Abstract
Accumulating evidence suggests that protein acetylation plays a major regulatory role in many facets of transcriptional control of metabolism. The enzymes that catalyze the addition and removal of acetyl moieties are the Histone Acetyl Transferases (HATs) and Histone Deacetylases (HDACs), respectively. A number of recent studies have uncovered novel mechanisms and contexts in which different HDACs play critical roles in metabolic control. Understanding the role of Class I and II HDACs in different metabolic programs during development, as well as in the physiology and pathology of the adult organism, will lead to novel therapeutics for metabolic disease. Here, we review the current understanding of how Class I and Class II HDACs contribute to metabolic control.
Keywords: HDAC, HAT, lipogenesis, gluconeogenesis, AMPK, HDAC3
Protein acetylation: a major regulatory mode of intracellular signaling
Recent advances have placed acetylation as a major regulatory mode of intracellular signaling. A number of studies have shown that acetylation and deacetylation is a dynamic process that occurs in a large fraction of the proteome [1–3]. Initially, the best-studied and most abundant examples of acetylation and deacetylation were those of lysine residues, on histones. Acetylation on the N-terminus lysine tail of histones leads to a decrease in a positive charge and hence decreased affinity to DNA [4]. In turn, this is thought to prime DNA for transcription, and facilitate RNA polymerase and transcription factors to bind to the relaxed chromatin, in the promoters of actively transcribed genes. Conversely, deacetylation of histones increases their affinity to DNA, with concomitant tightening of the chromatin and reduction of transcriptional activity. Due to the repressive role that histone deacetylation has on transcription, the HDACs are often referred to as transcriptional co-repressors. However, new findings suggest that Class I and II HDACs also deacetylate non-histone targets, and in some cases play an activating role in transcription. In lieu of these novel roles of HDACs regulating non-histone targets, several groups have suggested that perhaps renaming the HATs and HDACs to Lysine (K) Acetyl Transferases (KATs) or Lysine (K) Deacetylases (KDACs) will more appropriately reflect their function. For the purpose of this review, we will continue to refer of them by their original abbreviated names - HATs and HDACs. We review here recent findings of the function of Class I and Class II HDACs in control of cellular and organismal metabolism.
Structure and Function
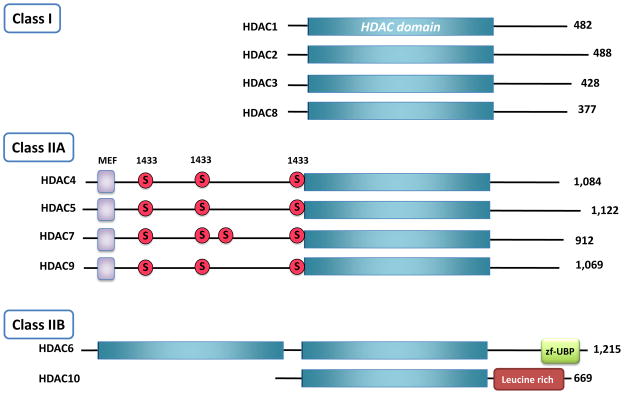
The HDACs are part of a large, evolutionarily conserved family of proteins, which dates back to prokaryotes [5, 6]. Since the discovery of the first HDACs [7, 8], eighteen distinct mammalian genes containing a deacetylase domain have been identified (Fig 1). These deacetylases are divided into two families, based on sequence similarity to their yeast orthologues, as well as co-factor dependence [9]. The first family of deacetylases can be further sub-divided into four subfamilies, containing Class I, Class IIa and IIb, and Class IV enzymes, that are grouped based on sequence homology to the S. cerevisiae deacetylases and which require zinc as a co-factor for enzymatic activity. The second family of deacetylases is often referred to as the Class III HDAC family, and its members are better known as the Sirtuin proteins. This class of deacetylases bears no sequence homology to the zinc dependant deacetylases, and requires NAD+ as a co-factor, and will not be discussed here.
Figure 1. The subclasses of the HDAC superfamily.
Class I HDACs are comprised almost entirely of a conserved deacetylase domain (shown in blue boxes). All Class IIA HDACs contain a long N-terminal adapter domain with myocyte enhancer factor 2 (MEF2) –binding sites (purple squares) and multiple phosphorylation sites (red circles) that are 14-3-3 chaperone protein binding sites, and a conserved deacetylase domain. HDAC6 is unique because it contains two deacetylase domains and a C-terminal zinc finger ubiquitin binding domain.
Class I HDACs
Class I HDACs (HDAC1, 2, 3 and 8) are homologous to budding yeast HDAC Rpd3. They are ubiquitously expressed with predominantly nuclear localization. Class I HDACs are almost entirely comprised of a conserved deacetylase domain (Fig 1) and have minimal N- and C- terminal domains. HDAC1 and 2, also referred to as the “canonical” HDACs, have strong enzymatic activity towards histones. In contrast to these conventional repressive effects on transcription, recent genome wide binding studies found these HDACs associated with active, as well as inactive chromatin [10]. Moreover, recent studies placed Class I HDACs on the map as key regulators of non-histone proteins, controlling deacetylation of transcription factors [11–16]. While first studied in depth for transcription factors such as p53 [17] and Stat3 [18], it has become clear that many, if not most, transcription factors and their co-regulators are controlled via acetylation. Like phosphorylation, in some instances acetylation can be activating for the target transcription factor whereas in others, this is an inactivation event. In addition to targeting transcription factors, Class I HDACs were also recently reported to deacetylate the AMP-activated Protein Kinase (AMPK), a central energy sensor with conserved roles in metabolism across all eukaryotes [19]. Similarly, HDAC8 was recently discovered as a major deacetylase of the cohesin subunit SMC3, whose proper deacetylation during anaphase is required to ensure proper regulation of sister chromatids [20].
HDAC1 and HDAC2 often have overlapping functions and predominantly act as part of transcriptional repressor multiprotein complexes, such as the Sin3 complex, the nucleosome remodeling and deacetylating (NuRD) complex, and the co-repressor for element-1-silencing transcription factor (CoREST) complex [21]. In addition to the Class I HDACs, some of the large repressor complexes also consist of other chromatin modifying enzymes. For example, the NURD complex contains lysine specific histone demethylases (LSD1), which allows the complex to serve several congruent functions in chromatin remodeling [10].
HDAC3 is usually found in a complex with two highly related hormone nuclear receptor co-repressors, namely Nuclear Receptor co-Repressor (NCoR1) and Silencing Mediator of Retinoic and Thyroid receptors (SMRT/NCoR2), and binding of NCor/SMRT to HDAC3 is required for its catalytic activity and recruitment to specific promoters, in vitro and in vivo [22–25]. Like HDAC1 and 2, HDAC3 was initially thought to solely act on histones and mediate transcriptional repression. However, recent studies suggest that HDAC3 may also have non-histone targets and does not always repress transcription. NCoR1/SMRT-bound HDAC3 can form a complex with the Class IIa HDACs [26], with HDAC3 being the major deacetylase activity in those complexes. Recent studies in mouse liver have shown that HDAC3/NCoR associates with circadian clock components, and control hepatic metabolism by repressing downstream target genes, in a circadian fashion [27]. It will be of interest to define the relative abundance of HDAC3 and the rest of the Class I HDACs in different transcriptional complexes, following different stimuli, as well as their subcellular and subnuclear localization, their promoter occupancy, and their relative roles on transcription, via effects on acetylation of histones vs. non-histone targets.
Class II HDACs
The mammalian Class II HDACs are most homologous to budding yeast HDA1 and are further subdivided into two classes – Class IIa (HDAC4, 5, 7, 9) and Class IIb (HDAC6 and HDAC10). Class IIa HDACs 4, 5, 7 and 9 have a very highly conserved deacetylase domain in their C-terminus (Fig. 1), and possess extensive N-terminus adapter domains that contain multiple conserved regulatory phosphorylation sites and protein binding domains[5, 28]. Distinct from the rest of the subclasses in the HDAC superfamily, the Class IIa HDACs can shuttle in and out of the nucleus, based on the phosphorylation status of few key serine residues in the N-terminus. Depending on the cell/tissue type and upstream stimulus, multiple kinase families including the Ca+/CaM-dependent protein kinases (CaMKs), Protein Kinase D (PKDs), and LKB1 dependent kinases of the AMPK family, can phosphorylate and regulate the localization of the Class IIa HDACs [29–34]. Indeed, phosphorylation on conserved serine residues within the N-terminus adapter domain promotes binding of the Class IIa HDACs to 14-3-3 adapter proteins, which promotes their nuclear export [35–38].
Class IIa HDACs also interact with the HDAC3/SMRT/NCoR complex [39–41]. In fact, the Class IIa HDACs are thought to bear minimal intrinsic deacetylase activity and any robust deacetylase activity has been attributed to their association with HDAC3 [42] [43]. Interestingly, evolutionary substitution of a key catalytic tyrosine residue (to histidine) that is conserved in all Class I HDACs, but not in vertebrate Class IIa HDACs, might account for their weak enzymatic activity, in conventional deacetylation assays [44]. Reversion of histidine to tyrosine, in mammalian HDAC4, increased deacetylase activity by 1,000 fold. This unveils a more specific role of vertebrate Class IIa deacetylases, which selects for low enzymatic activity and perhaps specificity towards largely undiscovered protein targets.
Recently, it has also been suggested that phosphorylation of the serine residues 278 and 279, located within the nuclear localization signal (NLS) of HDAC5, might regulate nuclear import/export. Mutating these serine residues to alanines redirects HDAC5 to the cytoplasm [45], thus decreasing its association with the HDAC3/NcoR1/SMRT complex, and indicating that these phosphorylation sites are important for nuclear import. Furthermore, serine 279 of HDAC5 can be phosphorylated by PKA, which in turn negatively regulates nuclear export [46].
Class IIb HDACs (HDAC6 and HDAC10) have less well-established functions, although HDAC6 is considered the major cytoplasmic deacetylase and is the only deacetylase from the superfamily that contains two deacetylase domains in addition to a ubiquitin binding domain at its C-terminus (Fig 1). HDAC6 is thought to regulate the deacetylation of α-tubulin, cortactin, chaperones, and IFNαR [47–52], and has recently been implicated in regulating autophagy as well as hepatic metabolism [53, 54]. Very little is currently known about the function of HDAC10 [55–58].
Role of the HDACs in physiology and metabolism
Transcriptional control of metabolism is a dynamic process that has been extensively studied over the past couple decades. Emerging evidence suggests that in addition to acetylation and deacetylation of histones, a number of transcription factors, co-activators, and repressors are also robustly controlled through acetylation and deacetylation.
Class I and II HDACS in cardiac and skeletal muscle physiology and metabolism
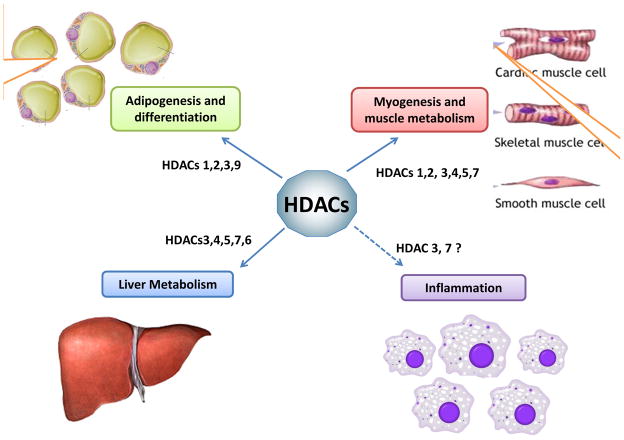
Recent findings have placed Class I HDACs in the heart of metabolic control of various tissues (see Figure 2). Hdac 1 and 2 were shown to play important but redundant roles in cardiac development and growth; complete genetic deletion of Hdac1 or Hdac2 led to embryonic or postnatal lethality respectively [59–61], however, inactivation of Hdac2 by lacZ mediated disruption produced viable mice, most likely attributed to the gene trap approach utilized [62]. Conditional deletion of both Hdac1 and Hdac2 in cardiomyocytes led to lethality shortly after birth, due to arrhythmia and inappropriate upregulation of calcium channels and contractile proteins [60]. Interestingly, even a single copy of Hdac1 or Hdac2 in the conditional mouse model was able to sustain mice through normal development. In adult mice, inhibition of Class I HDACs with apicidin derivative (API-D), an anti-parasitic agent and an HDAC inhibitor, prevented mice from getting cardiac hypertrophy by the thoracic aortic constriction pressure-overload model [63]. In addition, myocardial fibrosis was reduced by the HDAC inhibitors trichostatin A (TSA) and sodium valproate, in mice with left ventricular hypertrophy induced by aortic banding [64].
Figure 2. Class I and Class II HDACs are important in a number of metabolic tissues.
Deletion of both HDAC1 and 2 in mouse fibroblasts leads to a decrease in lipid accumulation following, adipogenic induction. Class IIa HDAC9 has also been implicated as a negative regulator in the control of adipogenesis. Class IIa HDACs are involved in myogenesis and deletion of multiple Class IIa HDACs in skeletal muscle de-repressess MEF2 targets resulting in an increase of slow myofibres. HDAC1 and 2 deletions in the myocardium result in lethality caused by dilated cardiomyopathy and arrhythmias. Class IIa HDAC5 and 9 have a role in suppressing cardiac growth in response to stress stimuli. Deletion of HDAC3 increases lipogenesis in the liver and causes severe hepatic steatosis. Class IIa HDACs regulate hepatic gluconeogenisis by recruitment of HDAC3, facilitating the deacetylation and activation of Foxo transcription factors during fasting. Other studies also implicate HDAC3 and HDAC7 in the control of inflammation.
Conditional deletion of Hdac3 under the control of the α-myosin heavy chain (α-MHC) promoter caused cardiac hypertrophy and re-programming of cardiomyocytes [65]. These mice had an increase in fatty acid uptake and oxidation, which led to significant myocardial lipid accumulation attributed to inappropriate increase in PPARα activity and PPARα target gene expression, suggesting that repression of this nuclear receptor by the NCoR/SMRT complex is lost in the absence of Hdac3. In a separate study, deletion of Hdac3 in the cardiac and skeletal muscle under the muscle creatine kinase (MCK) promoter, which allows for cardiac and skeletal muscle specific deletion postnatally, resulted in mice with no abnormalities, until challenged with a high fat diet (HFD) [66]. Upon switching to HFD, these mice presented with robust hypertrophic cardiomyopathy and heart failure, and a decrease in fatty acid metabolism genes, as well as a decrease in genes with functions in the electron transport chain and TCA cycle. These findings highlight the importance and differences in temporal regulations, even within the same gene and tissue. Furthermore, mice deficient in Hdac5 and Hdac9, Class IIa HDACs, presented with severe cardiac hypertrophy in response to cardiac stress [67, 68]. Collectively, these studies suggest that, despite redundant roles amongst some of the HDAC family members, the function of the Class I and II HDACs in developmental and postnatal transcriptional programs is quite complex.
The role of the Class IIa HDACs (HDAC4, 5, 7 and 9) in muscle physiology and metabolism has been well studied over the years. A number of studies have shown that Class IIa HDACs are regulated downstream of calcium signaling through the CaMK and PKD families, and more recently AMPK and its related kinases. Upon phosphorylation on conserved and specific residues within the N-terminus adaptor domain, the Class IIa HDACs bind to 14-3-3 scaffold proteins and are sequestered into the cytoplasm, where they are largely considered to be inactive. However, when dephosphorylated and nuclear, the Class IIa HDACs are thought to play a suppressive role on myogenesis and muscle fiber switch, via MEF2 (myocyte enhancer factor 2) specific repression. MEF2 transcription factors are believed to be key regulators for the oxidative, slow twitch (type I) myofibers. De-repression of MEF2 target genes downstream of Class IIa HDAC phosphorylation and calcium signaling allows for metabolic reprogramming. Consistent with this, genetic deletion of multiple Class IIa HDACs in skeletal muscle (due to some redundancy amongst different family members) promoted de-repression of MEF2 target genes and conversion of glycolytic fibers to oxidative fibers [69]. AMPK has been suggested to regulate the phosphorylation of Class IIa HDACs in myotubes allowing for MEF2-dependent derepression and induction of Glut4, providing one mechanism of how activated AMPK can enhance glucose uptake in the muscle [32]. Interestingly, another possible mode of regulation of MEF2 transcription factors could be through direct HDAC3 dependent deacetylation of MEF2 [12], which may be recruited to MEF2 by the Class IIa HDACs.
Recently, skeletal muscle specific deletion of NCoR1 in mice presented with a largely normal phenotype on a regular chow diet [70]. However, when challenged with a HFD, mice had increased muscle fiber size and exercise endurance, suggesting a suppressive role of NcoR1 in muscle reprogramming. Supporting this further, the NCoR1 KO mice on HFD had an increase in oxidative muscle metabolism, as well as mitochondrial quantity. These findings are consistent with the known inhibition by NCoR1 of nuclear receptors involved in this process, including PPARδ, and with the suppression of MEF2, which may be coordinately suppressed by NCoR1. Furthermore, MEF2 dependent genes were upregulated, which could be attributed to increased acetylation and activity of MEF2D transcription factor in the absence of NCoR1, due to destabilization of the NcoR1/SMRT/HDAC3 complex. Interestingly, NcoR1 mRNA was decreased by low glucose or high fatty acid levels in cells, paralleling its regulation by fasting and feeding and endurance exercise, in vivo. In addition to total NCoR1 levels being lowered under low energy conditions, the protein was localized in the nucleus in response to insulin, suggesting a crosstalk between classic insulin signaling and these transcriptional regulators [71]. Future studies are needed to dissect the molecular details of this crosstalk, in more depth. In addition, use of conditional NCoR1 mice can provide insight to what other transcription factors and nuclear receptors might be affected by to the loss of NCoR1, in different contexts.
Role of Class I and II HDACs in Adipogenesis
In a recent study, the canonical Class I HDACs 1 and 2 were shown to have a novel and unexpected role in the control of adipogenesis [71]. In this study, the authors utilized genetic deletion of both Hdac1 and Hdac2 in mouse embryonic fibroblasts and demonstrated a decrease in lipid accumulation following adipogenic induction of MEFs. Notably, deletion of each individual Class I HDAC did not have an effect on the differentiation process, supporting the notion that HDAC1 and HDAC2 have redundant functions in this cellular process.
Treatment of 3T3-L1 preadipocytes with the pan-HDAC inhibitors TSA, suberoylanilide hydroxamic acid (SAHA), or Scriptaid, led to a block in differentiation and adipogenesis, following induction [71]. The Class IIa specific inhibitor MC1568 was also recently shown to attenuate PPARγ-induced adipogenesis in 3T3-L1 cells, while the Class I-selective inhibitor MS275 blocked adipogenesis completely [72], consistent with other findings with TSA, SAHA and other HDAC inhibitors [73–75]. Interestingly, Class IIa HDAC9 has been implicated as a negative regulator in the control of adipogenesis [76]. In this study, out of the eleven HDACs examined, only HDAC9 mRNA was down-regulated during adipocyte differentiation. Interestingly, downregulation of HDAC9 happens relatively early and precedes the increase of expression of adipogenic genes during differentiation, suggesting that perhaps HDAC9 activity may need to be decreased in order for adipogenesis to proceed.
A recent report showed that deletion of NCoR1 in adipocytes leads to increased insulin sensitivity and reduced inflammatory response, in mice, despite excessive weight gain when challenged with a HFD [77]. These mice had a phenotype similar to the phenotype of mice treated with thiazolidinediode (TZD), a potent PPARγ agonist. Considering that HDAC3 may bind the NCoR1/SMRT co-repressor complex, it would be of interest to compare the phenotypes of HDAC3 adipose-specific knockout mice (KO) with the NCoR1 adipose-specific KO. Further genetic deletion analysis of single family members and combinations of the Class I and Class II HDACs in adipocytes in the intact mouse, will help us gain a better understanding the roles of the Class I HDACs in adipose tissues.
Role of Class I and II HDACs in liver metabolism
A number of recent studies have implicated a role for HDAC3 in the control of hepatic lipid metabolism. Conditional liver-specific KO of Hdac3 in adult mice resulted in severe hepatic steatosis with elevated expression of lipogenic enzymes [27, 78]. Genome-wide analysis of Hdac3 occupancy on lipogenic genes revealed a circadian binding pattern that inversely correlated with histone acetylation at these loci. Comparisons of Hdac3 cistromes to cistromes of its binding partner NCoR1 revealed high overlap, which was also shared with that of Rev-erbα, a nuclear receptor under circadian control, and a critical part of the core circadian clock machinery. The recruitment of HDAC3 to lipogenic gene loci, in liver, required Rev-erbα, as this binding and the circadian expression of these loci was lost when Rev-erbα was deleted. Strikingly, the binding of HDAC3 to genomic loci was extremely diurnal, with 99% binding observed during the day, when mice were not feeding. Collectively, the data suggest that Rev-Erbα recruits HDAC3 to lipogenic genes to repress their expression during the day (see Figure 3b). Whether HDAC3 catalytically acts on histones and/or additional targets in this context remains a subject for future studies.
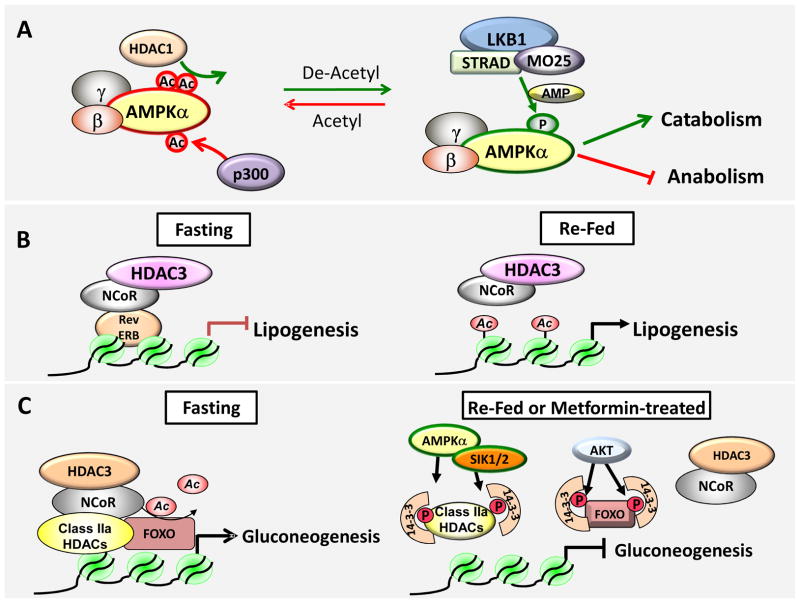
Figure 3. Molecular models for some of the metabolic processes regulated by Class I and II HDACs.
A) HDAC1 deacetylates AMPK and enhances its physical interaction with the upstream kinase LKB1. De-acetylated AMPK is more readily phosphorylated and activated. Activate AMPK positively regulates cellular processed that will replenish ATP in the cell and negatively regulates ones that are high in energy consumption. B) Rev-erbα recruits HDAC3 to lipogenic gene targets and globally regulates lipogenesis in the liver. HDAC3 recruitment to the genome displays a circadian rhythm and deletion of HDC3 in the liver leads to hepatic steatosis. C) Under fasting conditions, the hormone glucagon triggers dephosphorylation and nuclear recruitment of Class IIa HDACs, which in turn recruit the HDAC3/NCoR complex to deactylate FOXO family transcription factors. As acetylation inhibits FOXO binding to DNA, deacetylation of FOXO promotes its DNA binding and expression of its target genes. In the fed or metformin treated state, kinases of the AMPK family phosphorylate Class IIa HDACs and Akt phosphorylates FOXO, leading to 14-3-3 binding to both the Class IIa HDACs and FOXO, redirecting them in the cytoplasm and inactivating them.
The accumulation of hepatic lipids in HDAC3 liver specific KO mice fits with another study that used both inducible whole body KOs of HDAC3 in adult mice, and liver-specific KO starting during embryogenesis [78]. HDAC3 liver KO mice had lower fasting blood glucose and insulin levels, and an upregulation of a number of genes involved in lipid and fatty acid metabolism and cholesterol synthesis. The authors speculated that this may be due to increased activity of PPARγ [79], which was previously shown to recruit the HDAC3/NCoR1 complex [79, 80]. Inhibition with the PPARγ antagonist GW9662 in the HDAC3 null livers partially alleviated the lipid accumulation in these mice [78].
Interestingly, loss of Hdac3 in the liver activated the mammalian target of Rapamycin Complex 1 (mTORC1) pathway, and treatment of these mice with the mTORC1 inhibitor Rapamycin partially alleviated some of their fatty liver phenotype. Consistent with activation of such a pro-growth pathway, extended deletion of HDAC3 in the liver led to hepatocellular carcinoma (HCC) [81]. NCoR1 was down-regulated in tumors of these mice, suggesting that the heterotrimeric complex needs to stay intact to be functional. The cause of HCC was attributed to cumulative DNA damage [81], although deregulated liver metabolism and chronic stress from the fatty liver phenotype also could be considered a contributing factors.
A novel and unexpected role for the Class IIa HDACs in hepatic glucose metabolism was recently revealed. Treatment of mice with metformin or insulin, which respectively activate AMPK or related family members including Salt-Inducible Kinase 2 (SIK2), resulted in the phosphorylation of two key amino acids (Ser259 and Ser498 - using human HDAC5 numbering), in each of the Class IIa HDACs (Fig 1), and in nuclear exclusion of HDAC4, HDAC5, and HDAC7, in liver [34, 82]. Conversely, under fasted conditions in mouse liver, or in isolated hepatocytes treated with the fasting hormone glucagon, HDAC4, HDAC5, and HDAC7 underwent rapid dephosphorylation and nuclear accumulation (<15 minutes). To examine what genes the Class IIa HDACs regulates upon nuclear entry following glucagon treatment, unbiased microarray analysis was performed, revealing the unexpected finding that loss of these HDACs led to near complete loss of glucagon-induced expression of gluconeogenic genes. Surprisingly, the most HDAC-regulated gene on the entire whole genome microarray following glucagon or treatment with the cAMP agonist forskolin, was the catalytic subunit of Glucose 6 Phosphatase (G6pc). In this setting, the Class IIa HDACs appear to act as activators, a role opposite of their repressive function on MEF2 transcription factors. Further analysis demonstrated that Class IIa HDACs, through recruitment of HDAC3, regulated the acetylation and activity of the Foxo transcription factors, known inducers of the gluconeogenic transcription program [34]. Consistent with the recruitment of endogenous Class IIa HDACs to the promoters of the gluconeogenic genes G6pc and Pck1 in response to glucagon, the Class IIa HDACs were also required to recruit HDAC3 to these promoters (see Fig. 3c). In line with previous studies showing that acetylation of FOXO results in its inactivation [83, 84], hyperacetylation of FOXO1 in Class IIA HDAC shRNA expressing liver was accompanied by loss of FOXO target gene expression, like G6pc. As seen in the FOXO1 KO and G6PC liver-specific KO mice, mice bearing shRNAs that block Hdac4/5/7 expression in liver, showed increased glycogen accumulation and decreased blood glucose, in multiple murine models of metabolic syndrome (db/db, ob/ob, and C57Bl6 mice on a HFD). This phenocopies the human Glycogen Storage Disease Type I or Von Geirk’s disease, where G6PCc is inactivated due to mutations [85, 86]. Consistent with the finding that HDAC3 was the catalytic deacetylase regulating FOXO acetylation, a reduction of fasting blood glucose was also previous reported in the HDAC3 liver specific KO mice [78]. It remains to be seen what is the long-term consequences and phenotype of mice with liver specific deletions of the Class IIa HDACs. Interestingly, this mechanism was found to be conserved in fruit flies and the Drosophila orthologue dHDAC4 plays an important role of metabolic homeostasis of the fat body, in response to glucagon-like fly hormones [82]. Finally, though its connection to the above observations remains unclear, it was recently reported that HDAC6 may play a role in hepatic glucose metabolism as well [54].
Taken together, data on the role of Rev-ERB and HDAC3 in lipogenesis and on the role of Class IIa HDACs and HDAC3 in gluconeogenesis support the hypothesis that HDAC3 may only be bound to chromatin sites during fasting, whereas HDAC3 may associate with distinct activating and repressive transcriptional complexes during fasting. For example, for lipogenic genes and during the fasting period, Rev-Erb might recruit HDAC3 and NCoR to inhibit their expression. At the same time, Class IIa HDACs are activatively translocated into the nucleus by the fasting hormone glucagon where they recruit HDAC3/NCoR to deacetylate FOXO at gluconeogenic genes (see Fig 3c). One prediction of this model is that HDAC3 is at some promoters acting as a co-activator for gluconeogenic genes, while at the same time it is bound on promoters of lipogenic genes where it serves as a transcriptional repressor. Besides the presence and absence of Rev-ERB and the Class IIa HDACs, it will be important to define what other proteins are in the complexes that reside on the lipogenic and gluconeogenic promoters, and how fasting and feeding signals may regulate them.
As a final point, HDAC3’s binding partner NCoR1 has been reported to actively translocate to the nucleus of myocytes, following insulin treatment or refeeding [71], which was also observed in the livers of mice were the insulin-dependent mTORC1 pathway was genetically hyperactivated [70, 87]. At face value, if both signals are operational in liver at the same time, this would suggest that NCoR1 is shuttling into the nucleus under fed or insulin-stimulated conditions when the Class IIa HDACs may be shuttling out, though much further work is needed to fully investigate the regulation of HDAC3, NCor1, and Class IIa HDACs following distinct hormonal inputs in metabolic tissues.
It will be of great interest to also evaluate the role of Class I and Class IIa specific HDACs as targets for potential anti-diabetic therapies, using existing mouse models of diabetes. To this point, in spite of apparent lack of enzymatic activity, inhibitory compounds that directly bind to and induce degradation of Class IIa HDACs have been reported [88], and can be readily tested in models of metabolic dysfunction.
Role of Class I and II HDACs in Autophagy and other metabolic processes
Multiple recent reports suggest a role for both Class I and II HDACs in the regulation of autophagy [53, 89]. Conditional deletion of Hdac1 and Hdac2 in muscle led to myofiber degeneration and shared partial phenotypes with mice that are deficient in autophagy. Interestingly, deletion of both Hdac1/2 led to an increase in the levels of the polyubiquitin-binding protein p62 levels in skeletal muscle of neonatal animals, which was further enhanced when animals were fasted, implying a block in autophagy. The Class IIb HDAC6 has also been demonstrated to play a role in autophagy-mediated clearance of aggregated proteins and defective mitochondria [53, 90], though the specific target of its deacetylation in that process is not yet known. Notably however, HDAC6 is required for chloroqine-induced autophagy but not starvation-induced autophagy, suggesting a selectivity of HDAC6 for the “quality control” form of autophagy, which is basally required for the disposal of protein aggregates and damaged organelles [91]. Accordingly, the ubiquitin binding domain of HDAC6 (“zf-UBP” in Figure 1) is required for its ability to bind ubiquitinated protein aggregates and required for autophagosome-lysome fusion [86].
Another recent connection to autophagy control emerged from an unbiased RNAi screen, in which HDAC1 was discovered to be robustly controlling the acetylation of three lysine sites in the AMPK catalytic subunits AMPKa1 (PRKAA1) and AMPKa2 (PRKAA2) [17]. Deacetylation of the AMPK catalytic subunits led to their tighter association with the upstream kinase LKB1 that resulted in enhanced AMPK phosphorylation and activation [19] (see Fig 3a). This parallels previous studies showing acetylation control of the AMPK beta subunit Sip2 in budding yeast [92]. Considering that AMPK is a central regulator of metabolism and a key target of diabetes therapeutics [93–96] it would be interesting to further investigate whether the de-acetylation of AMPK in metabolic tissues is governed by HDAC1, and whether it is regulated by hormonal signals. Another core component of autophagy, which is activated by AMPK and inhibited by the mTORC1, is the autophagy kinase ULK1/Atg1, which itself was recently shown to be regulated by acetylation [97]. Indeed, a number of critical metabolic enzymes were shown to be regulated through reversible acetylation recently, including PEPCK [98], pyruvate kinase M2 [99] and a number of other metabolic enzymes [3, 10]. Whether Class I or II deacetylases control these events in vivo, remains to be investigated, though it is important to note that metabolic regulators beyond AMPK were found to be regulated by HDAC1 in the aforementioned study [19].
Finally, several reports have shown that HDAC3 complexed with NCoR1/SMRT plays a key role in macrophage activation, which is a major contributor to obesity induced inflammation [100–102]. Interestingly, while many early studies focused on the role of the HDAC3-NCor1 complex in the repression of Nuclear Receptors [103] and other transcription factors in macrophages [100], more recent studies suggest complex roles for HDAC3/NCor1 in the induction of a large percentage of inflammatory genes in macrophages through effects on interferon gene expression [104].
Concluding Remarks
Many recent studies have defined the role of acetylation and deacetylation as an abundant and dynamic process, integral to chromatin remodeling, as well as to the control of a great number of transcription factors and metabolic enzymes. Considering these crucial functions, it is necessary to further define the protein targets and functional roles each of the ten Class I and Class II HDACs play in metabolism during development, as well as in the physiology and pathology of adult tissues. The discovery that a vast percentage of metabolic enzymes are regulated by acetylation is particularly intriguing, though thus far most of these events have been suggested to be controlled by the Sirtuin/Class III HDAC family [105]. However, given recent studies discovering central metabolic regulators like AMPK as substrates for Class I HDACs [16], it will now be critical to re-examine how often Class I and II HDACs may control acetylation of non-histone targets. In addition, recent studies have uncovered a variety of new lysine modifications in histones, including lysine crotonylation, succinylation, and malonylation [106]. More recently, HDAC3 bound to NCor1 was reported to harbor decrotonylase activity in vitro [107], suggesting that the Class I and II HDACs may indeed hold novel activities and functions beyond what was imagined previously.
Defining the optimal therapeutic window for HDAC inhibitors in different disease states remains an urgent and ongoing area of investigation. It will be of great importance to determine if more specific HDAC inhibitors (HDACi) can be utilized for the treatment of metabolic disorders and the plethora of ailments associated with the metabolic syndrome [108]. In addition to the potential for harnessing HDAC modulation in the treatment of metabolic disease, it will be of great interest to explore what function these proteins might have in controlling metabolism in cancer cells, especially considering the re-emergence of cancer cell metabolism as a critical hallmark of tumorigenesis [109, 110] and the fact that two HDAC inhibitors were recently approved by the FDA as anti-cancer therapies [111–113]. Combining recent genetic dissections of HDAC function in mice with new biochemical and RNAi screens to decode their substrates, the next few years will no doubt reveal many new insights and new mysteries for this family of enzymes.
Acknowledgments
M.M.M. was supported T32CMG training grant to UCSD/Salk, as well as funding from the Rose Hills Foundation. Research in the lab of R.J.S. is supported by the Howard Hughes Medical Institute, NIH R01 DK080425, P01CA120964 and the American Diabetes Association Junior Faculty Award 1-08-JF-47. We thank the Leona and Harry Helmsley Charitable Trust for their generous support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kim SC, et al. Substrate and Functional Diversity of Lysine Acetylation Revealed by a Proteomics Survey. Molecular Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 2.Choudhary C, et al. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 3.Zhao S, et al. Regulation of Cellular Metabolism by Protein Lysine Acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton AL, et al. Enhanced Histone Acetylation and Transcription: A Dynamic Perspective. Molecular Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Haberland M, et al. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Ruijter AJ, et al. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taunton J, et al. A Mammalian Histone Deacetylase Related to the Yeast Transcriptional Regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 8.Grozinger CM, et al. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci U S A. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grozinger CM, Schreiber SL. Deacetylase Enzymes: Biological Functions Review and the Use of Small-Molecule Inhibitors. Chemistry & Biology. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L-f, et al. Duration of Nuclear NF-kB Action Regulated by Reversible Acetylation. Science. 2001;293:1653–1655. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 12.Gregoire S, et al. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiernan R, et al. Post-activation Turn-off of NF-κB-dependent Transcription Is Regulated by Acetylation of p65. The Journal of Biological Chemistry. 2003;278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 14.Canettieri G, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12:132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 15.Krämer OH, et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23:223–235. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Togi S, et al. HDAC3 influences phosphorylation of STAT3 at serine 727 by interacting with PP2A. Biochemical and Biophysical Research Communications. 2009;379:616–620. doi: 10.1016/j.bbrc.2008.12.132. [DOI] [PubMed] [Google Scholar]
- 17.Prives C, Manley JL. Why is p53 acetylated? Cell. 2001;107:815–818. doi: 10.1016/s0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- 18.Yuan ZL, et al. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y-y, et al. Functional dissection of lysine deacetylases reveals that HDAC1 and p300 regulate AMPK. Nature. 2012;482:251–255. doi: 10.1038/nature10804. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Deardorff MA, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012 doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Current Opinion in Genetics & Development. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 22.Li J, et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. The EMBO Journal. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guenther MG, et al. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon HG, et al. Purifcation and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. The EMBO Journal. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X-J, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nature Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischle W, et al. Enzymatic Activity Associated with Class II HDACs Is Dependent on a Multiprotein Complex Containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 27.Feng D, et al. A Circadian Rhythm Orchestrated by Histone Deacetylase 3 Controls Hepatic Lipid Metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang XJ, Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinsey TA, et al. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci U S A. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Passier R, et al. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J Clin Invest. 2000;105:1395–1406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vega RB, et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGee SL, et al. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 33.Berdeaux R, et al. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- 34.Mihaylova MM, et al. Class IIa Histone Deacetylases Are Hormone-Activated Regulators of FOXO and Mammalian Glucose Homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang AH, et al. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKinsey TA, et al. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao HY, et al. Mechanism for Nucleocytoplasmic Shuttling of Histone Deacetylase 7. The Journal of Biological Chemistry. 2001;276:47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 39.Kao HY, et al. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 40.Huang EY, et al. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2002;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 41.Downes M, et al. Identification of a nuclear domain with deacetylase activity. PNAS. 2000;97:10330–10335. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischle W, et al. Human HDAC7 Histone Deacetylase Activity Is Associated with HDAC3 in Vivo. The Journal of Biological Chemistry. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- 43.Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 44.Lahm A, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greco TM, et al. Nuclear Import of Histone Deacetylase 5 by Requisite Nuclear Localization Signal Phosphorylation. Phosphorylation Molecular & Cellular Proteomics. 2011;10.2:1–15. doi: 10.1074/mcp.M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ha CH, et al. PKA phosphorylates histone deacetylase 5 and prevents its nuclear export, leading to the inhibition of gene transcription and cardiomyocyte hypertrophy. PNAS. 2010;107:15467–15472. doi: 10.1073/pnas.1000462107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 48.Matsuyama A, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. The EMBO Journal. 2002;21 doi: 10.1093/emboj/cdf682. 6820-6868-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang X, et al. Acetylation-Dependent Signal Transduction for Type I Interferon Receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, et al. HDAC6 Modulates Cell Motility by Altering the Acetylation Level of Cortactin. Mol Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovacs JJ, et al. HDAC6 Regulates Hsp90 Acetylation and Chaperone-Dependent Activation of Glucocorticoid Receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 52.Kaluza D, et al. Class IIb HDAC6 regulates endothelial cell migration and angiogenesis by deacetylation of cortactin. The EMBO Journal. 2011;2011:4142–4156. doi: 10.1038/emboj.2011.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JY, et al. Disease-causing mutations in Parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winkler R, et al. Histone Deacetylase 6 (HDAC6) Is an Essential Modifier of Glucocorticoid-Induced Hepatic Gluconeogenesis. Diabetes. 2012;61:513–523. doi: 10.2337/db11-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kao HY, et al. Isolation and Characterization of Mammalian HDAC10, a Novel Histone Deacetylase. The Journal of Biological Chemistry. 2002;277:187–193. doi: 10.1074/jbc.M108931200. [DOI] [PubMed] [Google Scholar]
- 56.Fischer DD, et al. Isolation and Characterization of a Novel Class II Histone Deacetylase, HDAC10. The Journal of Biological Chemistry. 2002;277:6656–6666. doi: 10.1074/jbc.M108055200. [DOI] [PubMed] [Google Scholar]
- 57.Guardiola AR, Yao T-P. Molecular Cloning and Characterization of a Novel Histone Deacetylase HDAC10. The Journal of Biological Chemistry. 2002:277. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- 58.Tong JJ, et al. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain 2002. 2002;30:1114–1123. doi: 10.1093/nar/30.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression 2002. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kee HJ, Kook H. Roles and Targets of Class I and IIa Histone Deacetylases in Cardiac Hypertrophy. J of Biomed and Biotechn. 2011;2011 doi: 10.1155/2011/928326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trivedi CM, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3bold beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 63.Gallo P, et al. Inhibition of class I histone deacetylase with an apicidin derivative prevents cardiac hypertrophy and failure. Cardiovascular research. 2008;80:416–424. doi: 10.1093/cvr/cvn215. [DOI] [PubMed] [Google Scholar]
- 64.Kong Y, et al. Suppression of Class I and II Histone Deacetylases Blunts Pressure-Overload Cardiac Hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montgomery RL, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Z, et al. Diet-induced Lethality Due to Deletion of the Hdac3 Gene in Heart and Skeletal Muscle. J Biol Chem. 2011;286:33301–33309. doi: 10.1074/jbc.M111.277707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang CL, et al. Class II Histone Deacetylases Act as Signal-Responsive Repressors of Cardiac Hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang S, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potthoff MJ, et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamamoto H, et al. NCoR1 Is a Conserved Physiological Modulator of Muscle Mass and Oxidative Function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haberland M, et al. Redundant Control of Adipogenesis by Histone Deacetylases 1 and 2. J Biol Chem. 2010;285:14663–14670. doi: 10.1074/jbc.M109.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nebbioso A, et al. HDACs class II-selective inhibition alters nuclear receptor-dependent differentiation. J Mol Endocr. 2010;45:219–228. doi: 10.1677/JME-10-0043. [DOI] [PubMed] [Google Scholar]
- 73.Catalioto RM, et al. Chemically distinct HDAC inhibitors prevent adipose conversion of subcutaneous human white preadipocytes at an early stage of the differentiation program. Experimental Cell Research. 2009;2009:3267–3280. doi: 10.1016/j.yexcr.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 74.Burtona GR, et al. Microarray analysis of differentiation-specific gene expression during 3T3-L1 adipogenesis. Gene. 2004;329:167–187. doi: 10.1016/j.gene.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 75.Lagace DC, Nachtigal MW. Inhibition of Histone Deacetylase Activity by Valproic Acid Blocks Adipogenesis. J Biol Chem. 2004;279:18851–18860. doi: 10.1074/jbc.M312795200. [DOI] [PubMed] [Google Scholar]
- 76.Chatterjee TK, et al. Histone Deacetylase 9 Is a Negative Regulator of Adipogenic Differentiation. J Biol Chem. 2011;286:27836–27847. doi: 10.1074/jbc.M111.262964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li P, et al. Adipocyte NCoR Knockout Decreases PPARγ Phosphorylation and Enhances PPARγ Activity and Insulin Sensitivity. Cell. 2011;147:815–826. doi: 10.1016/j.cell.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Knutson SK, et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fajas L, et al. The Retinoblastoma-Histone Deacetylase 3 Complex Inhibits PPARγ and Adipocyte Differentiation. Dev Cell. 2002;3:903–910. doi: 10.1016/s1534-5807(02)00360-x. [DOI] [PubMed] [Google Scholar]
- 80.Guan HP, et al. Corepressors selectively control the transcriptional activity of PPARγ in adipocytes. Genes Dev. 2005;19:453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhaskara S, et al. Hdac3 Is Essential for the Maintenance of Chromatin Structure and Genome Stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang B, et al. A hormone-dependent module regulating energy balance. Cell. 2011;145:596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 84.Banks AS, et al. Dissociation of the Glucose and Lipid Regulatory Functions of FoxO1 by Targeted Knockin of Acetylation-Defective Alleles in Mice. Cell metabolism. 2011;14:587–597. doi: 10.1016/j.cmet.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salganik SV, et al. A detailed characterization of the adult mouse model of glycogen storage disease Ia. Lab Invest. 2009;89:1032–1042. doi: 10.1038/labinvest.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peng WT, et al. Generation of mice with a conditional allele for G6pc. Genesis. 2009;47:590–594. doi: 10.1002/dvg.20538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sengupta S, et al. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 88.Scognamiglioa A, et al. HDAC-class II specific inhibition involves HDAC proteasome-dependent degradation mediated by RANBP2. Biochimica et Biophysica Acta. 2008;1783:2030–2038. doi: 10.1016/j.bbamcr.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 89.Moresia V, et al. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. PNAS. 2012;109:1649–1654. doi: 10.1073/pnas.1121159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee JY, Yao TP. Quality control autophagy: A joint effort of ubiquitin, protein deacetylase and actin cytoskeleton. Autophagy. 2010:6. doi: 10.4161/auto.6.4.11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu JY, et al. Acetylation of Yeast AMPK Controls Intrinsic Aging Independently of Caloric Restriction. Cell. 2011;146:969–979. doi: 10.1016/j.cell.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kahn BB, et al. AMP-activated protein kinase: Ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 94.Zhang BB, et al. AMPK: an emerging drug target for diabetes and the metabolic syndrome. Cell Metab. 2009;9:407–416. doi: 10.1016/j.cmet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 95.Hardie DG, et al. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature cell biology. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin SY, et al. GSK3-TIP60-ULK1 Signaling Pathway Links Growth Factor Deprivation to Autophagy. Science. 2012;336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 98.Jiang W, et al. Acetylation Regulates Gluconeogenesis by Promoting PEPCK1 Degradation via Recruiting the UBR5 Ubiquitin Ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lv L, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ogawa S, et al. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1 dependent gene networks required for macrophage activation. PNAS. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghisletti S, et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Perissi V, et al. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genetics. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 103.Pascual G, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen X, et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiong Y, Guan KL. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J Cell Biol. 2012;198:155–164. doi: 10.1083/jcb.201202056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Madsen AS, Olsen CA. Profiling of Substrates for Zinc-dependent Lysine Deacylase Enzymes: HDAC3 Exhibits Decrotonylase Activity In Vitro. Angew Chem Int Ed Engl. 2012;51:9083–9087. doi: 10.1002/anie.201203754. [DOI] [PubMed] [Google Scholar]
- 108.Christensen DP, et al. Histone deacetylase (HDAC) inhibition as a novel treatment for diabetes mellitus. Mol Med. 2011;17:378–390. doi: 10.2119/molmed.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vander Heiden MG, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 111.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 112.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 113.Bolden JE, et al. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]