 46: 890–900 [DOI] [PubMed] [Google Scholar]
- Elias M., Drdova E., Ziak D., Bavlnka B., Hala M., Cvrckova F., Soukupova H., Zarsky V. (2003). The exocyst complex in plants. Cell Biol. Int. 27: 199–201 [DOI] [PubMed] [Google Scholar]
- Fang S., Jensen J.P., Ludwig R.L., Vousden K.H., Weissman A.M. (2000). Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275: 8945–8951 [DOI] [PubMed] [Google Scholar]
- Fölsch H., Pypaert M., Maday S., Pelletier L., Mellman I. (2003). The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J. Cell Biol. 163: 351–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Galan J.M., Peter M. (1999). Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96: 9124–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerges N.Z., Backos D.S., Rupasinghe C.N., Spaller M.R., Esteban J.A. (2006). Dual role of the exocyst in AMPA receptor targeting and insertion into the postsynaptic membrane. EMBO J. 25: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D.R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J.P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Göhre V., Spallek T., Häweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J.W., Robatzek S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Guichard A., McGillivray S.M., Cruz-Moreno B., van Sorge N.M., Nizet V., Bier E. (2010). Anthrax toxins cooperatively inhibit endocytic recycling by the Rab11/Sec15 exocyst. Nature 467: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Guo W. (2009). The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 21: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Shan L., Sheen J. (2007). The use of protoplasts to study innate immune responses. Methods Mol. Biol. 354: 1–9 [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M., Chang L., Hwang J., Chiang S.H., Saltiel A.R. (2003). The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature 422: 629–633 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Barber G.N. (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Ma Z., Barber G.N. (2009). STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461: 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov R., Gaude T. (2009). Endocytosis and endosomal regulation of the S-receptor kinase during the self-incompatibility response in Brassica oleracea. Plant Cell 21: 2107–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Langevin J., Morgan M.J., Sibarita J.B., Aresta S., Murthy M., Schwarz T., Camonis J., Bellaïche Y. (2005). Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev. Cell 9: 365–376 [DOI] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn J.A., Hsu P.C., Emr S.D. (2012). Ubiquitin and membrane protein turnover: From cradle to grave. Annu. Rev. Biochem. 81: 231–259 [DOI] [PubMed] [Google Scholar]
- Mao L., Takamiya K., Thomas G., Lin D.T., Huganir R.L. (2010). GRIP1 and 2 regulate activity-dependent AMPA receptor recycling via exocyst complex interactions. Proc. Natl. Acad. Sci. USA 107: 19038–19043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell J.M., Hoff T., Anderson R.G., Deegan D. (2011). Propagation, storage, and assays with Hyaloperonospora arabidopsidis: A model oomycete pathogen of Arabidopsis. Methods Mol. Biol. 712: 137–151 [DOI] [PubMed] [Google Scholar]
- McDowell J.M., Williams S.G., Funderburg N.T., Eulgem T., Dangl J.L. (2005). Genetic analysis of developmentally regulated resistance to downy mildew (Hyaloperonospora parasitica) in Arabidopsis thaliana. Mol. Plant Microbe Interact. 18: 1226–1234 [DOI] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y., Shiu S.H., Stone S.L., Salt J.N., Goring D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nicaise V., Roux M., Zipfel C. (2009). Recent advances in PAMP-triggered immunity against bacteria: Pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150: 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecenková T., Hála M., Kulich I., Kocourková D., Drdová E., Fendrych M., Toupalová H., Zársky V. (2011). The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J. Exp. Bot. 62: 2107–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F.G., Tör M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel M.A., Chong Y.T., Haasen K.E., Aldea-Brydges M.G., Stone S.L., Goring D.R. (2009). Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N., Prybylowski K., Petralia R.S., Chang K., Wang Y.X., Racca C., Vicini S., Wenthold R.J. (2003). NMDA receptor trafficking through an interaction between PDZ proteins and the exocyst complex. Nat. Cell Biol. 5: 520–530 [DOI] [PubMed] [Google Scholar]
- Scita G., Di Fiore P.P. (2010). The endocytic matrix. Nature 463: 464–473 [DOI] [PubMed] [Google Scholar]
- Shan L., He P., Li J., Heese A., Peck S.C., Nürnberger T., Martin G.B., Sheen J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Stone S.L., Arnoldo M., Goring D.R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Synek L., Schlager N., Eliás M., Quentin M., Hauser M.T., Zárský V. (2006). AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J. 48: 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush D.R., Maurice T., Roth D., Novick P. (1996). The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 15: 6483–6494 [PMC free article] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Trujillo M., Shirasu K. (2010). Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 13: 402–408 [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Pearce G., Ryan C.A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 103: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts:A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zárský V., Cvrcková F., Potocký M., Hála M. (2009). Exocytosis and cell polarity in plants - Exocyst and recycling domains. New Phytol. 183: 255–272 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]