Abstract
Smith-Lemli-Opitz (SLO) syndrome is an autosomal recessive disorder characterized by multiple congenital abnormalities and mental retardation. The condition is caused by the deficiency of 7-dehydrocholesterol reductase (DHCR7) which catalyzes the final step in cholesterol biosynthesis. Biochemical diagnosis is based on increased concentration of 7-dehydrocholesterol (7-DHC) in the patient serum. Both life expectancy and quality of life are severely affected by the disease. The estimated prevalence of SLO syndrome ranges between 1:20,000 and 1:40,000 among Caucasians. Although the mutational spectrum of the disease is wide, approximately 10 mutations are responsible for more than 80% of the cases. These mutations show a large interethnic variability. There are no mutation distribution data from Hungary to date. Thirteen patients were diagnosed with SLO syndrome in our laboratory. As first-line tests, serum 7-DHC and total cholesterol were measured and, in positive cases, molecular genetic analysis of the DHCR7 gene was performed. Complete genetic background of the disease could be identified in 12 cases. In 1 case only 1 mutation was detected in a heterozygote form. One patient was homozygous for the common splice site mutation c.964–1G>C, while all other patients were compound heterozygotes. One novel missense mutation, c.374A>G (p.Tyr125Cys) was identified.
Key Words: Cholesterol, 7-Dehydrocholesterol, DHCR7, Hungary, Mutation, SLO syndrome
Smith-Lemli-Opitz (SLO) syndrome is a monogenic, autosomal recessive, multiple congenital malformation syndrome which was first described by Smith et al. [1964]. The characteristic clinical features are mental retardation, behavior abnormalities, sleeping disturbances, failure to thrive, cardiac defects, facial dysmorphy, skeletal abnormalities (typical is the syndactyly of the 2nd and 3rd toe), genital and renal anomalies as well as photosensitivity [Kelley and Hennekam, 2000]. The clinical spectrum ranges from a mild disorder to severe appearance of the disease that often results in prenatal/neonatal death.
The disorder is caused by the deficiency of the DHCR7 enzyme which catalyzes the conversion of 7-dehydrocholesterol (7-DHC) to cholesterol in the cholesterol biosynthesis pathway [Tint et al., 1994]. Impaired function of the enzyme results in increased level of 7-DHC and decreased cholesterol level in plasma and tissues. Elevated concentration of 7-DHC biochemically confirms the diagnosis of SLO syndrome [Irons et al., 1993]. Cholesterol is an important component of the lipid rafts of cell membranes, it is the constituent of myelin, the precursor molecule of bile acids, steroid hormones and neurosteroids and plays an important role in the embryogenesis/morphogenesis as well [Kelley et al., 1996]. In SLO syndrome 7-DHC replaces cholesterol to a great extent in the sterol- and caveolin-rich membrane regions and disturbs cell signaling [Ren et al., 2011]. Very recently, it has been suggested that the neurological symptoms in SLO syndrome are associated with the oxysterols that are produced out of 7-DHC [Korade et al., 2010].
The DHCR7 cDNA was cloned in 1998 by 3 different groups [Fitzky et al., 1998; Wassif et al., 1998; Waterham et al., 1998]. The human DHCR7 gene localizes to chromosome 11q13 and spans 14,100 bp of genomic DNA [Fitzky et al., 1998]. It consists of 9 exons where the start codon is encoded by exon 3. The DHCR7 mRNA is 2,786 bp long with an open reading frame of 1,425 bp. No other disease is known to be associated with mutations of the DHCR7 gene; however, SNPs within or nearby the gene affect vitamin D metabolism [Wang et al., 2010; Berry and Hypponen, 2011].
SLO syndrome is relatively common in the Caucasian population with an estimated incidence between 1:20 000 and 1:40 000 [Tint et al., 1994] and a carrier frequency of 1:50 to 1:70 which may be the result of both heterozygote advantage [Porter, 2002] and genetic drift [Witsch-Baumgartner et al., 2008].
In the present study we investigated 13 patients from 12 families with clinically and biochemically confirmed SLO syndrome. Molecular genetic diagnosis was performed by standard methodology. In 1 case, where only 1 mutation was found using the standard protocol, we analyzed the promoter and the 3′ regions and the DHCR7 mRNA as well. The genetic background of the disease could be identified in 25/26 alleles.
Materials and Methods
Patients
Since 2008 we investigated 13 patients from different parts of Hungary with typical clinical features of SLO syndrome. The diagnosis of SLO syndrome was established by measurement of 7-DHC by UV spectrophotometry and further molecular genetic analysis of the DHCR7 was performed. Clinical information was available in each case and the clinical score was calculated according to Kelley and Hennekam [2000].
Clinical Features
Patient 1
This infant was born at 37 weeks of gestation by Cesarean section from the first pregnancy of her mother. The pregnancy was complicated by pre-eclampsia and diabetes. Her birth weight was 2,510 g. Observed anomalies were as follows: generalized, mild edema; generalized muscle hypotonia; microcephaly; microphthalmia; short palpebral fissure; cataract on the left side; low-set ears; hypoplastic tongue; gingival hypertrophy; on the right side completely, on the left side incompletely atretic choanae; micrognathia; on each extremity postaxial polydactyly; single transverse palmar crease; assymetric labia majora; missing labia minora; imperforate anus; and sacral pilonidal sinus. She died on the first day of life because of respiratory insufficiency. It is to be noted, that the mother has had 2 pregnancies since the birth of the abovementioned child and both terminated by spontaneous abortion in the 7th and 14th week of pregnancy, respectively.
Patient 2
This infant was born at 39 weeks of gestation by Cesarean section for breech presentation with multiple anomalies from the fifth pregnancy of his mother who had previously had 3 spontaneous abortions and thereafter gave birth to a healthy female infant. Perinatal asphyxia required a resuscitation after birth. Head circumference was 32.5 cm; body length and weight were 46 cm and 2,810 g, respectively. He presented with generalized muscular hypotonia and ambiguous genitalia (micropenis, bifid scrotum, perineoscrotal hypospadias, bilateral cryptorchidism). Cytogenetic investigation revealed a karyotype of 46,XY. Craniofacial abnormalities were microcephaly, small nose with broad, flat nasal bridge, anteverted nares, micrognathia, high-arched palate with cleft of the soft palate, and broad alveolar margins. Among skeletal abnormalities on both hands and on the right foot, postaxial polydactyly and bilateral, incomplete, soft tissue 2nd–3rd-toe syndactyly and a single transverse palmar crease were present. Other anomalies were bilateral cataract, cyst of the corpus pineale, enlarged lateral ventricles, and pyloric stenosis.
Patient 3
This patient is the brother of patient 2 and was born from the sixth pregnancy of his mother at 37 weeks of gestation by Cesarean section for breech presentation. Head circumference was 33.5 cm; body length and weight were 46 cm and 2,700 g, respectively. Available clinical data were less detailed than in the case of the older brother but phenotypically SLO syndrome was soon presumed. He presented with typical craniofacial features like micrognathia; high-arched palate with cleft of the soft palate; small nose with broad, flat nasal bridge; anteverted nares; with ambiguous genitalia (hypertrophic clitoris or micropenis, perineoscrotal hypospadias and bilateral cryptorchidism); postaxial polydactyly on the right hand; clinodactyly of the fifth finger on the left hand; bilateral, incomplete soft tissue 2nd–3rd-toe syndactyly; almost complete 4th–5th-toe syndactyly on the left foot and a single transverse palmar crease; vesicoureteral reflux and associated pyelectasis; muscular hypotonia; and adrenal hyperplasia.
Patient 4
This female patient was born at 35 weeks of gestation by Cesarean section from the first pregnancy of her mother. Birth weight and length were 2,000 g and 36 cm, respectively; head circumference was 31 cm. Multiple congenital abnormalities were observed: facial dysmorphia (micrognathia, cleft of the soft palate, to the cleft attached tongue), postaxial polydactyly on the extremities, incomplete 2nd–3rd- and complete 5th–6th-toe syndactyly, vertebral deformation, atrioventricular septal defect, open ductus Botalli, tricuspid insufficiency, and agenesis of the left kidney. The patient died at 12 days of age because of circulatory insufficiency. Resuscitation was unsuccessful. Postmortem examination revealed absence of lung lobulation and hypoplasia, hepatomegaly, bicornuate uterus, and pneumonia.
Patient 5
This female infant was born at 38 weeks of gestation. Her birth weight was 3,090 g. Minor anomalies were observed: bilateral incomplete 2nd–3rd-toe syndactyly; clinodactyly; on the right hand single transverse palmar crease; microcephaly; high-arched palate; bifid uvula; long philtrum; broad, flat nasal bridge; and mild hypertelorism.
Patient 6
This female patient was born at 38 weeks of gestation. Birth weight and length were 2,840 g and 52 cm, respectively; head circumference was 32 cm. Facial abnormalities were low-set ears, midline cleft of the hard palate, postaxial polydactyly on the right hand, bilateral 2nd–3rd-toe syndactyly, club foot on the left side, 11 ribs on the left side, right ventricular hypertrophy, tricuspid valve insufficiency, gastroesophageal reflux, feeding difficulties, dystopic kidney, and brain atrophy. She died at 1.5 years of age most likely because of an adverse reaction due to anesthesia.
Patient 7
This male patient was born from the first pregnancy of his mother at 39 weeks of gestation by Cesarean section because the amniotic fluid was stained with meconium. Anomalies observed were microcephaly, cleft palate, retrognathia, low-set ears, postaxial polydactyly, single transverse palmar crease, incomplete 2nd–3rd-toe syndactyly, micropenis, hypospadias, incomplete dysgenesis of corpus callosum, atrioventricular septal defect, and feeding difficulties.
Patient 8
This infant was born at 42 weeks of the first, uncomplicated gestation of his mother. His birth weight was 3,160 g. Minor anomalies were observed: micrognathia, epicanthal folds, short palpebral fissure, hypertelorism, high-arched palate, gingival hypertrophy, bilateral single transverse palmar crease, 2nd–3rd-toe syndactyly, far apart nipples, bifid scrotum, micropenis, as well as flexible joints and skin. Somatomental development was slow; at 8 months of age he died because of multiorgan failure of septic origin.
Patient 9
This female patient was born at 38 weeks of gestation. Birth weight was 2,710 g. Observed anomalies were: muscle hypotonia, micrognathia, epicanthal folds, broad nasal root, anteverted nares, incomplete 2nd–3rd-toe syndactyly, single transverse palmar crease, bifid uvula, feeding difficulties, and enlarged lateral ventricles.
Patient 10
This male patient was born at 36 weeks of gestation by Cesarean section because of pre-eclampsia. As a prenatal intrauterinal retardation had been diagnosed, amniocentesis was performed for chromosomal examination which revealed normal 46,XY karyotype. Observed abnormalities were as follows: intersex genitalia, hypospadias, hypertelorism, low-set ears, high-arched palate, on the right side postaxial polydactyly, 2nd–3rd-toe syndactyly, patent ductus arteriosus, mildly enlarged right lateral ventricle, and severe Hirschsprung disease.
Patient 11
This patient was born at 38 weeks of gestation, her mother having had 3 spontanous abortions and having given birth to a healthy son before. Birth weight was 2,350 g. Abnormalities were: postaxial polydactyly on all extremities, on the left side single transverse palmar crease, hypertonic extremities, limb shortness (upper arm, thigh), bilateral incomplete 2nd–3rd-toe syndactyly, high-arched palate, short neck, far apart nipples, intersex genitalia (karyotype 46,XY), pyloric stenosis, biliary cirrhosis, and atrioventricular septal defect. Pylorotomy did not solve the feeding difficulty so he needed parenteral nutrition. He died at 3 months of age after several infections.
Patient 12
This male patient was born at 35 weeks of gestation (complicated by diabetes mellitus and hypertension) by Cesarean section because of pre-eclampsia. Birth weight was 1,725 g. Anomalies observed were: inguinal hernia on the left side, bilateral incomplete 2nd–3rd-toe syndactyly, hypospadias, bifid scrotum, and atrial septal defect.
Patient 13
This female patient has got the mildest phenotype of all. Clinical signs are postaxial polydactyly, 2nd–3rd-toe syndactyly, atrial septal defect, mental retardation, scoliosis, strabismus, and hypacusis. She is 18 years old.
She had had an older sister who was born at 38 weeks of gestation in the state of acute asphyxia, her birth weight was 2,500 g. She was successfully resuscitated. Because of minor anomalies (no clinical data, after her mother's statement that as a newborn she looked like her younger sister) and congenital heart disease, Turner syndrome was suspected but chromosome analysis revealed a normal karyotype. Severe cardiovascular anomalies were observed: atrioventricular septal defect, open ductus arteriosus (Botalli), incomplete pulmonary vein transposition, absence of lung lobulation, and gallbladder agenesis. She developed cardiac decompensation, failed to thrive and died at 1 month of age.
Methods
Biochemical Measurements
Serum samples were used for 7-DHC measurement by the modified method of Honda et al. [1997]. Total cholesterol was measured by an enzymatic colometric method.
Analysis of the Coding Region of the DHCR7 Gene
DNA isolation from blood leukocytes was performed using QIAgen Blood Mini Kit (Qiagen) and molecular genetic analysis of DHCR7 was performed by amplifying exons 3–9 and the exon-flanking intron regions. PCR primers used were as described by Fitzky et al. [1998] except for exon 7 (e7F: 5′-GCT GGG CTC TCG CTA AGT AA-3′, e7R: 5′-GCA GTA GAT TAA GGT CAT GGG AAT-3′) and exon9a (e9aR: 5′-ATG TAG AAG TAG GGC AGC AGG TGG C-3′). For DNA sequencing the PCR products were cleaned using ultrafiltration microcolumns (Microcon YM-100, Millipore). Purified PCR products were sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies). Removal of unincorporated nucleotides was performed using gel filtration (DyeEx Kit, Qiagen). Capillary electrophoresis was performed on the ABI Prism 310 Genetic Analyzer (Life Technologies). Electropherograms of the sequenced PCR products were compared to the reference sequence of the DHCR7 gene (NG 012655.2).
When the mutations were found in the patient's sample, samples of the parents were analyzed to prove that the detected mutations are found in trans in the patient. In the case of patient 11, only the DNA samples of the parents were available.
Analysis of the Non-Coding Exons and Regulatory Sequences of the DHCR7 Gene
In 1 case (patient 13), where the abovementioned standard protocol was used, only 1 mutation was found in a heterozygote form. Therefore, the putative promoter region [Correa-Cerro and Porter, 2005] and the 3′ polyadenylation site were also investigated using the following primers: PM1F (5′-CCT GAC ACC CAT GTG TTT ACC-3′), PM1R (5′-CCG GAC TCG AGA TTG ACG-3′), PM2F (5′-AGC TGG GAT CCC GAA GAA G-3′), PM2R (5′-CAC GCC GCC TAC CCT CTA-3′), PM3-e1F (5′-GCA ATC GCT GAC ATC ATC C-3′), PM3-e1R (5′-CCT GTG AGT GGG CAC CTG-3′), e2F (5′-GAG CTT CTG CCC TCT CCT G-3′), e2R (5′-TCA ACG CTG TGA AGC CAT AG-3′), 3UTR1F (5′-CTA CAT GGC CAT CCT GCT G-3′), 3UTR1R (5′-CCC CAG GGA CAC TGA TTA GA-3′), 3UTR2F (5′-GTT CCT TGC TTT TGC CTT CA-3′) and 3UTR2R (5′-GGA GGG GGA TCT AGA GCT GA-3′).
Sequencing of the DHCR7 mRNA
In the same case the DHCR7 mRNA was also sequenced. Total RNA was extracted from EDTA-anticoagulated blood samples using Trizol reagent (Invitrogen) and reverse transcription was carried out using High Capacity cDNA kit (Life Technologies).
DHCR7 cDNA was amplified in 4 overlapping fragments using Pfu polymerase (Agilent Technologies). Following primers were used for the amplification: DHCR7mR1F (5′-TGA CAG AAC CGC ATC TCA A-3′), DHCR7mR1R (5′-CTC CTA CGT AGC CGG GTA GA-3′), DHCR7mR2F (5′-TAC CTT GTG GGT CAC CTT CC-3′), DHCR7mR2R (5′-ATT GGT CAC ATG GCT GTG G-3′), DHCR7mR3F (5′-GAT CGG GAA GTG GTT TGA CT-3′), DHCR7mR3R (5′-GCC ACC CGG AAG ATG TAG TA-3′), DHCR7mR4F (5′-GAC TGT GTC TGG CTG CCT TA-3′) and DHCR7mR4R (5′-AGG ATG GCC ATG TAG ATG ATG-3′).
Assessing the Pathogenicity of the Novel Variant
For the possible deleterious effect of the p.Tyr125Cys mutation, a SIFT analysis was performed [Ng and Henikoff, 2001, 2002]. Alignment analysis of DHCR7 proteins were carried out using ClustalW [Larkin et al., 2007]. The presence of the c.374A>G (p.Tyr125Cys) mutation in healthy individuals was tested by SnaBI enzyme digestion of PCR-amplified products. Exon 5 of the DHCR7 gene was amplified in a 250-bp PCR product. In homozygous wild-type samples, digestion with SnaBI yielded a 121-bp and a 129-bp restriction fragment while this SnaBI site is lost in the case of the c.374A>G mutation.
Results and Discussion
Both cholesterol and 7-DHC levels were measured in the same serum samples of the suspected patients. Extremely elevated serum 7-DHC levels were measured in all patients, ranging from 87 mg/l to 302 mg/l. Table 1 shows a representative measurement in the patients that was performed before any therapy was started (i.e. cholesterol supplementation). Cholesterol levels were generally low, in 8 patients lower than the lower limit of their age-specific reference range. Cholesterol/7-DHC ratio was abnormal in all patients (not shown).
Table 1.
Genetic background, 7-DHC and cholesterol levels in the Hungarian SLO patients
| Patients | Genotype | Effect on protein level | Severity scorea | 7-DHC, mg/1 (<0.15 mg/l)b | Cholesterol, mM (0–1 years: >1.3 mM; >1 years: 2.8–5.2 mM)b |
|---|---|---|---|---|---|
| 1 | [c.964-lG>C] + [c.964-lG>C] | frameshift | 55-S | 109 | 0.31 |
| 2 | [c.964-lG>C] + [c.1190C>T] | frameshift/p.Ser397Leu | 50-T | 302 | 1.4 |
| 3 | [c.964-lG>C] + [c.1190C>T] | frameshift/p.Ser397Leu | 40-T | 253 | 1.08 |
| 4 | [c.964-lG>C] + [c.1190C>T] | frameshift/p.Ser397Leu | 55-S | 174 | 0.58 |
| 5 | [c.452G>A] + [c.740C>T] | p.Trp151 */p.Ala247Val | 10-M | 130 | 3.47 |
| 6 | [c.976G>T] + [c.374A>G] | p.Val326Leu/p.Tyrl25Cys | 40-T | 205 | 2.1 |
| 7 | [c.730G>A] + [c.976G>T] | p.Gly244Arg/p.Val326Leu | 40-T | 155 | 0.72 |
| 8 | [c.326T>C] + [c.452G>A] | p.Leul09Pro/p.Trpl51* | 30-T | 126 | 0.77 |
| 9 | [c.1295A>G] + [c.1328G>A] | p.Tyr432Cys/p.Arg443His | 15-M | 217 | 2.0 |
| 10 | [c.452G>A] + [c.976G>T] | p.Trpl51*/p.Val326Leu | 20-T | 156 | 1.57 |
| 11 | [c.725G>A] + [c.452G>A] | p.Arg242His/p.Trpl51* | 55-S | 215 | 0.8 |
| 12 | [c.452G>A] + [c.1295A>G] | p.Trpl51*/p.Tyr432Cys | 20-T | 274 | 1.47 |
| 13 | [c.452G>A] + [?] | p.Trpl51*/? | 15-M | 87 | 2.44 |
M = Mild; S = severe; T = typical.
Normal reference values are shown for comparison.
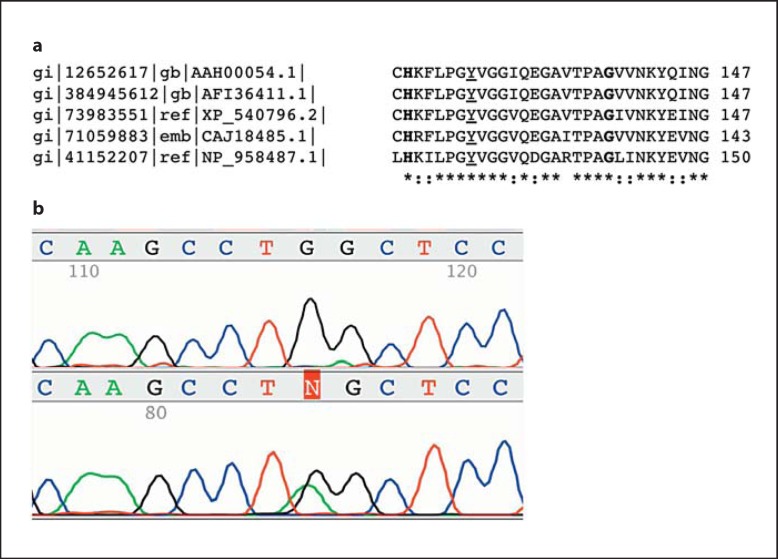
In 12 of the 13 patients, establishing the molecular diagnosis of SLO syndrome was successful. Genotypes of the patients are shown in table 1. In addition to the known nonsense, missense and splicing mutations, 1 previously unidentified mutation, c.374A>G (p.Tyr125Cys) was detected. The pathogenicity of this mutation is supported by the following facts: (1) the affected residue is phylogenetically highly conserved in human, dog, zebrafish, mouse, and rhesus (fig. 1a); (2) the Grantham difference is 194 and (3) the affected residue does not tolerate the tyrosine to cysteine missense mutation according to the SIFT prediction. In addition, this was the only candidate mutation detected in trans with another known SLO-causing mutation, c.976G>T (p.Val326Leu) in the affected patient. The closest amino acid positions that have been a subject to pathogenic mutation leading to SLO syndrome are 119 (p.His119Leu [Waterham et al., 1998]) and 138 (p.Gly138Val [Waye et al., 2005]) (fig. 1a). In the case of the c.374A>G (p.Tyr125Cys) mutation, RFLP analysis of exon 5 of the DHCR7 gene was performed to exclude the possibility of being a polymorphism in the Hungarian population. The results showed that this mutation was not present in 50 healthy individuals representing 100 alleles.
Fig. 1.
a Amino acid sequence alignment of DHCR7 orthologs (AAH00054.1: human, AFI36411.1: rhesus, XP 540796.2: dog, CAJ18485.1: mouse, NP 958487.1: zebrafish). The affected tyrosine is underlined. The 2 closest known sites affected by pathogenic missense mutations are in bold. b The lack of the allele harboring p.Trp151* in cDNA analysis suggests the presence of NMD. Upper panel: cDNA, lower panel: gDNA.
As the elevated 7-DHC levels are specific for SLO [Porter, 2008] and can distinguish between SLO and the 2 similar rare disorders of cholesterol biosynthesis, lathosterolosis [Brunetti-Pierri et al., 2002] and desmosterolosis [Andersson et al., 2002], the diagnosis that was based on the clinical symptoms and the measurement of serum 7-DHC could be established in patient 13. In this unique case, a 3-tier molecular genetic analysis approach was used. First-tier analysis that involved sequencing of the coding exons revealed 1 previously described pathogenic mutation in a heterozygous form, namely c.452G>A (p.Trp151*). In the second tier of molecular testing, sequence analysis of the non-coding exons and the putative promoter as well as the 3′ untranslated region (462 nucleotides downstream from the last codon) of the gene was performed. This analysis did not reveal any potentially responsible genetic alteration. Next, cDNA sequencing was performed in the patient's sample. This analysis led to the following conclusions: first, no other mutation that affects the coding sequence could be detected which suggests that rather mRNA quantity than quality might be affected by the other, unidentified alteration. Second, the analysis of the DNA and the RNA level confirmed the presence of nonsense-mediated mRNA decay (NMD) as the mRNA derived from blood leukocytes lacks the p.Trp151* allele as judged by the sequencing (fig. 1b). NMD affecting the nonsense mutations in the DHCR7 gene has been shown before [Correa-Cerro et al., 2005].
The fact that this patient's phenotype lies at the upper end of the extremely wide clinical spectrum of SLO syndrome makes these findings even more puzzling. It is known from the literature that in some SLO cases only 1 mutation can be found and sequence analysis of all exons and all exon-intron boundaries detects about 96% of all mutations [Witsch-Baumgartner et al., 2001a]. Porter [2008] suggests that, at least in some of those cases, the second allele is not expressed. What we found is somewhat different as, according to the NMD, the other, non-nonsense allele is the only that is expressed, at least in the blood leukocytes. We cannot exclude the possibility of a mutation present in some uncharacterized distant or deep intronic regulatory motifs that might modulate (i.e. decrease but not completely abolish) the expression level of the DHCR7 protein. The presence of another, tissue-specific promoter at the more important synthesis site(s) cannot be excluded either. This speculation, however, cannot explain the strikingly different phenotype that was observed in the sister of the patient (although biochemical and genetic tests were not performed in that case). In the case of SLO syndrome, the development of a sensitive assay system that is capable to measure the activity of DHCR7, similarly to the one that had been used before [Shefer et al., 1995] rather to quantitate its substrate, would be of great importance and might give answer for the obvious question: how can the clinical spectrum be so wide? We believe that from the genetic point of view the wide range of symptoms in addition to the possible genetic modifiers [Witsch-Baumgartner et al., 2004] might be attributed to the enzyme activity, i.e. 2 null alleles are often associated with spontaneous abortion or very severe phenotype [Witsch-Baumgartner et al., 2000] and some residual activity is needed to survive the prenatal period.
Before this study, only 1 report of an SLO patient from Hungary was published [Szabó et al., 2009].
Mutations that cause SLO syndrome show an interesting geographical prevalence rate (table 2). The literally most common c.964–1G>C mutation that is probably of British origin [Witsch-Baumgartner et al., 2001b] and frequently detected in the Mediterraneum [Witsch-Baumgartner et al., 2005] but almost absent in patients of Slavic ethnicity [Kozák et al., 2000; Witsch-Baumgartner et al. 2001b] was found to be the second most frequent in our data set, being present in 4 alleles. On the other hand, 6 p.Trp151* alleles were identified in our patients. This mutation seems to occur frequently in Poland, Germany and Austria, Czech and Slovak Republics as well as in Hungary but to be very rare in Great Britain, Spain and Italy (table 2). The other typically Eastern-European p.Val326Leu allele [Witsch-Baumgartner et al., 2001b] shows a very similar distribution pattern. It was detected in 3 disease-causing alleles and, together with the p.Trp151* and c.964–1G>C were responsible for the 54% of unrelated pathogenic mutations in Hungary. These data show that the allelic heterogeneity is significantly different in Europe, similarly to other monogenic diseases [Dörk et al., 2000; Ivady et al., 2011]. Testing for 7 DHCR7 mutations would result in 59% detection sensitivity in Great Britain while this would be enough to reach 83% in Poland and Spain or even 100% in the case of Czech and Slovak patients. Hungarian mutation frequency distribution lies between the abovementioned data as testing for 5 mutations results in a detection sensitivity of 71%, which is very similar to Austria and Germany, where testing for 6 mutations results in 68% mutation detection sensitivity.
Table 2.
Prevalence (%) of the most common SLO-causing DHCR7 gene mutations in Europe
| Country (no. of alleles): | Great Britain (44) | Poland (30) | Germany and Austria (44) | Italy (20) | Spain (40) | Czech and Slovak Republics (20) | Hungary (24) | |
|---|---|---|---|---|---|---|---|---|
| p.Trp151* | 2.3 | 33.3 | 18.2 | 5.0 | 2.5 | 50.0 | 25.0 | |
| p.Val326Leu | 2.3 | 23.3 | 18.2 | 0 | 0 | 25.0 | 12.5 | |
| p.Arg352Trp | 0 | 13.3 | 6.8 | 5.0 | 2.5 | 0 | 0 | |
| p.Leu157Pro | 0 | 6.7 | 2.3 | 0 | 0 | 5.0 | 0 | |
| c.964–1G>C | 34.1 | 3.3 | 20.5 | 20.0 | 30.0 | 5.0 | 16.7 | |
| p.Arg446Gln | 0 | 3.3 | 0 | 0 | 7.5 | 5.0 | 0 | |
| p.Arg404Cys | 9.1 | 0 | 0 | 5.0 | 0 | 0 | 0 | |
| p.Thr93Met | 6.8 | 0 | 0 | 45.0 | 23.0 | 0 | 0 | |
| p.Gly410Ser | 2.3 | 0 | 2.3 | 0 | 7.5 | 10.0 | 0 | |
| p.Phe302Leu | 2.3 | 0 | 0 | 0 | 10.0 | 0 | 0 | |
| p.Ser397Leu | 0 | 0 | 0 | 0 | 0 | 0 | 8.3 | |
| p.Tyr432Cys | 0 | 0 | 0 | 0 | 0 | 0 | 8.3 | |
| Sum above | 59.1 | 83.3 | 68.2 | 80.0 | 83.0 | 100.0 | 70.8 | |
| Reference1 | a | a | a | b | b | c | this study | |
In all but 1 family, the pathogenic combinations were either nonsense-missense, missense-missense or splicing (c.964–1G>C)-missense mutations, which means that a residual enzyme activity cannot be excluded. In the only case where 2 proven null alleles were present (c.964–1G>C), the first affected child died at the first day after delivery and the second and third pregnancies ended in spontaneous abortions at week 7 and 14 of the pregnancy, respectively. Molecular analysis of the second fetus had not been performed, but in the case of the other 2, homozygosity for the c.964–1G>C mutation could be confirmed. Taken together, our results support the previous findings that the presence of 2 severe null alleles – with very few exceptions – is likely to be incompatible with life and the phenotype of the affected children depends on the genetic basis, the presence of modifiers and the environmental situation (for example vitamin intake).
References
- Andersson HC, Kratz L, Kelley R. Desmosterolosis presenting with multiple congenital anomalies and profound developmental delay. Am J Med Genet. 2002;113:315–319. doi: 10.1002/ajmg.b.10873. [DOI] [PubMed] [Google Scholar]
- Berry D, Hypponen E. Determinants of vitamin D status: focus on genetic variations. Curr Opin Nephrol Hypertens. 2011;20:331–336. doi: 10.1097/MNH.0b013e328346d6ba. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Corso G, Rossi M, Ferrari P, Balli F, et al. Lathosterolosis, a novel multiple-malformation/mental retardation syndrome due to deficiency of 3beta-hydroxysteroid-delta5-desaturase. Am J Hum Genet. 2002;71:952–958. doi: 10.1086/342668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Cerro LS, Porter FD. 3beta-hydroxysterol delta7-reductase and the Smith-Lemli-Opitz syndrome. Mol Genet Metab. 2005;84:112–126. doi: 10.1016/j.ymgme.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Correa-Cerro LS, Wassif CA, Waye JS, Krakowiak PA, Cozma D, et al. DHCR7 nonsense mutations and characterisation of mRNA nonsense mediated decay in Smith-Lemli-Opitz syndrome. J Med Genet. 2005;42:350–357. doi: 10.1136/jmg.2004.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörk T, Macek M, Jr, Mekus F, Tümmler B, Tzountzouris J, Casals T, et al. Characterization of a novel 21-kb deletion, CFTRdele2,3 (21 kb), in the CFTR gene: a cystic fibrosis mutation of Slavic origin common in Central and East Europe. Hum Genet. 2000;106:259–268. doi: 10.1007/s004390000246. [DOI] [PubMed] [Google Scholar]
- Fitzky BU, Witsch-Baumgartner M, Erdel M, Lee JN, Paik YK, et al. Mutations in the delta7-sterol reductase gene in patients with the Smith-Lemli-Opitz syndrome. Proc Natl Acad Sci USA. 1998;95:8181–8186. doi: 10.1073/pnas.95.14.8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Batta AK, Salen G, Tint GS, Chen TS, Shefer S. Screening for abnormal cholesterol biosynthesis in the Smith-Lemli-Opitz syndrome: rapid determination of plasma 7-dehydrocholesterol by ultraviolet spectrometry. Am J Med Genet. 1997;68:288–293. doi: 10.1002/(sici)1096-8628(19970131)68:3<288::aid-ajmg8>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Irons M, Elias ER, Salen G, Tint GS, Batta AK. Defective cholesterol biosynthesis in Smith-Lemli-Opitz syndrome. Lancet. 1993;341:1414. doi: 10.1016/0140-6736(93)90983-n. [DOI] [PubMed] [Google Scholar]
- Ivady G, Madar L, Nagy B, Gonczi F, Ajzner E, et al. Distribution of CFTR mutations in Eastern Hungarians: relevance to genetic testing and to the introduction of newborn screening for cystic fibrosis. J Cyst Fibros. 2011;10:217–220. doi: 10.1016/j.jcf.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Kelley RI, Hennekam RC. The Smith-Lemli-Opitz syndrome. J Med Genet. 2000;37:321–335. doi: 10.1136/jmg.37.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Roessler E, Hennekam RC, Feldman GL, Kosaki K, et al. Holoprosencephaly in RSH/Smith-Lemli-Opitz syndrome: does abnormal cholesterol metabolism affect the function of sonic hedgehog? Am J Med Genet. 1996;66:478–484. doi: 10.1002/(SICI)1096-8628(19961230)66:4<478::AID-AJMG22>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Korade Z, Xu L, Shelton R, Porter NA. Biological activities of 7-dehydrocholesterol-derived oxysterols: implications for Smith-Lemli-Opitz syndrome. J Lipid Res. 2010;51:3259–3269. doi: 10.1194/jlr.M009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozák L, Francová H, Hrabincová E, Procházková D, Jüttnerová V, et al. Smith-Lemli-Opitz syndrome: molecular-genetic analysis of ten families. J Inherit Metab Dis. 2000;23:409–412. doi: 10.1023/a:1005616321794. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD. Malformation syndromes due to inborn errors of cholesterol synthesis. J Clin Invest. 2002;110:715–724. doi: 10.1172/JCI16386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet. 2008;16:535–541. doi: 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- Ren G, Jacob RF, Kaulin Y, Dimuzio P, Xie Y, et al. Alterations in membrane caveolae and BKCa channel activity in skin fibroblasts in Smith-Lemli-Opitz syndrome. Mol Genet Metab. 2011;104:346–355. doi: 10.1016/j.ymgme.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer S, Salen G, Batta AK, Honda A, Tint GS, et al. Markedly inhibited 7-dehydrocholesterol-delta 7-reductase activity in liver microsomes from Smith-Lemli-Opitz homozygotes. J Clin Invest. 1995;96:1779–1785. doi: 10.1172/JCI118223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DW, Lemli L, Opitz JM. A Newly Recognized Syndrome of Multiple Congenital Anomalies. J Pediatr. 1964;64:210–217. doi: 10.1016/s0022-3476(64)80264-x. [DOI] [PubMed] [Google Scholar]
- Szabó GP, Oláh AV, Kozak L, Balogh E, Nagy A, et al. A patient with Smith-Lemli-Opitz syndrome: novel mutation of the DHCR7 gene and effects of therapy with simvastatin and cholesterol supplement. Eur J Pediatr. 2009;169:121–123. doi: 10.1007/s00431-009-0987-z. [DOI] [PubMed] [Google Scholar]
- Tint GS, Irons M, Elias ER, Batta AK, Frieden R, et al. Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N Engl J Med. 1994;330:107–113. doi: 10.1056/NEJM199401133300205. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassif CA, Maslen C, Kachilele-Linjewile S, Lin D, Linck LM, et al. Mutations in the human sterol delta7-reductase gene at 11q12-13 cause Smith-Lemli-Opitz syndrome. Am J Hum Genet. 1998;63:55–62. doi: 10.1086/301936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Wijburg FA, Hennekam RC, Vreken P, Poll-The BT, et al. Smith-Lemli-Opitz syndrome is caused by mutations in the 7-dehydrocholesterol reductase gene. Am J Hum Genet. 1998;63:329–338. doi: 10.1086/301982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Krakowiak PA, Wassif CA, Sterner AL, Eng B, et al. Identification of nine novel DHCR7 missense mutations in patients with Smith-Lemli-Opitz syndrome (SLOS) Hum Mutat. 2005;26:59. doi: 10.1002/humu.9346. [DOI] [PubMed] [Google Scholar]
- Witsch-Baumgartner M, Fitzky BU, Ogorelkova M, Kraft HG, Moebius FF, et al. Mutational spectrum in the delta7-sterol reductase gene and genotype-phenotype correlation in 84 patients with Smith-Lemli-Opitz syndrome. Am J Hum Genet. 2000;66:402–412. doi: 10.1086/302760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witsch-Baumgartner M, Löffler J, Utermann G. Mutations in the human DHCR7 gene. Hum Mutat. 2001a;17:172–182. doi: 10.1002/humu.2. [DOI] [PubMed] [Google Scholar]
- Witsch-Baumgartner M, Ciara E, Löffler J, Menzel HJ, Seedorf U, et al. Frequency gradients of DHCR7 mutations in patients with Smith-Lemli-Opitz syndrome in Europe: evidence for different origins of common mutations. Eur J Hum Genet. 2001b;9:45–50. doi: 10.1038/sj.ejhg.5200579. [DOI] [PubMed] [Google Scholar]
- Witsch-Baumgartner M, Gruber M, Kraft HG, Rossi M, Clayton P, et al. Maternal apo E genotype is a modifier of the Smith-Lemli-Opitz syndrome. J Med Genet. 2004;41:577–584. doi: 10.1136/jmg.2004.018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witsch-Baumgartner M, Clayton P, Clusellas N, Haas D, Kelley RI, et al. Identification of 14 novel mutations in DHCR7 causing the Smith-Lemli-Opitz syndrome and delineation of the DHCR7 mutational spectra in Spain and Italy. Hum Mutat. 2005;25:412. doi: 10.1002/humu.9328. [DOI] [PubMed] [Google Scholar]
- Witsch-Baumgartner M, Schwentner I, Gruber M, Benlian P, Bertranpetit J, et al. Age and origin of major Smith-Lemli-Opitz syndrome (SLOS) mutations in European populations. J Med Genet. 2008;45:200–209. doi: 10.1136/jmg.2007.053520. [DOI] [PubMed] [Google Scholar]