FoxO6 is enriched in the adult hippocampus, yet little is known about the role of FoxO factors in cognition. Salih et al. now show that FoxO6 knockout mice learn normally but are impaired in contextual fear conditioning and novel object recognition. Disrupting FoxO6 in the adult hippocampus leads to memory consolidation defects. Furthermore, FoxO6 mutant mice exhibit decreased dendritic spine density in hippocampal neurons. FoxO6 may thus promote memory consolidation by coordinating hippocampal neuronal connectivity.
Keywords: consolidation, FoxO transcription factors, hippocampus, insulin signaling, learning and memory, synaptic function
Abstract
The FoxO family of transcription factors is known to slow aging downstream from the insulin/IGF (insulin-like growth factor) signaling pathway. The most recently discovered FoxO isoform in mammals, FoxO6, is highly enriched in the adult hippocampus. However, the importance of FoxO factors in cognition is largely unknown. Here we generated mice lacking FoxO6 and found that these mice display normal learning but impaired memory consolidation in contextual fear conditioning and novel object recognition. Using stereotactic injection of viruses into the hippocampus of adult wild-type mice, we found that FoxO6 activity in the adult hippocampus is required for memory consolidation. Genome-wide approaches revealed that FoxO6 regulates a program of genes involved in synaptic function upon learning in the hippocampus. Consistently, FoxO6 deficiency results in decreased dendritic spine density in hippocampal neurons in vitro and in vivo. Thus, FoxO6 may promote memory consolidation by regulating a program coordinating neuronal connectivity in the hippocampus, which could have important implications for physiological and pathological age-dependent decline in memory.
The FoxO family of Forkhead transcription factors has attracted a lot of interest because of its conserved role in the regulation of aging downstream from the insulin and insulin-like growth factor (IGF) signaling pathway (Kenyon 2005). Mutations that attenuate the insulin/IGF signaling pathway—and thereby activate FoxO—extend life span in worms, flies, and mice (Partridge and Bruning 2008). Expression of FoxO family members can extend the life span of invertebrates (Henderson and Johnson 2001; Libina et al. 2003; Giannakou et al. 2004; Hwangbo et al. 2004). Mammals have four FoxO isoforms (FoxO1, FoxO3, FoxO4, and FoxO6) (Salih and Brunet 2008). Single-nucleotide polymorphisms in two of these isoforms (FoxO1 and FoxO3) have recently been associated with exceptional longevity in humans (Willcox et al. 2008; Flachsbart et al. 2009; Li et al. 2009; Pawlikowska et al. 2009; Soerensen et al. 2010; Kleindorp et al. 2011). Thus, the FoxO family of transcription factors appears to play a conserved “prolongevity” role from worms to humans.
The most recently identified FoxO family member, FoxO6, is unique in that its mRNA is expressed predominantly in the CNS in mammals (Jacobs et al. 2003; Hoekman et al. 2006), whereas FoxO1, FoxO3, and FoxO4 are expressed relatively ubiquitously (Furuyama et al. 2000; Biggs et al. 2001; Hoekman et al. 2006). FoxO6 mRNA is particularly highly expressed in the hippocampus (Jacobs et al. 2003; Hoekman et al. 2006), a region important for learning and memory. Like other FoxO family members, FoxO6 is negatively regulated by the insulin/IGF signaling pathway in cells. Phosphorylation of FoxO6 in response to insulin/IGF signaling attenuates FoxO6-dependent transcription, although it does not affect FoxO6 nuclear localization (Jacobs et al. 2003; van der Heide et al. 2005). Together, these observations raise the possibility that FoxO6 plays an important role in the hippocampus in conditions where insulin/IGF levels are low.
Relatively few studies have explored the mode of action of FoxO transcription factors in the nervous system. During development, the FoxO family is required to establish neuronal polarity, and the ectopic expression of FoxO6 can rescue the neuronal polarity defects due to deficiency of the FoxO family (de la Torre-Ubieta et al. 2010). In adults, FoxO family members play an important role in neural stem cell self-renewal and fate (Paik et al. 2009; Renault et al. 2009) and in neuronal apoptosis following epileptic seizures (Shinoda et al. 2004). At the organismal level, the deletion of FoxO1 or FoxO3 results in reduced anxiety- and depression-like behaviors, respectively (Polter et al. 2009). Interestingly, the insulin/IGF pathway, which regulates FoxO transcription factors, has been found to impact learning and memory in physiological or pathological conditions such as Alzheimer's disease (Cohen et al. 2009; Freude et al. 2009; Killick et al. 2009; Chen et al. 2011). However, the importance and mode of action of the hippocampal-enriched FoxO6 in cognitive behavior are not known.
While the role in learning and memory of prolongevity transcription factors such as FoxO is largely unexplored, it is well known that the regulation of gene expression is critical for proper cognitive function and reward behavior (Flavell and Greenberg 2008). For example, the activity-dependent transcription factor CREB plays an important role in regulating a gene expression program important for synaptic function (Tao et al. 1998), synaptic plasticity (Barco et al. 2002), and reward and addiction behaviors (Carlezon et al. 1998; Larson et al. 2011). Furthermore, MEF2, another activity-dependent transcription factor, is important for synaptic function (Flavell et al. 2006; Shalizi et al. 2006), learning and memory (Barbosa et al. 2008), and behavioral plasticity in response to drugs of abuse (Pulipparacharuvil et al. 2008). Several additional transcriptional regulators, including histone acetylases and deacetylases (Alarcon et al. 2004; Fischer et al. 2007; Peleg et al. 2010), histone methyltransferases (Maze et al. 2010; Covington et al. 2011), and chromatin-binding proteins like MeCP2 (Moretti et al. 2006; Zhou et al. 2006; Chahrour et al. 2008; Deng et al. 2010; Cohen et al. 2011; Goffin et al. 2012), also play an essential role in the regulation of gene expression, synaptic plasticity, and cognitive behaviors. However, the role in cognition of transcription factors that are regulated by insulin and IGF, such as FoxO factors, is much less clear. Furthermore, the gene expression program regulated by FoxO6 has never been characterized. Whether FoxO6-regulated genes overlap with genes controlled by other transcriptional regulators that play a role in synaptic and cognitive function is not known.
To investigate the role of the FoxO6 transcription factor in vivo, we generated mice with a deletion in the FoxO6 gene. FoxO6 mutant mice were viable and outwardly normal but demonstrated a significant impairment of contextual and object memory consolidation. Interestingly, disruption of FoxO6 activity in the CA1 region of the adult hippocampus was sufficient to lead to defects in memory consolidation. FoxO6 mutant mice also showed irregular theta wave frequency electroencephalography oscillations in the hippocampus as they explored novel objects, indicating that FoxO6 is required for neuronal synchronization. A genome-wide approach revealed that the gene expression program regulated by FoxO6 following novel object learning in the hippocampus was enriched for genes involved in synaptic function. Consistently, FoxO6 deficiency resulted in decreased dendritic spine density in the CA1 region of the hippocampus. Taken together, our findings show that FoxO6—a transcription factor that belongs to a family known to extend life span downstream from insulin/IGF signaling—promotes memory consolidation and modulates synaptic function, which has important implications for preventing the age-dependent decline in cognitive function.
Results
FoxO6 is expressed predominantly in the CA1 and CA3 regions of the hippocampus
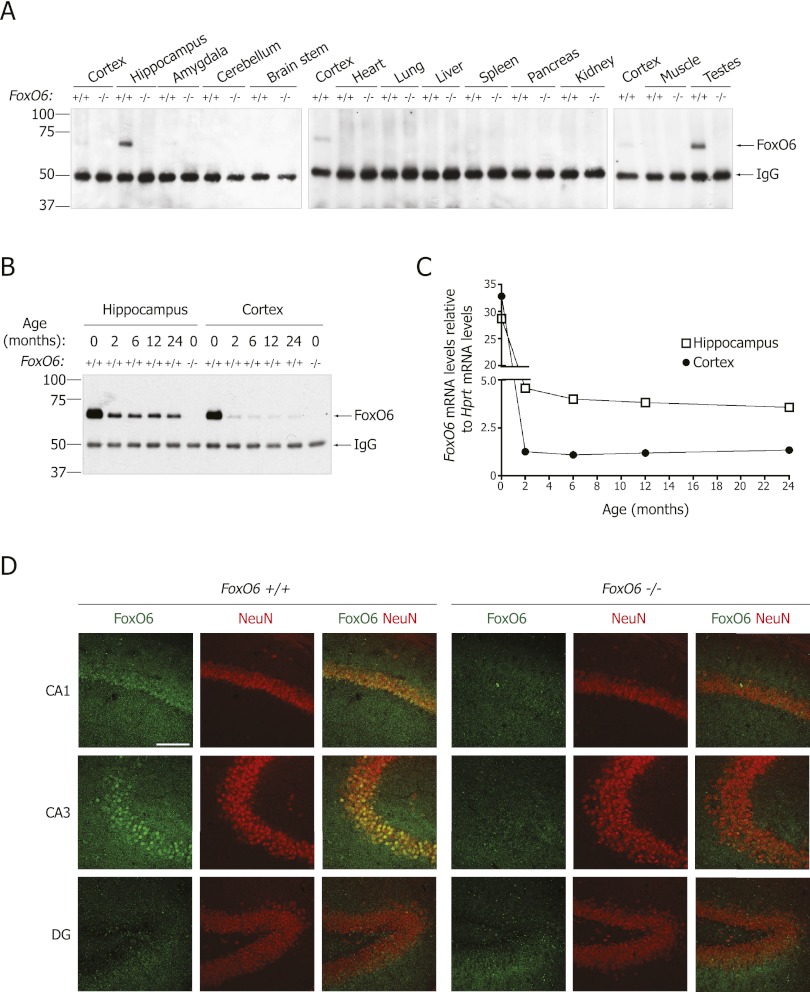
To examine FoxO6 protein expression in the adult brain, we generated two different antibodies to mouse FoxO6. Both antibodies were specific to FoxO6, as they recognized a protein of the correct molecular weight (65 kDa) in wild-type mouse tissues but not in tissues from FoxO6-null mice (Fig. 1A; Supplemental Fig. 1). We assessed FoxO6 protein levels in tissues from adult mice by immunoprecipitating FoxO6 with one of the antibodies and immunoblotting with the other antibody. FoxO6 protein was expressed abundantly in the adult hippocampus and at lower levels in the cortex and amygdala (Fig. 1A). FoxO6 protein was not detectable in the cerebellum and brain stem or in the nonneuronal tissues we tested, except for the testis (Fig. 1A). The expression pattern of the FoxO6 protein in adult mice was consistent with that of FoxO6 mRNA (Jacobs et al. 2003; Hoekman et al. 2006).
Figure 1.
FoxO6 is expressed predominantly in the CA1 and CA3 regions of the hippocampus in adult mice. (A) FoxO6 protein is highly expressed in the hippocampus of the adult mouse brain. Tissues from 8-wk-old FoxO6 mutant (−/−) and wild-type (+/+) siblings were tested by immunoprecipitation with an antibody to full-length FoxO6 and Western blotting with an antibody to an N-terminal FoxO6 peptide. (B,C) FoxO6 protein (B) and mRNA (C) are expressed abundantly in the hippocampus and cortex at birth and decrease while becoming enriched in the hippocampus relative to the cortex in adult mice. (D) FoxO6 protein is expressed in the nuclei of neurons of the pyramidal cell layer of the CA1 and CA3 hippocampal fields of the adult mouse brain but not in the dentate gyrus (DG). Immunohistochemistry on coronal brain sections from 2.5-mo-old FoxO6 mutant and wild-type siblings with antibodies to full-length FoxO6 and the neuronal marker NeuN. Bar, 100 μm.
We determined whether FoxO6 expression was affected by age in the hippocampus and cortex of mice. Both FoxO6 protein (Fig. 1B) and mRNA (Fig. 1C) decreased between birth and adulthood (2 mo of age) but then stayed relatively constant through old age (24 mo of age) (Fig. 1B,C). Interestingly, while FoxO6 protein and mRNA were expressed at similar levels in the hippocampus and the cortex at birth, FoxO6 was preferentially expressed in the hippocampus relative to the cortex in adult mice (Fig. 1B,C).
Immunohistochemistry with FoxO6 antibodies revealed that in the adult hippocampus, FoxO6 was expressed in the pyramidal cell layers of the CA1 and CA3 fields but not in the dentate gyrus (Fig. 1D). Furthermore, FoxO6 was localized in the nucleus of pyramidal neurons (Fig. 1D). These findings indicate that FoxO6 protein is predominantly expressed in neurons of the CA1 and CA3 regions of the hippocampus—two regions that are important for learning and memory in adult mice (Nakashiba et al. 2008; Hunsaker et al. 2009; Burgess and O'Keefe 2011).
FoxO6-null mice are viable and outwardly normal
To investigate the role of FoxO6 in vivo, we generated FoxO6-null mice by substituting the first coding exon of FoxO6 with a neomycin cassette using homologous recombination in embryonic stem (ES) cells (Supplemental Fig. 1A). Two independently targeted ES cell clones gave rise to male chimeric founders that transmitted the FoxO6-null allele to F1 offspring. Southern blots with three independent probes confirmed accurate homologous recombination in F1 progeny (Supplemental Fig. 1B–D). Western blots and RT-qPCR confirmed that full-length FoxO6 was not expressed in the FoxO6-null mice (Fig. 1A,B; Supplemental Fig. 1E), although we cannot rule out the possibility that short C-terminal fragments of FoxO6 are still being expressed. The protein levels of the other FoxO isoforms (FoxO1, FoxO3, and FoxO4) were not obviously increased in various brain regions of FoxO6 mutant mice (Supplemental Fig. 1F), suggesting that other FoxO isoforms do not compensate for FoxO6 loss, at least at the protein level and in these brain regions. FoxO6 mutant mice were born viable, and their body weight was normal to adulthood (Supplemental Fig. 2A). The brain of FoxO6 mutant mice had a normal weight and an overall normal structure (Supplemental Fig. 2B; data not shown). There were no obvious defects in the hippocampus of FoxO6 mutant mice, as shown by the neuronal stain NeuN (Fig. 1D) and cresyl violet staining (Supplemental Fig. 2C).
FoxO6 mutant mice displayed normal levels of thigmotaxis (an anxiety-related “wall-hugging” tendency innate to rodents) (Supplemental Fig. 3A,B) and normal levels of rearing (Supplemental Fig. 3C) in an open field arena. FoxO6 mutant mice showed slightly decreased locomotion (Supplemental Fig. 3D) and increased self-directed grooming (Supplemental Fig. 3E). FoxO6 mutant mice had normal motor coordination, as both the FoxO6 mutant mice and wild-type siblings maintained their balance for the same amount of time on a one-trial accelerating RotaRod assay (Supplemental Fig. 3F) and displayed no differences for interlimb coordination using the CatWalk apparatus (Supplemental Fig. 3G). The neuromuscular capacity of the FoxO6 mutant mice was also normal (Supplemental Fig. 3H,I). These findings indicate that FoxO6 is dispensable for overall survival, brain development, and motor coordination.
FoxO6 is necessary for consolidation of contextual and object recognition memory
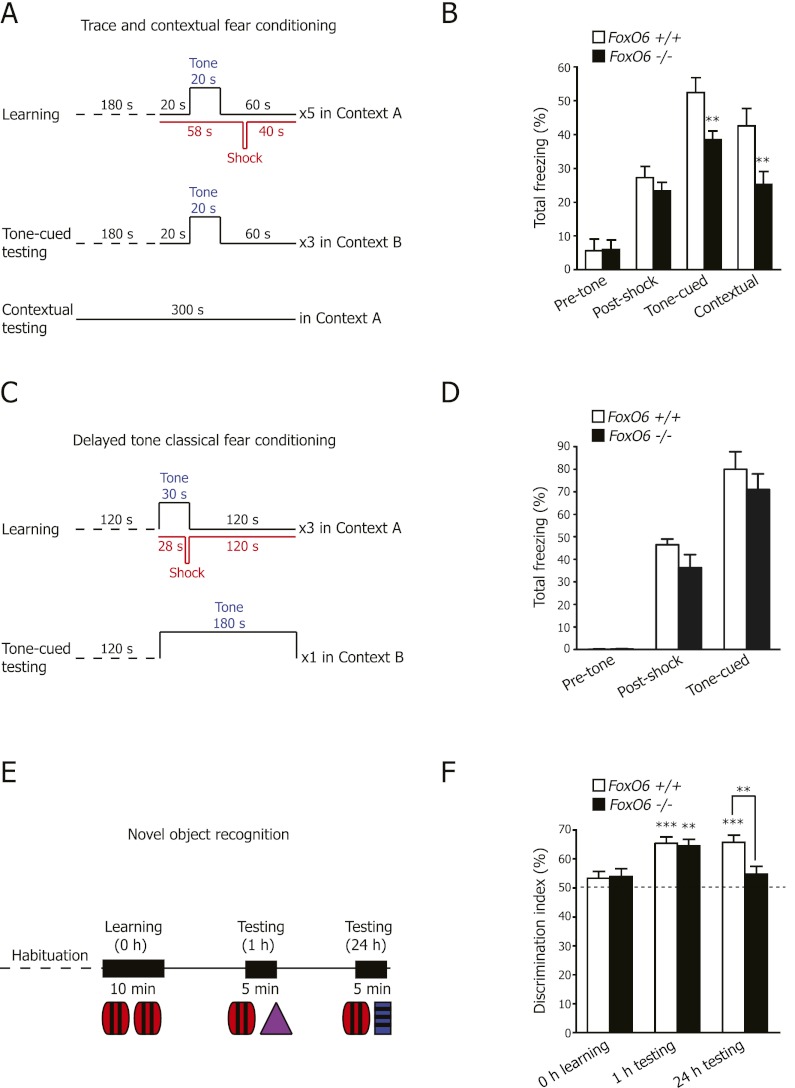
Given the selective expression of FoxO6 in the CA1 and CA3 regions of the adult hippocampus, we examined the behavior of FoxO6 mutant mice in hippocampal-dependent learning and memory. We first used contextual fear conditioning, a robust associative learning task that requires a functional hippocampus and amygdala (Kim and Fanselow 1992; Chowdhury et al. 2005; Salehi et al. 2009). In this test, a mild electric foot shock (unconditional stimulus) is paired to a specific context and a tone cue trace (conditional stimulus) (Fig. 2A). Both FoxO6-mutant and wild-type mice displayed increased freezing levels in response to the cycles of tone and foot shock (post-shock) (Fig. 2B), suggesting that both groups of mice learned the task comparably. Interestingly, when the FoxO6 mutant mice were tested for tone-cued memory 24 h later, they froze significantly less compared with their wild-type counterparts (tone-cued) (Fig. 2B), suggesting that the FoxO6 mutant mice have a defect in memory consolidation. Furthermore, FoxO6 mutant mice also froze significantly less compared with their wild-type counterparts when mice were tested for contextual memory 48 h after the training phase (contextual) (Fig. 2B), confirming a memory consolidation defect in FoxO6 mutant mice. The decreased freezing level displayed by the FoxO6 mutants during the contextual memory tests was not due to an impaired sensation of pain or impaired hearing and vision, as the FoxO6 mutant mice responded normally in a hot plate nociception assay (Supplemental Fig. 4A) and in a startle response assay (Supplemental Fig. 4B) and were able to find a visible platform in a swimming pool (Supplemental Fig. 4C). Together, these results indicate that the consolidation of fear memories is impaired in the FoxO6 mutant mice. The contextual fear-conditioning test that we used is thought to involve the hippocampus and the amygdala. The addition of a silent time interval between the tone and the foot shock (Fig. 2A) is thought to make the consolidation of this memory more hippocampus-dependent (Chowdhury et al. 2005). Furthermore, FoxO6 mutant mice did not exhibit significant defects in a classical fear-conditioning test (Fig. 2C,D), which is dependent on the amygdala (Han et al. 2009). These results suggest that the contextual fear memory defects of FoxO6 mutant mice are dependent on the hippocampus.
Figure 2.
FoxO6 mutant mice have impaired expression of contextual and recognition memories. (A) Scheme to test contextual learning and memory using trace tone-cued and contextual fear conditioning. Learning (day 1), tone-cued memory testing (day 2), and contextual memory testing (day 3). (B) FoxO6 mutant mice are significantly impaired in the consolidation of both trace tone-cued and contextual fear memories. Results are expressed as the percentage of time for which freezing behavior was shown prior to receiving the shock (pretone), during the learning stage (post-shock), during tone-cued memory testing (tone-cued), and during contextual memory testing (contextual). Mean ± SEM. n = 16–19 mice (4- to 7-mo-old males) per genotype. P < 0.0001 for the stage of the fear-conditioning test, two-way repeated measures ANOVA with genotype and stage of the fear conditioning as the factors, F3,99 = 64.29; (**) P < 0.01 FoxO6 wild-type versus null, unpaired Student's t-test. (C) Scheme to test auditory (tone) fear learning and memory using delayed tone fear conditioning. Learning (day 1) and tone-cued memory testing (day 2). (D) FoxO6 mutant mice are not impaired in the consolidation of tone-cued fear memories. Results are expressed as the percentage of time for which freezing behavior was shown prior to receiving the shock (pretone), during the learning stage (post-shock), and during tone-cued memory testing (tone-cued). Mean ± SEM. n = 7–9 mice (2- to 3-mo-old males) per genotype. P < 0.0001 for the stage of the fear-conditioning test, two-way repeated measures ANOVA with genotype and stage of the fear conditioning as the factors, F2,28 = 163.6. (E) Scheme to test object learning and memory using the novel object recognition task. (F) FoxO6 mutant mice could learn the novel object recognition task (1-h testing), but FoxO6 mutant mice are significantly impaired in the consolidation or retrieval of object recognition memories at 24 h. The discrimination index (percent) is shown for the time spent exploring the novel object versus the total time spent exploring both objects. Mean ± SEM. n = 20–21 mice (2- to 5 mo-old males) per genotype. P < 0.0001 for the stage of the novel object recognition test, two-way repeated measures ANOVA with genotype and stage of the test as the factors, F2,78 = 13.21. FoxO6 mutant and wild-type mice both show a preference for the novel object at 1 h. (**) P < 0.01; (***) P < 0.001 for 0 h versus 1 h, paired Student's t-test. Wild-type mice, but not FoxO6 mutant mice, show a preference for the novel object at 24 h. (***) P < 0.001 for FoxO6 wild-type 0 h versus 24 h, paired Student's t-test; (**) P < 0.01 for FoxO6 mutant versus wild-type at 24 h, unpaired Student's t-test.
We next tested FoxO6 mutant mice for learning and memory using the novel object recognition task, a test that has the advantage of not exposing the mice to overly stressful stimuli and that is thought to involve the hippocampus (Clark and Martin 2005; Bevins and Besheer 2006). This test involves mice exploring and learning two identical objects in a small arena for 10 min (Fig. 2E). Both FoxO6 mutants and wild-type siblings explored these original objects similarly (Supplemental Fig. 4D). One hour later, one of the objects was replaced with a novel object, and the percentage of time spent exploring the novel object was assessed. Both FoxO6 mutants and wild-type siblings displayed a comparable and significant preference for the novel object 1 h after the learning period (1-h testing) (Fig. 2F). These results indicate that FoxO6 mutant mice did not have impaired interest in the objects and that the FoxO6 mutant mice could learn the novel object recognition task normally. In contrast, when mice were tested for memory 24 h after the learning period by substituting the novel object for a different novel object, FoxO6 mutant mice no longer displayed a significant preference for the novel object, whereas wild-type mice did (24 h testing) (Fig. 2F). This result suggests that memory consolidation is defective in FoxO6-null mice, even in a situation of relatively low stress levels that is unlikely to involve the amygdala. In contrast, FoxO6-null mice do not have global defects in all hippocampus-dependent memory tasks, since FoxO6 mutant mice did not exhibit significant defects in spatial memory in the Morris water maze (Supplemental Fig. 5). Collectively, these results argue for a role of FoxO6 in circuits/networks of the hippocampus involved in contextual and object memory but not spatial memory.
Local and acute disruption of FoxO6 function in the CA1 region of the hippocampus is required for memory consolidation in adult wild-type mice
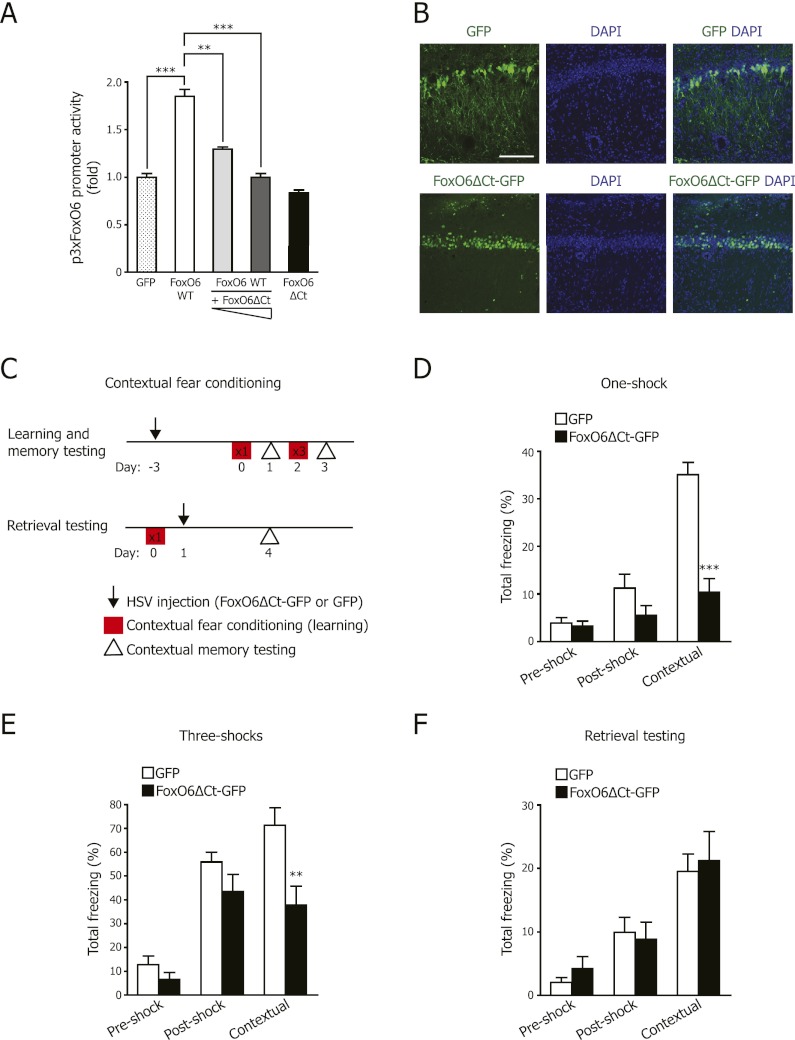
To test whether the memory deficits in FoxO6-null mice are due to the specific action of FoxO6 in adult mice, we used a viral approach to disrupt FoxO6 function locally and acutely in the CA1 region of the dorsal hippocampus in adult mice. We used replication-defective herpes simplex virus (HSV) because HSV is naturally neurotropic (Fink et al. 1996). To disrupt FoxO6 function, we generated HSV encoding a fusion between GFP and a truncated form of FoxO6 with a deletion of the C-terminal transactivation domain (FoxO6ΔCt-GFP) (Supplemental Fig. 6A). We verified that FoxO6ΔCt-GFP was transcriptionally inactive using a luciferase reporter gene under the control of FoxO6-binding sites (Fig. 3A). Importantly, FoxO6ΔCt-GFP acted as a dominant negative for wild-type FoxO6 function in cells (Fig. 3A). When overexpressed in cultured cells, FoxO6ΔCt-GFP also inhibited the activity of FoxO3 and FoxO4 (Supplemental Fig. 6B). However, FoxO4 is not expressed at detectable levels in the CA1 (Hoekman et al. 2006), and FoxO3 protein is expressed at lower levels in the CA1 than in the CA3 and the dentate gyrus (V Renault and A Brunet, unpubl.), so FoxO6ΔCt-GFP expression in the CA1 region is likely to affect mostly FoxO6 (and, to some extent, FoxO3). Stereotactic microinjection of HSV expressing FoxO6ΔCt-GFP or GFP alone as a control produced robust bilateral transgene expression that was localized to the CA1 region of the dorsal hippocampus of adult wild-type mice (Fig. 3B; Supplemental Fig. 6C), with no detectable expression in other regions, including the amygdala (Supplemental Fig. 6C). FoxO6ΔCt-GFP is localized in the nucleus of CA1 neurons (Fig. 3B). Together, these data indicate that stereotactic injection of HSV in the adult hippocampus leads to the relatively specific expression of a dominant-negative form of FoxO6 in the CA1 region of the hippocampus.
Figure 3.
Disrupting FoxO6 function locally and acutely in the CA1 region of the dorsal hippocampus impairs the consolidation of contextual fear memories. (A) FoxO6 with a deletion of the C-terminal region acts as a dominant negative. Luciferase assays in U2OS cells using constructs expressing wild-type FoxO6-GFP (FoxO6 WT) alone or with increasing amounts of FoxO6ΔCt-GFP (FoxO6ΔCt) and a luciferase reporter driven by three consensus FoxO6-binding sites (p3xFoxO6). Results are normalized to renilla and to GFP construct alone. Mean ± SEM from three independent experiments performed in triplicate. (**) P < 0.01; (***) P < 0.001, one-way ANOVA with the Bonferroni post hoc test. (B) HSV vectors expressing FoxO6ΔCt-GFP or GFP were microinjected into the CA1 region of the hippocampus in adult wild-type mice. FoxO6ΔCt-GFP was predominantly in the nucleus of neurons from the CA1 pyramidal cell layer. Counterstained with the nuclear marker DAPI. Bar, 100 μm. (C) Scheme to test contextual learning and memory in adult wild-type mice injected with HSV expressing FoxO6ΔCt-GFP or GFP. Contextual fear conditioning was performed with one or three foot shocks as indicated in the squares. (D) Wild-type mice expressing FoxO6ΔCt-GFP are significantly impaired in contextual fear memory following one foot shock. Mice were microinjected with viral vectors expressing FoxO6ΔCt-GFP or GFP in the CA1 region of the hippocampus, and contextual fear conditioning was performed 3 d following surgery. Results are expressed as the percentage of time for which freezing behavior was shown prior to receiving the shock (preshock), during the learning stage (post-shock), and during contextual memory testing (contextual). Mean ± SEM. n = 7 mice (2- to 3-mo-old) per viral construct. P < 0.0001 for the stage of the fear-conditioning test, two-way repeated measures ANOVA with viral construct and stage of the fear conditioning as the factors, F2,24 = 38.26; (***) P < 0.001 FoxO6ΔCt-GFP versus GFP, unpaired Student's t-test. (E) Adult mice injected with FoxO6ΔCt-GFP are significantly impaired in stronger contextual fear memory (in response to three foot shocks). Results are expressed as in D. Mean ± SEM. n = 7 mice (2- to 3-mo-old) per viral construct. P < 0.0001 for the stage of the fear-conditioning test, two-way repeated measures ANOVA with viral construct and stage of the fear conditioning as the factors, F2,24 = 56.95; (**) P < 0.01 FoxO6ΔCt-GFP versus GFP, unpaired Student's t-test. (F) Adult mice injected with FoxO6ΔCt-GFP after learning and consolidation can retrieve contextual fear memory normally. The learning phase of the contextual fear conditioning was performed on wild-type mice; 1 d later, the mice were microinjected with viral vectors expressing FoxO6ΔCt-GFP or GFP in the CA1 region of the hippocampus, and contextual fear conditioning was performed 3 d following surgery. Results are expressed as in D. Mean ± SEM. n = 6–7 mice (2- to 3-mo-old) per viral construct. P < 0.0001 for the stage of the fear-conditioning test, two-way repeated measures ANOVA with viral construct and stage of the fear conditioning as the factors, F2,22 = 20.72.
To investigate the importance of FoxO6 activity in learning and memory in adult mice, we stereotactically injected FoxO6ΔCt-GFP or GFP viruses and, 3 d later, conducted contextual fear conditioning in which a specific context was paired with a single foot shock (0.5 mA) (Fig. 3C) and then with three foot shocks (0.5 mA spaced 1 min apart) (Fig. 3C). Twenty-four hours after training, memory was tested by returning the mice to the original context. We used this simplified contextual fear-conditioning paradigm instead of the trace tone-cued and contextual procedure that we used in Figure 2 because this experiment was conducted at a different site. The simplified contextual fear-conditioning paradigm has been shown to assess hippocampal-dependent processes (Kim and Fanselow 1992; Wang et al. 2009). Mice expressing FoxO6ΔCt-GFP or GFP alone displayed similar levels of freezing both before and immediately after the single foot shock (Fig. 3D) or three foot shocks (Fig. 3E) as well as similar reactivity to the shocks (Supplemental Fig. 6D,E), indicating that disrupting FoxO6 function does not impair learning. Interestingly, expression of FoxO6ΔCt-GFP in the CA1 region of the hippocampus resulted in significantly decreased freezing levels 24 h after training compared with expression of GFP alone after a single foot shock (Fig. 3D) or after three foot shocks (Fig. 3E), indicating that memory consolidation is impaired in mice expressing FoxO6ΔCt-GFP. Importantly, mice microinjected with FoxO6ΔCt-GFP after fear conditioning still exhibit high levels of freezing (Fig. 3F; see below), indicating that this construct is not simply inducing nonspecific lesioning of hippocampal circuits. These results suggest that FoxO6 activity (and perhaps FoxO3 activity) is required in the CA1 region of the adult hippocampus for memory consolidation. HSV infects excitatory neurons rather than inhibitory neurons (Cole et al. 2012). Thus, the fact that injecting HSV expressing FoxO6ΔCt-GFP affects memory consolidation suggests that this function of FoxO6 is mediated by its action in excitatory neurons. It is also worth noting that the defects in memory consolidation of mice with acute FoxO6 dominant-negative expression are more pronounced than those of the chronic FoxO6 knockout. This could be due to slight differences in the contextual fear-conditioning assays, a possible compensation of chronic FoxO6 loss by other family members, and/or the fact that the FoxO6ΔCt-GFP can affect other FoxO family members that are expressed in the CA1 region (e.g., FoxO3).
We examined whether disrupting FoxO6 function affected retrieval of a contextual fear memory. Mice were first trained by contextual fear conditioning and stereotactically injected 24 h later with vectors expressing FoxO6ΔCt-GFP or GFP. The mice were then tested for contextual memory 3 d after the virus injection (Fig. 3C). Mice microinjected with FoxO6ΔCt-GFP virus showed freezing levels similar to mice microinjected with control GFP virus in this memory retrieval paradigm (Fig. 3F). These findings suggest that mice expressing FoxO6ΔCt in the hippocampus have intact retrieval of contextual fear memories that have already been consolidated. Taken together, these findings indicate that attenuating FoxO6 function in the CA1 region of the dorsal hippocampus disrupts the consolidation of contextual fear memories in adult mice without affecting learning or memory retrieval.
FoxO6 is necessary for neuronal synchronization upon novel object learning in the hippocampus
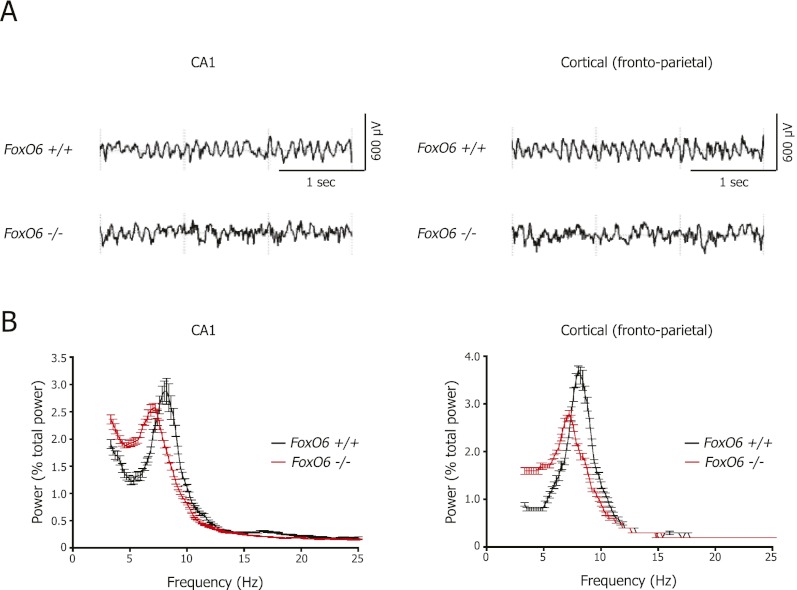
The synchronization of neuronal circuits is a crucial component in processing and storing the information upon learning for the consolidation of memories. To test whether FoxO6 is required for global synchronization of neuronal circuits in vivo, we measured theta rhythms in electroencephalogram (EEG) signals from the CA1 field of the hippocampus and from the cortex (frontoparietal) in FoxO6 mutant mice in response to novel objects (Fig. 4). Theta rhythms are oscillations in the frequency range of 6–12 Hz and represent the synchronization of the neuronal network when the hippocampus is engaged by activities such as exploration (Buzsaki 2002). As expected, wild-type mice displayed robust theta rhythms in the hippocampus during the active exploration of novel objects (Fig. 4A, left panel). In contrast, FoxO6 mutant mice showed irregular theta rhythms in the hippocampus (Fig. 4A, left panel). The averaged power spectra confirmed that the theta frequency oscillations in the hippocampus of FoxO6 mutant mice were irregular during the active exploration of novel objects (Fig. 4B, left panel). Theta frequency oscillations were also irregular in the frontoparietal cortex (Fig. 4A,B, right panels). The irregularity of the theta waves in the hippocampus and between the hippocampus and cortex of FoxO6 mutant mice suggests that neuronal network synchronization is impaired in FoxO6 mutants. These results raise the possibility that in the absence of FoxO6, the information being acquired during object exploration may not be correctly encoded into neuronal connections during memory consolidation.
Figure 4.
FoxO6 is necessary for neural network synchronicity in the hippocampus. (A) Representative spectrograms of EEG recordings from the hippocampal CA1 area (left panels) and the frontoparietal cortex region (right panels) for wild-type (top) and FoxO6 mutant (bottom) siblings. (B) Theta frequency oscillations are irregular in the FoxO6 mutant mice within the hippocampus and the frontoparietal cortex region. Normalized and averaged power spectra of EEG recordings from the hippocampal CA1 area (left panel) and the frontoparietal cortex region (right panel) for FoxO6 wild-type and mutant siblings. The X-axis represents the EEG frequency scale for 0.2-Hz frequency bins from 2 to 25 Hz. Mean ± SEM. n = 7–8 mice (3- to 4-mo-old males) per genotype. Peak theta frequency for wild-type siblings at 7.9 ± 0.2 Hz and for FoxO6 mutant mice at 7.0 ± 0.3 Hz in the CA1 region (P < 0.05 for FoxO6 mutant vs. wild-type mice, unpaired Student's t-test). Peak theta frequency for wild-type at 8.1 ± 0.1 Hz and for FoxO6 mutant mice at 7.3 ± 0.2 Hz in the frontoparietal cortex region (P < 0.01 for FoxO6 mutant vs. wild-type mice; unpaired Student's t-test).
FoxO6 is required for the expression of a program of genes involved in synaptic function in response to novel object learning
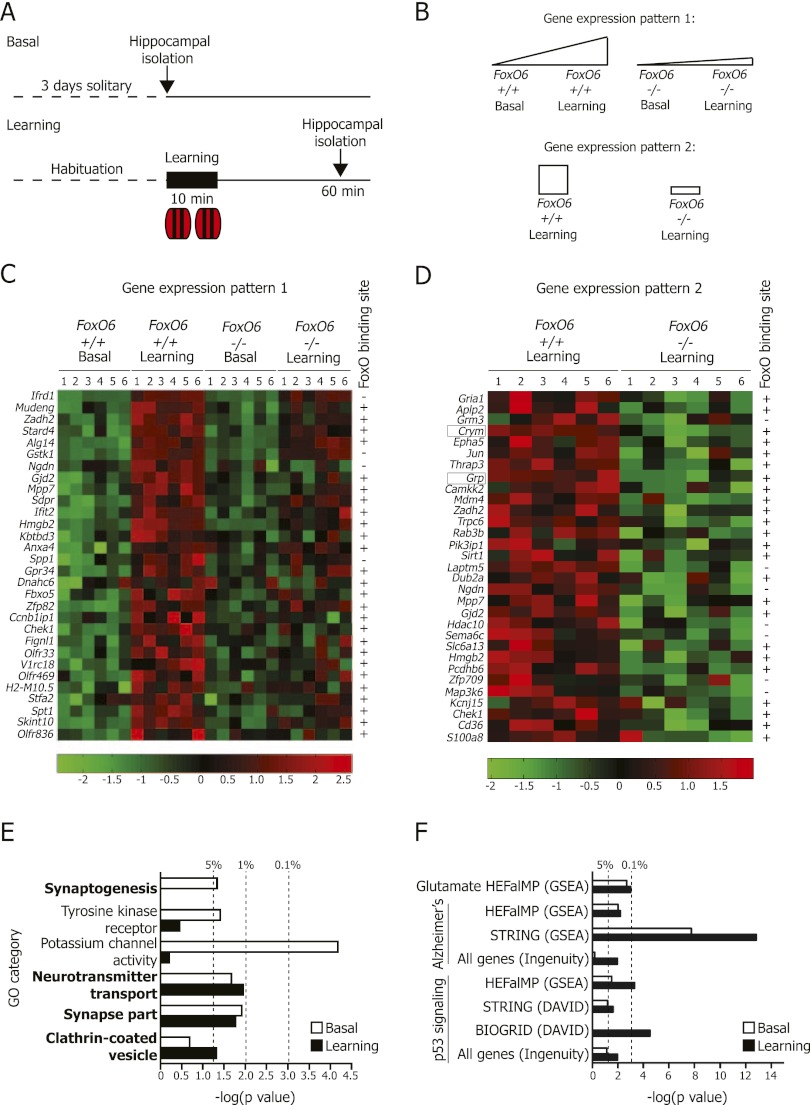
To determine the mechanisms underlying the regulation of memory consolidation by FoxO6, we used a genome-wide microarray approach to compare genes differentially expressed in the hippocampus of adult FoxO6 mutant versus wild-type mice before and after novel object learning (Fig. 5A; Supplemental Table 1). We first focused on genes that were up-regulated significantly following novel object learning in wild-type mice compared with FoxO6 mutant mice (gene expression pattern 1 in Fig. 5B). Interestingly, 176 genes were up-regulated significantly following object learning in wild-type mice compared with FoxO6 mutant mice (P < 0.05 for interaction between the factors genotype and learning, two-way ANOVA; P < 0.05 for wild-type sibling basal vs. learning, one-way ANOVA) (Fig. 5C; Supplemental Table 1). We investigated whether genes induced by learning in wild-type mice compared with FoxO6 mutant siblings were enriched for specific gene categories using gene ontology (GO) analysis. Genes induced by learning in only wild-type mice showed significant enrichment for genes involved in cognition (P = 1.9 × 10−4, modified Fisher's exact test), indicating that FoxO6 is required for the expression of a program of genes coordinating cognitive function.
Figure 5.
FoxO6 is necessary for the expression of a program of genes involved in synaptic function in response to novel object learning. (A) Scheme to test the hippocampal gene expression of the FoxO6 mutant and wild-type mice under basal conditions (basal) or after a novel object learning task. n = 6 mice (8- to 9-wk-old males) per genotype per condition. (B) The differentially expressed genes in the hippocampus of adult FoxO6 mutant versus wild-type mice before and after object learning were divided into two gene expression patterns: (1) genes significantly induced by learning in wild-type mice relative to FoxO6 mutant mice and (2) genes down-regulated in FoxO6 mutant mice after learning. (C,D) Heat map of selected genes significantly up-regulated by object learning in the hippocampus of wild-type mice relative to FoxO6 mutant mice (C) or significantly decreased in the hippocampus of FoxO6 mutant mice compared with wild-type siblings following object learning (D). P < 0.05 for interaction between genotype and learning, two-way ANOVA; P < 0.05 for wild-type siblings basal versus learning, one-way ANOVA; P < 0.05 for FoxO6 mutant versus wild-type, one-way ANOVA. The entire set of genes with this expression profile is presented in Supplemental Table 1. Colors represent the Z score of the expression level for each gene (red is high expression, and green is low expression). (+) Presence of a consensus FoxO-binding site in the gene promoters (5 kb upstream of or downstream from the transcriptional start site). (E) Genes regulated by FoxO6 after novel object learning are enriched for genes involved in synaptic function. Genes differentially expressed in the FoxO6 mutant mice under basal (white bars) or learning conditions (black bars) were compared with GO categories using DAVID version 6.7 (P < 0.05 for FoxO6 mutant vs. wild-type; one-way ANOVA). GO category information is presented in Supplemental Table 1. (Dashed lines) P = 0.05, P = 0.01, and P = 0.001, modified Fisher's exact test. (F) Genes regulated by FoxO6 are enriched for genes involved in glutamate signaling, Alzheimer's disease, and the p53 signaling pathway. Genes differentially expressed in the FoxO6 mutant mice under basal (white bars) or learning conditions (black bars) (P < 0.05 for FoxO6 mutant vs. wild-type, one-way ANOVA) were compared. (Dashed lines) P = 0.05 and P = 0.001.
To gain further insight into the mechanistic role of FoxO6 in memory consolidation, we examined the ensemble of genes that were expressed at significantly lower levels in FoxO6-null mice compared with wild-type mice after object learning (gene expression pattern 2 in Fig. 5B) and before object learning (Supplemental Table 1). We found that 601 genes were expressed at significantly lower levels in FoxO6 mutant mice compared with wild-type siblings following object learning, and 411 genes were expressed at significantly lower levels in FoxO6 mutant mice compared with wild-type siblings before object learning (P < 0.05 for FoxO6 mutant vs. wild-type, one-way ANOVA) (Fig. 5D; Supplemental Table 1). Genes down-regulated in FoxO6 mutant mice compared with wild-type mice after learning include the gastrin-releasing polypeptide (Grp) (P = 4.3 × 10−5 for FoxO6 mutant vs. wild-type after learning, one-way ANOVA) and μ-crystallin (Crym) (P = 1.9 × 10−4 for FoxO6 mutant vs. wild-type after learning, one-way ANOVA) (Fig. 5D; Supplemental Table 1). Grp modulates synaptic plasticity and memory formation (Shumyatsky et al. 2002; Roesler et al. 2006, 2009), and Crym is highly expressed in hippocampal and cortical neurons, where it is thought to play a role in neuronal specification (Arlotta et al. 2005; Lein et al. 2007; Suzuki et al. 2007). We independently confirmed that Grp and Crym were significantly down-regulated in FoxO6 mutant mice compared with wild-type mice using RT-qPCR (Supplemental Fig. 7A,B).
To gain a global understanding of the FoxO6 program, we analyzed whether genes regulated by FoxO6 before and after learning were enriched for specific categories using GO analysis, gene set enrichment analysis (GSEA), and protein analysis through evolutionary relationships (PANTHER). FoxO6-regulated genes before learning show significant enrichment for genes involved in synaptogenesis and synapse organization (Fig. 5E; Supplemental Fig. 7D), suggesting that FoxO6 is required for the expression of a program of genes regulating synapse formation under basal conditions. Interestingly, the genes regulated by FoxO6 upon learning show enrichment for genes involved in neurotransmitter transport and release, synaptic transmission, clathrin-coated vesicles, and tight junction regulation (Fig. 5E; Supplemental Fig. 7E). Furthermore, FoxO6-regulated genes comprise a number of genes that are directly involved in synapses and dendritic spines (e.g., myosin VI, Myo6 [Osterweil et al. 2005], and Park7 [Sheng et al. 2013]). This unbiased analysis of microarray data suggests that FoxO6 regulates a program of genes involved in synapse function.
To understand better the gene pathways involved in learning and memory in response to FoxO6, we performed further bioinformatic analyses of the microarray data using additional tools, such as GSEA; the Database for Annotation, Visualization, and Integrated Discovery (DAVID); Ingenuity; human experimental/functional mapper (HEFalMp); BIOGRID; and STRING. This deeper analysis highlighted that the FoxO6 program was enriched for genes involved in three main pathways: glutamate signaling, Alzheimer's disease, and the p53 pathway (Fig. 5F; Supplemental Figs. 7F, 8–12). The glutamate signaling pathway is known to be involved in synapse function (Malenka and Bear 2004; Watkins and Jane 2006; Lu et al. 2009; Arendt et al. 2010). FoxO6 regulates a large number of genes involved in this glutamate pathway (e.g., the ionotropic AMPA glutamate receptor 1 [Gria1], the ionotropic NMDA glutamate receptor subunits 2C and 2D [Grin2c and Grin2d], and cAMP-dependent protein kinase regulatory subunit type 1 α [Prkar1a]) (Supplemental Figs. 8A, 10). Some of these genes could underlie the memory consolidation defects of FoxO6 mutant mice. Gria1 (also known as GluR1) is key for synaptic transmission and plasticity (Zamanillo et al. 1999; Lu et al. 2009). Furthermore, turning off NMDA receptor activity resulted in defects in memory consolidation (Shimizu et al. 2000). Interestingly, pathways regulated by FoxO6 were also enriched for genes involved in Alzheimer's disease (Supplemental Figs. 8B, 11), a pathology characterized by age-dependent progressive memory defects (Guerreiro and Hardy 2011; Walsh and Teplow 2012). Thus, genes involved in Alzheimer's disease (e.g., the amyloid precursor-like protein [Aplp2], low-density lipoprotein receptor-related protein 8 [Lrp8], and Gria1) could underlie, at least in part, the defect in memory consolidation of FoxO6-null mice. Most unexpected was the p53 pathway, with genes such as Tp53 itself; the related isoform Tp73, which is known to have a role in cognitive function (Wetzel et al. 2008); and a variety of DNA repair pathway genes (Supplemental Figs. 9, 12). These data suggest that DNA repair and stress response mechanisms play an important role in memory consolidation, perhaps by protecting the cells in which synaptic connections have been strengthened.
FoxO6-regulated genes show enrichment for the activity-dependent MEF2 transcription factor-binding sites
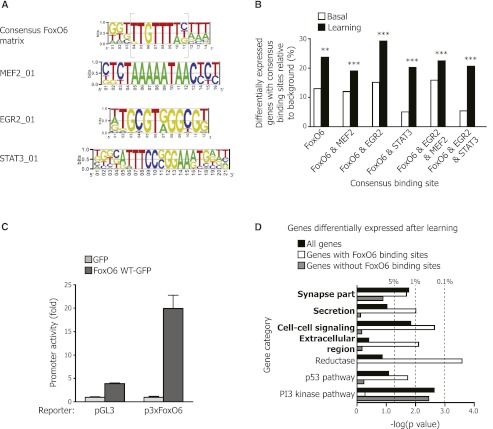
To analyze more specifically the potential direct targets of the FoxO6 transcription factor, we searched for putative FoxO-binding sites in the promoters of the genes regulated by FoxO6 both before and after learning. We found that several promoters contained FoxO-binding sites and that these binding sites formed the following matrix: (G/T)(G/T)(G/T)TTGTTT(A/T)(T/C)(A/T)(A/T)(A/T) (Fig. 6A,B). This matrix is similar to the canonical FoxO-binding sequence (TTGTTTAC) (Furuyama et al. 2000) but has additional requirements on the 5′ and 3′ ends and may represent a more specific “FoxO6 consensus matrix.” A luciferase reporter gene driven by this FoxO6 consensus matrix was up-regulated by FoxO6 expression in cultured neurons (Fig. 6C), indicating that this consensus matrix allows FoxO6-dependent expression. Analysis of the potential direct targets of FoxO6 revealed that they were enriched for genes involved in the synapse compartment, secretion, and cell–cell signaling (Fig. 6D), suggesting that one direct function of this transcription factor may be to regulate synaptic function and/or signaling.
Figure 6.
The promoters of genes that are positively regulated by FoxO6 after novel object learning contain consensus binding sites for FoxO and for the activity-dependent transcription factor MEF2. (A) Consensus matrix for FoxO6-binding sites constructed by aligning the promoter sequences from the genes differentially expressed in the FoxO6 mutant and wild-type mice both in basal conditions (basal) and after object learning (learning). The known FoxO-binding site is shown in gray brackets. Consensus matrices for MEF2_01-, EGR2_01-, and STAT3_01-binding sites from the TRANSFAC database shown to co-occur with the consensus FoxO6 matrix are represented. (B) Proportion of genes down-regulated in FoxO6 mutant versus wild-type mice containing the consensus binding sites described in A in basal conditions (white bars) and after learning (black bars). (**) P < 0.01; (***) P < 0.001, log rank test. (C) The consensus FoxO6 matrix drives FoxO6-dependent transcription in cultured neurons. Luciferase assays in primary cultures of cerebellar granule neurons (CGNs) transfected with constructs expressing a control luciferase reporter (pGL3), a luciferase reporter driven by three consensus FoxO6-binding sites (p3xFoxO6), and a construct to ectopically express FoxO6-GFP. Results are normalized to renilla. Mean ± SD of a representative experiment performed in triplicate. (D) Genes regulated by FoxO6 after novel object learning and containing FoxO-binding sites are enriched for genes involved in the synapse compartment, secretion, and cell–cell signaling. All genes differentially expressed in the FoxO6 mutant mice after learning (black bars), the subset of genes containing the FoxO-binding site (white bars), or the subset of genes without a FoxO-binding site (gray bars) were compared with gene categories using DAVID version 6.7 (P < 0.05 for FoxO6 mutant vs. wild-type, one-way ANOVA). DAVID category information is presented in Supplemental Table 1. (Dashed lines) P = 0.05, P = 0.01, and P = 0.001, modified Fisher's exact test.
We next asked whether binding motifs for other transcription factors co-occurred with the FoxO6-binding matrix in the promoters of genes regulated by FoxO6 upon learning. Interestingly, unbiased analysis revealed that the promoters of FoxO6-regulated genes were significantly enriched for consensus binding motifs for neuronal activity-dependent transcription factors such as MEF2 and EGR (Fig. 6A,B; Supplemental Table 1). The MEF2 family of transcription factors is known to regulate a program of genes that suppress excitatory synapse numbers and function (Flavell et al. 2006, 2008; Barbosa et al. 2008). The observation that the promoters of FoxO6-regulated genes also contain MEF2- and EGR-binding motifs, coupled with the enrichment of FoxO6-regulated genes in molecular signatures for synaptic function, raises the possibility that one mechanism by which FoxO6 regulates consolidation of memory upon object learning is by controlling synaptic number and/or function. Interestingly, the genes involved in the p53 pathway were more enriched in FoxO6 direct genes (Fig. 6D), suggesting that stress resistance and DNA repair responses may be key for the function of FoxO6 in hippocampal neurons.
FoxO6 regulates dendritic spine density in vitro and in vivo
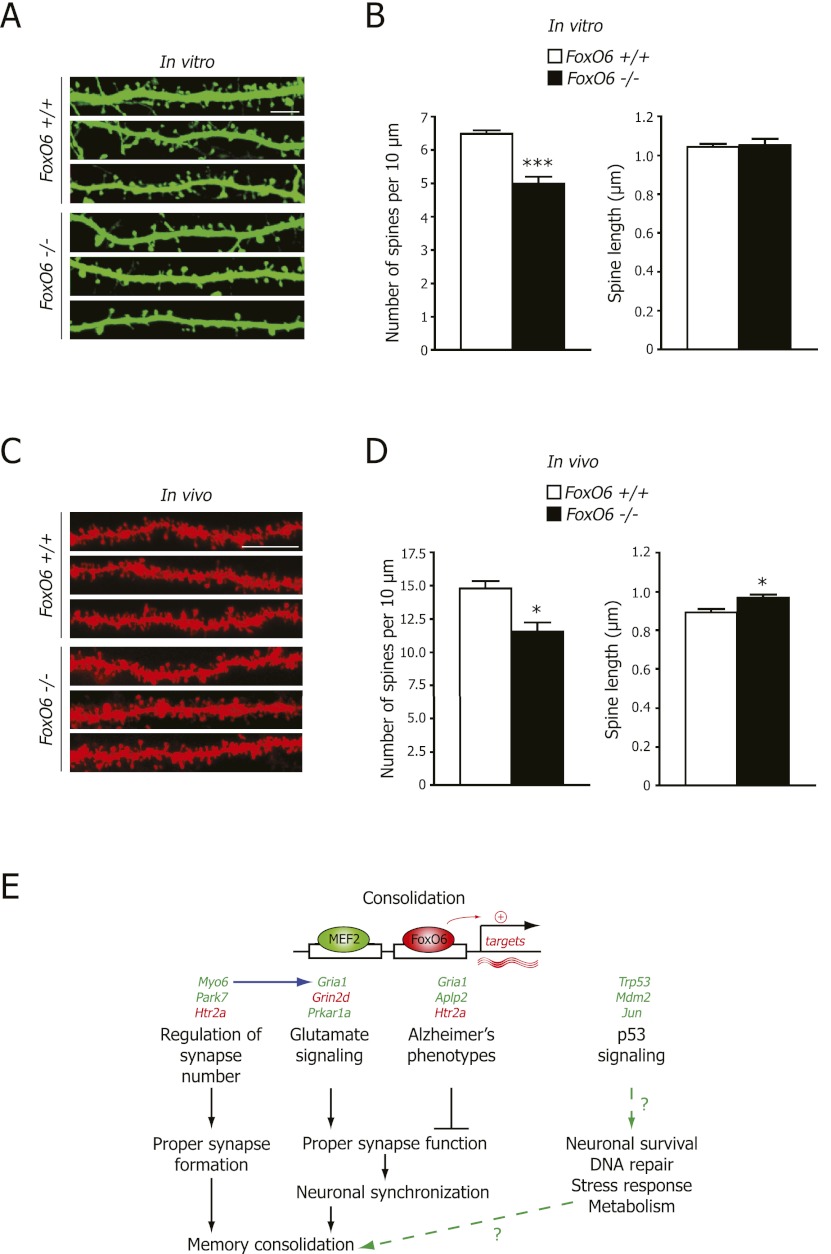
Our unbiased analysis of the microarray data revealed that a striking number of FoxO6 target genes are involved in synaptogenesis, dendritic spines, and excitatory synapse signaling. These observations raise the possibility that FoxO6 controls synapse number and/or function. To test whether FoxO6 is important for synaptic number, we assessed the number of dendritic spines. Dendritic spines are considered to be a good approximation of synapse number because they generally form one excitatory synaptic connection with a presynaptic axonal bouton (Shepherd and Harris 1998). To assess spine number in FoxO6 mutant neurons, we cultured hippocampal neurons from FoxO6 mutant and wild-type embryonic mice (Brewer et al. 1993) and transfected these neurons with a construct expressing GFP at 10–12 d in culture. We then quantified the number of spines relative to dendritic length 8 d later (Fig. 7A,B) as well as the length and width of the spines (Fig. 7A,B; Supplemental Fig. 13A). Hippocampal neurons from FoxO6 mutant mice showed a significant decrease in dendritic spine density in culture (Fig. 7A,B). The length and width of the dendritic spines were not significantly different between FoxO6 mutant and wild-type hippocampal neurons in culture (Fig. 7A,B; Supplemental Fig. 13A). In addition, the length of dendrites and axons of the cultured hippocampal neurons were similar in FoxO6 mutant mice and wild-type siblings (Supplemental Fig. 13C,D). These findings indicate that FoxO6 deficiency results in decreased spine number in cultured hippocampal neurons.
Figure 7.
FoxO6 deficiency leads to decreased spine density in vitro and in vivo. (A) The dendrites and spines from cultured embryonic hippocampal neurons transfected with GFP were visualized after 18–20 d by staining with an antibody against GFP. Representative images of spines in FoxO6 mutant and wild-type neurons are shown. Bar, 5 μm. (B) Spine density is decreased in hippocampal neurons from FoxO6 mutant animals compared with wild-type siblings, but spine length is not affected. (Left panel) Quantification of spine number per length of dendrite of neurons prepared as in A. (Right panel) Quantification of dendritic spine length of neurons prepared as in A. Mean ± SEM. n = 7–8 mice per genotype. Spine density: 125 neurons analyzed in total; spine length: eight–10 neurons per mouse and 50 spines per neuron, resulting in 2950–3200 spines per genotype. (***) P < 0.001 for FoxO6 mutant versus wild-type mice, unpaired Student's t-test. (C) The dendrites and spines in the stratum radiatum of the CA1 region were visualized by sporadically labeling hippocampal neurons by diolistic delivery of the fluorescent carbocyanine dye DiI. Representative images are shown. Bar, 10 μm. (D) Spine density is decreased in vivo in hippocampal CA1 pyramidal neurons from adult FoxO6 mutants compared with wild-type siblings, and spine length is increased in the FoxO6 mutant hippocampal neurons. (Left panel) Quantification of spine number per length of dendrite of neurons visualized as in C. (Right panel) Quantification of dendritic spine length of neurons visualized as in C. Mean ± SEM. n = 4 mice (4- to 4.5-mo-old) per genotype. Spine density: Eight to 10 neurons per mouse were analyzed by counting the number of spines in 30- to 160-μm lengths of dendrite with two to five replicates per neuron; spine length: eight to 10 neurons per mouse and 50 spines per neuron, resulting in 1600–1650 spines per genotype. (*) P < 0.05 for FoxO6 mutant versus wild-type mice, unpaired Student's t-test. (E) Model of FoxO6 action. Upon learning, FoxO6 would induce the expression of genes that coordinate proper synaptic number (e.g., Myo6 and Park7), glutamate signaling (e.g., Gria1 and Prkar1a), and p53 signaling (e.g., Trp53 and Jun) in the hippocampus, which in turn would allow proper synapse formation/function and neuronal stress responses, which are required for the consolidation of contextual and object memories. Accordingly, genes dysregulated in Alzheimer's disease are also regulated by FoxO6 (e.g., Gria1 and Aplp2). (Green) Genes down-regulated in FoxO6 mutant mice; (red) genes up-regulated in FoxO6 mutant mice.
To determine whether adult FoxO6 mutant mice also had dendritic spine defects in vivo, we visualized dendritic spines using diolistic delivery of the fluorescent carbocyanine dye DiI into CA1 pyramidal neurons of brain slices from adult mice (Fig. 7C). Consistent with the in vitro results, FoxO6 mutant mice had a significant decrease in dendritic spine density in the CA1 region of the hippocampus compared with wild-type siblings (Fig. 7D). Dendritic spine length was significantly increased in the CA1 neurons of the adult FoxO6 mutant hippocampus in vivo compared with wild-type siblings (Fig. 7D), but spine width was not affected (Supplemental Fig. 13B). Overall, FoxO6 loss results in a reduction of spine number both in vitro and in vivo. The quantitative differences in spine number or spine length between in vitro and in vivo experiments are likely due to culture conditions (e.g., growth factors, oxygen levels, etc.). Together with our microarray data, these results suggest that FoxO6 promotes synapse formation and/or maintenance and may affect synaptic function.
Collectively, our findings suggest the following model for FoxO6 function in memory consolidation (Fig. 7E): Upon learning, FoxO6 would induce the expression of genes that orchestrate proper synaptic number and function, DNA repair, and stress responses to lead to correct neuronal connectivity in the hippocampus, which in turn would be required for the consolidation of the contextual and object memories. This model is consistent with the dependence of memory consolidation on de novo transcription (Abel and Lattal 2001).
Discussion
Our results show that FoxO6—a transcription factor that belongs to a family involved in slowing aging downstream from the insulin/IGF/PI3K pathway—plays a crucial role in memory consolidation. One mechanism by which FoxO6 regulates memory consolidation may be by coordinating a gene expression program that controls synaptic number and function as well as neuronal connectivity in the hippocampus after learning. Before this study, very little was known about the role of FoxO6 and, more generally, about the family of insulin/IGF-dependent FoxO transcription factors in cognitive processes. Because FoxO transcription factors are required for an extended life span in a range of organisms, our findings may also provide insights into the molecular mechanisms underlying age-dependent cognitive decline in mammals.
Region and time of action of FoxO6 in memory consolidation
FoxO6 does not appear to be necessary for learning because FoxO6 mutant mice performed similarly to wild-type siblings during the learning stages of the contextual fear-conditioning and novel object recognition tasks. Similarly, disruption of FoxO6 using a viral construct to express a dominant-negative form of FoxO6 in the CA1 region of the adult hippocampus does not significantly affect the learning phase of contextual fear conditioning. However, memory was impaired in FoxO6 mutant mice in both contextual fear conditioning and novel object recognition, suggesting either a defect in the mechanisms of actual memory retrieval or a defect in memory consolidation. Consolidation is important for the transfer of the information from the hippocampus to other brain regions and is thought to require waves of transcription and translation (Abel and Lattal 2001). Our viral experiments with acute expression of a dominant-negative form of FoxO6 in the adult hippocampus revealed that memory was only impaired when the dominant-negative FoxO6 virus was injected before learning and consolidation of a contextual fear-conditioning task but not once the mice had learned and consolidated the task. These observations suggest that FoxO6 regulates memory consolidation but not retrieval.
Our observation that FoxO6 mutant mice have impaired consolidation of contextual and object recognition memories but normal consolidation of spatial memories suggests that FoxO6 may regulate specific neural circuits. Contextual memory formation is known to require the trisynaptic pathway of the hippocampus (involving the CA3, the CA1, and the entorhinal cortex), whereas spatial memory formation requires the monosynaptic pathway (involving the CA1 field and entorhinal cortex) (Nakashiba et al. 2008; Hunsaker et al. 2009; Burgess and O'Keefe 2011). Thus, FoxO6 may play a more prominent role in the trisynaptic pathway than in the monosynaptic pathway. FoxO6 is expressed at detectable levels in the amygdala and may also function in the amygdala to contribute to the expression of fear memories. In addition, it is also possible that low-level expression of FoxO6 in other tissues (e.g., immune system, adipose tissue, liver, muscle, intestine, hypothalamus) (Kim et al. 2011) could affect hippocampal-dependent behavior in a non-cell-autonomous manner by affecting the systemic environment. However, FoxO6 mutant mice do not show defects in a classical fear-conditioning paradigm (which is dependent on the amygdala). Furthermore, disruption of FoxO6 in the CA1 region of the adult hippocampus impairs memory consolidation. Together, these observations strongly suggest that FoxO6 regulates memory by acting mainly in the hippocampus, although we cannot completely rule out the possibility that FoxO6 also modulates behavior by acting in other tissues.
The finding that the disruption of FoxO6 activity in the adult hippocampus using a viral approach impairs the consolidation of contextual fear memories suggests that FoxO6 acts in adult circuits for memory consolidation. However, other aspects of the FoxO6 mutant phenotype that are evident in adulthood may still arise during development. For example, dendritic spine density is decreased in embryonic hippocampal neurons from FoxO6-null mice, suggesting that FoxO6 loss results in a reduced number of synapses during development. At the molecular level, the genes regulated by FoxO6 before object learning (basal) are enriched for genes involved in synapse formation, and at least one FoxO6-regulated gene, Crym, which is thought to play a role in neuronal specification and differentiation (Arlotta et al. 2005; Lein et al. 2007; Suzuki et al. 2007), is also differentially regulated by FoxO6 in neonatal mice (Supplemental Fig. 7C). However, the reduced spine density of FoxO6-null mice does not appear to have overall pervasive effects, as FoxO6 mutant animals develop normally and have normal learning in independent tests (contextual fear conditioning and novel object recognition).
FoxO6 regulates a gene expression program involved in synapse function
Our genome-wide analysis of gene expression in the hippocampus of FoxO6 mutant and wild-type mice both before and after a novel object learning task revealed that FoxO6 regulates a program of genes involved in synaptic function upon learning. Before our study, the target genes for FoxO6 were not known, apart from Pak1 in the context of knockdown of other FoxO isoforms (de la Torre-Ubieta et al. 2010). The FoxO6 gene expression program mostly includes genes involved in synapse formation and glutamate signaling. Accordingly, several FoxO6 target genes are found to also be dysregulated in Alzheimer's disease. Interestingly, the FoxO6 gene expression program also includes genes involved at multiple levels in the p53 pathway, which is normally involved for the stress response of cycling cells. In neurons, this pathway may serve to coordinate a stress response/DNA repair pathway and perhaps protect cells in which proper synaptic connections have occurred. It is highly unlikely that a single target gene or a single cellular function mediates FoxO6 function in memory consolidation. Instead, FoxO6 more likely orchestrates the expression of a program of genes and coordinates complementary aspects of synapse function and neuronal connectivity.
Intriguingly, there is a significance co-occurrence of potential consensus binding motifs for FoxO6 and for MEF2, EGR, and/or STAT transcription factors in the promoters of genes down-regulated in FoxO6 mutant mice following learning. Thus, FoxO6 may cooperate with activity-dependent transcription factors such as MEF2 to transactivate specific programs of gene expression following neuronal activity that may be required for memory consolidation. In that regard, it is interesting to note that, similar to FoxO6 mutant mice, Egr-1-null mice are defective in the retrieval of object recognition memories 24 h following novel object training, but their 1-h performance is not impaired (Jones et al. 2001). Furthermore, like FoxO6 mutant mice, mice deficient for Mef2c have defects in contextual memory (Barbosa et al. 2008). However, in contrast to FoxO6, the MEF2 family of transcription factors negatively regulates synapse number (Flavell et al. 2006; Barbosa et al. 2008; Pulipparacharuvil et al. 2008). Thus, it is possible that FoxO6 and MEF2 share similar subsets of genes, but that these transcription factors regulate genes in an opposing manner or a different time frame. FoxO6 and MEF2 may also negatively regulate each other or respond differentially to neuronal activity.
Role of FoxO6 compared with other members of the FoxO signaling pathway
FoxO isoforms share high similarity (>98%) in their DNA-binding domain (Jacobs et al. 2003), suggesting that they may have overlapping gene expression programs. In contrast, the other domains (e.g., transactivation and regulation) show much less similarity between FoxO family members, raising the possibility that FoxO isoforms are regulated differently in the nervous system. FoxO isoforms also have different patterns of expression in the hippocampus, with FoxO6 being enriched in the CA1 and CA3 regions, FoxO3 being more highly expressed in the dentate gyrus and CA3 regions than in the CA1, FoxO1 being enriched in the dentate gyrus, and FoxO4 being expressed at very low levels in the brain. Until this study, very little was known about the program of gene expression and the role of other FoxO family members in cognition. To date, mice with a specific deletion of FoxO1 in the brain and mice null for FoxO3 have been shown to have decreased anxiety-like and decreased depression-like behaviors using open field assessment, the elevated plus maze, and the forced swim test (Polter et al. 2009). However, the role of FoxO1 and FoxO3 in learning and memory has never been tested. The difference in expression patterns suggests that FoxO isoforms may have specific and perhaps complementary roles in cognition. On the other hand, FoxO family members may have overlapping roles, which may mask a number of phenotypes in FoxO6 mutant mice. For example, the FoxO family as a whole is necessary for the establishment of axo–dendritic polarity in developing neurons, but knocking down individual FoxO isoforms does not lead to neuronal polarity defects (de la Torre-Ubieta et al. 2010). While our study has identified a specific role for FoxO6 in the regulation of synapse formation that cannot be compensated by the remaining FoxO isoforms, it does not exclude the possibility that FoxO6 may have other functions when other FoxO family members are deficient.
FoxO transcription factors are positively regulated by the phosphatase PTEN. Human patients with inherited PTEN mutations display brain disorders, including autism spectrum disorder and mental retardation (Butler et al. 2005). Specific deletion of Pten in the mature neurons of the cerebral cortex and hippocampus in mice recapitulates phenotypic aspects of human patients with PTEN deletions, such as abnormal social interactions and learning deficits in some memory tests (Kwon et al. 2006). However, the Pten mutant mice showed no defect in context- and cue-dependent fear conditioning (Kwon et al. 2006), which contrasts with the contextual memory consolidation defect observed in the FoxO6 mutant mice. The social interaction phenotype of FoxO6 mutant mice is not yet known. The differences in phenotypes between mice with deletions in the Pten and FoxO6 genes may be due to the fact that PTEN regulates other FoxO family members or that PTEN also controls other pathways in addition to FoxO, including GSK3 and mTOR/S6K, that may have dominant effects on social behavior and cognition.
The paradoxical roles of the insulin/IGF signaling pathway in cognition
The insulin/IGF signaling pathway has paradoxical roles in cognition. Activating the insulin/IGF pathway—which is known to inhibit FoxO factors—is important for memory retention and synaptogenesis (O'Kusky et al. 2000; Aleman and Torres-Aleman 2009; Chen et al. 2011). On the other hand, reducing the insulin/IGF pathway—which is known to activate FoxO factors—is important for extending longevity and preventing symptoms of Alzheimer's disease (Cohen et al. 2009; Freude et al. 2009; Killick et al. 2009). It is possible that lowering the levels of insulin/IGF (leading to high FoxO activity) is beneficial for overall life span and some aspects of cognition, such as memory consolidation. It is also possible that lowering insulin/IGF signaling “sensitizes” the organism to signals that transiently activate PI3K signaling. The capacity to retain transient waves of insulin/IGF/PI3K signaling activity (leading to FoxO activity oscillations) may be needed during memory retention and retrieval (Lin et al. 2001; Opazo et al. 2003; Chen et al. 2005; Horwood et al. 2006). Indeed, recent studies have also shown that injection of an IGF family member, IGF2, into the hippocampus after training enhanced memory (Chen et al. 2011) and that injection of IGF2 after extinction trials facilitated extinction of memory (Agis-Balboa et al. 2011). Finally, it is also possible that the positive effects on memory retention of insulin/IGF are mediated by other targets of the insulin/IGF pathway (i.e., mTOR, GSK3, etc.) (Chen et al. 2011).
Finally, FoxO6 mutant mice share several phenotypes with prematurely aged animals. Indeed, aged rats have defects in memory consolidation/retrieval after 24 h (Blalock et al. 2003) but can learn the novel object recognition task, similar to FoxO6 mutant mice. In addition, theta frequency waves become irregular with advancing age in rodents (Colas et al. 2005). Endogenous FoxO6 activity may decrease during aging or during age-dependent neurodegenerative diseases. An age-dependent decline in FoxO6 activity could contribute to part of the cognitive decline seen with age in healthy adults or in pathological conditions. Teasing apart the importance of the insulin/IGF–FoxO pathway in longevity and cognitive functions will have critical implications for understanding the mechanisms underlying the maintenance of cognitive function with advancing age.
Materials and methods
Animals
Mice were housed under standard conditions with food and water ad libitum in the Stanford University Animal Facility. All animal care and procedures were in accordance with the policies set forth by the Stanford Animal Care and Use Committee. Aged C57BL/6J male mice at 6, 12, and 24 mo of age were obtained from the National Institute on Aging. Young equivalent C57BL/6J mice were obtained from the Jackson Laboratory. Mice obtained from external sources were allowed to acclimatize for at least a week before being used for experiments. The FoxO6-null and wild-type siblings were generated from mating two FoxO6 heterozygous parents to avoid potential maternal problems that may have arisen from using FoxO6-null mothers. FoxO6-null mice were maintained on a mixed C57BL/6 and 129sv strain unless stated otherwise.
Antibodies
Antibodies to full-length mouse FoxO6 were generated by injection of a FoxO6-GST fusion protein into rabbits, and the antibodies were purified by affinity (Quality Controlled Biochemicals). The antibodies to a FoxO6 peptide were generated against the N-terminal peptide of mouse FoxO6 (MAAKLRAHQVDVKKC) (Princeton Biomolecules). This FoxO6 peptide was conjugated to the keyhole limpet hemocyanin (KLH) carrier protein and injected into rabbits (Covance). The antibodies were purified by affinity using the same peptide. The antibodies to NeuN (clone A60, MAB377), β-actin, and GADPH were obtained from Millipore, Novus Biological, and Abcam, respectively.
Immunoprecipitation and Western blotting
Mouse tissues were sonicated in RIPA buffer (1% [v/v] Triton X-100, 1% [w/v] sodium deoxycholate, 0.1% [w/v] SDS, 0.15 M NaCl, 20 mM Tris-HCl at pH 7.4, 2 mM EDTA at pH 8.0, 50 mM NaF, 40 mM β-glycerophosphate, 1 mM EGTA at pH 8.0, 2 mM sodium orthovanadate, protease inhibitor tablet [Roche], 0.055 U of aprotinin [Sigma], phosphatase inhibitor cocktail 1 and 2 [1:100; Sigma], 1 mM PMSF). Tissues were homogenized using 9–12 W of power in 30-sec pulses using a sonicator. Five-hundred micrograms of protein homogenate was incubated with 4 μg of full-length FoxO6 antibody overnight at 4°C. Protein-A agarose beads (Sigma) were added to each sample, and proteins were eluted by boiling in 3× Laemmli buffer (without β-mercaptoethanol). Protein samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were incubated with the peptide FoxO6 antibody (1:1000), and the primary antibody was visualized using 200 ng mL−1 HRP-conjugated protein-A (Sigma) and enhanced chemiluminescence (ECL, Amersham).
Immunohistochemistry on mouse brain sections
Mice were intracardially perfused with ice-cold PBS (pH 7.4) containing 10 U mL−1 heparin, followed by ice-cold 4% (w/v) paraformaldehyde (PFA) in PBS. The brain was harvested, post-fixed in 4% PFA for 48 h at 4°C, and then incubated in 30% sucrose in PBS for 48 h at 4°C. The brains were embedded in O.C.T. Tissue-Tek (Sakura) at −80°C. Coronal brain sections (30 μm) were collected using a microtome (Microm). Antigen retrieval was performed using a 30-min incubation in 10 mM sodium citrate (pH 6.0) containing 0.05% (v/v) Triton X-100 at 80°C. Nonspecific binding sites were blocked in 3% normal donkey serum (Equitech Bio) in tris-buffered saline (TBS; pH 7.4) with 0.3% Triton X-100 (TBS-TDS). Sections were incubated with the primary antibodies (full-length FoxO6 antibody, 1:250; NeuN antibody, 1:600), in TBS-TDS containing donkey serum (Jackson ImmunoResearch) overnight at 4°C. Consequent incubations with a secondary biotinylated donkey anti-rabbit antibody (1:500; Jackson ImmunoResearch), then streptavidin coupled to Fluorescein-DFAT (1:400; Jackson ImmunoResearch) and a secondary donkey anti-mouse antibody coupled to Texas red (1:400; Jackson ImmunoResearch) were each performed in TBS-TDS overnight at 4°C. The sections were mounted using Vectashield mountant containing DAPI (Vector Laboratories). Fluorescent images were captured with a Zeiss LSM 510 confocal laser scanning microscope equipped with a Ti:Sapphire laser (Cell Sciences Imaging Facility, Stanford University, CA).
Fear conditioning
The trace tone-cued fear-conditioning experiment with FoxO6 mutant and wild-type siblings used two different contexts, A and B, and was performed according to Saxe et al. (2006) with the modification described in Salehi et al. (2009) using a fear-conditioning chamber from Coulbourn Instruments. On the first day, mice were individually placed in context A and, after 3 min, received five tone–shock pairing cycles (tone is referred to as a conditional stimulus [CS], and the shock is referred to as an unconditional stimulus [US]). The shock (0.5 mA, 50 Hz, 2 sec) was delivered after 18 sec from the end of the tone (70 dB, 2 kHz, 20 sec). On the second day, mice were placed in context B (containing a novel olfactory scent, novel floor texture, and novel visual cues) for 3 min and were subsequently presented with three tone cycles without any shocks. On the third day, mice were placed in the original context A for 5 min without any CS and US. Freezing was defined as the complete lack of motion for a minimum of 0.75 sec, and the percent of freezing in each period of time was reported.
The delayed-tone classical fear-conditioning experiment with FoxO6 mutant and wild-type siblings used two different contexts, A and B, and was performed according to Han et al. (2009) using a fear-conditioning chamber from Coulbourn Instruments. On the first day, mice were individually placed in context A and, after 2 min, received three tone–shock pairing cycles. The shock (0.5 mA, 50 Hz, 2 sec) was delivered during the last 2 sec of the tone (85 dB, 2.8 kHz, 30 sec). On the second day, mice were placed in context B (containing a novel olfactory scent, novel floor texture, and novel visual cues) for 2 min and were subsequently presented with 180 sec of tone without any shocks. Freezing was defined as the complete lack of motion for a minimum of 0.75 sec, and the percent of freezing in each period of time was reported.
Novel object recognition
The novel object recognition test was performed on two independent cohorts of mice according to Nilsson et al. (2007). One cohort consisted of mice on the mixed C57BL/6J-129sv background, and the second cohort consisted of mice that had been backcrossed to the C57BL/6J strain for at least four generations. Mice were individually habituated to an open arena (50 × 50 × 50 cm) for three consecutive days. On the fourth day, two identical objects (orange barrels with vertical black stripes) were placed into the arena, and the animals were allowed to explore for 10 min. During the memory testing sessions, after delays of 1 h and 24 h, the animals were placed back into the arena with a clean copy of the familiar object used during training and a novel object of similar dimensions but with a different shape and color. Mice were allowed to explore freely for 5 min during the memory testing sessions. Digital video tracking (using an infrared camera and vplsi Viewpoint software) of body movements and nose position was used to quantify the locomotor activity and the exploratory activity around the objects (2.5-cm zone around the objects). The discrimination index (DI)—i.e., the ratio of the time spent exploring the novel object over the time spent exploring the two objects—was used to represent memory. The DI was calculated for each animal for 5-min windows and averaged among the groups of mice by genotype. To evaluate memory, comparisons were made between the two testing sessions and the learning session for a given genotype. The DI during learning was also calculated over 10 min to ensure that the mice had equally explored the two identical objects. Mice that did not explore the objects were excluded from further analysis (two wild-type mice and one FoxO6 mutant mouse).
Viral constructs
Murine FoxO6 with a deletion of the C terminus at amino acid 389 (FoxO6ΔCt) was fused to GFP, subcloned into the HSV amplicon (pHSV), and packaged using a replication-defective helper virus as previously described (Han et al. 2009). The pHSV amplicon containing only GFP was used as a control. Virus was purified on a sucrose gradient, pelleted, and resuspended in 10% sucrose. The average titer of the recombinant virus stocks was typically 5.0 × 107 infectious units per milliliter.
Stereotactic injection of virus in the hippocampus
For HSV injection in the hippocampus, mice were housed under standard conditions with food and water ad libitum at the Hospital for Sick Children (Toronto, Canada). Animal care and procedures were approved by the Hospital for Sick Children Animal Care and Use Committee. Male and female adult mice at 2–3 mo of age of an F1 hybrid strain (C57BL/6NTac × 129S6/SvEvTac) were used. Mice were group-housed (three to five mice per cage), on a 12-h light/dark cycle. For the surgeries, mice were pretreated with atropine sulfate (0.1 mg kg−1 i.p.), anesthetized with chloral hydrate (400 mg kg−1 i.p.), and placed in a stereotaxic frame. The skin was retracted and holes were drilled in the skull bilaterally above the dorsal hippocampus (rostrocaudal, −2.30 mm; mediolateral, ±1.5 mm; dorsoventral, −1.7 mm vs. Bregma) according to Paxinos and Franklin (2001). Bilateral microinjections of the viruses (2.0 μL) were delivered into the CA1 region over 20 min through glass micropipettes. Micropipettes were left in place an additional 10 min to ensure diffusion of the viruses. Because transgene expression using this viral system typically peaks 3–4 d following infusion (Barrot et al. 2002), behavioral experiments were conducted 3 d following surgery.
Contextual fear conditioning on stereotactically injected mice
The experiments with recombinant HSV infections of the wild-type adult hippocampus were performed with the apparatus and behavioral procedures that have been previously described (Wang et al. 2009). Contextual fear-conditioning experiments were conducted in a windowless room containing four conditioning chambers. Each conditioning context consisted of a stainless steel conditioning chamber (31 × 24 × 21 cm; Med Associates) containing a stainless steel shock grid floor. Shock grid bars (3.2 mm diameter) were spaced 7.9 mm apart, and the grid floor was positioned over a stainless steel drop pan. The front, top, and back of the chamber were made of clear acrylic, and the two sides were made of modular aluminum. Mouse freezing behavior was monitored via four overhead cameras. Freezing was assessed using an automated scoring system (Actimetrics), which digitized the video signal at 4 Hz and compared movement frame by frame to determine the amount of freezing.
During learning, mice were placed in the context for 3 min and were presented with one unsignaled foot shock (0.5 mA, 2 sec) starting at 2 min. After the foot shock, mice remained in the context for an additional minute and then were returned to their home cage. Twenty-four hours later, mice were placed back into the same context for a total of 5 min, and freezing was monitored. After a further 24 h, mice were then retrained under stronger conditions by placing the mice in the context for 5 min and presenting three foot shocks (0.5 mA, 2 sec) at 2 min, 3 min, and 4 min. Contextual fear memory was assessed again 24 h later by placing the animals back in the context for 5 min. Contextual fear-conditioning experiments were conducted during the light phase. No gender differences were observed.
In addition to measuring freezing as a measure of memory formation, an additional series of parameters was measured to confirm that the behavioral effects observed were not due to factors other than memory deficits. First, baseline activity prior to any tone or shock on the learning day can be a useful measure of general activity (such as in the open field). Activity during this period, for example, can readily detect the effects of hippocampal lesions or psychostimulant drugs (Anagnostaras et al. 1999; Shuman et al. 2009). Secondly, the gross motor reactivity to the shock, known as the activity burst or unconditional response, during the actual 2-sec shock can be used as a measure of shock reactivity or pain sensitivity (DeLorey et al. 1998; Anagnostaras et al. 2000; Shuman et al. 2009). Finally, the post-shock freezing response can be an indicator of the animal's ability to learn, as it is thought to reflect a conditional (i.e., learned) response to the context rather than an unconditional response to the shock (Fanselow 1986).
The placement and extent of the viral infection for each mouse were determined following the behavioral experiments (see below) using GFP fluorescence by an experimenter unaware of the behavioral data. Two hours after the final behavioral experiment, mice were perfused with 4% (w/v) PFA, and their brains were harvested and post-fixed in 4% PFA overnight at 4°C. The following day, coronal brain sections (40 μm) were collected and mounted on gelatin-coated slides for fluorescent confocal microscopy (LSM710, Zeiss). Mice were classified as “hits” and included in the subsequent data analysis if they had robust bilateral transgene expression in the CA1 region of the dorsal hippocampus.
In vivo cortical and local EEG recordings in behaving mice
FoxO6 mutant and wild-type siblings were equipped for frontoparietal and hippocampal EEG recording (Colas et al. 2008). Cortical EEG electrodes (0.9 mm diameter) were placed in epidural position over the right hemisphere: frontal, +1.5 mm versus Bregma; parietal, −1.5 mm versus Bregma; both, 1 mm lateral versus midline. Hippocampal electrodes (0.28 mm diameter) were placed within the CA1 layer of the left hemisphere (rostrocaudal, −1.90 mm; mediolateral, +1.0 mm; dorsoventral, −1.6 mm). After 1 wk of recovery, the mice were habituated to the recording system for an additional week. Recording sessions were conducted in a 50 × 30 cm plexiglass box including objects for 10 min. The EEG signals were collected using a commercial amplifier (Embla, model A10), analog-to-digital-converted with a 200-Hz sampling rate. Remote video viewing was used to ensure that the mice were actively involved in exploratory behavior. The signals were subjected to spectral analysis using a fast Fourier transform (3-sec windows, 1024-Hz resolution) yielding power spectra with a 0.2-Hz resolution (PRANA suite from Phytools). Individual power spectra were normalized against total power in the 3- to 50-Hz band, and spectrograms were expressed as the percentage of power against total power per 0.2-Hz frequency bin. The resulting individual spectrograms were averaged for FoxO6 mutant and wild-type groups.
RNA extraction
mRNA samples were collected from the hippocampus from FoxO6 mutant and wild-type siblings before (basal) or after novel object learning. The cohort of basal mice was housed individually for at least 3 d prior to tissue harvesting. The mice used to collect hippocampal samples after the object learning task were handled daily for at least 7 d in the procedure room, prior to the object learning task. Twenty-four hours before the object learning task, each mouse was individually habituated to the empty novel object arena for 10 min. On the days of the novel object learning task and the empty arena habituation, the mice were transferred to the procedure room and acclimatized for 60 min. For the novel object learning task, the mice were allowed to explore two identical and novel objects for 10 min in a 70 × 70 × 70-cm black plastic arena with a white PVC vinyl material on the base. A video tracking system (Videotrack automated behavioral analysis system; Viewpoint Life Science, Inc.) was used to record mouse location and running path with time. After the object exploration, mice were transferred to an empty cage and were euthanized after 60 min. The brains were dissected and incubated in ice-cold RNAlater (Ambion) for 24 h at 4°C. The brains were then rapidly frozen in O.C.T. Tissue-Tek (Sakura) at −80°C. Coronal brain sections (300 μm) were made using a microtome (Microm), and the hippocampus was finely dissected and collected into RNA lysis buffer (Ambion) and homogenized for 30 sec using a rotar-stat homogenizer. Total RNA was extracted using the RNAqueous column (Ambion) following the manufacturer's protocol.
Microarray analysis
Microarray hybridization was performed at the Stanford PAN facility. Briefly, 250 ng of total RNA was reverse-transcribed to cDNA, and cDNA was hybridized to the Mouse Gene 1.0 ST array (Affymetrix). A background adjustment and normalization with RMA (robust multiarray analysis) (Irizarry et al. 2003) was performed. The determination of differentially expressed probes was tested using significance analysis of microarrays (SAM) (Tusher et al. 2001), rank products (Breitling et al. 2004), and ANOVA. Probes were mapped to known genes using the Array Information Library Universal Navigator (AILUN) (Chen et al. 2007), using annotation from the mouse Mm9 genome build. Fold change is the ratio of the log2 of expression (FoxO6-null relative to wild type, or novel object learning task over basal conditions). For functional analyses, the expression profiles were analyzed for common functional themes and similarity to previously generated molecular signatures using GSEA (Subramanian et al. 2005), GO category analysis using DAVID version 6.7 (Dennis et al. 2003), PANTHER (Thomas et al. 2003), and Ingenuity (Ingenuity Systems, Inc.).
For the transcription factor-binding site analysis, the HugoOnce procedure was applied to identify one representative probe set for each gene probed on the array (http://r2.amc.nl). Differential gene expression between FoxO6 wild-type and null mice under basal conditions or after novel object learning was calculated using a one-way ANOVA test on log2 transformed RMA-Sketch values (P < 0.01). To generate a representation of genes expressed in the mouse hippocampus, a data set from the Gene Expression Omnibus (GEO) database containing the profiles of 20 wild-type mouse hippocampi (GSE10784) was used, and MAS 5.0 normalization (Affymetrix) to generate absent/present (A/P) calls for each gene was applied. If a gene was called “present” within all 20 wild-type hippocampi, the gene was then included for further analysis. Promoter regions were defined as 2.5 kb upstream of and 500 base pairs (bp) downstream from the transcriptional start site for each gene. TRANSFAC version 11.1 was used as the source of transcription factor motifs for all analyses (http://www.biobase.de). The CREAD package and the Storm program were used to search for occurrences of transcription factor-binding sites within promoters (Smith et al. 2006). For Storm searches, P < 0.0001 was used as a match threshold against whole-genome intergenic sequence hit tables constructed for the Mm8 genome assembly using CREAD with parameters -gapsize = 6 and -wordsize = 8. To determine whether a motif was significantly overrepresented in a particular set of genes (foreground), a background set of genes (composed of any genes that pass the hippocampal filter described above but are not found in the foreground set) was built that contained at least five times the number of sequences and drawn at random from the filtered set. Both sets of genes were then searched using identical Storm criteria and input motifs. Each motif was scored by performing a proportion test using the prop.test function in R (version 2.6.1). To determine whether two or more motifs were significantly co-occurring, the same test was applied only to promoters that contained one or more instances of each motif queried in the combination. To build a consensus, sequences identified by these matrices were aligned using the ClustalX program. Sequence LOGOs were generated using the STAMP program (http://www.benoslab.pitt.edu/stamp). The consensus FoxO6 matrix and all Perl scripts written to implement these procedures are available on request.
To compare the FoxO6 expression profiles to specific gene signatures, a variety of tools were used: HEFalMp (Huttenhower et al. 2009), STRING (von Mering et al. 2003), and BioGRID (Stark et al. 2006). The HEFalMp database was queried for a specific gene in relation to a biological process or for a biological process (or disease) in relation to all genes. For both types of queries, we used the recommended cutoffs: 0.5 for relationship confidence score and 0.05 for approximate P-value. Several gene sets were generated based on the following queries: Tp53 in relation to DNA damage (309 genes), glutamate signaling in relation to learning (930 genes), and Alzheimer's disease-related genes (1486 genes). These gene sets were then tested for enrichment of FoxO6-regulated genes using GSEA (Subramanian et al. 2005). Multiple hypotheses testing correction was applied to final GSEA results using the method of Benjamini and Hochberg (1995). Using the STRING and BioGRID databases, we identified the FoxO6-regulated genes that form pairwise interactions in order to reveal the function of the pool of interacting genes. The functional enrichment of the interacting FoxO6-regulated genes was determined by using GSEA (Subramanian et al. 2005) and DAVID (Dennis et al. 2003; Huang et al. 2009). Ingenuity was also used to highlight FoxO6-regulated genes in pathway diagrams.
Luciferase assays
The consensus FoxO6 matrix identified from the promoters of genes differentially expressed in the microarray analysis (Fig. 6A) was used to create a FoxO6-responsive luciferase reporter. Three tandem repeats of the consensus FoxO6-binding site were cloned into the NheI and BglII sites of the pGL3-basic luciferase vector (Promega) by ligation of the following oligonucleotides: 5′-CTAGCGGTTTGTTTTCTTTGATAGGTTTGTTTTCTTTGATAGGTTTGTTTTCTTTA-3′ and 5′-GATCTAAAGAAAACAAACCTATCAAAGAAAACAAACCTATCAAAGAAAACAAACCG-3′.
U2OS human osteosarcoma cell lines were cultured in McCoy's 5A modified medium supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin/glutamine (P/S/Q) (Invitrogen) at 37°C in 5% CO2 and 95% humidity. U20S cells were plated at a density of 7.5 × 104 cells per milliliter in 24-well plates. The next day, each well of cells was transfected using Lipofectamine LTX (Invitrogen) with 200 ng of a luciferase reporter construct driven by three tandem repeats of the FoxO6 consensus binding motif (p3xFoxO6) and 100 ng of a renilla luciferase reporter construct (pRL0). For the constructs to be tested, 50 ng of each was transfected: wild-type FoxO6-GFP (FoxO6 WT-GFP) or FoxO6ΔCt-GFP in the pEGFP-N1 vector. To test for dominant-negative activity of FoxO6ΔCt-GFP, 50 ng of FoxO6 WT-GFP was spiked with 50 or 150 ng of the FoxO6ΔCt-GFP construct. For the transfection of each well of cells, the total amount of DNA was made up to 850 ng with empty pEGFP-N1 vector. Fifty-two hours after transfection, the cells were lysed, and the luciferase and renilla luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's protocol.
Cerebellar granule neurons (CGNs) were cultured and transfected 2 d after plating using a modified calcium phosphate method as described (Shalizi et al. 2006; de la Torre-Ubieta et al. 2010), with 500 ng of the p3xFoxO6 construct or the empty pGL3-basic luciferase construct and 50 ng of a renilla luciferase reporter construct (pRL-TK). To test the FoxO6 dependency of the p3xFoxO6 construct, 500 ng of wild-type FoxO6-GFP (FoxO6 WT-GFP) in the pEGFP-N1 vector or the pEGFP-N1 vector alone was also transfected. Two days later, the CGNs were subjected to dual-luciferase assays (Promega).
Neuron culture, transfections, and immunocytochemistry
Primary cultures of hippocampal neurons were prepared from individual embryonic day 18 (E18) to E19 mouse embryos as described (Brewer et al. 1993). Hippocampi from each embryo were dissected and processed individually. Neurons were plated on poly-L-lysine/laminin-coated coverslips at a density of 1000 (spine morphology) or 200 (axonal and dendritic length) cells per square millimeter in Neurobasal/B27 medium containing 2% FBS. The next day and every third day, one-half of the volume was replaced with Neurobasal/B27 medium without serum. For neuronal morphology analyses, neurons were transfected at 1 d in vitro (DIV1) with a modified calcium phosphate protocol (Konishi et al. 2002; de la Torre-Ubieta et al. 2010), fixed at DIV5, and subjected to immunocytochemistry with the GFP antibody together with an antibody to the dendritic marker MAP2 (Sigma) and the axonal marker Tau-1 (Chemicon). In all transfections, the anti-apoptotic protein Bcl-xl was coexpressed to rule out potential effects of cell survival on neuronal morphology. The expression of Bcl-xl had no effect on the morphology of neurons (Gaudilliere et al. 2004; de la Torre-Ubieta et al. 2010). Individual images of GFP-labeled DIV5 primary hippocampal neurons prepared from FoxO6 mutant or wild-type mice were captured in a blinded manner on a Nikon eclipse TE2000 epifluorescence microscope using a digital CCD camera (Diagnostic Instruments). Axons and dendrites were identified by morphology and the presence of Tau-1 and MAP2, respectively. Axonal and dendritic length was quantified by tracing using the Spot imaging software (Diagnostic Instruments). Total length per neuron is the length of the axon or dendrite including all of its branches summed together for each neuron.
For spine analyses, neurons were transfected after DIV10 with the pEGFP-N1 plasmid (Clonetech) using the Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's guidelines and fixed at DIV18–DIV20 in 4% (w/v) PFA and 4% (w/v) sucrose. Following fixation, neurons were subjected to immunocytochemistry with an antibody to GFP (Molecular Probes), and nuclei were stained with Hoechst 33258. To characterize dendritic spine morphology, individual 1024 × 1024 z-stacks (0.5-μm depth intervals) of GFP-labeled primary hippocampal neurons were captured on an Olympus Fluoview confocal laser scanning microscope with a 60× objective in a blinded manner. Spine density was quantified by counting the number of spines in three independent 50- to 100-μm dendritic segments of eight to 10 neurons per animal. Spine length and width were quantified by tracing individual spines as described (Sala et al. 2001). Fifty spines per neuron from eight to 10 neurons per animal were quantified. FoxO6 loss did not affect neuronal survival.
In vivo labeling of hippocampal neurons using diolistics
A diolistic approach was used to label individual neurons in FoxO6 mutant and wild-type littermate adult hippocampi (Gan et al. 2000; O'Brien and Lummis 2006). Briefly, coronal hippocampal sections were fixed for 10 min in 4% PFA and then incubated for 1 h in 30% sucrose at 4°C before diolistic delivery of DiI-coated tungsten particles using a Helios Gene Gun (Bio-Rad). Sections were incubated overnight at room temperature in 4% PFA and 30% sucrose, stained with Hoechst 33258, and mounted for analysis. Individual z-stacks of stratum radiatum dendrites of hippocampal CA1 pyramidal neurons that were labeled by DiI were captured and analyzed in a blinded manner as above.
Acknowledgments
We thank R. Malenka, U. Haditsch, T. Palmer, R. Tsien, A. West, T. Wyss-Coray, A. Citri, and the members of the Brunet laboratory for helpful discussions and critical reading of the manuscript. We thank P. Thekkat and J. White for help with mouse colony management, U. Haditsch and J. Ramunas for help with the behavioral experiments, and J. Koster for the use of the R2 platform. We thank B. Benayoun for help with bioinformatic analyses. This work was supported by NIH grant R01 (AG026648), a Klingenstein fellowship, an AFAR/Pfizer grant, and a NARSAD grant (to A. Brunet). D.A.M.S. was supported by a Stanford University Dean's fellowship. A. Bonni was supported by NIH grant NS041021. D.V.M. was supported by NIH grant MH065541 and the G. Harold and Leila Y. Mathers Charitable Foundation. A.A.M. and A.J.B. were supported by the National Library of Medicine (T15 LM007033 and R01 LM009719, respectively). D.A.M.S. and A. Brunet conceived and planned the study, and D.A.M.S. performed and analyzed experiments. A.J.R., C.J.C., and S.A.J. designed and performed the experiments to test the role of FoxO6 in adult mice (Fig. 3B–F; Supplemental Fig. 6C–E). D.C. planned and performed the novel object recognition tests and the EEG studies (Figs. 2E,F, 4; Supplemental Fig. 4D). K.D. performed the luciferase assays in Supplemental Figure 6B. L.T.U. and A. Bonni designed and performed the studies on spines and on the luciferase reporter driven by FoxO6-binding sites in neurons (Figs. 6C, 7A–D; Supplemental Fig. 13A–D). R.P.Z. helped with RT-qPCR studies (Supplemental Fig. 7A–C). A.A.M. and A.J.B. helped with microarray analysis (Fig. 5E; Supplemental Fig. 7D–F). D.U. helped with pathway analysis (Fig. 5F; Supplemental Figs. 8, 9). E.E.S. helped with promoter analysis (Fig. 6A,B,D). M.S. helped with the contextual and classical fear conditioning (Fig. 2A–D; Supplemental Figs. 3G,I, 4A–C) and the Morris water maze (Supplemental Fig. 5). D.V.M. provided intellectual contribution and helped with interpretation of synaptic data. D.A.M.S. and A. Brunet wrote the paper. All authors discussed the results and commented on the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.208926.112.
References
- Abel T, Lattal KM 2001. Molecular mechanisms of memory acquisition, consolidation and retrieval. Curr Opin Neurobiol 11: 180–187 [DOI] [PubMed] [Google Scholar]
- Agis-Balboa RC, Arcos-Diaz D, Wittnam J, Govindarajan N, Blom K, Burkhardt S, Haladyniak U, Agbemenyah HY, Zovoilis A, Salinas-Riester G, et al. 2011. A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J 30: 4071–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A 2004. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42: 947–959 [DOI] [PubMed] [Google Scholar]
- Aleman A, Torres-Aleman I 2009. Circulating insulin-like growth factor I and cognitive function: Neuromodulation throughout the lifespan. Prog Neurobiol 89: 256–265 [DOI] [PubMed] [Google Scholar]
- Anagnostaras SG, Maren S, Sage JR, Goodrich S, Fanselow MS 1999. Scopolamine and Pavlovian fear conditioning in rats: Dose-effect analysis. Neuropsychopharmacology 21: 731–744 [DOI] [PubMed] [Google Scholar]
- Anagnostaras SG, Josselyn SA, Frankland PW, Silva AJ 2000. Computer-assisted behavioral assessment of Pavlovian fear conditioning in mice. Learn Mem 7: 58–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt KL, Royo M, Fernandez-Monreal M, Knafo S, Petrok CN, Martens JR, Esteban JA 2010. PIP3 controls synaptic function by maintaining AMPA receptor clustering at the postsynaptic membrane. Nat Neurosci 13: 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD 2005. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45: 207–221 [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, et al. 2008. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci 105: 9391–9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Alarcon JM, Kandel ER 2002. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell 108: 689–703 [DOI] [PubMed] [Google Scholar]
- Barrot M, Olivier JD, Perrotti LI, DiLeone RJ, Berton O, Eisch AJ, Impey S, Storm DR, Neve RL, Yin JC, et al. 2002. CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proc Natl Acad Sci 99: 11435–11440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Bevins RA, Besheer J 2006. Object recognition in rats and mice: A one-trial non-matching-to-sample learning task to study ‘recognition memory.' Nat Protoc 1: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Biggs WH III, Cavenee WK, Arden KC 2001. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm Genome 12: 416–425 [DOI] [PubMed] [Google Scholar]
- Blalock EM, Chen KC, Sharrow K, Herman JP, Porter NM, Foster TC, Landfield PW 2003. Gene microarrays in hippocampal aging: Statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci 23: 3807–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P 2004. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ 1993. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res 35: 567–576 [DOI] [PubMed] [Google Scholar]
- Burgess N, O'Keefe J 2011. Models of place and grid cell firing and theta rhythmicity. Curr Opin Neurobiol 21: 734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, et al. 2005. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet 42: 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G 2002. Theta oscillations in the hippocampus. Neuron 33: 325–340 [DOI] [PubMed] [Google Scholar]
- Carlezon WA Jr, Thome J, Olson VG, Lane-Ladd SB, Brodkin ES, Hiroi N, Duman RS, Neve RL, Nestler EJ 1998. Regulation of cocaine reward by CREB. Science 282: 2272–2275 [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320: 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Garelick MG, Wang H, Lil V, Athos J, Storm DR 2005. PI3 kinase signaling is required for retrieval and extinction of contextual memory. Nat Neurosci 8: 925–931 [DOI] [PubMed] [Google Scholar]
- Chen R, Li L, Butte AJ 2007. AILUN: Reannotating gene expression data automatically. Nat Methods 4: 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DY, Stern SA, Garcia-Osta A, Saunier-Rebori B, Pollonini G, Bambah-Mukku D, Blitzer RD, Alberini CM 2011. A critical role for IGF-II in memory consolidation and enhancement. Nature 469: 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury N, Quinn JJ, Fanselow MS 2005. Dorsal hippocampus involvement in trace fear conditioning with long, but not short, trace intervals in mice. Behav Neurosci 119: 1396–1402 [DOI] [PubMed] [Google Scholar]
- Clark RE, Martin SJ 2005. Interrogating rodents regarding their object and spatial memory. Curr Opin Neurobiol 15: 593–598 [DOI] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, et al. 2009. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell 139: 1157–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, et al. 2011. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron 72: 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas D, Cespuglio R, Sarda N 2005. Sleep wake profile and EEG spectral power in young or old senescence accelerated mice. Neurobiol Aging 26: 265–273 [DOI] [PubMed] [Google Scholar]
- Colas D, Valletta JS, Takimoto-Kimura R, Nishino S, Fujiki N, Mobley WC, Mignot E 2008. Sleep and EEG features in genetic models of Down syndrome. Neurobiol Dis 30: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, Vetere G, Pekar T, Ross PJ, Neve RL, et al. 2012. MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat Neurosci 15: 1255–1264 [DOI] [PubMed] [Google Scholar]
- Covington HE III, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, Dietz DM, Lobo MK, Ghose S, Mouzon E, et al. 2011. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 71: 656–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Gaudilliere B, Yang Y, Ikeuchi Y, Yamada T, DiBacco S, Stegmuller J, Schuller U, Salih DA, Rowitch D, et al. 2010. A FOXO–Pak1 transcriptional pathway controls neuronal polarity. Genes Dev 24: 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorey TM, Handforth A, Anagnostaras SG, Homanics GE, Minassian BA, Asatourian A, Fanselow MS, Delgado-Escueta A, Ellison GD, Olsen RW 1998. Mice lacking the β3 subunit of the GABAA receptor have the epilepsy phenotype and many of the behavioral characteristics of Angelman syndrome. J Neurosci 18: 8505–8514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE 2010. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci 13: 1128–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3 doi: 10.1186/gb-2003-4-5-p3 [PubMed] [Google Scholar]
- Fanselow MS 1986. Conditioned fear-induced opiate analgesia: A competing motivational state theory of stress analgesia. Ann N Y Acad Sci 467: 40–54 [DOI] [PubMed] [Google Scholar]
- Fink DJ, DeLuca NA, Goins WF, Glorioso JC 1996. Gene transfer to neurons using herpes simplex virus-based vectors. Annu Rev Neurosci 19: 265–287 [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH 2007. Recovery of learning and memory is associated with chromatin remodelling. Nature 447: 178–182 [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A 2009. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci 106: 2700–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME 2008. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci 31: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME 2006. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freude S, Hettich MM, Schumann C, Stohr O, Koch L, Kohler C, Udelhoven M, Leeser U, Muller M, Kubota N, et al. 2009. Neuronal IGF-1 resistance reduces Aβ accumulation and protects against premature death in a model of Alzheimer's disease. FASEB J 23: 3315–3324 [DOI] [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N 2000. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 349: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan WB, Grutzendler J, Wong WT, Wong RO, Lichtman JW 2000. Multicolor ‘DiOlistic' labeling of the nervous system using lipophilic dye combinations. Neuron 27: 219–225 [DOI] [PubMed] [Google Scholar]
- Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A 2004. A CaMKII–NeuroD signaling pathway specifies dendritic morphogenesis. Neuron 41: 229–241 [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L 2004. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305: 361. [DOI] [PubMed] [Google Scholar]
- Goffin D, Allen M, Zhang L, Amorim M, Wang IT, Reyes AR, Mercado-Berton A, Ong C, Cohen S, Hu L, et al. 2012. Rett syndrome mutation MeCP2 T158A disrupts DNA binding, protein stability and ERP responses. Nat Neurosci 15: 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Hardy J 2011. Alzheimer's disease genetics: Lessons to improve disease modelling. Biochem Soc Trans 39: 910–916 [DOI] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, Bontempi B, Neve RL, Frankland PW, Josselyn SA 2009. Selective erasure of a fear memory. Science 323: 1492–1496 [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol 11: 1975–1980 [DOI] [PubMed] [Google Scholar]
- Hoekman MF, Jacobs FM, Smidt MP, Burbach JP 2006. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns 6: 134–140 [DOI] [PubMed] [Google Scholar]
- Horwood JM, Dufour F, Laroche S, Davis S 2006. Signalling mechanisms mediated by the phosphoinositide 3-kinase/Akt cascade in synaptic plasticity and memory in the rat. Eur J Neurosci 23: 3375–3384 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hunsaker MR, Tran GT, Kesner RP 2009. A behavioral analysis of the role of CA3 and CA1 subcortical efferents during classical fear conditioning. Behav Neurosci 123: 624–630 [DOI] [PubMed] [Google Scholar]
- Huttenhower C, Haley EM, Hibbs MA, Dumeaux V, Barrett DR, Coller HA, Troyanskaya OG 2009. Exploring the human genome with functional maps. Genome Res 19: 1093–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M 2004. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429: 562–566 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jacobs FM, van der Heide LP, Wijchers PJ, Burbach JP, Hoekman MF, Smidt MP 2003. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem 278: 35959–35967 [DOI] [PubMed] [Google Scholar]
- Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S 2001. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci 4: 289–296 [DOI] [PubMed] [Google Scholar]
- Kenyon C 2005. The plasticity of aging: Insights from long-lived mutants. Cell 120: 449–460 [DOI] [PubMed] [Google Scholar]
- Killick R, Scales G, Leroy K, Causevic M, Hooper C, Irvine EE, Choudhury AI, Drinkwater L, Kerr F, Al-Qassab H, et al. 2009. Deletion of Irs2 reduces amyloid deposition and rescues behavioural deficits in APP transgenic mice. Biochem Biophys Res Commun 386: 257–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Fanselow MS 1992. Modality-specific retrograde amnesia of fear. Science 256: 675–677 [DOI] [PubMed] [Google Scholar]
- Kim DH, Perdomo G, Zhang T, Slusher S, Lee S, Phillips BE, Fan Y, Giannoukakis N, Gramignoli R, Strom S, et al. 2011. FoxO6 integrates insulin signaling with gluconeogenesis in the liver. Diabetes 60: 2763–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleindorp R, Flachsbart F, Puca AA, Malovini A, Schreiber S, Nebel A 2011. Candidate gene study of FOXO1, FOXO4, and FOXO6 reveals no association with human longevity in Germans. Aging Cell 10: 622–628 [DOI] [PubMed] [Google Scholar]
- Konishi Y, Lehtinen M, Donovan N, Bonni A 2002. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol Cell 9: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF 2006. Pten regulates neuronal arborization and social interaction in mice. Neuron 50: 377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EB, Graham DL, Arzaga RR, Buzin N, Webb J, Green TA, Bass CE, Neve RL, Terwilliger EF, Nestler EJ, et al. 2011. Overexpression of CREB in the nucleus accumbens shell increases cocaine reinforcement in self-administering rats. J Neurosci 31: 16447–16457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445: 168–176 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, et al. 2009. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet 18: 4897–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C 2003. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115: 489–502 [DOI] [PubMed] [Google Scholar]
- Lin CH, Yeh SH, Lin CH, Lu KT, Leu TH, Chang WC, Gean PW 2001. A role for the PI-3 kinase signaling pathway in fear conditioning and synaptic plasticity in the amygdala. Neuron 31: 841–851 [DOI] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA 2009. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron 62: 254–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Bear MF 2004. LTP and LTD: An embarrassment of riches. Neuron 44: 5–21 [DOI] [PubMed] [Google Scholar]
- Maze I, Covington HE III, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, et al. 2010. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327: 213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY 2006. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci 26: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Young JZ, McHugh TJ, Buhl DL, Tonegawa S 2008. Transgenic inhibition of synaptic transmission reveals role of CA3 output in hippocampal learning. Science 319: 1260–1264 [DOI] [PubMed] [Google Scholar]
- Nilsson M, Hansson S, Carlsson A, Carlsson ML 2007. Differential effects of the N-methyl-d-aspartate receptor antagonist MK-801 on different stages of object recognition memory in mice. Neuroscience 149: 123–130 [DOI] [PubMed] [Google Scholar]
- O'Brien JA, Lummis SC 2006. Diolistic labeling of neuronal cultures and intact tissue using a hand-held gene gun. Nat Protoc 1: 1517–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kusky JR, Ye P, D'Ercole AJ 2000. Insulin-like growth factor-I promotes neurogenesis and synaptogenesis in the hippocampal dentate gyrus during postnatal development. J Neurosci 20: 8435–8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo P, Watabe AM, Grant SG, O'Dell TJ 2003. Phosphatidylinositol 3-kinase regulates the induction of long-term potentiation through extracellular signal-related kinase-independent mechanisms. J Neurosci 23: 3679–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterweil E, Wells DG, Mooseker MS 2005. A role for myosin VI in postsynaptic structure and glutamate receptor endocytosis. J Cell Biol 168: 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, et al. 2009. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5: 540–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Bruning JC 2008. Forkhead transcription factors and ageing. Oncogene 27: 2351–2363 [DOI] [PubMed] [Google Scholar]
- Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, et al. 2009. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell 8: 460–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KB 2001. The mouse brain in stereotaxic coordinates, 2nd ed. Academic Press, San Diego [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. 2010. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328: 753–756 [DOI] [PubMed] [Google Scholar]
- Polter A, Yang S, Zmijewska AA, van Groen T, Paik JH, Depinho RA, Peng SL, Jope RS, Li X 2009. Forkhead box, class O transcription factors in brain: Regulation and behavioral manifestation. Biol Psychiatry 65: 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, et al. 2008. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron 59: 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. 2009. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5: 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler R, Luft T, Oliveira SH, Farias CB, Almeida VR, Quevedo J, Dal Pizzol F, Schroder N, Izquierdo I, Schwartsmann G 2006. Molecular mechanisms mediating gastrin-releasing peptide receptor modulation of memory consolidation in the hippocampus. Neuropharmacology 51: 350–357 [DOI] [PubMed] [Google Scholar]
- Roesler R, Valvassori SS, Castro AA, Luft T, Schwartsmann G, Quevedo J 2009. Phosphoinositide 3-kinase is required for bombesin-induced enhancement of fear memory consolidation in the hippocampus. Peptides 30: 1192–1196 [DOI] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M 2001. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31: 115–130 [DOI] [PubMed] [Google Scholar]
- Salehi A, Faizi M, Colas D, Valletta J, Laguna J, Takimoto-Kimura R, Kleschevnikov A, Wagner SL, Aisen P, Shamloo M et al. 2009. Restoration of norepinephrine-modulated contextual memory in a mouse model of Down syndrome. Sci Transl Med 1: 7ra17 doi: 10.1126/scitranslmed.3000258 [DOI] [PubMed] [Google Scholar]
- Salih DA, Brunet A 2008. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 20: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe MD, Battaglia F, Wang JW, Malleret G, David DJ, Monckton JE, Garcia AD, Sofroniew MV, Kandel ER, Santarelli L, et al. 2006. Ablation of hippocampal neurogenesis impairs contextual fear conditioning and synaptic plasticity in the dentate gyrus. Proc Natl Acad Sci 103: 17501–17506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A 2006. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311: 1012–1017 [DOI] [PubMed] [Google Scholar]
- Sheng C, Heng X, Zhang G, Xiong R, Li H, Zhang S, Chen S 2013. DJ-1 deficiency perturbs microtubule dynamics and impairs striatal neurite outgrowth. Neurobiol Aging 34: 489–498 [DOI] [PubMed] [Google Scholar]
- Shepherd GM, Harris KM 1998. Three-dimensional structure and composition of CA3 → CA1 axons in rat hippocampal slices: Implications for presynaptic connectivity and compartmentalization. J Neurosci 18: 8300–8310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu E, Tang YP, Rampon C, Tsien JZ 2000. NMDA receptor-dependent synaptic reinforcement as a crucial process for memory consolidation. Science 290: 1170–1174 [DOI] [PubMed] [Google Scholar]
- Shinoda S, Schindler CK, Meller R, So NK, Araki T, Yamamoto A, Lan JQ, Taki W, Simon RP, Henshall DC 2004. Bim regulation may determine hippocampal vulnerability after injurious seizures and in temporal lobe epilepsy. J Clin Invest 113: 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman T, Wood SC, Anagnostaras SG 2009. Modafinil and memory: Effects of modafinil on Morris water maze learning and Pavlovian fear conditioning. Behav Neurosci 123: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumyatsky GP, Tsvetkov E, Malleret G, Vronskaya S, Hatton M, Hampton L, Battey JF, Dulac C, Kandel ER, Bolshakov VY 2002. Identification of a signaling network in lateral nucleus of amygdala important for inhibiting memory specifically related to learned fear. Cell 111: 905–918 [DOI] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Xuan Z, Zhang MQ 2006. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc Natl Acad Sci 103: 6275–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Christensen K, McGue M, Stevnsner T, Bohr VA, Christiansen L 2010. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell 9: 1010–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M 2006. BioGRID: A general repository for interaction datasets. Nucleic Acids Res 34: D535–D539 doi: 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Suzuki N, Mori J, Oshima A, Usami S, Hashizume K 2007. Micro-crystallin as an intracellular 3,5,3′-triiodothyronine holder in vivo. Mol Endocrinol 21: 885–894 [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME 1998. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20: 709–726 [DOI] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, et al. 2003. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 31: 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heide LP, Jacobs FM, Burbach JP, Hoekman MF, Smidt MP 2005. FoxO6 transcriptional activity is regulated by Thr26 and Ser184, independent of nucleo-cytoplasmic shuttling. Biochem J 391: 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B 2003. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res 31: 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DM, Teplow DB 2012. Alzheimer's disease and the amyloid β-protein. Prog Mol Biol Transl Sci 107: 101–124 [DOI] [PubMed] [Google Scholar]
- Wang SH, Teixeira CM, Wheeler AL, Frankland PW 2009. The precision of remote context memories does not require the hippocampus. Nat Neurosci 12: 253–255 [DOI] [PubMed] [Google Scholar]
- Watkins JC, Jane DE 2006. The glutamate story. Br J Pharmacol 147: S100–S108 doi: 10.1038/sj.bjp.0706444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel MK, Naska S, Laliberte CL, Rymar VV, Fujitani M, Biernaskie JA, Cole CJ, Lerch JP, Spring S, Wang SH, et al. 2008. p73 regulates neurodegeneration and phospho-tau accumulation during aging and Alzheimer's disease. Neuron 59: 708–721 [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD 2008. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci 105: 13987–13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanillo D, Sprengel R, Hvalby O, Jensen V, Burnashev N, Rozov A, Kaiser KM, Koster HJ, Borchardt T, Worley P, et al. 1999. Importance of AMPA receptors for hippocampal synaptic plasticity but not for spatial learning. Science 284: 1805–1811 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. 2006. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52: 255–269 [DOI] [PMC free article] [PubMed] [Google Scholar]