The targeting of proteins to specific locations within a cell is a dynamic process that requires sophisticated sorting mechanisms. This work shows that Arabidopsis KEEP ON GOING plays a central role in regulating movement of proteins from the plasma membrane to the central vacuole and in the secretion of defense proteins to the extracellular space.
Abstract
In plants, the trans-Golgi network and early endosomes (TGN/EE) function as the central junction for major endomembrane trafficking events, including endocytosis and secretion. Here, we demonstrate that the KEEP ON GOING (KEG) protein of Arabidopsis thaliana localizes to the TGN/EE and plays an essential role in multiple intracellular trafficking processes. Loss-of-function keg mutants exhibited severe defects in cell expansion, which correlated with defects in vacuole morphology. Confocal microscopy revealed that KEG is required for targeting of plasma membrane proteins to the vacuole. This targeting process appeared to be blocked at the step of multivesicular body (MVB) fusion with the vacuolar membrane as the MVB-associated small GTPase ARA6 was also blocked in vacuolar delivery. In addition, loss of KEG function blocked secretion of apoplastic defense proteins, indicating that KEG plays a role in plant immunity. Significantly, KEG was degraded specifically in cells infected by the fungus Golovinomyces cichoracearum, suggesting that this pathogen may target KEG to manipulate the host secretory system as a virulence strategy. Taking these results together, we conclude that KEG is a key component of TGN/EE that regulates multiple post-Golgi trafficking events in plants, including vacuole biogenesis, targeting of membrane-associated proteins to the vacuole, and secretion of apoplastic proteins.
INTRODUCTION
The post-Golgi endomembrane system in eukaryotic cells consists of a complex collection of organelles involved in modifying and trafficking membranes and proteins (Hanton et al., 2007; Hwang and Robinson, 2009; Murphy et al., 2009; Scheuring et al., 2011). In plant cells, there are only two types of well-described endosomal compartments, the trans-Golgi network/early endosome (TGN/EE) and the multivesicular body (MVB), which is sometimes referred to as the prevacuolar compartment (PVC). The TGN/EE is a highly dynamic endomembrane organelle derived from the trans-most Golgi stacks and functions as a major hub for both endocytic and secretory pathways, including receiving endocytosed materials from the plasma membrane (PM) and biosynthetic cargo molecules from the Golgi and sorting them to the PM, cell exterior, or vacuoles (Dettmer et al., 2006; Otegui et al., 2006; Hanton et al., 2007; Lam et al., 2007; Reichardt et al., 2007; Chow et al., 2008; Toyooka et al., 2009; Viotti et al., 2010; Kang et al., 2011; Reyes et al., 2011; Scheuring et al., 2011). The TGN/EE also plays an essential role in transmitting signals initiated by diverse receptor families that regulate processes, such as growth and pathogen resistance (Robatzek et al., 2006; Geldner et al., 2007; Geldner and Robatzek, 2008; Murphy et al., 2009).
Proteins associated with the TGN/EE are required for proper vacuole biogenesis, cell expansion, senescence, and responses to various hormones and environmental stresses. For example, VHA-a1, a TGN/EE-localized vacuolar-type H+-ATPase, modulates cell expansion and salt sensitivity and functions to maintain proper Golgi morphology (Dettmer et al., 2006; Brüx et al., 2008; Krebs et al., 2010). The TGN/EE resident protein ECHIDNA (ECH) is an Arabidopsis thaliana ortholog of yeast Tlg2p-vesicle protein 23 and is required for cell expansion, protein secretion, and TGN integrity (Gendre et al., 2011). VACUOLAR PROTEIN SORTING45 (VPS45), a member of the Sec1p family from Arabidopsis, interacts with the SYP41/SYP61/VTI12 SNARE complex at the TGN/EE to regulate vacuolar protein sorting and plays a potential role in autophagy (Bassham et al., 2000; Surpin et al., 2003; Zouhar et al., 2009).
Several TNG/EE-localized proteins in Arabidopsis have been implicated in secretion and defense against pathogens. For example, mutations in the TGN/EE protein HopM interactor7 compromise resistance to Pseudomonas syringae (Nomura et al., 2006, 2011). HopM interactor7 is an ADP ribosylation factor guanine nucleotide exchange factor protein and has been shown to modulate vesicle trafficking and polar deposition of callose in response to infection by bacterial pathogens (Nomura et al., 2006,Nomura et al., 2011). Similarly, SYP42 and SYP43, TGN/EE-localized Q-SNARE proteins, play important but redundant roles in secretion, vacuolar transport, and resistance to a fungal pathogen (Aoyama and Chua, 1997; Brocard et al., 2002; Uemura et al., 2012).
The KEG protein of Arabidopsis localizes mainly to the TGN/EE and appears to be multifunctional based on its unique collection of domains consisting of a RING finger E3 ligase domain, a kinase domain, nine Ankyrin repeats, and 12 HERC2-like repeats (Stone et al., 2006; Gu and Innes, 2011). Previously, KEG has been implicated in regulation of abscisic acid signaling via degradation of the transcription factor ABSCISIC ACID INSENSITIVE5 (ABI5) (Stone et al., 2006; Liu and Stone, 2010). However, little is known about the function of KEG at the subcellular level. Here, we demonstrate that KEG plays a central role in vacuolar biogenesis and the endomembrane trafficking system, including regulation of protein secretion and targeting of membrane proteins to the vacuole for degradation. In addition, we found that KEG is selectively degraded in cells infected with a powdery mildew fungus, suggesting that KEG plays a role in plant immunity.
RESULTS AND DISCUSSION
KEG Is Essential for Cell Expansion
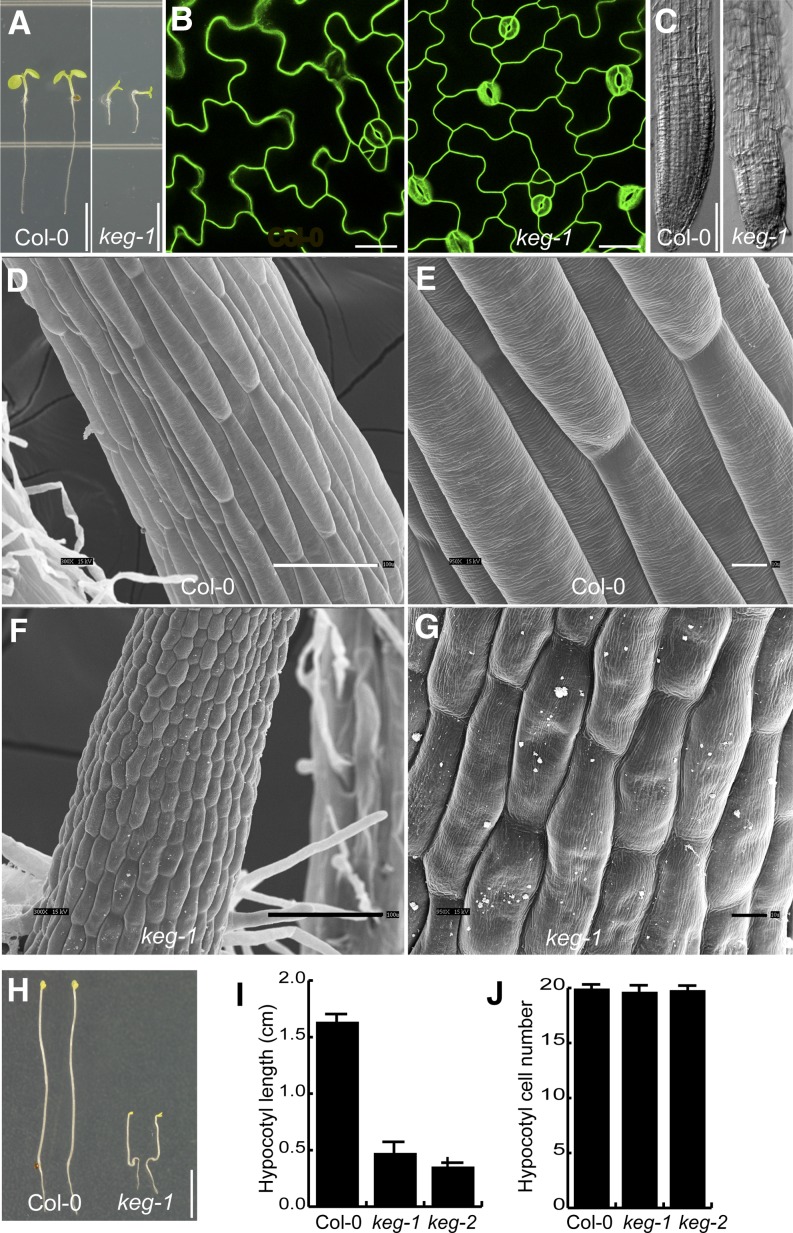
The T-DNA insertion mutants keg-1 and keg-2 show a severely stunted growth phenotype and are seedling lethal (Figure 1A; Stone et al., 2006; Supplemental Table 1 online for list of primers used to genotype mutants). This phenotype has been proposed to result, in part, from enhanced abscisic acid signaling due to the accumulation of ABI5 (Stone et al., 2006). However, regulation of ABI5 levels cannot be the only function of KEG, as overaccumulation of ABI5 alone is not sufficient to induce growth arrest and loss-of-function mutations in ABI5 only partially restore seedling viability to the keg-1 mutant (Brocard et al., 2002; Lopez-Molina et al., 2003; Stone et al., 2006). To gain additional insight into the underlying causes of growth inhibition in keg mutant plants, we performed confocal laser scanning microscopy (CLSM) and scanning electron microscopy analyses on keg mutant seedlings. CLSM analysis of cotyledon epidermal cells revealed that keg-1 seedlings have smaller cells with smaller lobes than wild-type Columbia-0 seedlings (Figure 1B). Reduced cell size in keg-1 seedlings was also apparent in root epidermal cells, with the cell pattern appearing irregular at the root tip (Figure 1C). Scanning electron microscopy analyses of hypocotyl cells revealed severe anisotropic cell expansion defects in the keg-1 mutant, with apparent alterations in cell surface texture, suggesting defects in cell wall structure (Figures 1D to 1G). These analyses were also performed on keg-2 mutant seedlings, with identical results, establishing that these defects are due to loss of KEG function and not a second site mutation (data not shown). Hypocotyl elongation of etiolated keg-1 and keg-2 seedlings was reduced by ∼75% compared with the wild type, whereas hypocotyl cell number was the same (Figures 1H to 1J; Stone et al., 2006), indicating that loss of KEG function causes a defect in cell expansion rather than cell division.
Figure 1.
Arabidopsis keg Mutants Are Defective in Cell Expansion.
(A) Five-day-old seedlings grown on half-strength MS. Col-0, Columbia-0. Bars = 5 mm.
(B) Cotyledon epidermal cells of 5-d-old seedlings expressing the PM marker BRI1-sYFP. Bars = 25 μm.
(C) Root tips of 5-d-old seedlings. Bars = 100 μm.
(D) to (G) Scanning electron micrographs of hypocotyls of 5-d-old seedlings grown in the presence of 1% Suc. Bars = 100 μm in (D) and (F) and 10 μm in (E) and (G).
(H) Seven-day-old etiolated seedlings. Bar = 5 mm.
(I) Hypocotyl length of 7-d-old etiolated seedlings. Data represent means ± sd (n = 30).
(J) Hypocotyl cell number of 7-d-old etiolated seedlings. Data represent means ± sd (n = 30).
[See online article for color version of this figure.]
Vacuole Development Is Compromised in keg Mutants
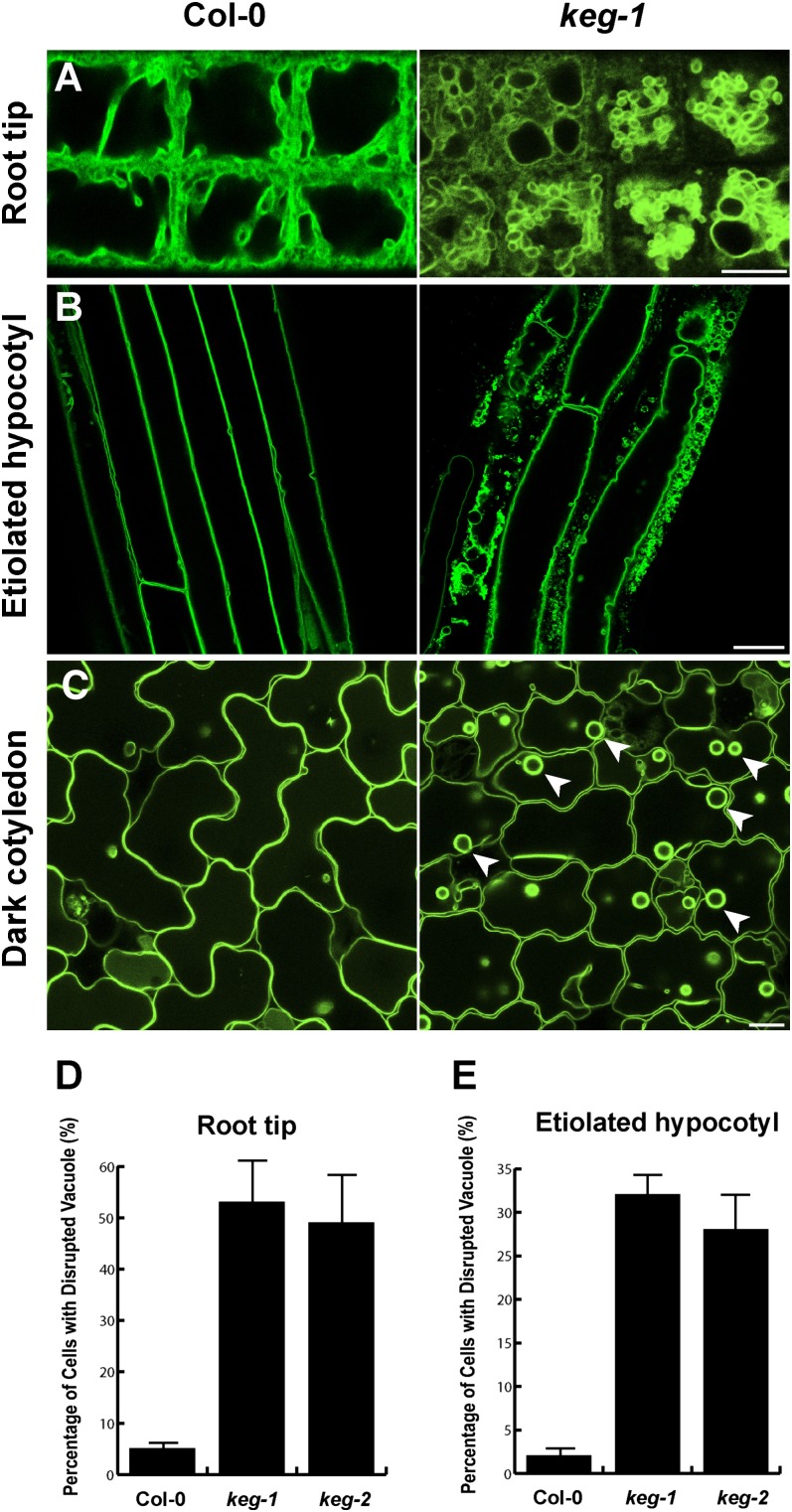
Cell expansion in plants is dependent on turgor pressure provided by the central vacuole. We thus investigated whether the cell expansion defect in keg mutant seedlings could be related to a defect in vacuole structure. Using the fluorescent tonoplast marker γ-TIP-mCherry (Nelson et al., 2007) to image vacuoles, we observed alterations in vacuole structure in the root elongation zone, hypocotyl, and cotyledon epidermal cells of keg-1 mutant seedlings (Figure 2; see Supplemental Figure 1 online). Approximately half of the cells in the root elongation zone of keg-1 seedlings contained multiple smaller vacuoles rather than a single large central vacuole (Figures 2A and 2D), indicating a defect in vacuole biogenesis. Similarly, vacuoles in hypocotyl cells of etiolated keg-1 seedlings were severely disrupted, with accumulation of numerous vesicles and aberrant compartments (Figures 2B and 2E). Tonoplasts in keg-1 cotyledon and hypocotyl epidermal cells were relatively intact, but with a more ruffled appearance. In addition, many bulb-like structures were observed in keg-1 epidermal cells (Figure 2C, arrowheads). Although the mechanism by which these bulbs are formed is not clear (Saito et al., 2002), their appearance in the keg-1 mutant is consistent with defects in membrane fusion processes. In sum, these observations suggest that the defect in cell expansion in keg seedlings is caused at least in part by defects in vacuole formation and function.
Figure 2.
The Development of Central Vacuoles Is Compromised in keg Mutants.
(A) Confocal microscopy images of vacuoles in the root elongation zone of 5-d light-grown 35S:γ-TIP-mCherry transgenic seedlings. Col-0, Columbia-0.
(B) Confocal microscopy images of vacuoles in hypocotyl of 7-d dark-grown 35S:γ-TIP-mCherry transgenic seedlings.
(C) Confocal microscopy images of vacuoles in cotyledons of 7-d dark-grown 35S:γ-TIP-mCherry transgenic seedlings.
(D) Percentage of cells with fragmented vacuoles in the root elongation zone shown in (A). Data represent means ± sd (n = 30).
(E) Percentage of cells with disrupted vacuoles in etiolated hypocotyl shown in (B). Data represent means ± sd (n = 30).
Bars = 10 μm.
[See online article for color version of this figure.]
KEG Localizes to the TGN/EE in Arabidopsis
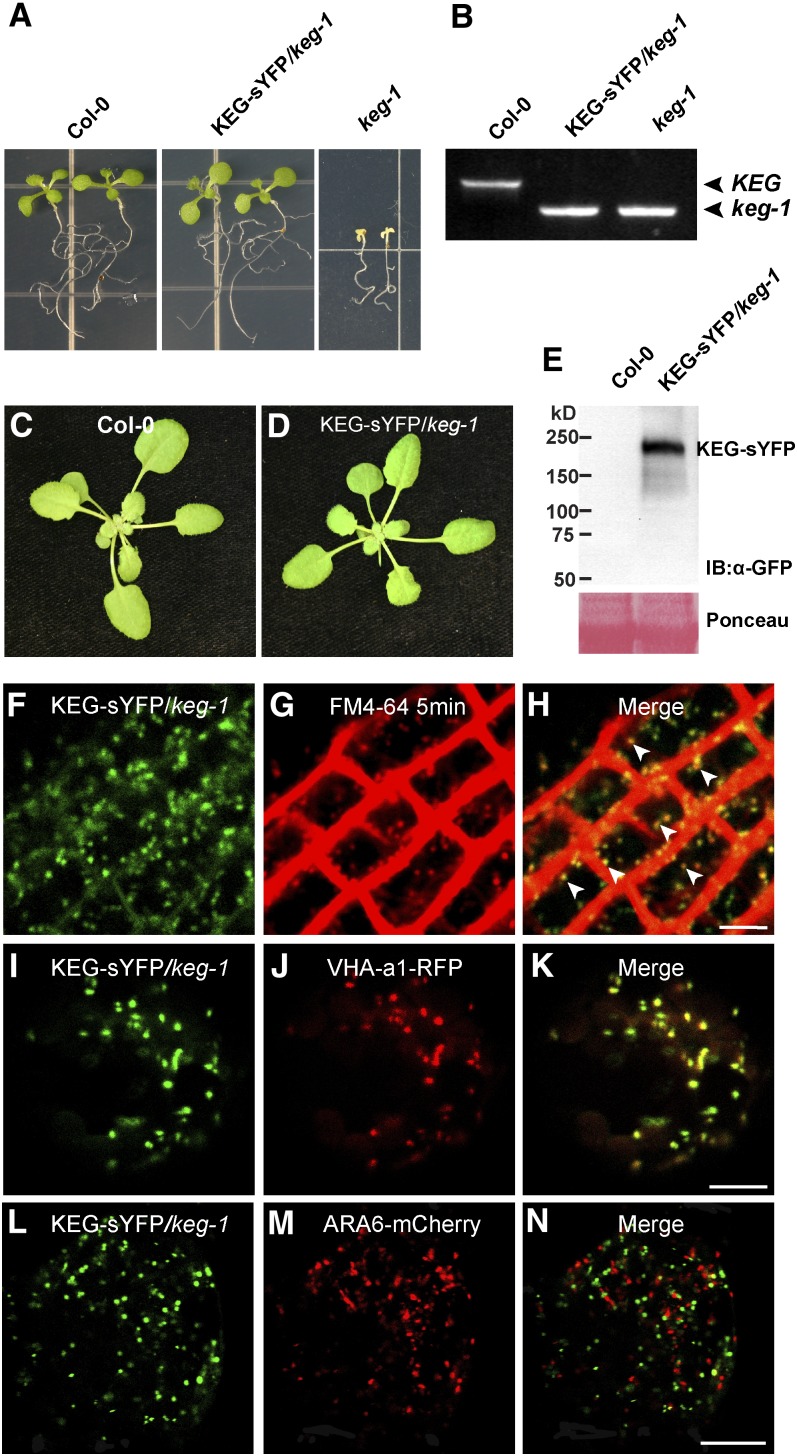
Previously, we reported that KEG localizes to TGN/EE vesicles defined by the Arabidopsis syntaxin protein SYP61 when transiently overexpressed in Nicotiana benthamiana (Gu and Innes, 2011). To confirm that KEG localizes to the TGN/EE in Arabidopsis, we examined KEG subcellular localization in transgenic Arabidopsis lines. KEG fused with super yellow fluorescent protein (KEG-sYFP) driven by its native promoter cannot be detected by conventional fluorescence microscopy (Gu and Innes, 2011), and overexpression of KEG under a steroid-inducible promoter causes massive cell death in transgenic Arabidopsis (Wawrzynska et al., 2008). We thus transformed keg-1 mutant Arabidopsis with KEG-sYFP driven by a 35S promoter and conducted an extensive screen for transformants that expressed KEG-sYFP at a detectable level and that appeared morphologically wild type. We obtained several such lines in a homozygous keg-1 mutant background, indicating that KEG-sYFP is functional and that its subcellular distribution must at least partially overlap with that of the endogenous KEG (Figures 3A to 3E).
Figure 3.
KEG-sYFP Fully Complements keg-1 Mutant Phenotypes and Localizes to the TGN/EE.
(A) Ten-day old seedlings of wild-type plants, 35S:KEG-sYFP transgenic keg-1 plants, and nontransgenic keg-1 plants grown on half-strength MS plates. Col-0, Columbia-0.
(B) KEG genotypes of seedlings shown in (A) analyzed by PCR.
(C) and (D) Three-week-old wild-type plants and 35S:KEG-sYFP transgenic keg-1 plants.
(E) KEG-sYFP protein levels in the plants shown in (C) and (D).
(F) to (H) Cells in the root elongation zone of 35S:KEG-sYFP transgenic keg-1 plants stained with the endocytic tracer FM4-64 for 5 min.
(I) to (N) Protoplasts prepared from 35S:KEG-sYFP transgenic keg-1 plants transfected with 35S:VHA-a1-RFP or 35S:ARA6-mCherry. Images were taken 16 h after transfection.
Bars = 5 μm.
In root tip cells of KEG-sYFP transgenic lines, KEG-sYFP localized to intracellular punctate structures that were quickly (<10 min) labeled by the lipophilic dye FM4-64 after the dye’s internalization from the PM (Figures 3F to 3H), indicating that KEG-sYFP localizes to early endosomes. To confirm the identity of these endosomes, we prepared protoplasts from leaves of 35S:KEG-sYFP lines and transfected them with the TGN/EE marker VHA-a1-RFP (for red fluorescent protein) (Dettmer et al., 2006) and the MVB/PVC marker ARA6-mCherry (Ueda et al., 2004; Haas et al., 2007). We observed that KEG-sYFP colocalized almost completely with VHA-a1-RFP but not with ARA6-mCherry (Figures 3I to 3N), confirming that KEG associates with the TGN/EE.
When KEG-sYFP was transiently overexpressed in N. benthamiana in the presence of latrunculin B, which disrupts actin filaments and, thus, TGN/EE movements, we observed a weak PM signal (see Supplemental Movie 1 and Supplemental Figure 2 online). We were unable to see a PM signal in transgenic Arabidopsis plants treated with latrunculin B, but this is likely due to the significantly lower expression level of KEG-sYFP in Arabidopsis. The PM localization in N. benthamiana was enhanced when we expressed a mutant form of KEG lacking E3 ubiquitin ligase activity (Liu and Stone, 2010) (see Supplemental Figure 2 online). Although we cannot rule out that the PM localization of KEG in N. benthamiana is an artifact of overexpression, these data suggest that KEG localization may be regulated in part by its E3 ligase activity.
Loss of KEG Function Does Not Affect Formation of MVBs, Golgi, or Endoplasmic Reticulum
Arabidopsis mutants containing mutations in genes encoding TGN/EE components, such as VHA-a1, ECH, VPS45, and the SYP4 group, share phenotypes with keg mutants in terms of reduced seedling viability and defects in cell expansion (Brüx et al., 2008; Zouhar et al., 2009; Gendre et al., 2011; Uemura et al., 2012). Functional studies of these TGN/EE-associated proteins have shown that they play roles in maintaining endomembrane organelle integrity (Bassham et al., 2000; Surpin et al., 2003; Brüx et al., 2008; Zouhar et al., 2009; Viotti et al., 2010; Gendre et al., 2011; Kim and Bassham, 2011; Uemura et al., 2012). To investigate whether KEG is also necessary for maintaining the structural integrity of major endomembrane organelles beside the vacuole, we generated transgenic keg seedlings constitutively expressing VHA-a1-RFP to label the TGN/EE (Dettmer et al., 2006), GmManI49-mCherry to label Golgi (Nelson et al., 2007), ARA6-sYFP to label MVB/PVC (Ueda et al., 2001,Ueda et al., 2004; Haas et al., 2007), and HDEL-GFP (for green fluorescent protein) to label endoplasmic reticulum (ER) (Nelson et al., 2007). Surprisingly, our CLSM analyses revealed no significant differences between the wild type and the keg-1 mutant in terms of size, shape, number, or movement of these organelles in various tissues (see Supplemental Figures 3 to 5 online). In addition, keg-1 seedlings responded the same as wild-type seedlings to the pharmacological trafficking inhibitors Brefeldin A (BFA) and wortmannin (Wm) with regards to aggregation of VHA-a1-RFP and dilation of ARA6-sYFP–labeled vesicles (see Supplemental Figures 3D, 3H, 4D, and 4H online), further validating the identity of TGN/EEs and MVB/PVCs in keg-1 mutants. Finally, ultrastructure analyses using transmission electron microscopy demonstrated that endomembrane organelles maintained regular structural morphologies in the keg-1 mutant and did not display gross defects (see Supplemental Figures 3 to 5 online). Taken together, these data indicate that KEG is required for proper vacuole development but not for development of TGN/EE, Golgi, MVB/PVC, or ER.
KEG Regulates Vacuolar Transport of PM Proteins
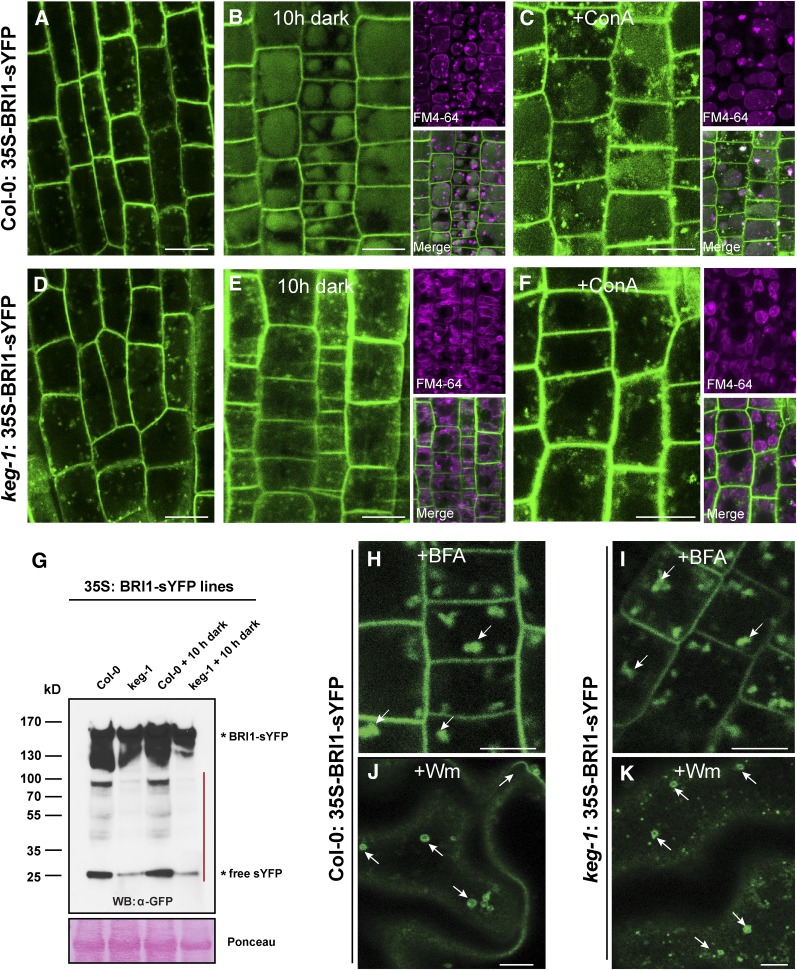
An important functional role of many TGN/EE-associated proteins is regulating intracellular trafficking processes, such as vacuolar protein transport, protein secretion, and autophagy (Bassham et al., 2000; Surpin et al., 2003; Brüx et al., 2008; Zouhar et al., 2009; Viotti et al., 2010; Gendre et al., 2011; Kim and Bassham, 2011; Uemura et al., 2012). To test if KEG plays a role in endomembrane trafficking pathways, we constructed transgenic plants constitutively expressing the brassinosteroid receptor BRI1-sYFP, a well-studied protein that cycles between the PM and TGN/EE (Geldner et al., 2007; Kleine-Vehn et al., 2008; Viotti et al., 2010). As shown in Figure 4, BRI1-sYFP localized to both the PM and endosomal vesicles in both wild-type and keg-1 mutant seedlings with no apparent difference (Figures 4A and 4D). Treatment with BFA induced similar intracellular aggregates in both wild-type and keg-1 mutant seedlings (Figures 4H and 4I), suggesting that KEG is not required for endocytosis of BRI1. Following endocytosis, BRI-1 is targeted for degradation in the vacuole, which can be visualized by fluorescence accumulation in vacuole lumens when plants are incubated in the dark or treated with concanamycin A (ConA), an inhibitor of vacuolar H+-ATPases (Tamura et al., 2003; Kleine-Vehn et al., 2008). As shown in Figure 4B, we observed a clear sYFP signal within vacuoles of wild-type plants after 10 h of dark incubation. By contrast, sYFP was almost undetectable in vacuoles of the keg-1 and keg-2 mutant under the same conditions (Figure 4E; see Supplemental Figure 6 online). We quantified the total BRI1-YFP fluorescence signal in both the PM and in the vacuoles using NIH ImageJ and found vacuolar fluorescence in keg-1 cells was reduced 2.7-fold relative to the wild type, while BRI1-sYFP fluorescence in the PM did not significantly differ (see Supplemental Table 2 online). Consistent with the observed reduction in BRI1-sYFP fluorescence in the vacuoles of keg-1 seedlings, we also observed reduced levels of free sYFP relative to full-length BRI-sYFP, indicating a reduced rate of BRI1-sYFP degradation (Figure 4G). Treatment with ConA yielded a weaker vacuolar BRI1-sYFP accumulation compared with dark treatment in wild-type plants and induced intracellular aggregates, which is a known side effect of ConA on BRI1 trafficking (Dettmer et al., 2006) (Figure 4C). ConA induced similar intracellular aggregates of BRI1-sYFP in keg-1 seedlings, but no vacuolar accumulation (Figure 4F). We also analyzed the vacuolar transport of PIP2A-mCherry, a PM aquaporin protein (Cutler et al., 2000; Nelson et al., 2007; Kleine-Vehn et al., 2008), and observed a similar effect of keg-1 on vacuolar accumulation (see Supplemental Figure 7 online), demonstrating that KEG is required for targeting of multiple PM proteins to the vacuole. These latter data also demonstrate that the reduced BRI1-sYFP fluorescence observed in keg-1 vacuoles is not an artifact caused by potential pH differences between keg-1 and wild-type vacuoles, which would affect sYFP fluorescence.
Figure 4.
KEG Regulates Vacuolar Transport of BRI1.
(A) and (D) Confocal microscopy images of root cells of wild-type (A) and keg-1 mutant (D) 35S:BRI1-sYFP transgenic seedlings grown under a 16-h/8-h light/dark cycle for 5 d and imaged near the end of the light cycle. Col-0, Columbia-0.
(B) and (E) The above seedlings were transferred to the dark ∼6 h into the light cycle on the sixth day and imaged 10 h later. These seedlings were also stained with the lipophilic dye FM4-64 to reveal vacuole structures.
(C) and (F) Seedlings treated with 0.5 μM ConA for 10 h.
(G) Total protein was extracted from 5-d-old 35S:BRI1-sYFP transgenic seedlings and immunoblotted with anti-GFP antibody. The red line indicates BRI1-sYFP degradation products.
(H) and (I) Seedlings treated with 100 μg/mL BFA for 1 h. Arrows indicate BFA compartments.
(J) and (K) Seedlings treated with 33 μM Wm for 1 h. Arrows indicate ring-like structures formed by MVB dilation.
Bars = 10 μm.
Vacuolar degradation of PM receptors through the endocytic pathway plays an important role in quenching receptor activities and thus fine-tuning signal output (Robatzek et al., 2006; Geldner et al., 2007; Robatzek, 2007; Kleine-Vehn et al., 2008; Reyes et al., 2011). We thus tested whether the lack of vacuolar degradation in keg mutants affected signaling output by the BRI1 receptor. We found that keg mutant seedlings were more sensitive to brassinolides in terms of root growth inhibition and hypocotyl elongation (see Supplemental Figure 8 online), suggesting that KEG indeed contributes to downregulation of BRI1 signaling, presumably through affecting its vacuolar degradation.
To investigate at what step vacuolar trafficking of BRI1 is impaired in keg mutants, we asked whether BRI1 is sorted to MVBs and whether it is subsequently internalized into intraluminal vesicles (ILVs) of MVBs before delivery to the lytic vacuole. To answer the first question, we treated BRI1-sYFP transgenic plants with the PI3 kinase inhibitor Wm, which induces dilation of MVBs, producing a ring-like appearance in Arabidopsis due to homotypic fusion events (Tse et al., 2004; Jaillais et al., 2006; Lam et al., 2007; Silady et al., 2008; Wang et al., 2009). We observed clear ring-like structures labeled by BRI1-sYFP in cotyledon cells of both wild-type and keg-1 mutant seedlings treated with Wm (Figures 4J and 4K). This result indicates that loss of KEG does not affect BRI1 sorting to the MVB/PVC, although it is possible that some of the ring-like structures are derived from the TGN/EE (Wang et al., 2009). We therefore reasoned that the vacuolar trafficking defect of BRI1 likely occurs after BRI1 is sorted to MVBs.
During the maturation of MVBs, internalized PM proteins are sequestrated to ILVs of MVBs by the ESCRT complex (for endosomal sorting complexes required for transport). Mutations in plant ESCRT-related proteins, such as CHMP1A and CHMP1B, disrupt invagination from the MVB limiting membrane and thus reduce formation of ILVs. As a result, PM cargo cannot be delivered into the vacuole lumen but instead accumulates at the vacuole membrane upon MVB fusion with the vacuole (Spitzer et al., 2009). We therefore assessed whether BRI1-sYFP accumulated at the vacuolar membrane in dark- or ConA-treated keg-1 seedlings (Figures 4E and 4F). However, no obvious accumulation of BRI1-sYFP was observed. Instead, there was an increase in punctate signals in keg-1 seedlings compared with the wild type after dark incubation, suggesting that BRI1-containing endosomes may be accumulating in the dark rather than fusing with the vacuole.
KEG Is Required for Trafficking of ARA6-Associated MVBs to the Lytic Vacuole
A previous electron microscopy study showed that BRI1 can be detected inside MVBs (Viotti et al., 2010), suggesting that BRI1 transits through MVBs on its way to the vacuole. To confirm this prediction, we transiently coexpressed BRI1-sYFP and ARA6-mCherry in both Arabidopsis protoplasts and N. benthamiana epidermal cells. As shown in Supplemental Figure 9 online, intracellular BRI1-sYFP puncta mostly colocalized with ARA6-mCherry–labeled endosomes in both systems, indicating that after endocytosis, BRI1 passes through ARA6-positive MVBs before fusing with the vacuole.
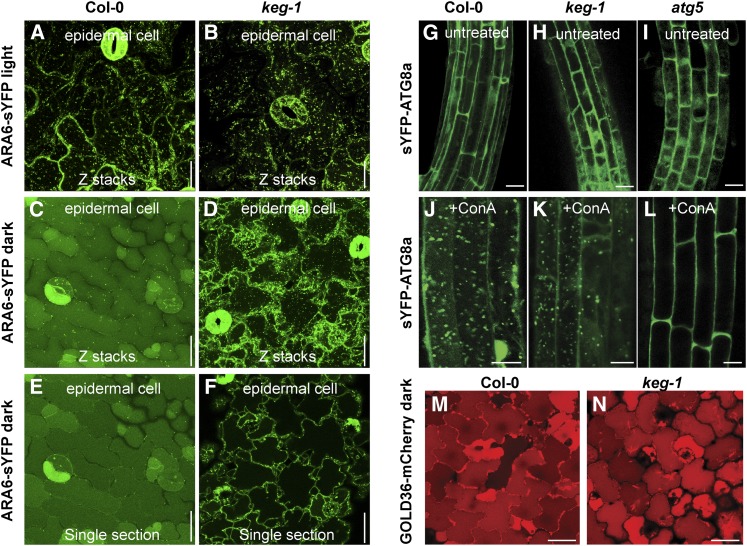
Given that BRI1-containing endosomes accumulated in the keg-1 mutant in the dark, we hypothesized that KEG is required for fusion of MVBs with the vacuole. To test this hypothesis, we examined MVB behavior in 35S:ARA6-sYFP transgenic plants incubated in the dark. ARA6 belongs to the Rab GTPase family and is associated with MVBs through N-terminal acylation (Ueda et al., 2001). In wild-type ARA6-sYFP seedlings, incubation in the dark induced accumulation of a strong fluorescent signal in the vacuolar lumen and a reduction in the number of intracellular puncta (presumptive MVBs) relative to light-grown seedlings (Figure 5). Although the precise mechanism remains unclear, these data demonstrate that ARA6 is deposited into the vacuolar lumen following MVB fusion with the vacuolar membrane in wild-type seedlings. In keg-1 and keg-2 mutant seedlings, by contrast, no fluorescence was detected in the vacuolar lumen of dark-grown seedlings, and the number of intracellular puncta did not decrease relative to light-grown plants, resulting in many more puncta than observed in dark-grown wild-type seedlings (Figure 5; see Supplemental Figure 6 online). Quantification of these puncta in cotyledon epidermal cells of dark-grown seedlings revealed that keg-1 cells contained over 60% more fluorescent puncta than wild-type seedlings (see Supplemental Table 3 online). Taken together, we conclude that KEG is required for transport of ARA6-associated MVBs into vacuoles, which thus affects vacuolar transport of PM proteins that depend on MVBs as carriers.
Figure 5.
KEG Is Required for Vacuolar Targeting of ARA6-Associated MVBs but Not for Vacuolar Targeting of Autophagosomes or the Soluble Cargo Protein GOLD36/MVP1.
(A) to (D) Confocal microscopy images of cotyledon epidermal cells of wild-type ([A] and [C]) and keg-1 ([B] and [D]) transgenic seedlings expressing ARA6-sYFP grown in the light ([A] and [B]) or dark for 7 d ([C] and [D]). Images are Z-stacks of five single sections. Col-0, Columbia-0.
(E) and (F) Single sections from (C) and (D).
(G) to (I) Cytosolic distribution of sYFP-ATG8a in root cells of 5-d-old transgenic seedlings grown on half-strength MS media.
(J) to (L) Visualization of autophagosomes inside vacuole lumens of root cells after incubation of sYFP-ATG8a transgenic seedlings with 0.5 μM ConA for 10 h.
(M) and (N) Vacuolar accumulation of GOLD36/MVP1-mCherry in cotyledon cells of 7-d-old wild-type and keg-1 seedlings grown under dark conditions.
Bars = 10 μm.
[See online article for color version of this figure.]
KEG Is Not Required for Vacuolar Trafficking of Soluble Cargo Proteins, nor for Autophagy
Because KEG appeared to regulate trafficking of MVBs to the vacuole, we asked whether other vesicle trafficking pathways involving vacuolar targeting were compromised in the keg-1 mutant. The first pathway we examined was macroautophagy, which is a major component of the cellular recycling system. Macroautophagy initiates in the cytosol with the de novo assembly of autophagosomes, which are double-membrane vesicles that engulf cytosolic macromolecules, including mitochondria and chloroplasts, and delivers them to the vacuole for disassembly. Similar to MVBs, fusion of autophagosomes with the vacuolar membrane results in release of internal vesicles to the lumen of the vacuole. This transport process can be visualized in roots of transgenic plants expressing an autophagosome membrane component GFP-Autophagy Deficient8a (GFP-ATG8a) by the appearance of fluorescent puncta inside the vacuolar lumen in the presence of ConA, which stabilizes autophagic bodies inside the vacuole (Thompson et al., 2005; Chung et al., 2010; Suttangkakul et al., 2011). A 10-h ConA treatment induced clear vacuole luminal accumulation of autophagosomes labeled by sYFP-ATG8a in both wild-type and keg-1 transgenic seedlings, but not in the autophagy deficient mutant atg5 (Figures 5G to 5L), indicating that KEG is not required for either autophagosome formation or their subsequent fusion with the vacuole.
The second pathway we examined was vacuolar transport of a biosynthetic cargo protein, GOLGI DEFECTS 36/MODIFIED VACUOLE PHENOTYPE 1 (GOLD36/MVP1). GOLD36 is a member of the myrosinase superfamily that resides in the vacuole but is synthesized in the ER and is subsequently transported to the vacuole via an unknown route (Agee et al., 2010; Marti et al., 2010). In contrast with ARA6-sYFP and BRI1-sYFP, steady state accumulation of GOLD36/MVP1-mCherry was not affected by the keg-1 mutation in transgenic plants grown in the dark (Figures 5M and 5N). This finding suggests that delivery of at least some soluble cargo proteins to the vacuole may differ in mechanism from delivery of PM proteins. Taken together, KEG appears to specifically regulate vacuolar transport mediated by ARA6-defined MVBs but not by autophagosomes or GOLD36/MVP1-associated vacuolar transport carriers.
Accumulating evidence indicates that MVBs are formed directly from the TGN/EE through a process of gradual maturation (Ueda et al., 2004; Niemes et al., 2010a,Niemes et al., 2010b; Kang et al., 2011; Scheuring et al., 2011). For example, formation of ESCRT-mediated intraluminal vesicles appears to initiate in the TGN/EE (Scheuring et al., 2011). Because KEG is localized primarily to TGN/EEs and is required for vacuolar transport of MVBs, we hypothesize that during MVB maturation, KEG modifies and regulates MVB-associated proteins, which in turn regulate a subsequent trafficking event, which could be associated with targeting, tethering, docking, or fusion with the vacuole.
KEG Is Required for Apoplastic Protein Secretion
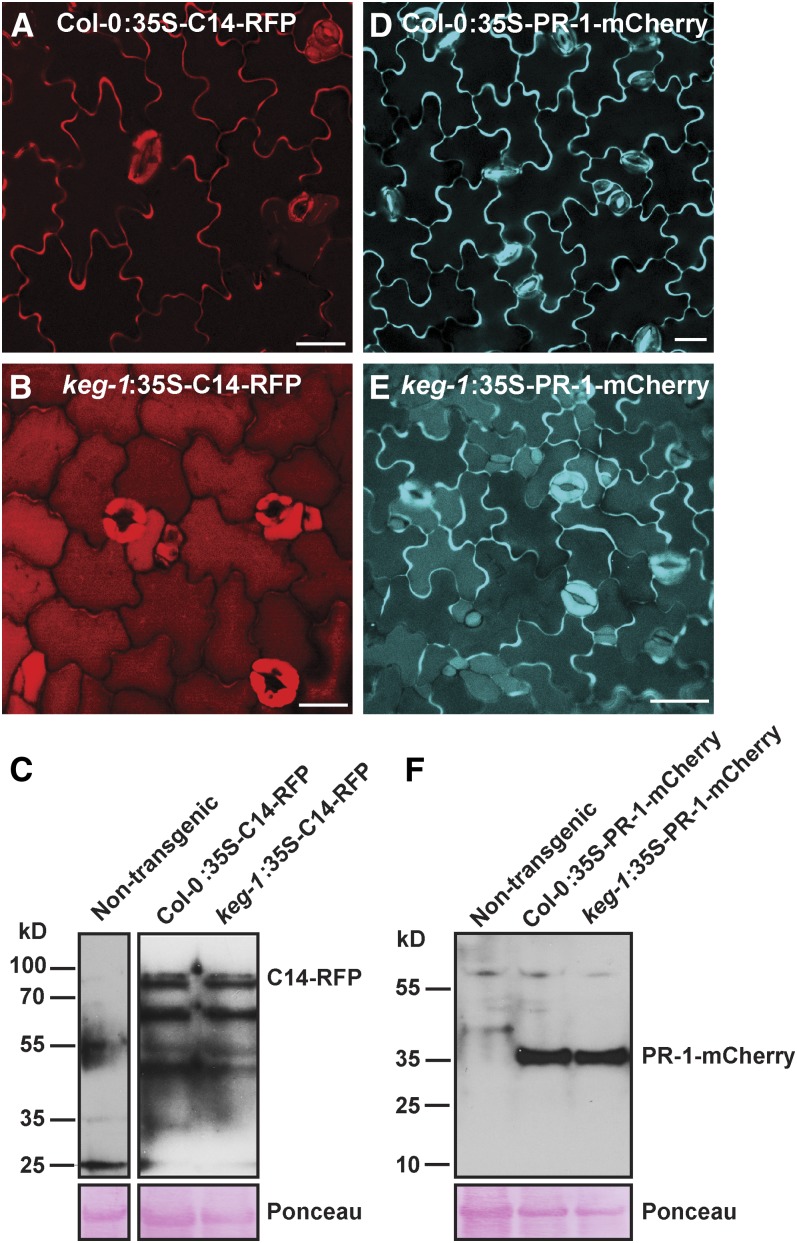
The above data suggested that KEG plays a role in regulating trafficking of TGN/EE-derived vesicles. Since the TGN/EE produces vesicles that are destined for the PM and the vacuole (Viotti et al., 2010; Kang et al., 2011), we also investigated whether KEG is required for secretion of apoplastic proteins. For these analyses, we generated transgenic Arabidopsis lines expressing two different apoplastic defense proteins, tomato (Solanum lycopersicum) C14-Red Fluorescent Protein (C14-RFP; Bozkurt et al., 2011) and PR1-mCherry (van Loon et al., 2006), in wild-type and keg-1 mutant backgrounds and analyzed the steady state accumulation of each in the epidermis of cotyledons. C14-RFP and PR1-mCherry accumulated to high levels in the apoplastic space of wild-type seedlings, with only a weak signal detected in the vacuolar lumen (Figures 6A and 6D). In keg-1 and keg-2 seedlings, by contrast, the C14-RFP signal was almost entirely in the vacuolar lumen with only trace amounts in the apoplast (Figure 6B; see Supplemental Figure 6 online). The PR1-mCherry signal in keg-1 seedlings appeared in both the apoplast and vacuolar lumen, but the vacuolar signal was much stronger than in wild-type seedlings (Figure 6E). Of particular note is that PR1-mCherry fluorescence was quite bright in the central opening of stomata in wild-type seedlings and very faint in keg-1 seedlings (Figures 6D and 6E), suggesting that PR1 accumulates at a potential entry point for pathogens in wild-type plants. Immunoblot analyses using anti-RFP and anti-mCherry antibodies against total protein revealed no differences in terms of protein accumulation pattern or degradation patterns between wild-type and keg-1 transgenic plants (Figures 6C and 6F), suggesting that the mislocalization of these proteins in the keg-1 mutant is not due to protein degradation and that the majority of the observed signal is derived from full-length proteins.
Figure 6.
KEG Is Required for Efficient Secretion of Apoplastic Defense Proteins.
(A) and (B) Confocal microscopy images of cotyledon epidermal cells of 5-d-old 35S:C14-RFP transgenic plants. Col-0, Columbia-0.
(C) Anti-RFP immunoblot against total proteins from 35S:C14-RFP transgenic seedlings.
(D) and (E) Confocal microscopy images of cotyledon epidermal cells of 5-d-old 35S:PR-1-mCherry transgenic plants.
(F) Anti-mCherry immunoblot against total proteins from 35S:PR-1-mCherry transgenic seedlings.
Bars = 10 μm.
[See online article for color version of this figure.]
The above data indicate that KEG is required for proper secretion of C14 and PR-1. However, we did not observe any defects in the PM delivery of BRI1, PIN1, and AUX1 in the keg-1 mutant (Figure 4D; see Supplemental Figure 10 online); thus, KEG may be required specifically for secretion of proteins into the apoplastic space, suggesting that vesicles delivering secreted proteins may differ from those delivering PM-localized proteins.
KEG Is Specifically Degraded during Infection by a Powdery Mildew Fungus
The failure of keg-1 mutant plants to secrete defense proteins suggested that KEG may play a role in pathogen defense. This supposition is also supported by our previous work in which we showed that a missense mutation in the C-terminal HERC2-like repeats of KEG (keg-4) can suppress enhanced disease resistance1–mediated enhanced resistance to the fungal pathogen Golovinomyces cichoracearum (causal agent of powdery mildew on Arabidopsis; Wawrzynska et al., 2008). G. cichoracearum is a host adapted pathogen that colonizes the leaf surface of Arabidopsis and establishes intimate contact with host cells by forming feeding structures called haustoria in which the host cell PM and fungal cell PM are brought into close proximity. Haustoria are the sites at which fungal effector proteins are secreted to manipulate the metabolism and defense responses of host cells and where the plant directs defense proteins; thus, haustoria are sites of greatly elevated endomembrane trafficking (Panstruga and Dodds, 2009; Leborgne-Castel et al., 2010).
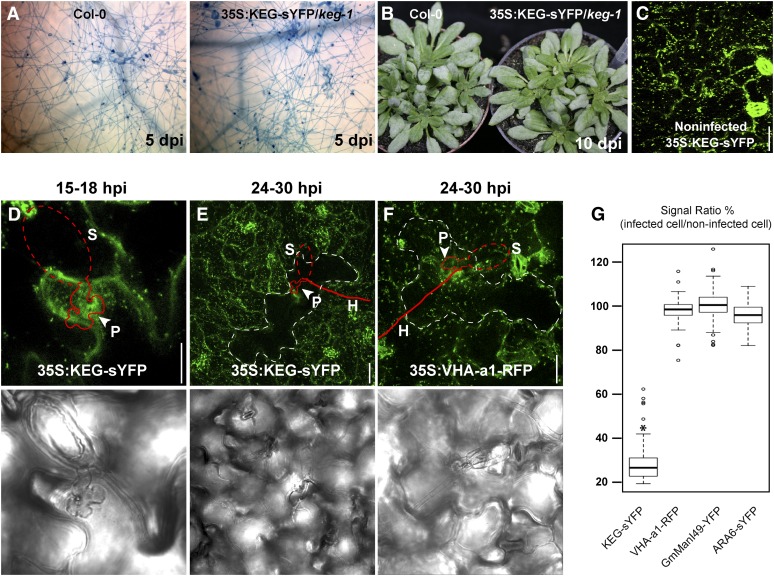
To investigate whether KEG plays a role in haustorial-associated endomembrane trafficking, we tracked the subcellular dynamics of KEG during G. cichoracearum infection using 35S:KEG-sYFP transgenic lines. As controls, we inoculated three organelle marker lines, VHA-a1-RFP (TGN/EE), ARA6-sYFP (MVB/PVC), and GmManI49-YFP (Golgi), in parallel. Growth of G. cichoracearum on the 35S:KEG-sYFP lines was indistinguishable from that on wild-type plants in terms of fungal hyphae growth and macroscopic symptoms (powder production; Figures 7A and 7B). Subcellularly, KEG-sYFP was associated with broadly distributed vesicles in epidermal cells prior to infection (Figure 7C). However, ∼15 to 18 h postinoculation (hpi), KEG became localized to fungal penetration sites (Figure 7D), suggesting that KEG plays a role in defense during early infection stages. Significantly, ∼24 to 30 hpi, when haustoria mature, the KEG-sYFP signal, but not the VHA-a1-RFP signal, became depleted specifically in infected cells (Figures 7E and 7F). Quantification of fluorescence signal revealed that the KEG-sYFP signal was downregulated more than 70% in infected cells versus noninfected cells, but the fluorescence signal of other markers remained unchanged after pathogen infection (Figure 7G). These data demonstrate that TGN/EEs accumulate around fungal penetration sites and, most significantly, that KEG is specifically targeted for degradation following the maturation of fungal haustoria. Degradation of KEG would then inhibit secretion of defense proteins, favoring further growth of the pathogen.
Figure 7.
KEG Is Specifically Degraded in Powdery Mildew Infected Cells.
(A) Trypan blue staining of G. cichoracearum fungal hyphae on Arabidopsis leaves 5 d postinoculation (dpi). Col-0, Columbia-0.
(B) Fungal growth on 4-week-old plants at 10 d postinoculation.
(C) Distribution of KEG-sYFP in uninfected epidermal cells of transgenic Arabidopsis plants.
(D) Localization of KEG-sYFP to the penetration site of G. cichoracearum at 15 to 18 hpi.
(E) and (F) Specific degradation of KEG-sYFP (E) but not VHA-a1-RFP labeled TGN/EE (F) in G. cichoracearum infected cells at 24 to ∼30 hpi. White dashed lines outline cells penetrated by the fungus. Arrowheads indicate pathogen penetration sites, and red lines outline the penetrating fungus as observed by standard light microscopy in the bottom panels. H, hyphae; P, penetration site; S, spore. Each image is a Z-stack of six sections.
(G) Ratio of fluorescence signal in pathogen infected cells versus adjacent noninfected cells. The total fluorescence signal in each of 70 infected cells was measured for each transgenic line using NIH ImageJ and compared with the total fluorescence in an equal-sized field taken from adjacent noninfected cells. Results are provided as means with 25th and 75th percentiles (box) and range (whiskers). Statistical outliers are shown as circles. Asterisk denotes significantly different value from the three controls (P < 0.001; F test).
Bars = 20 μm.
Because KEG is degraded following haustoria formation, we speculate that it is targeted by a fungal effector protein. In this context, it is noteworthy that an oomycete effector protein (Avrblb2 from Phytophthora infestans) has recently been identified that inhibits secretion of defense proteins (Bozkurt et al., 2011). Avrblb2 promotes vacuolar accumulation of C14 in N. benthamiana (Bozkurt et al., 2011), reminiscent of the effect of losing KEG function in Arabidopsis. Additionally, degradation of KEG would be expected to lead to accumulation of proteins in the cytoplasm due to inhibition of vacuolar targeting, which might also benefit the pathogen.
A Model for KEG’s Role in Regulating Protein Trafficking
Our data demonstrate that KEG plays a role in trafficking of TGN/EE-derived vesicles to both the vacuole and PM. KEG contains a functional RING E3 ubiquitin ligase domain and a kinase domain (Stone et al., 2006), making it plausible that KEG regulates vesicle trafficking via ubiquitination and/or phosphorylation of other TGN/EE-associated proteins. The E3 ligase activity of KEG is essential for KEG function as the KEGAA RING domain mutant only partially complements the keg-1 mutation, with plants producing a few true leaves, but remaining severely dwarfed and sterile (Liu and Stone, 2010). By contrast, kinase activity is not essential for KEG function, as a kinase inactive KEG variant (Lys176Arg) can fully complement the keg-1 mutation (Liu and Stone, 2010). Ubiquitination is a well-documented mechanism for regulating trafficking of membrane proteins in yeast and animals, with both endocytosis and sorting to MVBs being controlled by ubiquitination (Katzmann et al., 2001; Mukhopadhyay and Riezman, 2007; Lauwers et al., 2009; Wollert and Hurley, 2010). Targeting of PM proteins to the vacuole in plants also appears to require ubiquitination. For example, the borate transporter BOR1 of Arabidopsis is ubiquitinated in response to excess borate, and blockage of ubiquitination (by mutation of the target site on BOR1) blocks translocation of BOR1 from an early endosomal compartment to MVBs (Kasai et al., 2011). Similarly, ubiquitination of the Arabidopsis iron transporter IRT1 is required for its endocytosis and subsequent targeting to the vacuole (Barberon et al., 2011). The essential role of ubiquitination in regulating protein trafficking in plants is also demonstrated by mutations in the Arabidopsis deubiquitinating enzyme AMSH3. AMSH3 has been shown to interact with classical ESCRT-III components and is required for the formation of a central lytic vacuole, proper sorting of vacuolar cargo proteins, and trafficking of the PM protein PIN2 to the vacuole (Isono et al., 2010; Katsiarimpa et al., 2011).
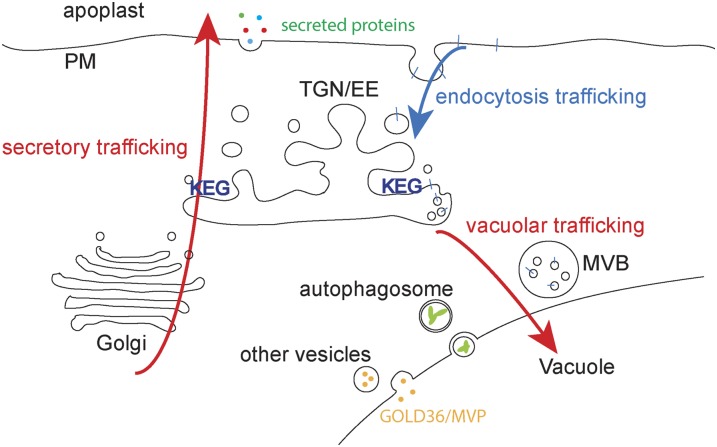
We propose that KEG functions as a TGN/EE resident E3 ligase to ubiquitinate either the proteins being sorted (e.g., PM proteins being targeted to the vacuole) and/or other proteins involved in directing traffic (e.g., members of the ESCRT complex) (Figure 8). In the case of PM proteins being targeted to the vacuole, this ubiquitination is required for targeting of MVBs containing these PM proteins to the vacuole. In the case of defense proteins targeted to the apoplast, this ubiquitination would be necessary to direct the vesicles containing these proteins to the PM as opposed to the vacuole. KEG does not appear to regulate trafficking of proteins that do not pass through the TGN/EE, however, as the keg-1 mutation did not affect autophagy, nor delivery of GOLD36/MVP. Initial endocytosis of PM proteins also does not appear to require KEG, even though KEG itself may partially localize to the PM. Proteins with the structure of KEG (i.e., containing an N-terminal RING domain followed by a kinase domain, ankyrin repeats, and HERC2-like repeats) appear to be unique to plants (Stone et al., 2006), suggesting that regulation of endomembrane trafficking in plants has evolved additional complexities relative to yeast and mammals.
Figure 8.
Model Showing Endomembrane Trafficking Processes Regulated by KEG.
KEG resides on the TGN/EE where it regulates secretion of defense proteins from the Golgi to the apoplast and trafficking of PM proteins through MVBs to the vacuole (red arrows). KEG does not appear to play a role in endocytosis from the PM (blue arrow), in the formation of autophagosomes or their delivery to the vacuole, nor in the delivery of at least some soluble cargo proteins to the vacuole, such as GOLD36/MVP1.
METHODS
Plant Growth Conditions
Arabidopsis thaliana seeds were surface sterilized with 50% (v/v) bleach and 0.1% Triton X-100. Two days after stratification at 4°C, seeds were germinated and grown on half-strength Murashige and Skoog (MS) medium containing 0.8% agar without added sugar (except where indicated) under a photoperiod of 16 h light and 8 h dark at 22°C and a light intensity of ∼100 µE m−2. For pathogen assays, 7-d-old seedlings were transferred from half-strength MS plates to MetroMix 360 soil (Sun Gro Horticulture) and grown under a photoperiod of 9 h light and 15 h dark at 22°C and a light intensity of ∼150 µE m−2 for another 3 weeks before inoculation. Nicotiana benthamiana plants used for transient protein expression were grown under the same short-day conditions, except that seeds were planted directly into MetroMix 360. For dark-grown conditions, seeds were stratified on agar in the dark for 2 d, exposed to light for 4 h before being wrapped in a double layer of aluminum foil, and then germinated and grown for 7 d at 22°C. For hypocotyl growth measurements, visualization of ARA6-sYFP, γ-TIP-mCherry, and GOLD36/MVP1-mCherry, images were recorded immediately after removal from the dark. Assessment of dark-induced BRI1-sYFP and PIP2A-mCherry vacuolar degradation was performed as previously described (Kleine-Vehn et al., 2008). In brief, seedlings were grown on half-strength MS agar as described above for 5 d, and on the sixth day, seedlings were transferred to the dark ∼6 h into the light cycle and incubated for 10 h in the dark and then imaged.
Pathogen Assays
Golovinomyces cichoracearum strain UCSC1 was maintained on hypersusceptible Arabidopsis pad4-2 mutant plants. Plants were inoculated between 3 and 4 weeks of age by gently brushing the leaves of diseased plants onto healthy plants to pass the conidia (asexual spores). Trypan blue staining of fungal hyphae was performed as previously described (Frye and Innes, 1998).
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
The RING E3 ubiquitin ligase domain of KEG was mutated using a QuikChange II site-directed mutagenesis kit (Agilent Technologies) using primers described in Liu and Stone (2010) to generate C29A/H31A amino acid substitutions (KEGAA). cDNA clones of ATG8a, BRI1, ARA6, PR-1, and GOLD36/MVP1 and DNA constructs of 35S:γ-TIP-mCherry, 35S:GmManI49-mCherry and 35S:PIP2A-mCherry (Nelson et al., 2007) were provided by the ABRC at Ohio State University. Fusion of fluorescent proteins to KEG, KEGAA, BRI1, ATG8a, ARA6, PR-1, and GOLD36/MVP1 was accomplished using a multisite Gateway cloning strategy (Invitrogen) as described previously (Gu and Innes, 2011). For stable transformation of Arabidopsis, we used 35S promoter–driven pEarleyGate100 (Earley et al., 2006) as the destination vector in BRI1-sYFP, PR-1-mCherry, and GOLD36/MVP1-mCherry transgenic lines and pMDC32 (Qi and Katagiri, 2009) as the destination vector in KEG-sYFP, ARA6-sYFP, and sYFP-ATG8a transgenic lines. For transient expression in N. benthamiana, we used pEarleyGate100 as destination vector for ARA6-mCherry, BRI1-sYFP, and VHA-a1-RFP and steroid-inducible promoter pTA7002 (Aoyama and Chua, 1997) as destination vector for KEG-sYFP, KEGAA-sYFP, and KEG-mCherry.
Plasmids were transformed into Agrobacterium tumefaciens strain GV3101 (pMP90) by electroporation with selection on Luria-Bertani plates containing 50 μg/mL kanamycin sulfate (Sigma-Aldrich) and 20 μg/mL gentamycin (Gibco). For generating 35S:γ-TIP-mCherry, 35S:GmManI49-mCherry, 35S:PIP2A-mCherry, 35S:BRI1-sYFP, 35S:ARA6-sYFP, 35S:KEG-sYFP, 35S:GOLD36-mCherry, 35S:PR-1-mCherry, and 35S:sYFP-ATG8a transgenic lines, Arabidopsis plants were transformed using the floral dip method (Clough and Bent, 1998). As keg-1 (Salk_049542) and keg-2 (Salk_018105) homozygous plants are seedling lethal, heterozygous plants were used for transformation. T1 generation plants containing the transgene were selected either by growing on half-strength MS with 0.8% agar and 30 μg/mL hygromycin B (Sigma-Aldrich) or by spraying 1-week-old seedlings with 300 μM BASTA (Finale) three times in 2-d intervals. Transgenic T1 plants were then genotyped for the presence of keg-1 or keg-2 mutations using PCR. T2 generation plants homozygous for keg-1 and keg-2 were then identified by seedling morphology and used for microscopy, with their wild-type-appearing siblings were used as controls. 35S:HEDL-GFP, 35S:VHA-a1-RFP, 35S:C14-RFP, pPIN1-PIN1-GFP, and pAUX1-AUX1-GFP transgenic lines were generated through genetic crosses with keg-1 and keg-2 heterozygous T-DNA insertion lines. Primers used for creating plasmid constructs and genotyping are listed in Supplemental Table 1 online.
Transient Protein Expression
For transient protein expression in N. benthamiana, Agrobacterium GV3101 (pMP90) strains transformed with indicated constructs were grown and prepared for transient expression as previously described (Wroblewski et al., 2005). Briefly, Agrobacterium cultures were resuspended in water at an OD600 of 0.8. Suspensions were mixed in equal ratios for coexpression. Bacterial suspension mixtures were infiltrated using a needleless syringe into expanding leaves of 4-week-old N. benthamiana plants. For 35S-driven constructs, CLSM was performed 2 d after infiltration. For steroid-inducible constructs, leaves were sprayed with 50 mM dexamethasone (Sigma-Aldrich) 40 h after injection, and microscopy imaging was performed 24 h after hormone application. For transient protein expression in Arabidopsis protoplasts, protoplasts were prepared from 35S:KEG-sYFP transgenic plants following a previously published procedure (Sheen et al., 1995), and 10 μg of either 35S:VHA-a1-RFP or 35S:ARA6-mCherry plasmid was used for transfection. CLSM was performed 18 to ∼24 h after transfection.
Drug Treatments
Solutions of BFA (Invitrogen; 100 μg/mL), Wm (Sigma-Aldrich; 30 μM), or latrunculin B (Calbiochem; 25 μM) were used to treat Arabidopsis seedlings or N. benthamiana leaves, and images were taken 1 h after exposure to BFA or Wm. ConA (0.5 μM) was added in half-strength MS plates on which plants were grown for indicated times. To stain membranes with FM4-64 dye (Invitrogen), whole Arabidopsis seedlings were incubated in 2 μM FM4-64 for 5 min, washed with water three times, and incubated in the dark at room temperature for the indicated times.
CLSM
To image fluorescent protein fusions in live cells, CLSM was performed using a Leica SP5 AOBS inverted confocal microscope (Leica Microsystems) equipped with ×20 numerical aperture 0.7 and ×63 numerical aperture 1.3 water objectives. GFP fluorescence (excited by the 488-nm argon laser) was detected using a custom 495- to 550-nm band-pass emission filter, sYFP fluorescence (excited by the 514-nm argon laser) was detected using a custom 522- to 545-nm band-pass emission filter, and mCherry and RFP fluorescence (excited using 561-nm He-Ne laser) was detected using a custom 595- to 620-nm band-pass emission filter. To obtain three-dimensional images and movies, a series of Z-stack images were collected and processed by three-dimensional image analysis software (IMARIS 7.0; Bitplane Scientific Software). Quantification of total fluorescence in individual cells was accomplished using NIH ImageJ (Schneider et al., 2012).
Transmission and Scanning Electron Microscopy
Seedlings were fixed with 2% glutaraldehyde and 4% paraformaldehyde in 100 mM sodium phosphate buffer, pH 7.2. Samples were postfixed with 2% (w/v) osmium tetroxide, dehydrated using ethanol, and embedded in Spurr’s resin (Electron Microscopy Sciences). Thin sections were stained with 3% uranyl acetate followed by 0.4% lead citrate. The samples were imaged with a JEM-1010 transmission electron microscope (JEOL). For scanning electron microscopy, fixed samples were dehydrated and then dried with a critical point dryer (CPD-030; Balzers). Samples were coated with platinum/palladium using an ion coater (E5100 SEM Coating Unit; Polaron) and observed with a JSM-5800LV scanning electron microscope (JEOL).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: KEG (At5g13530), ATG8a (At4g21980), BRI1 (At4g39400), ARA6 (At3g54840), GOLD36/MVP1 (At1g54030), VHA-a1 (At2g28520), γ-TIP (At2g36830), PR-1 (At2g14610), PIP2A (At3g53420), PIN1 (At1g73590), and AUX1 (At2g38120). C14 can be found under tomato accession number T06416.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Confocal Microscopy of Tonoplast Marker γ-TIP-mCherry in Cotyledon and Hypocotyl Cells of Transgenic Seedlings.
Supplemental Figure 2. Mutation of the KEG RING E3 Ubiquitin Ligase Domain Enhances Plasma Membrane Localization of KEG.
Supplemental Figure 3. CLSM and TEM Micrographs of TGN/EE and Golgi Structures in Wild-Type and keg-1 Mutant Seedlings.
Supplemental Figure 4. CLSM and TEM Micrographs of MVBs in Wild-Type and keg-1 Mutant Seedlings.
Supplemental Figure 5. CLSM and TEM Micrographs of ER Structure in Wild-Type and keg-1 Mutant Seedlings.
Supplemental Figure 6. The keg-2 Mutation Affects Protein Vacuolar Transport and Apoplast Secretion the Same Way as the keg-1 Mutation.
Supplemental Figure 7. Transport of the PM Aquaporin Protein PIP2A-mCherry to the Vacuole Is Inhibited in the keg-1 Mutant.
Supplemental Figure 8. Enhanced Brassinolide Responses in keg Mutant Seedlings.
Supplemental Figure 9. Colocalization of BRI1-sYFP and ARA6-mCherry in Arabidopsis and N. benthamiana.
Supplemental Figure 10. Delivery of Plasma Membrane Proteins PIN1 and AUX1 to the Plasma Membrane Is Not Affected by the keg-1 Mutation.
Supplemental Table 1. Primers Used in This Work.
Supplemental Table 2. Fluorescence Quantification of BRI1-sYFP in Vacuoles and Plasma Membranes of keg-1 and Wild-Type Arabidopsis Seedlings.
Supplemental Table 3. Quantification of ARA6-Labeled MVBs in Cotyledon Epidermal Cells of Dark-Grown Seedlings.
Supplemental Movie 1. Transient Expression of KEG-sYFP in an N. benthamiana Epidermal Cell in the Presence of Latrunculin B.
Acknowledgments
We thank Karin Schumacher, Glenn Hicks, Sid Shaw, and Sophien Kamoun for sharing published research materials. We also thank Sid Shaw and Xuhong Yu for insightful discussion. We acknowledge the Indiana University Light Microscopy Imaging Center for access to the Leica SP5 confocal microscope and the Indiana Molecular Biology Institute for access to scanning and transmission electron microscopes. The ABRC at Ohio State University provided cDNA clones and seed for Arabidopsis T-DNA insertion lines and transgenic marker lines. We acknowledge the National Institute of General Medical Sciences of the National Institutes of Health for funding (Grant R01 GM063761 to R.W.I.).
AUTHOR CONTRIBUTIONS
Y.G. and R.W.I. designed the research and wrote the article. Y.G. performed all of the experiments.
Glossary
- TGN/EE
trans-Golgi network/early endosome
- MVB
multivesicular body
- PVC
prevacuolar compartment
- PM
plasma membrane
- CLSM
confocal laser scanning microscopy
- ER
endoplasmic reticulum
- BFA
Brefeldin A
- Wm
wortmannin
- sYFP
to be defined
- RFP
red fluorescent protein
- ConA
concanamycin A
- ILV
intraluminal vesicle
- hpi
hours postinoculation
- MS
Murashige and Skoog
- GFP
green fluorescent protein
References
- Agee A.E., et al. (2010). MODIFIED VACUOLE PHENOTYPE1 is an Arabidopsis myrosinase-associated protein involved in endomembrane protein trafficking. Plant Physiol. 152: 120–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C., Sanderfoot A.A., Kovaleva V., Zheng H., Raikhel N.V. (2000). AtVPS45 complex formation at the trans-Golgi network. Mol. Biol. Cell 11: 2251–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt T.O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., Cano L.M., Jones A.M., Huitema E., van der Hoorn R.A., Kamoun S. (2011). Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA 108: 20832–20837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard I.M., Lynch T.J., Finkelstein R.R. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüx A., Liu T.Y., Krebs M., Stierhof Y.D., Lohmann J.U., Miersch O., Wasternack C., Schumacher K. (2008). Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth inhibition in Arabidopsis. Plant Cell 20: 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.M., Neto H., Foucart C., Moore I. (2008). Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20: 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Frye C.A., Innes R.W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Hyman D.L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Robatzek S. (2008). Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 147: 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D., Oh J., Boutté Y., Best J.G., Samuels L., Nilsson R., Uemura T., Marchant A., Bennett M.J., Grebe M., Bhalerao R.P. (2011). Conserved Arabidopsis ECHIDNA protein mediates trans-Golgi-network trafficking and cell elongation. Proc. Natl. Acad. Sci. USA 108: 8048–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Innes R.W. (2011). The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol. 155: 1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas T.J., Sliwinski M.K., Martínez D.E., Preuss M., Ebine K., Ueda T., Nielsen E., Odorizzi G., Otegui M.S. (2007). The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton S.L., Matheson L.A., Chatre L., Rossi M., Brandizzi F. (2007). Post-Golgi protein traffic in the plant secretory pathway. Plant Cell Rep. 26: 1431–1438 [DOI] [PubMed] [Google Scholar]
- Hwang I., Robinson D.G. (2009). Transport vesicle formation in plant cells. Curr. Opin. Plant Biol. 12: 660–669 [DOI] [PubMed] [Google Scholar]
- Isono E., Katsiarimpa A., Müller I.K., Anzenberger F., Stierhof Y.D., Geldner N., Chory J., Schwechheimer C. (2010). The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Rollin C., Gaude T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109 [DOI] [PubMed] [Google Scholar]
- Kang B.H., Nielsen E., Preuss M.L., Mastronarde D., Staehelin L.A. (2011). Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Kasai K., Takano J., Miwa K., Toyoda A., Fujiwara T. (2011). High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 286: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A., Anzenberger F., Schlager N., Neubert S., Hauser M.T., Schwechheimer C., Isono E. (2011). The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell 23: 3026–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D.J., Babst M., Emr S.D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155 [DOI] [PubMed] [Google Scholar]
- Kim S.J., Bassham D.C. (2011). TNO1 is involved in salt tolerance and vacuolar trafficking in Arabidopsis. Plant Physiol. 156: 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M., Beyhl D., Görlich E., Al-Rasheid K.A., Marten I., Stierhof Y.D., Hedrich R., Schumacher K. (2010). Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. USA 107: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.K., Siu C.L., Hillmer S., Jang S., An G., Robinson D.G., Jiang L. (2007). Rice SCAMP1 defines clathrin-coated, trans-golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Jacob C., André B. (2009). K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leborgne-Castel N., Adam T., Bouhidel K. (2010). Endocytosis in plant-microbe interactions. Protoplasma 247: 177–193 [DOI] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti L., Stefano G., Tamura K., Hawes C., Renna L., Held M.A., Brandizzi F. (2010). A missense mutation in the vacuolar protein GOLD36 causes organizational defects in the ER and aberrant protein trafficking in the plant secretory pathway. Plant J. 63: 901–913 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. (2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Murphy J.E., Padilla B.E., Hasdemir B., Cottrell G.S., Bunnett N.W. (2009). Endosomes: A legitimate platform for the signaling train. Proc. Natl. Acad. Sci. USA 106: 17615–17622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Niemes S., Labs M., Scheuring D., Krueger F., Langhans M., Jesenofsky B., Robinson D.G., Pimpl P. (2010a). Sorting of plant vacuolar proteins is initiated in the ER. Plant J. 62: 601–614 [DOI] [PubMed] [Google Scholar]
- Niemes S., Langhans M., Viotti C., Scheuring D., San Wan Yan M., Jiang L., Hillmer S., Robinson D.G., Pimpl P. (2010b). Retromer recycles vacuolar sorting receptors from the trans-Golgi network. Plant J. 61: 107–121 [DOI] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y.N., Imboden L.A., Chang J.H., He S.Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10774–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Herder R., Schulze J., Jung R., Staehelin L.A. (2006). The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18: 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga R., Dodds P.N. (2009). Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science 324: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Katagiri F. (2009). Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J. 57: 932–944 [DOI] [PubMed] [Google Scholar]
- Reichardt I., Stierhof Y.D., Mayer U., Richter S., Schwarz H., Schumacher K., Jürgens G. (2007). Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr. Biol. 17: 2047–2053 [DOI] [PubMed] [Google Scholar]
- Reyes F.C., Buono R., Otegui M.S. (2011). Plant endosomal trafficking pathways. Curr. Opin. Plant Biol. 14: 666–673 [DOI] [PubMed] [Google Scholar]
- Robatzek S. (2007). Vesicle trafficking in plant immune responses. Cell. Microbiol. 9: 1–8 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito C., Ueda T., Abe H., Wada Y., Kuroiwa T., Hisada A., Furuya M., Nakano A. (2002). A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J. 29: 245–255 [DOI] [PubMed] [Google Scholar]
- Scheuring D., Viotti C., Krüger F., Künzl F., Sturm S., Bubeck J., Hillmer S., Frigerio L., Robinson D.G., Pimpl P., Schumacher K. (2011). Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J., Hwang S., Niwa Y., Kobayashi H., Galbraith D.W. (1995). Green-fluorescent protein as a new vital marker in plant cells. Plant J. 8: 777–784 [DOI] [PubMed] [Google Scholar]
- Silady R.A., Ehrhardt D.W., Jackson K., Faulkner C., Oparka K., Somerville C.R. (2008). The GRV2/RME-8 protein of Arabidopsis functions in the late endocytic pathway and is required for vacuolar membrane flow. Plant J. 53: 29–41 [DOI] [PubMed] [Google Scholar]
- Spitzer C., Reyes F.C., Buono R., Sliwinski M.K., Haas T.J., Otegui M.S. (2009). The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Williams L.A., Farmer L.M., Vierstra R.D., Callis J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M., Zheng H., Morita M.T., Saito C., Avila E., Blakeslee J.J., Bandyopadhyay A., Kovaleva V., Carter D., Murphy A., Tasaka M., Raikhel N. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555 [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K., Goto Y., Asatsuma S., Koizumi M., Mitsui T., Matsuoka K. (2009). A mobile secretory vesicle cluster involved in mass transport from the Golgi to the plant cell exterior. Plant Cell 21: 1212–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Uemura T., Kim H., Saito C., Ebine K., Ueda T., Schulze-Lefert P., Nakano A. (2012). Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc. Natl. Acad. Sci. USA 109: 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon L.C., Rep M., Pieterse C.M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Viotti C., et al. (2010). Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynska A., Christiansen K.M., Lan Y., Rodibaugh N.L., Innes R.W. (2008). Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol. 148: 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Hurley J.H. (2010). Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464: 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski T., Tomczak A., Michelmore R. (2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3: 259–273 [DOI] [PubMed] [Google Scholar]
- Zouhar J., Rojo E., Bassham D.C. (2009). AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 149: 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]