Abstract
The NF-κB transcription factor is a central mediator of inflammatory and innate immune signaling pathways. Activation of NF-κB is achieved by K63-linked polyubiquitination of key signaling molecules which recruit kinase complexes that in turn activate the IκB kinase (IKK). Ubiquitination is a highly dynamic process and is balanced by deubiquitinases that cleave polyubiquitin chains and terminate downstream signaling events. The A20 deubiquitinase is a critical negative regulator of NF-κB and inflammation, since A20-deficient mice develop uncontrolled and spontaneous multi-organ inflammation. Furthermore, specific polymorphisms in the A20 genomic locus predispose humans to autoimmune disease. Recent studies also indicate that A20 is an important tumor suppressor that is inactivated in B-cell lymphomas. Therefore, targeting A20 may form the basis of novel therapies for autoimmune disease and lymphomas.
Keywords: A20, inflammation, NF-κB, ubiquitin
Introduction
The NF-κB transcription factor is ubiquitously expressed and is activated by diverse immune stimulatory ligands, including interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α), bacterial lipopolysaccharide (LPS) and antigenic peptides. NF-κB forms various combinations of homo- and heterodimers containing p50 (NF-κB1), p52 (NF-κB2), RelA (p65), RelB and c-Rel.1 NF-κB subunits each contain an approximately 300-amino-acid domain located in the amino (N)-terminus known as the Rel homology domain. The Rel homology domain confers DNA binding, nuclear localization and dimerization of NF-κB proteins. Under resting conditions, NF-κB is held inactive as a latent complex in the cytoplasm by the ankyrin repeat domain containing inhibitor IκBα as well as other IκB family members.2 In response to immune-activating stimuli or stress, IκBα is phosphorylated by the IκB kinase (IKK) complex, which consists of two catalytically active kinases, IKKα and IKKβ, and a regulatory subunit IKKγ (NEMO).3 Phosphorylation of IκBα on serines 32 and 36 trigger its ubiquitination and proteasome-dependent degradation thus allowing bound NF-κB dimers to translocate to the nucleus and activate gene expression. The transcriptional activity of NF-κB must be tightly regulated to curb persistent NF-κB activation that may harm the host. IκBα is itself an NF-κB target gene and contributes to the termination of NF-κB signaling in a negative feedback loop.4 A number of other negative regulatory mechanisms, some of which will be described later in this review, also function to inhibit NF-κB and maintain homeostasis.
It has become clear that ubiquitination of key signaling molecules plays a central role in the activation of IKK and NF-κB. Ubiquitin is covalently attached to lysine residues on protein substrates and either triggers proteasomal degradation, protein trafficking, signal transduction or the DNA damage response. Ubiquitination involves a three-step enzymatic reaction catalyzed by three classes of proteins: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases.5 The E1 enzyme activates ubiquitin via adenosine triphosphate and forms a thioester linkage between the catalytic cysteine of the E1 and the carboxy (C)-terminal glycine of ubiquitin. The activated ubiquitin is then transferred to a specific E2 ubiquitin-conjugating enzyme forming an E2 ubiquitin thioester. The E3 ubiquitin ligase then conjugates ubiquitin to a lysine residue in the substrate in the final step of ubiquitination. There are over 600 E3 ubiquitin ligases in humans that are classified into different groups, such as homologous to E6-associated protein C-terminus (HECT) and really interesting new gene (RING). The E3 ligase is thought to confer specificity to ubiquitination and directly contacts the substrate, although the mechanism of ubiquitin transfer differs among the E3 ligase groups. For example, HECT E3 ligases directly transfer the ubiquitin to substrates, whereas RING ligases function as scaffolds and coordinate transfer together with the E2 enzyme.6
Polyubiquitination of proteins occurs when ubiquitin molecules are linked to one another via internal lysine residues to form chains. There are seven potential lysine (K6, K11, K27, K29, K33, K48 and K63) residues in ubiquitin that may participate in chain linkage formation.7 In addition, ubiquitin molecules may be assembled into linear chains whereby the carboxy and amino termini of ubiquitin molecules are linked.8 Linear polyubiquitination has emerged as an important regulator of NF-κB activation by multiple stimuli although the precise role of linear ubiquitin chains in these pathways is not well understood.9 An E3 ligase complex termed LUBAC (linear ubiquitin chain assembly complex) containing two RING finger proteins, HOIL-1L and HOIP, and a third subunit known as SHARPIN, catalyzes linear polyubiquitin chains and is important for NF-κB activation in multiple inflammatory pathways.8, 10
In addition to linear polyubiquitination, both K48- and K63-linked polyubiquitin chains also play important roles in NF-κB signaling. K48-linked polyubiquitin chains trigger proteasomal degradation of substrates, whereas K63-linked polyubiquitin chains generally regulate nonproteolytic functions such as receptor trafficking, kinase activation, signal transduction and the DNA damage response.5, 7 The E2 enzyme Ubc13, together with Uev1a, is specific for the synthesis of K63-linked polyubiquitin chains, although UbcH5 may also function as an E2 for K63-linked ubiquitination.11, 12 The roles of other ubiquitin linkages such as K6, K11, K27, K29, or K33 are poorly understood, although K11-, K27- and K29-linked polyubiquitin chains may also promote protein degradation.13, 14
The proinflammatory cytokine TNF is a potent activator of NF-κB. TNF binding to TNF receptor 1 (TNFR1) triggers trimerization of the receptor and recruitment of the signaling molecule TRADD to the death domain of the receptor. TRADD serves as a platform for binding to the adaptor molecule RIP1 and the E3 ligases TRAF2, cIAP1/2 and LUBAC.15 RIP1 then becomes modified with K63-linked and linear polyubiquitin chains which function as scaffolds to recruit ubiquitin-binding domain protein complexes.16, 17 The ubiquitin-binding adaptors TAB2/TAB3 and NEMO recruit the kinases TAK1 and IKK, respectively, to ubiquitinated RIP1 to enable TAK1 to phosphorylate, and hence activate IKKβ.18, 19, 20 A similar paradigm exists in other NF-κB pathways, such as IL-1R/TLR4 signaling.
IL-1β or LPS binding to IL-1R or TLR4, respectively, results in the rapid association of the adaptor molecule Myd88 with the receptors and subsequent recruitment of the kinases IRAK1 and IRAK4.21 Activation and phosphorylation of IRAK kinases leads to the recruitment of the E3 ubiquitin ligase TRAF6 which undergoes K63-linked autoubiquitination and serves as a scaffold to recruit TAK1 and IKK kinases through binding to TAB2/3 and NEMO, respectively.21 TAK1 is subsequently activated and triggers IKK/NF-κB and mitogen-activated protein kinase/c-Jun N-terminal kinase activation.22
Ubiquitination can be reversed by deubiquitinating enzymes (DUBs) or deubiquitinases that cleave ubiquitin molecules and therefore disassemble polyubiquitin chains from the substrate. The human genome contains nearly 100 DUBs, subdivided into five families based on sequence and structural similarity: ubiquitin-specific proteases, ubiquitin C-terminal hydrolases, ovarian tumor proteases (OTUs), Machado–Joseph disease protein domain proteases and JAMM/MPN domain-associated metallopeptidases (JAMMs).23 The ubiquitin carboxy-terminal hydrolase, ubiquitin-specific protease, OTU and Josephin DUBs function as cysteine proteases, and JAMMs are zinc metalloproteases.24, 25 Many DUBs harbor ubiquitin-binding motifs, such as the zinc finger ubiquitin-specific protease domain, the ubiquitin-interacting motif and the ubiquitin-associated domain.26 These motifs likely coordinate the recognition and the recruitment of ubiquitinated substrates to ensure specificity and tight control.
Given the central role of ubiquitination in NF-κB signaling pathways, it is not surprising that DUBs play a critical role in the inhibition of NF-κB pathways. The two most prominent and best studied DUBs that regulate NF-κB are cylindromatosis (CYLD) and A20.27 Both CYLD and A20 are key inhibitors of NF-κB and are important for homeostatic control of NF-κB and inflammation. The role of CYLD in the inhibition of NF-κB has been reviewed elsewhere,28 and the main focus of this review article will be on A20 inhibition of NF-κB.
A20 inhibition of NF-κB signaling
A20 (also known as TNFAIP3) was first discovered in 1990 as a cytokine-induced gene in human umbilical vein endothelial cells.29 It was also found that multiple NF-κB activating stimuli induce A20 expression via NF-κB sites in the A20 promoter.30, 31 Although A20 was initially described as an inhibitor of TNF-induced cell death,32 subsequent studies showed that overexpression of A20 inhibited NF-κB activation in response to different stimuli.33, 34 In the TNFR pathway, A20 was found to interact with the E3 ligase TRAF2 and inhibit TRAF2-induced NF-κB activation.34 Similarly, A20 also inhibited NF-κB activation triggered by RIP1 overexpression.35 These initial studies suggested that A20 may be targeting upstream receptor proximal molecules to regulate NF-κB signaling. Indeed, A20 also interacted with the E3 ligase TRAF6 and inhibited IL-1-induced NF-κB activation at the level of TRAF6.36
Mice lacking A20 were generated shortly thereafter and provided important insight into the biological role of A20. A20-deficient mice died prematurely as a result of spontaneous multi-organ inflammation and cachexia.37 These mice also exhibited a profound defect in resolving acute inflammatory responses and succumbed to exposure to sublethal doses of LPS.37 Mechanistic studies performed in A20-deficient mouse embryonic fibroblasts (MEFs) indicated a defect in the termination of TNF-induced IKK and NF-κB activation.37 Thus, A20 is an important negative feedback regulator of NF-κB essential for immune homeostasis.
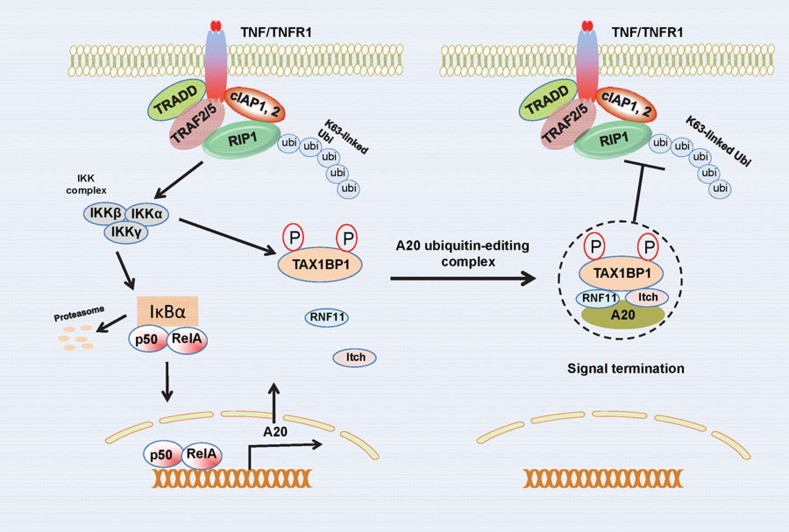
Despite the important insight into A20 function from initial studies with A20-deficient mice, it was still yet to be elucidated how A20 inhibited NF-κB. A20 was subsequently found to contain an already defined previously OTU DUB domain embedded in its N-terminus.38, 39 Overexpression of A20, but not a catalytically inactive DUB mutant, inhibited both RIP1 and TRAF6 ubiquitination.39, 40 A20 also contains seven C-terminal zinc finger domains, one of which functions as an E3 ligase. Taken together, A20 functions as a ubiquitin-editing enzyme in the TNFR pathway by first cleaving K63-linked polyubiquitin chains on RIP1 followed by catalysis of K48-linked chains via the fourth zinc finger (ZnF4) to trigger proteasomal degradation of RIP1 (Figure 1).39 The DUB and E3 ligase activities likely function in a sequential manner, although the degradation of RIP1 does not occur until several hours after TNF stimulation, at which time NF-κB signaling is largely inhibited.
Figure 1.
Mechanisms of NF-κB inhibition by A20 in the TNFR1 signaling pathway. A20 is induced by NF-κB in the TNFR1 pathway and functions in a negative feedback loop. The A20 adaptor molecule TAX1BP1 is phosphorylated on Ser593 and Ser624 by IKKα which nucleates the A20 ubiquitin-editing complex and is essential for interactions between TAX1BP1, Itch, RNF11 and A20. The A20 ubiquitin-editing complex inhibits RIP1 K63-linked polyubiquitination to terminate NF-κB signaling downstream of TNFR1. IKK, IκB kinase; RNF11, ring finger protein 11; TAX1BP1, Tax1-binding protein 1; TNFR1, TNF receptor 1.
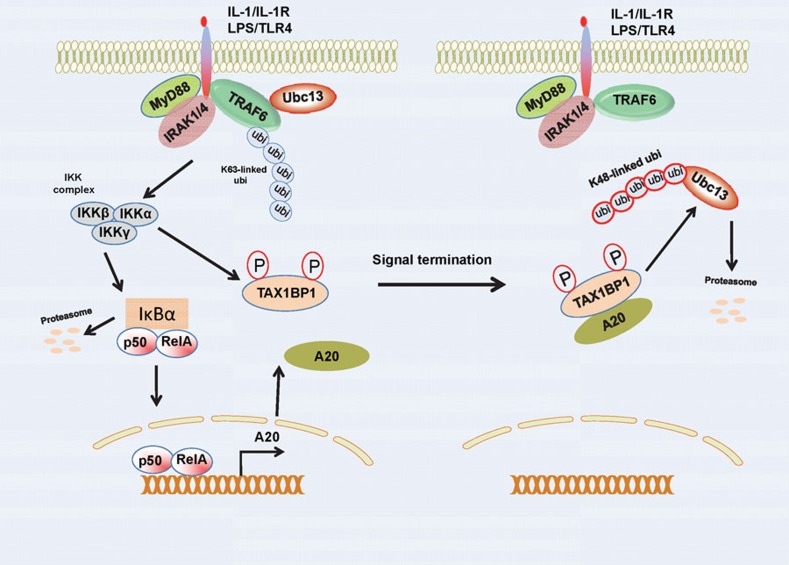
Thus far, the ubiquitin-editing function of A20 has only been described in the TNFR pathway. In the IL-1R/TLR4 pathways, A20 blocks TRAF6 ubiquitination, although it does not trigger its degradation, in contrast to the TNFR pathway where RIP1 is degraded. Therefore, A20 either inhibits IL-1R/TLR4 solely via its DUB domain, or alternative mechanisms exist. We have described such an alternative mechanism that occurs independently of the catalytic function of A20, whereby A20 targets E3 ligases and disrupts the interactions between E2:E3 ubiquitin enzyme complexes.41 A20 is recruited to the E3 ligases TRAF6, TRAF2 and cIAP1/2, and disrupts their binding with the E2 enzymes Ubc13 and UbcH5c, and furthermore triggers the ubiquitination and degradation of the E2 enzymes at later time points after stimulation (4–6 h) (Figure 2).41 Therefore, it appears that A20 utilizes distinct mechanisms to inhibit NF-κB activation in different pathways.
Figure 2.
Mechanisms of NF-κB inhibition by A20 in IL-1R/TLR4 signaling pathways. A20 is induced by NF-κB downstream of IL-1R/TLR4. TAX1BP1 cooperates with A20 to disrupt the interactions between the E3 ligase TRAF6 and the E2 enzymes Ubc13 and UbcH5c upon IL-1R/TLR4 stimulation. At later times, A20 conjugates K48-linked polyubiquitin chains on the E2 enzymes to trigger their proteasomal degradation. TAX1BP1, Tax1-binding protein 1.
A20 contains seven zinc finger domains, one of which (ZnF4) appears to function as an E3 ligase domain.39 Interestingly, structural studies have revealed that ZnF4 interacts with mono-ubiquitin and K63-linked polyubiquitin chains.42 Therefore, ZnF4 may participate in the recognition of ubiquitinated A20 substrates. The zinc finger domains of A20 may also be involved in targeting A20 to a membrane-associated compartment where it targets certain substrates such as TRAF2 for lysosomal degradation.43 This function of A20 was restricted to zinc fingers 6–7, although redundancy with adjacent zinc fingers was observed, consistent with other studies showing that some of the A20 zinc fingers are functionally redundant.44
Recently, a study by the Chen group reported that A20 inhibits IKK by a noncatalytic mechanism. When overexpressed in 293 cells, A20 inhibits TNF-induced IKK activation without reducing RIP1 ubiquitination.45 The A20 DUB mutant (C103A) inhibited IKK activation as effectively as wild-type A20 in this system. A20 inhibition of IKK was dependent on A20 binding to NEMO which was induced by K63-linked polyubiquitin chains.45 The binding of A20 with NEMO occurred rapidly upon IL-1 stimulation (2–5 min) in HeLa cells.45 Therefore, the polyubiquitin chains that serve to activate IKK also serve a dual function in its negative regulation by the recruitment of A20 to NEMO. The interaction of A20 with NEMO blocked the phosphorylation of IKK by TAK1. Interestingly, A20 zinc fingers 4 and 7 interacted with ubiquitin and were important for the inhibition of NF-κB consistent with prior studies.42 How can these results be reconciled with previous studies that clearly showed that A20 functions upstream of IKK?39, 41 Why would A20 need to inhibit multiple targets in the same pathway, particularly those downstream of its other well established targets? It is important to note that A20 is not expressed (or expressed at very low levels) in most cell types but is induced within 30–60 min upon NF-κB activation. Therefore, caution must be taken upon overexpression studies with A20 or the use of transformed cell lines where the expression of A20 may be dysregulated. The findings by Chen and colleagues suggest that A20 forms a complex with NEMO within minutes of ligand binding to the receptor, leading to inhibition of IKK via a noncatalytic mechanism. Therefore, this model may be more relevant in T and B lymphocytes where the basal levels of A20 are high.46 Taken together, it has become increasingly clear that A20 may inhibit NF-κB independently of its deubiquitinase activity, although additional studies are clearly needed to further test and refine the different models of A20 inhibition of NF-κB.
A20 regulation of innate and adaptive immunity
As mentioned earlier, Tnfaip3–/– mice develop spontaneous multi-organ inflammation that leads to premature lethality.37 Tnfaip3–/– mice on a Rag1–/– background still developed spontaneous inflammation, indicating that adaptive lymphocytes were not essential for the uncontrolled inflammatory response,37 although hematopoietic cells were clearly involved.47 Signals from TNF were similarly dispensable for the inflammation in Tnfaip3–/– mice, and A20 potently inhibited TLR signaling in macrophages.40 Consistent with these observations, homeostatic TLR signaling dependent on the Myd88 adaptor was dysregulated in A20-deficient mice.47 The homeostatic Myd88-dependent signals were derived from commensal intestinal bacteria since treatment of Tnfaip3–/– mice with broad spectrum antibiotics reduced systemic inflammation and prevented cachexia.47 A20 is therefore a critical regulator of immune homeostasis and TLR signaling in vivo.
Conditional A20 knockout mice have revealed important and surprising roles of A20 in different tissues. Mice lacking A20 in B cells (Tnfaip3fl/flCD19-Cre) developed autoimmune disease similar to systemic lupus erythematosus and exhibited autoantibody production.48, 49, 50 B cells deficient in A20 were hyper-responsive to B cell-activating stimuli and had much greater activation of NF-κB which led to enhanced proliferation and survival.48 Tnfaip3fl/flCD19-Cre mice had increased levels of proinflammatory cytokines such as IL-6 which contributed to a loss of B-cell tolerance coupled with an expansion of myeloid and effector T cells.48, 49 Thus, A20 expressed in the B-cell compartment is essential for B-cell homeostasis and the prevention of autoimmunity.
Deletion of A20 in dendritic cells (DCs) and myeloid cells also results in autoimmunity. Mice lacking A20 in DCs (Tnfaip3fl/flCD11c-Cre) did not develop severe multi-organ inflammation observed in Tnfaip3–/– mice, but rather exhibited splenomegaly, lymphadenopathy and autoimmune diseases such as colitis and spondyloarthritis.51 These mice also displayed characteristic features of autoimmunity such as autoantibody production and nephritis.52 A20-deficient DCs spontaneously matured, were hyper-responsive to TLR ligands and produced greater amounts of cytokines such as IL-6 and TNF.51, 52 Knockdown of A20 with small interfering RNA in DCs also enhanced DC maturation and activation.53 The loss of A20 in DCs also has a profound effect on other immune cell populations, and their activation since A20-deficient DCs trigger the expansion of myeloid and lymphoid cells and the aberrant activation of T and B lymphocytes.51, 52 Tnfaip3fl/flCD11c-Cre mice developed spontaneous inflammatory bowel disease that was mediated by activated T lymphocytes.51 Young (2–3 months of age) Tnfaip3fl/flCD11c-Cre mice were also more susceptible to dextran sodium sulfate-induced colitis.51 A20 also plays a role in human inflammatory bowel disease, since genome wide association studies have identified single-nucleotide polymorphisms in human A20 associated with Crohn's disease.51 Indeed, mucosal biopsies from Crohn's disease patients revealed aberrantly low A20 expression, suggesting that polymorphisms that decrease A20 expression may predispose individuals to Crohn's disease.54
Further insight into the role of A20 in preventing inflammatory bowel disease has been obtained by the generation of mice with specific deletion of A20 in intestinal epithelial cells (Tnfaip3fl/flVillin-Cre).55 These mice did not develop spontaneous colitis, but were more susceptible to dextran sodium sulfate-induced colitis.55 Since Tnfaip3fl/flVillin-Cre mice on a TNFR1-deficient background developed less severe colitis, it appears that a key function of A20 in enterocytes is to restrict TNFR signaling.55 However, intestinal epithelial cells lacking A20 were more sensitive to TNF-dependent apoptosis, resulting in impaired intestinal barrier function and subsequent commensal bacteria infiltration and systemic inflammation.55 Therefore, A20 is a key anti-apoptotic protein in intestinal epithelial cells that maintains epithelial barrier integrity and homeostasis. A20 also promotes intestinal epithelial barrier integrity and maintains homeostasis by a distinct mechanism involving deubiquitination of the tight junction protein occludin.56
Mice lacking A20 specifically in the myeloid compartment (Tnfaip3fl/flLysM-Cre) also developed autoimmune disease, although in this case the mice had spontaneous destructive polyarthritis.57 Tnfaip3fl/flLysM-Cre mice exhibited elevated serum levels of inflammatory cytokines TNF, IL-1β, IL-6 and MCP-1 that was also mirrored in joint tissue.57 A20-deficient peritoneal macrophages displayed enhanced LPS-induced IκBα degradation and TNF secretion.57 The autoimmunity and pathology in Tnfaip3fl/flLysM-Cre mice was dependent on IL-6 and the TLR4-Myd88 pathway, but independent of TNF.57 In humans, genome wide association studies have identified variants of the A20 locus, as well as other NF-κB regulatory genes such as TRAF1 and CD40 that are associated with rheumatoid arthritis.58
A20 has also been deleted in keratinocytes by crossing Tnfaip3fl/fl mice with transgenic mice expressing Cre under the control of the keratin 14 (K14) gene promoter (Tnfaip3fl/flK14-Cre).59 Interestingly, these mice do not develop pronounced skin inflammation but exhibit keratinocyte hyperproliferation and developmental abnormalities such as disheveled hair and abnormal ectodermal appendages59 Thus, the predominant function of A20 in keratinocytes is to oppose NF-κB in the ectodysplasin receptor pathway that regulates appendage development and skin homeostasis.59
A20 also regulates NF-κB signaling by the pattern recognition receptor NOD2 (nucleotide-binding oligomerization domain containing 2) which recognizes muramyl dipeptide, a peptidoglycan constituent. A20 inhibits NOD2 signaling by deubiquitinating the adaptor RIP2 (also known as RICK) to block downstream NF-κB signaling.60, 61 Macrophages lacking A20 show increased muramyl dipeptide-induced RIP2 K63-linked polyubiquitination and downstream NF-κB activation.61 Thus, A20 targets RIP2 ubiquitination to restrict NOD2-dependent signaling.
Although A20 is inducible by proinflammatory cytokines in most cell types, the regulation of A20 differs in lymphocytes. T lymphocytes express high basal levels of A20 that diminish upon T-cell activation.46 Nevertheless, A20 inhibits antigen receptor signaling to NF-κB in both T and B lymphocytes downstream of protein kinase C. Carma1 (also known as CARD11), Bcl10 and MALT1 form a signaling complex in T and B cells known as ‘CBM' that relays signals from protein kinase C to IKK and NF-κB activation.62 MALT1 undergoes K63-linked polyubiquitination by TRAF6 in response to TCR stimulation and this event is antagonized by A20 via its DUB domain.63 MALT1 is a paracaspase and mutually regulates A20 by cleavage at Arginine 439 to impair its NF-κB inhibitory function.64 Therefore, the balance of A20 inactivation and MALT1 ubiquitination likely determines the threshold of TCR-induced NF-κB activation. The regulation of MALT1 by A20 has largely been studied in T lymphocytes although A20 clearly is a negative regulator of BCR-induced NF-κB activation.49
Regulation of A20 by accessory proteins
In vitro studies conducted with recombinant A20 have indicated that A20 does not efficiently hydrolyze K63-linked polyubiquitin chains, but instead has a preference for K48-linked polyubiquitin chains.65 Therefore, A20 may rely on accessory proteins to provide specificity for K63-linked chains in vivo. One such protein is TAX1BP1 (Tax1-binding protein 1), a molecule that was identified in yeast two-hybrid screens as an interacting protein of human T cell leukemia virus type 1 (HTLV-1) Tax, A20 and TRAF6.66, 67, 68 Initial studies with TAX1BP1 suggested that it cooperates with A20 to inhibit cell death, although the mechanisms were unclear.67 Tax1bp1–/– mice were generated and showed an impairment in resolving acute and homeostatic inflammation, although the inflammation was not as severe as Tnfaip3–/– mice and was mainly restricted to the heart and skin.69 Tax1bp1–/– macrophages and fibroblasts activated NF-κB persistently in response to proinflammatory cytokines indicating a defect in the termination of NF-κB signaling.69, 70 TAX1BP1 inhibits NF-κB upstream of IKK since TRAF6 and RIP1 K63-linked ubiquitination were more pronounced in LPS and TNF-stimulated Tax1bp1–/– MEFs.41, 70 TAX1BP1 was also essential for interactions between A20 and its substrates TRAF6 and RIP1 in the IL-1R/TLR4 and TNFR pathways respectively.69, 70 Therefore, TAX1BP1 serves as a bona fide adaptor molecule for A20, consistent with previous studies that show that TAX1BP1 interacts with A20 and mediates its anti-apoptotic function.67
In addition to TAX1BP1, there are also other components of the A20 ubiquitin-editing complex that are essential for the function of A20. Itch (also known as AIP4) and RNF11 (ring finger protein 11) are E3 ligases that interact with each other and also both regulate A20.71 Itch contains several WW domains, a modular domain with conserved tryptophan residues that mediates interactions with proline-rich peptide-motifs 72 Itch interacts with TAX1BP1 in a TNF and IL-1-dependent manner via two ‘PPXY' motifs (where P=Proline, X=any amino acid and Y=tyrosine) located within the zinc finger motifs in TAX1BP1.73 Thus, TAX1BP1 recruits Itch to the A20 complex where Itch somehow facilitates TAX1BP1 and A20 with the binding and/or degradation of substrates. Itch-deficient MEFs phenocopy Tax1bp1–/– MEFs since NF-κB signaling is persistent upon LPS or TNF stimulation.73 The E3 ligase activity of Itch is also essential for NF-κB inhibition and Itch is important for the TNF-induced degradation of RIP1 which was impaired in Itch-deficient MEFs.73 Notably, Itchy mice (Itch knockout mice) also exhibit spontaneous inflammation and succumb to uncontrolled pulmonary inflammation.74
The RING domain E3 ligase RNF11 regulates tumor growth factor-β signaling by interactions with multiple signaling pathway components.75, 76 As mentioned earlier, Itch interacts with RNF11 and also ubiquitinates RNF11, suggesting that Itch may regulate RNF11 function.77 RNF11 was also shown to interact with A20, TAX1BP1 and NEMO by yeast two-hybrid screening suggesting its potential involvement in NF-κB signaling.78 Indeed, silencing of RNF11 by RNAi potentiated NF-κB activation indicating its role as a negative regulator of NF-κB.79 RNF11 interacts with TAX1BP1 and Itch upon TNF stimulation and is required for targeting of A20 to RIP1.79 The ‘PPXY' and RING domains of RNF11 are both essential for inhibition of NF-κB, although it remains unclear what the exact substrates of RNF11 are in NF-κB signaling. Regardless, RNF11 plays an important role in the termination of NF-κB signaling by regulating A20 in the context of a ubiquitin-editing complex. Generation of RNF11-deficient mice will be important to confirm its role as a regulator of A20 and NF-κB.
A number of other A20 interacting proteins have also been shown to inhibit NF-κB activation. ABIN1 (A20 binding inhibitor of NF-κB 1) was originally identified in a yeast two-hybrid screen as an A20-interacting protein which inhibits NF-κB when overexpressed.35, 80 There is evidence that ABIN1 recruits A20 to NEMO as a mechanism to inhibit NF-κB.81 However, MEFs derived from ABIN1 knockout mice did not have any major defects in NF-κB regulation, suggesting that ABIN1 may have a tissue-specific role in NF-κB inhibition or there may be redundancy with other ABIN molecules (ABIN2 and ABIN3).82 It is also possible that ABIN1 regulates other TLR-associated inflammatory pathways such as CCAAT/enhancer-binding protein β.83 Regardless, ABIN1 appears to be an important negative regulator of TLR signaling, since ABIN1-deficient mice developed a progressive lupus-like inflammatory disease and died prematurely.83 A similar phenotype was observed in ABIN1 knock-in mice with a disrupted ubiquitin-binding domain.84, 85
The 14–3–3 proteins also interact with A20 and may modulate the localization of A20 via chaperone activity.86, 87 However, 14–3–3 does not appear to regulate NF-κB suggesting that binding to A20 is for a distinct purpose.87 YMER (also known as CCDC50) was also reported to interact with A20, inhibit NF-κB and interact with ubiquitin via a ubiquitin-binding domain.88 However, YMER-deficient mice will be needed to firmly establish its role as an A20 and NF-κB regulator.
Since A20 inducibly interacts with a number of accessory proteins (TAX1BP1, Itch, RNF11) to assemble the A20 ubiquitin-editing complex upon TNF or IL-1 stimulation, it is likely that a common signaling protein in these pathways regulates the assembly of this complex. Indeed, we recently reported that IKK phosphorylates TAX1BP1 on two serine residues to trigger the nucleation of the A20 ubiquitin-editing complex (Figure 1).89 Surprisingly, the IKKα (but not IKKβ) subunit of IKK was essential for TAX1BP1 phosphorylation and the termination of TNF- and IL-1-induced NF-κB signaling.89 This finding is consistent with prior studies performed with IKKα-deficient mice and Ikkα–/– macrophages, showing that IKKα was an essential negative regulator of inflammation and canonical NF-κB signaling.90, 91 Exactly how TAX1BP1 phosphorylation nucleates the A20 ubiquitin-editing complex is unclear, but it is reasonable to predict that a conformational change is triggered by the phosphorylation. IKK also directly phosphorylates A20, although it is the IKKβ subunit that phosphorylates A20 on Ser381.92 A20 phosphorylation promotes NF-κB inhibition although the mechanism is unclear.
The role of A20 in cancer
NF-κB is persistently activated in most cancers and confers a survival advantage to cancer cells leading to resistance to chemotherapy and radiation therapies. It is now clear that genetic loss of key NF-κB inhibitors such as A20 may predispose to certain lymphoid malignancies. Indeed, A20 is commonly deleted in several subtypes of B-cell lymphomas (about 40%) including marginal zone lymphoma, diffuse large B-cell lymphoma, follicular lymphoma, MALT lymphoma and Hodgkin lymphoma.93, 94, 95, 96, 97 Not surprisingly, persistent NF-κB activation is a hallmark of all of these malignancies. A20 is inactivated in lymphomas by either point mutations, deletions or epigenetic inactivation by promoter methylation.96 Loss of A20 has also been found in Sezary syndrome, a cutaneous T-cell lymphoma.98 The paracaspase MALT1, which cleaves A20, may serve as a therapeutic target for B-cell lymphomas, since inhibitors of MALT1 catalytic activity suppress NF-κB activation and rescue A20 cleavage in DLBC lymphomas of the activated B-cell type.99 Whether MALT1 inhibitors will have efficacy for the treatment of tumors genetically inactivated for A20 is unclear.
In the context of oncogenic virus infection (Epstein–Barr virus) in Hodgkin lymphoma, much fewer mutations in A20 were observed suggesting that the virus may have a mechanism to inactivate A20.97 In fact, this may be a common feature for oncogenic viruses, since the HTLV-1 Tax oncoprotein blocks TAX1BP1 phosphorylation to inhibit A20 function.89 Viral inactivation of A20 further underscores its important tumor suppressor role in lymphoid cells.
The role of A20 in solid tumors is more complicated and is likely tissue- and context-specific, since A20 may be oncogenic in certain solid tumors such as breast carcinoma and glioma. In these cancers, the anti-apoptotic function of A20 may have been usurped in the pathway to tumorigenesis. A20 is overexpressed in aggressive breast carcinomas lacking expression of the estrogen receptor or progesterone receptor or in high-grade tumor samples.100 A20 protects against cell death in the breast cancer cell line MCF-7 treated with tamoxifen.100 A20 is similarly overexpressed in gliomas and also appears to function as an oncogene. RNAi-mediated knockdown of A20 in glioma cells reduced proliferation, promoted cell cycle arrest and triggered cell death.101 Interestingly, A20 was also overexpressed in glioma stem cells and played a role in cell survival, self-renewal and tumorigenesis.102 Although A20 may potentially serve as a therapeutic target in breast cancer or glioma, systemic inhibition of A20 may have deleterious consequences on immune homeostasis as we have learned from studies with knockout mice.
Concluding remarks and outstanding questions
In conclusion, the A20 deubiquitinase is a key negative regulator of NF-κB signaling that is essential for maintaining immune homeostasis and downregulating inflammation. However, it is important to consider that in addition to A20, there are a number of other DUBs that regulate NF-κB, most notably CYLD.103 Since A20 and CYLD share a number of substrates such as RIP1, what are the key determinants for specificity? It is likely that adaptor molecules, such as TAX1BP1 for A20, play a major role in mediating specificity for individual DUBs. In fact, there may be multiple adaptor and/or effector molecules that assemble into unique ubiquitin-editing complexes containing both DUBs and E3 ligases that are specific for certain substrates or pathways. In support of this notion, a recent study demonstrated that the E3 ligase Itch also cooperates with CYLD to downregulate TAK1 ubiquitination and inflammation.104 Therefore, Itch appears to be a key component of both A20 and CYLD complexes.
In addition to unique DUB-specific adaptor molecules and complexes, DUBs may also hydrolyze distinct polyubiquitin chains (i.e., K11 vs. K63) on substrates. Therefore, multiple DUBs may target the same substrate, but have specificity for different ubiquitin chains. Finally, it is also important to consider that DUBs may regulate NF-κB via distinct temporal and spatial differences. Indeed, CYLD is thought to primarily inhibit basal NF-κB activation, whereas in most cell types A20 inhibits activation-induced NF-κB.103
Despite the intense research on A20 and the impressive progress made in the last decade on its mechanisms of action, certain key questions remain. In addition to its well-ascribed functions as a DUB and ubiquitin-editing enzyme, there is also evidence that A20 may inhibit NF-κB via non-catalytic mechanisms.41, 45 For example, A20 disrupts the binding between E2 and E3 ubiquitin enzyme complexes to terminate IL-1R/TLR4 signaling.41 As mentioned earlier, a recent study suggests that A20 may also directly inhibit IKK through a noncatalytic mechanism dependent on ubiquitin binding via zinc finger domains.45 Therefore, what are the precise roles of the A20 OTU domain and the C-terminal zinc fingers in NF-κB and cell death inhibition? The zinc fingers in A20 have been shown to regulate E3 ligase activity, cellular localization and binding to ubiquitin and adaptor molecules. Which of these functions is most important for inhibition of NF-κB? Further studies are also necessary to determine if ubiquitin editing and/or noncatalytic inhibition of IKK or ubiquitin enzyme complexes by A20 may serve tissue-specific roles. A20 complexes and associated adaptor molecules may also differ depending on the cell type. Finally, although A20 is an important tumor suppressor in B-cell lymphomas, its role in other cancers seems to be highly tissue and context-specific, probably relating to whether the dominant function of A20 in a particular tissue is to inhibit NF-κB or apoptosis. The answers to the questions posed above may aid in the development of future therapies that modulate A20 expression and/or function in autoimmune diseases or cancer.
Acknowledgments
The laboratory of E W Harhaj is supported by NIH grant nos. PO1CA128115, RO1CA135362 and RO1GM083143.
References
- Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-κB activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skånland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Chastagner P, Israel A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE. 2008;3:e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Dixit VM. Ubiquitin-mediated regulation of TNFR1 signaling. Cytokine Growth Factor Rev. 2008;19:313–324. doi: 10.1016/j.cytogfr.2008.04.014. [DOI] [PubMed] [Google Scholar]
- O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-κB-independent cell-death switch in TNF signaling. Curr Biol. 2007;17:418–424. doi: 10.1016/j.cub.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, et al. TAB2 and TAB3 actvate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Akira S. Toll-like receptor signaling. J Biol Chem. 2003;278:38105–38108. doi: 10.1074/jbc.R300028200. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Ninomiya-Tsuji J, Qian Y, Matsumoto K, Li X. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phosphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol Cell Biol. 2002;22:7158–7167. doi: 10.1128/MCB.22.20.7158-7167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Harhaj EW, Dixit VM. Deubiquitinases in the regulation of NF-κB signaling. Cell Res. 2011;21:22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-κB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opipari AW, Jr, Boguski MS, Dixit VM. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J Biol Chem. 1990;265:14705–14708. [PubMed] [Google Scholar]
- Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by κB elements. J Biol Chem. 1992;267:17971–17976. [PubMed] [Google Scholar]
- Laherty CD, Perkins ND, Dixit VM. Human T cell leukemia virus type I Tax and phorbol 12-myristate 13-acetate induce expression of the A20 zinc finger protein by distinct mechanisms involving nuclear factor κB. J Biol Chem. 1993;268:5032–5039. [PubMed] [Google Scholar]
- Opipari AW, Jr, Hu HM, Yabkowitz R, Dixit VM. The A20 zinc finger protein protects cells from tumor necrosis factor cytotoxicity. J Biol Chem. 1992;267:12424–12427. [PubMed] [Google Scholar]
- Jaattela M, Mouritzen H, Elling F, Bastholm L. A20 zinc finger protein inhibits TNF and IL-1 signaling. J Immunol. 1996;156:1166–1173. [PubMed] [Google Scholar]
- Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-κB activation. Proc Natl Acad Sci USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyninck K, de Valck D, Vanden Berghe W, van Criekinge W, Contreras R, Fiers W, et al. The zinc finger protein A20 inhibits TNF-induced NF-κB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-κB-inhibiting protein ABIN. J Cell Biol. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyninck K, Beyaert R. The cytokine-inducible zinc finger protein A20 inhibits IL-1-induced NF-κB activation at the level of TRAF6. FEBS Lett. 1999;442:147–150. doi: 10.1016/s0014-5793(98)01645-7. [DOI] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, et al. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PC Ovaa H, Hamon M, Kilshaw PJ, Hamm S, Bauer S, et al. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J. 2004;378:727–734. doi: 10.1042/BJ20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- Shembade N, Ma A, Harhaj EW. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Li L, Soetandyo N, Wang Q, Ye Y. The zinc finger protein A20 targets TRAF2 to the lysosomes for degradation. Biochim Biophys Acta. 2009;1793:346–353. doi: 10.1016/j.bbamcr.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkenberg M, van Huffel S, Heyninck K, Beyaert R. Functional redundancy of the zinc fingers of A20 for inhibition of NF-κB activation and protein-protein interactions. FEBS Lett. 2001;498:93–97. doi: 10.1016/s0014-5793(01)02504-2. [DOI] [PubMed] [Google Scholar]
- Skaug B, Chen J, Du F, He J, Ma A, Chen ZJ. Direct, Noncatalytic Mechanism of IKK Inhibition by A20. Mol Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari M, Wolf FW, Seldin MF, O'Shea KS, Dixit VM, Turka LA. Lymphoid expression and regulation of A20, an inhibitor of programmed cell death. J Immunol. 1995;154:1699–1706. [PubMed] [Google Scholar]
- Turer EE, Tavares R, Hitotsumatsu O, Advincula R, Lee BL, Shifrin N, et al. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J Exp Med. 2008;205:451–464. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares RM, Turer EE, Liu CL, Advincula R, Scapini P, Rhee L, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Vahl JC, Kumar D, Heger K, Bertossi A, Wójtowicz E, et al. B cells lacking the tumor suppressor TNFAIP3/A20 display impaired differentiation and hyperactivation and cause inflammation and autoimmunity in aged mice. Blood. 2011;117:2227–2236. doi: 10.1182/blood-2010-09-306019. [DOI] [PubMed] [Google Scholar]
- Hovelmeyer N, Reissig S, Xuan NT, Adams-Quack P, Lukas D, Nikolaev A, et al. A20 deficiency in B cells enhances B-cell proliferation and results in the development of autoantibodies. Eur J Immunol. 2011;41:595–601. doi: 10.1002/eji.201041313. [DOI] [PubMed] [Google Scholar]
- Hammer GE, Turer EE, Taylor KE, Fang CJ, Advincula R, Oshima S, et al. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat Immunol. 2011;12:1184–1193. doi: 10.1038/ni.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, van Loo G, Waelput W, de Prijck S, Muskens F, Sze M, et al. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity. 2011;35:82–96. doi: 10.1016/j.immuni.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Song XT, Evel-Kabler K, Rollins L, Aldrich M, Huang XF, Chen SY. A20 is an antigen presentation attenuator, and its inhibition overcomes regulatory T cell-mediated suppression. Nat Med. 2008;14:258–265. doi: 10.1038/nm1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenescu R, Bruno ME, Rogier EW, Stefka AT, McMahan AE, Wright TB, et al. Signature biomarkers in Crohn's disease: toward a molecular classification. Mucosal Immunol. 2008;1:399–411. doi: 10.1038/mi.2008.32. [DOI] [PubMed] [Google Scholar]
- Vereecke L, Sze M, Mc Guire C, Rogiers B, Chu Y, Schmidt-Supprian M, et al. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J Exp Med. 2010;207:1513–1523. doi: 10.1084/jem.20092474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej LE, Jacques P, Maelfait J, Verheugen E, Kool M, Sze M, et al. TNFAIP3 maintains intestinal barrier function and supports epithelial cell tight junctions. PLoS ONE. 2011;6:e26352. doi: 10.1371/journal.pone.0026352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matmati M, Jacques P, Maelfait J, Verheugen E, Kool M, Sze M, et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens S, Lefebvre S, Gilbert B, Sze M, Devos M, Verhelst K, et al. Keratinocyte-specific ablation of the NF-κB regulatory protein A20 (TNFAIP3) reveals a role in the control of epidermal homeostasis. Cell Death Differ. 2011;18:1845–1853. doi: 10.1038/cdd.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Núñez G, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonska M, Lin X. NF-κB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 2011;21:55–70. doi: 10.1038/cr.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwel M, Welteke V, Oeckinghaus A, Baens M, Kloo B, Ferch U, et al. A20 negatively regulates T cell receptor signaling to NF-κB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-κB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Peleraux A, Thebault S, Dick J, Lemasson I, Devaux C, et al. CREB-2, a cellular CRE-dependent transcription repressor, functions in association with Tax as an activator of the human T-cell leukemia virus type 1 promoter. J Virol. 1998;72:8332–8337. doi: 10.1128/jvi.72.10.8332-8337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Valck D, Jin DY, Heyninck K, van de Craen M, Contreras R, Fiers W, et al. The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene. 1999;18:4182–4190. doi: 10.1038/sj.onc.1202787. [DOI] [PubMed] [Google Scholar]
- Ling L, Goeddel DV. T6BP, a TRAF6-interacting protein involved in IL-1 signaling. Proc Natl Acad Sci USA. 2000;97:9567–9572. doi: 10.1073/pnas.170279097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iha H, Peloponese JM, Verstrepen L, Zapart G, Ikeda F, Smith CD, et al. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J. 2008;27:629–641. doi: 10.1038/emboj.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Harhaj NS, Liebl DJ, Harhaj EW. Essential role for TAX1BP1 in the termination of TNF-α-, IL-1- and LPS-mediated NF-κB and JNK signaling. EMBO J. 2007;26:3910–3922. doi: 10.1038/sj.emboj.7601823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitching R, Wong MJ, Koehler D, Burger AM, Landberg G, Gish G, et al. The RING-H2 protein RNF11 is differentially expressed in breast tumours and interacts with HECT-type E3 ligases. Biochim Biophys Acta. 2003;1639:104–112. doi: 10.1016/j.bbadis.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- Shembade N, Harhaj NS, Parvatiyar K, Copeland NG, Jenkins NA, Matesic LE, et al. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- Matesic LE, Copeland NG, Jenkins NA. Itchy mice: the identification of a new pathway for the development of autoimmunity. Curr Top Microbiol Immunol. 2008;321:185–200. doi: 10.1007/978-3-540-75203-5_9. [DOI] [PubMed] [Google Scholar]
- Subramaniam V, Li H, Wong M, Kitching R, Attisano L, Wrana J, et al. The RING-H2 protein RNF11 is overexpressed in breast cancer and is a target of Smurf2 E3 ligase. Br J Cancer. 2003;89:1538–1544. doi: 10.1038/sj.bjc.6601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi P, Seth A. RNF11 is a multifunctional modulator of growth factor receptor signalling and transcriptional regulation. Eur J Cancer. 2005;41:2549–2560. doi: 10.1016/j.ejca.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Santonico E, Belleudi F, Panni S, Torrisi MR, Cesareni G, Castagnoli L. Multiple modification and protein interaction signals drive the Ring finger protein 11 (RNF11) E3 ligase to the endosomal compartment. Oncogene. 2010;29:5604–5618. doi: 10.1038/onc.2010.294. [DOI] [PubMed] [Google Scholar]
- Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, et al. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-κB signalling. EMBO J. 2009;28:513–522. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-κB activation, ABIN-1. FEBS Lett. 2003;536:135–140. doi: 10.1016/s0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A. ABIN-1 binds to NEMO/IKKγ and co-operates with A20 in inhibiting NF-κB. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wu R, High AA, Slaughter CA, Finkelstein D, Rehg JE, et al. A20-binding inhibitor of NF-κB (ABIN1) controls Toll-like receptor-mediated CCAAT/enhancer-binding protein beta activation and protects from inflammatory disease. Proc Natl Acad Sci USA. 2011;108:E998–E1006. doi: 10.1073/pnas.1106232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Löhr F, et al. Ubiquitin binding mediates the NF-κB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- Nanda SK, Venigalla RK, Ordureau A, Patterson-Kane JC, Powell DW, Toth R, et al. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J Exp Med. 2011;208:1215–1228. doi: 10.1084/jem.20102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenz C, Dixit VM. 14-3-3 proteins associate with A20 in an isoform-specific manner and function both as chaperone and adapter molecules. J Biol Chem. 1996;271:20029–20034. doi: 10.1074/jbc.271.33.20029. [DOI] [PubMed] [Google Scholar]
- de Valck D, Heyninck K, van Criekinge W, Vandenabeele P, Fiers W, Beyaert R. A20 inhibits NF-κB activation independently of binding to 14-3-3 proteins. Biochem Biophys Res Commun. 1997;238:590–594. doi: 10.1006/bbrc.1997.7343. [DOI] [PubMed] [Google Scholar]
- Bohgaki M, Tsukiyama T, Nakajima A, Maruyama S, Watanabe M, Koike T, et al. Involvement of Ymer in suppression of NF-κB activation by regulated interaction with lysine-63-linked polyubiquitin chain. Biochim Biophys Acta. 2008;1783:826–837. doi: 10.1016/j.bbamcr.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Shembade N, Pujari R, Harhaj NS, Abbott DW, Harhaj EW. The kinase IKKα inhibits activation of the transcription factor NF-κB by phosphorylating the regulatory molecule TAX1BP1. Nat Immunol. 2011;12:834–843. doi: 10.1038/ni.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Q, Bottero V, Estepa G, Morrison L, Mercurio F, et al. Enhanced NF-κB activation and cellular function in macrophages lacking IκB kinase 1 (IKK1) Proc Natl Acad Sci USA. 2005;102:12425–12430. doi: 10.1073/pnas.0505997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutti JE, Abbott DW, Zhou AY, Sprott KM, Asara JM, Hahn WC. IKKβ Phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-κB pathway. Mol Cell Biol. 2007;27:7451–7461. doi: 10.1128/MCB.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- Honma K, Tsuzuki S, Nakagawa M, Tagawa H, Nakamura S, Morishima Y, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114:246–275. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- Chanudet E, Huang Y, Ichimura K, Dong G, Hamoudi RA, Radford J, et al. A20 is targeted by promoter methylation, deletion and inactivating mutation in MALT lymphoma. Leukemia. 2010;24:483–487. doi: 10.1038/leu.2009.234. [DOI] [PubMed] [Google Scholar]
- Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun FC, Grabarczyk P, Möbs M, Braun FK, Eberle J, Beyer M, et al. Tumor suppressor TNFAIP3 (A20) is frequently deleted in Sezary syndrome. Leukemia. 2011;25:1494–1501. doi: 10.1038/leu.2011.101. [DOI] [PubMed] [Google Scholar]
- Ferch U, Kloo B, Gewies A, Pfänder V, Düwel M, Peschel C, et al. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell JA, Ghayad S, Ben-Larbi S, Dumontet C, Mechti N, Cohen PA. A20/TNFAIP3, a new estrogen-regulated gene that confers tamoxifen resistance in breast cancer cells. Oncogene. 2007;26:4656–4667. doi: 10.1038/sj.onc.1210269. [DOI] [PubMed] [Google Scholar]
- Guo Q, Dong H, Liu X, Wang C, Liu N, Zhang J, et al. A20 is overexpressed in glioma cells and may serve as a potential therapeutic target. Expert Opin Ther Targets. 2009;13:733–741. doi: 10.1517/14728220903045018. [DOI] [PubMed] [Google Scholar]
- Hjelmeland AB, Wu Q, Wickman S, Eyler C, Heddleston J, Shi Q, et al. Targeting A20 decreases glioma stem cell survival and tumor growth. PLoS Biol. 2010;8:e1000319. doi: 10.1371/journal.pbio.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N, Zeng M, Sinha I, Polin L, Wei WZ, Rathinam C, et al. The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat Immunol. 2011;12:1176–1183. doi: 10.1038/ni.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]