Abstract
Because of the high risk of recurrence in high-grade serous ovarian carcinoma (HGS-OvCa), the development of outcome predictors could be valuable for patient stratification. Using the catalog of The Cancer Genome Atlas (TCGA), we developed subtype and survival gene expression signatures, which, when combined, provide a prognostic model of HGS-OvCa classification, named “Classification of Ovarian Cancer” (CLOVAR). We validated CLOVAR on an independent dataset consisting of 879 HGS-OvCa expression profiles. The worst outcome group, accounting for 23% of all cases, was associated with a median survival of 23 months and a platinum resistance rate of 63%, versus a median survival of 46 months and platinum resistance rate of 23% in other cases. Associating the outcome prediction model with BRCA1/BRCA2 mutation status, residual disease after surgery, and disease stage further optimized outcome classification. Ovarian cancer is a disease in urgent need of more effective therapies. The spectrum of outcomes observed here and their association with CLOVAR signatures suggests variations in underlying tumor biology. Prospective validation of the CLOVAR model in the context of additional prognostic variables may provide a rationale for optimal combination of patient and treatment regimens.
Introduction
High-grade serous ovarian carcinoma (HGS-OvCa) accounts for 60%–80% of the approximately 26,000 women diagnosed with epithelial ovarian carcinoma in the US annually (1, 2). Known risk determinants for the development of ovarian carcinoma include BRCA1/BRCA2 mutations, family history, nulliparity, oral contraceptive use, tubal ligation, pregnancy, and lactation (1, 3). A common treatment regimen consists of tumor debulking, followed by administration of platinum and taxane-based chemotherapy (4). The advanced stage at which most patients present, combined with the high rate of relapse, results in a 5-year survival rate less than 40% (5, 6). Identification of nonresponders and patients with primary platinum resistance (recurrence less than 6 months after last chemotherapy cycle) is an important step toward achieving greater life expectancy for serous ovarian carcinoma patients (7). Gene expression profiles have been established that are associated with overall survival (8, 9), debulking status (10), and response to platinum therapy (11–15). Despite those encouraging developments, no biomarkers for prediction of response to therapy are yet in clinical use.
Gene expression–based outcome predictors have had the greatest impact in breast cancer, where gene signature–based assays of recurrence, chemotherapy efficacy, and metastasis were developed with the potential to guide therapy decisions (16–18). Predictors of prognosis have been developed for other cancers, but have not necessarily led to changes in clinical practice. In addition to predicting survival, the potential of prognostic classifiers lies in the ability to recognize categories of patients that are more likely to respond to particular therapies. For example, the Mesenchymal expression subtype of glioblastoma is being investigated in relation to response to angiogenesis inhibitors such as bevacizumab (19).
In a recent report from The Cancer Genome Atlas (TCGA) Research Network, 489 cases of HGS-OvCa were analyzed using copy number, expression and methylation arrays, and exonic sequencing of more than 18,500 genes (20). The HGS-OvCa genome was shown to harbor more somatic copy number alterations and fewer gene mutations than glioblastoma, which was also studied by TCGA (21). Consistent with a recent report (22), 96.5% of ovarian cases studied by TCGA contained a mutation in TP53 while also harboring a large number of copy number alterations, with the major target genes including CCNE1, MYC, TERT, and NF1. The TCGA analysis additionally described 4 expression subtypes of HGS-OvCa. Based on the expression of marker genes, these subtypes were termed “Differentiated,” “Immunoreactive,” “Mesenchymal,” and “Proliferative” (23). A supervised prognostic signature based on 193 genes developed using 215 TCGA expression profiles was able to classify a validation set of 524 expression profiles into a group with a better predicted prognosis (median survival, approximately 48 months) and a group with a worse predicted prognosis (median survival, approximately 36 months).
Here, we expanded the description of the 4 recently identified signatures of HGS-OvCa using the full catalog of TCGA genomic and clinical data. We integrated subtype and prognostic classifiers defined in a set of 489 expression profiles into a prognostic framework termed “Classification of Ovarian Cancer” (CLOVAR) and tested its accuracy in predicting outcome on a dataset of 879 publicly available expression profiles of HGS-OvCa.
Results
Gene expression profiles from individual HGS-OvCa tumor samples exhibit features of multiple subtype signatures.
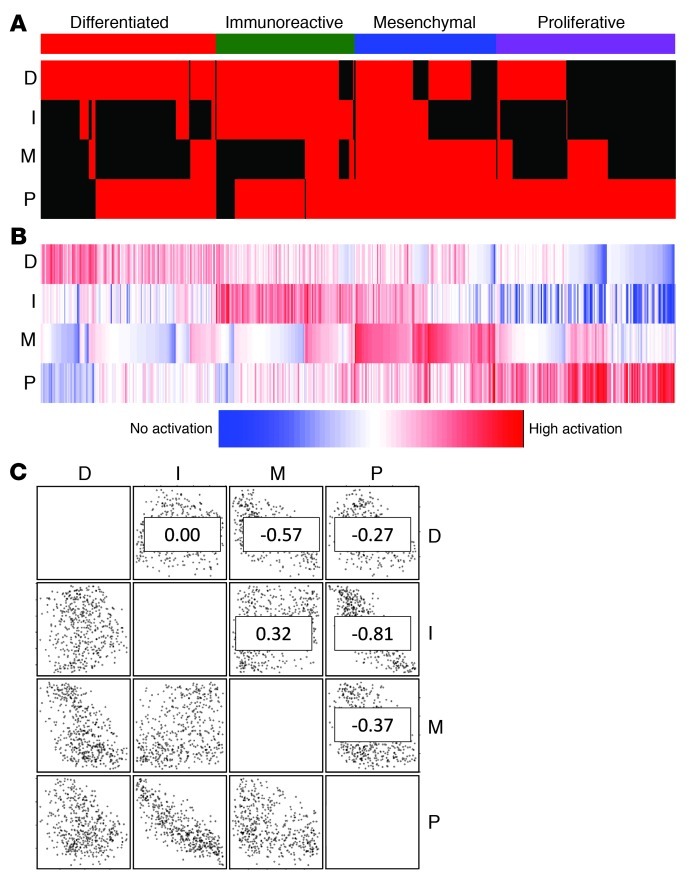
It was previously reported that a portion of ovarian tumor samples are characterized by high numbers of infiltrating T lymphocytes or stromal cells and that such tumor samples can be recognized by expression profiling (23, 24). Using immunohistochemistry, we identified an increased number of samples with infiltrating T lymphocytes in the Immunoreactive group (42% [n = 19] vs. 16% [n = 80]; P < 0.01, Fisher exact test), whereas desmoplasia, associated with infiltrating stromal cells, was found more often in the Mesenchymal cluster, with borderline significance (27% [n = 26] vs. 12% [n = 63]; P = 0.07, Fisher exact test). To investigate the possibility that tumor samples expressing the Immunoreactive and Mesenchymal signatures were additionally expressing the non–infiltrating cell–associated Differentiated or Proliferative signature, we used single-sample gene set enrichment analysis (ssGSEA) to assess gene set activation scores for all 489 tumor samples. ssGSEA is a rank-based comparison of the expression levels of genes in the gene set with all other genes in an expression profile. In this analysis, expression profiles clustering with, for instance, the Differentiated group will have a high Differentiated signature score. Signatures of the 4 subtypes were described previously (20). To define ssGSEA score cutoffs for inclusion in the set of samples expressing a subtype signature, we first assessed the silhouette width for each clustered profile. The silhouette width is defined as the ratio between the smallest correlation between a profile and all other profiles within its cluster and the largest correlation between that profile and all other profiles outside its cluster. The minimum ssGSEA score among samples that defined the original clusters and that clustered with a positive silhouette width to establish the ssGSEA score was used as cutoff value for inclusion into a subtype set. In this analysis, 82% of the 489 expression profiles were assigned to at least 2 subtypes (Figure 1 and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI65833DS1). For comparison, a similar analysis on our previously reported subtypes of glioblastoma (25) assigned 24% of expression profiles to more than 1 class (Supplemental Figure 1). Every possible signature combination was observed in at least 1 tumor sample, and negative correlations were observed between the Immunoreactive and Proliferative signatures (r2 = –0.81) and between the Differentiated and Mesenchymal signatures (r2 = –0.57). The overlap in gene signature scores suggests that HGS-OvCa does not consist of mutually exclusive expression subtypes, but that each tumor sample is represented by multiple signatures at different levels of activation. This pattern may reflect a higher level of homogeneity than is seen in other tumor types, such as glioblastoma and breast cancer.
Figure 1. HGS-OvCa signature ssGSEA scores. ssGSEA scores for 489 HGS-OvCa tumor samples were generated using 4 previously described gene expression signatures (20): differentiated (D), immunoreactive (I), mesenchymal (M), and proliferative (P).
(A) Binary scores indicating whether a tumor sample activates the gene signature. Each column represents 1 sample; each row represents 1 gene signature. Red, activated; black, not activated. (B) Raw gene set activation scores. Each column represents 1 sample; each row represents 1 gene signature. (C) Scatter plots and r2 values (numbers within plots) for all gene set combinations.
Characterization of gene expression signatures.
To assess whether the 4 HGS-OvCa subtype expression signatures were associated with specific genomic aberrations, we investigated whether gene set activation scores were significantly increased in altered versus nonaltered samples. Genes with alterations in more than 3% of samples were included in the analysis (copy number alterations, n = 489; mutation events, n = 316). After Benjamini-Hochberg correction of gene set score t test P values for multiple testing, no gene mutation or copy number deletion resulted in a significant change of gene set score. Copy number amplifications of 367 genes, mapping to 13 different commonly amplified regions, were found to be associated with a significant increase of activation levels of at least 1 of the 4 ovarian cancer signatures (Supplemental Table 2). To gain further insight into the possible target of amplification for the 13 amplification regions, we calculated Pearson correlation coefficients among 279 amplified genes with matching expression data and gene set activation scores. Of 11 genes with matching expression data at 1q32.1, LAD1, which regulates stability of epithelial layer stability with mesenchyme, showed a correlation with the Differentiated gene set that was distinctly higher than other genes. When linking 105 genes at the 19q13 locus with expression levels to the Proliferative signature, we found NOTCH3 to be among the most highly correlated. NOTCH3 inactivation in samples expressing the Proliferative signature may provide a therapeutic opportunity (26). The 12q14 gene whose expression correlated most highly with the Proliferative gene set was HMGA2, which suggests a role for this chromatin-modifying gene that has previously been related to progression in lung adenocarcinoma (27). Amplification of the transcription factor MYC was associated with a significant difference in Differentiated and Immunoreactive gene set scores, but correlation was much higher with Differentiated than with Immunoreactive signature scores.
The gene signatures were further characterized by correlating signature activation scores to the activation scores of 7,565 gene sets from the Molecular Signatures Database ( http://www.broadinstitute.org/msigdb/; ref. 28) and gene expression levels (see Supplemental Methods).
Predicting outcome using an integrated signature classifier.
We previously described a transcriptional signature of 193 genes predictive of overall survival, which was developed using univariate Cox regression analysis on the expression profiles of 215 HGS-OvCa samples (20). In order to maximize the discovery power of the TCGA dataset, we used all 481 TCGA HGS-OvCa–derived expression profiles with matching clinical data, rather than the 215 profiles used in our previous work (20). To generate the new signature, we selected the 100 genes whose expression was most correlated (good prognosis genes, n = 47), or anticorrelated (poor prognosis genes, n = 53), with survival (Supplemental Table 3). We chose a signature of 100 genes to prevent sensitivity to overfitting of the data and to permit analysis of new samples with a medium-throughput expression assay. We named this signature the “Classification of Ovarian Cancer” (CLOVAR) survival signature. Gene set enrichment analysis of the 100-gene CLOVAR survival signature, which includes genes such as RB1, NFKBIB, and RXRB, showed an overlap between the signature and Molecular Signatures Database gene sets such as BENPORATH_MYC_MAX_TARGETS and LY_AGING_OLD_UP (Supplemental Table 4). Of the 100 genes in the CLOVAR survival signature, 36 were included from our recently reported 193-gene survival signature (20).
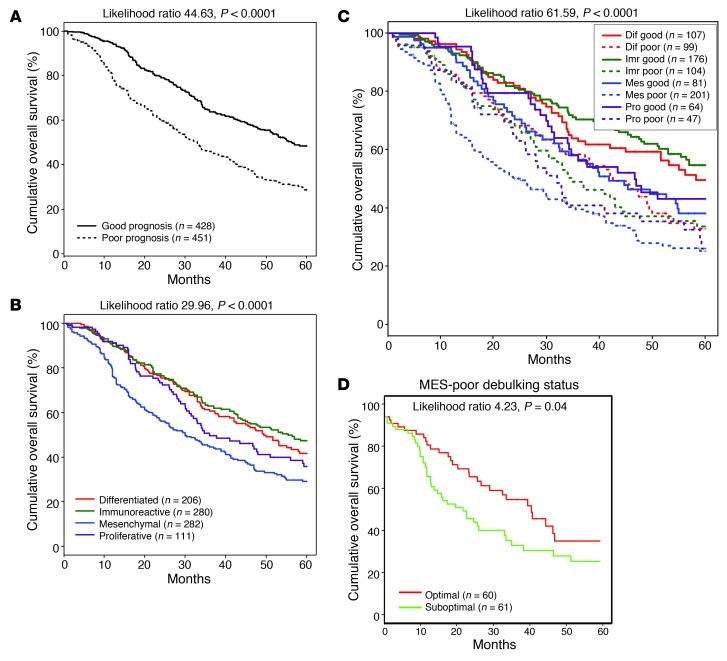
To verify the prognostic power of the CLOVAR survival signature, we used ssGSEA to analyze 64 TCGA expression profiles not included in the training dataset and 815 expression profiles with matching survival annotation from 6 published studies (9, 10, 23, 29–31). CLOVAR survival signature ssGSEA scores were calculated for each of the 7 validation datasets. The validation cohort included data generated on 5 different expression platforms, and the scale of the ssGSEA scores was dependent on the total number of genes included in the expression platform used. To make ssGSEA scores comparable between different expression platforms, scores were converted to a [0,1] scale within each dataset (see Methods). The validation set was stratified into CLOVAR poor prognosis or CLOVAR good prognosis groups using a cutoff score of 0.5. The difference in survival between the 2 prognostic groups was highly statistically significant (P < 0.0001; Figure 2A).
Figure 2. Survival per CLOVAR subtype, CLOVAR survival class, and double classifiers.
Kaplan Meier curves for CLOVAR survival class (A), survival per CLOVAR subtype (B), and double classification of samples by both CLOVAR subtype and CLOVAR survival signature (C). Data represent survival capped at 60 months for 879 samples in the validation set. (D) A further significant difference in survival was observed between optimally and suboptimally debulked CLOVAR Mesenchymal/CLOVAR poor prognosis patients.
A significant difference in recurrence-free survival was observed between patients in the TCGA cohort whose tumor samples expressed either the Immunoreactive or the Mesenchymal signature (Supplemental Figure 2). Next, we set out to verify this survival characteristic in the validation cohort. As reducing the size of the signature allows for the use of medium-high throughput gene expression quantification systems, we first reduced the 4 subtype signatures from 800 genes to gene signatures that, combined, contained 100 genes. We refer to these herein as CLOVAR subtype signatures. The CLOVAR subtype signatures classified the TCGA cohort into the 4 groups used to define the signatures with a cross-validation error rate of 8%, which indicates that the original clustering was captured by the reduced signatures. In order to determine whether validation cohort samples expressed the subtype signatures, we assessed ssGSEA scores of CLOVAR subtype signatures in the TCGA expression dataset. The minimum ssGSEA score among core samples was used to establish the ssGSEA score used as a cutoff value for inclusion into a subtype set. Using these ssGSEA cutoff values, tumor samples in the validation cohort were identified as expressing the CLOVAR Immunoreactive or CLOVAR Mesenchymal signatures. Cases expressing both signatures were assigned to a class according to the higher of the 2 scores. Those samples that did not express one of the signatures related to infiltrating cells were assigned to the CLOVAR Proliferative, CLOVAR Differentiated, CLOVAR Immunoreactive, or CLOVAR Mesenchymal class depending on the highest normalized gene set score for the 4 CLOVAR gene set signatures. Kaplan-Meier analysis revealed a highly significant difference in survival among the 4 validation set expression groups established by this approach (P < 0.0001; Figure 2B).
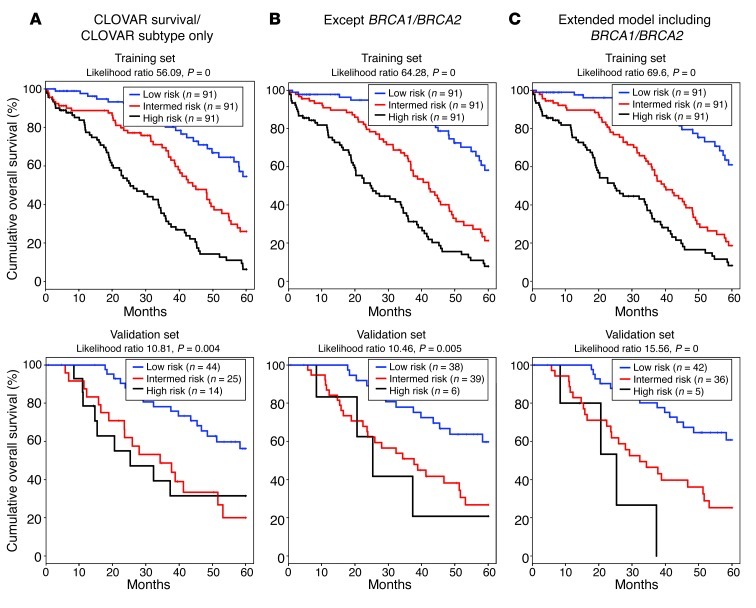
To combine the CLOVAR survival and CLOVAR subtype signatures for outcome prediction, we established a multiple covariate model using the TCGA dataset and including 3 signature scores as variables: CLOVAR survival, CLOVAR Immunoreactive, and CLOVAR Mesenchymal. In this model, CLOVAR survival was the dominant variable (Supplemental Table 5). Based on this model with CLOVAR survival and CLOVAR subtype as covariates, we computed risk scores of patients in training and validation sets. Patients in each set were then classified into good, intermediate, and poor outcome groups according the risk scores, using tertiles of risk scores for the TCGA set as cutoffs. We used this model to compute risk scores and classified the validation set (n = 879) into good, intermediate, and poor outcome groups, which showed 5-year survival of 55%, 38%, and 29%, respectively. Next, we tested whether including additional prognostic factors could improve the predictions. CLOVAR survival, CLOVAR Immunoreactive, and CLOVAR Mesenchymal scores, stage, grade, residual disease, and age annotation were available on 428 TCGA patients, which were used to calibrate a model (Supplemental Table 5). Using this extended model, we classified 328 patients from the validation set for which these parameters were available. The 5-year survival of the good, intermediate, and poor outcome groups using the extended model was 60%, 39%, and 27%, respectively. Lastly, we tested whether addition of “presence of BRCA1/BRCA2 germline mutations” to the model could further inform prognostic classification. BRCA1/BRCA2 germline data were available for 273 patients in the TCGA set and 83 patients in the validation set. The 5-year survival of the good, intermediate, and poor outcome groups in the validation set using the extended model including BRCA1/BRCA2 germline mutations was 61%, 25%, and 0%, respectively. To compare the performance of the 3 models, we computed likelihoods for the TCGA and validation sets using high/intermediate/low classification as a covariate, only including samples with nonmissing data on all variables (TCGA, n = 273; validation set, n = 83; Figure 3). Both in the TCGA set, which was used to train the models, and in the validation set, the extended model combined with BRCA1/BRCA2 germline mutation status showed the highest likelihood, suggesting most accurate performance in outcome prediction. Although the comparison may be hampered by the specific set of cases for which the correct annotation was available, this result suggests that a model combining CLOVAR survival, CLOVAR Immunoreactive, and CLOVAR Mesenchymal scores and BRCA1/BRCA2 germline mutation status provides optimal outcome predictions.
Figure 3. Survival analysis of multiple covariate outcome predictions.
A multiple covariate proportional hazards model was trained using (A) only CLOVAR survival and CLOVAR subtype signatures; (B) CLOVAR survival and CLOVAR subtype signatures, stage, grade, age, and residual disease; or (C) CLOVAR survival and CLOVAR subtype signatures, stage, grade, age, residual disease, and BRCA1/BRCA2 germline mutations. Models were trained on a training dataset (TCGA samples; n = 273) and tested on the validation set (n = 83), including all samples with nonmissing data on all variables. The difference in outcome of the 3 predicted groups was tested using Kaplan-Meier survival analysis.
As BRCA1/BRCA2 germline mutation status was only available for 9.4% of samples in the validation set, we sought to further optimize outcome prediction through combination of the CLOVAR survival and CLOVAR subtype signatures. We performed Kaplan-Meier survival analysis between samples with CLOVAR poor prognosis and CLOVAR good prognosis survival signatures within each of the CLOVAR Immunoreactive, CLOVAR Mesenchymal, CLOVAR Proliferative, and CLOVAR Differentiated subtypes. Importantly, the CLOVAR survival classifier predicted outcome with high significance within each of the 4 CLOVAR subtype groups (Figure 2C). The worst outcome was found in patients with tumors classified as both CLOVAR Mesenchymal and CLOVAR poor prognosis, which represented 23% of the validation set and showed a median overall survival of 23 months. The hazard ratio for death within 5 years of diagnosis for this group compared with the remainder of the sample cohort was 1.95 (95% confidence interval, 1.59 to 2.40), and the hazard ratio for recurrence within 12 months was 1.66 (95% confidence interval, 1.26 to 2.18) (Supplemental Table 6). Of the CLOVAR Mesenchymal/CLOVAR poor prognosis cases, 63% were determined to be resistant to platinum therapy, defined as having tumor recurrence within 6 months after the end of platinum therapy (Supplemental Table 1 and Supplemental Figure 3). All other samples — those not classified as CLOVAR Mesenchymal/CLOVAR poor prognosis — showed a platinum resistance rate of 23%. To refine the classification, we sought to understand those CLOVAR Mesenchymal/CLOVAR poor prognosis patients that had a relatively favorable outcome, with greater than 12 months progression-free survival (PFS). Germline BRCA1/BRCA2 mutations are associated with increased response rates to chemotherapy and overall survival (32). However, germline BRCA1/BRCA2 mutations were not found more frequently among tumor samples predicted as CLOVAR Mesenchymal/CLOVAR poor prognosis with greater than 12 months PFS compared with those CLOVAR Mesenchymal/CLOVAR poor prognosis samples with less than 12 months PFS. Similarly, such patients were not more likely to have been optimally debulked, were not younger, and did not receive a different treatment regimen compared with patients for which there was concordance between molecular prediction and poor clinical outcome. Interestingly, the amount of residual tumor after surgical debulking was independently related to prognosis in the CLOVAR Mesenchymal/CLOVAR poor prognosis, CLOVAR Immunoreactive/CLOVAR poor prognosis, and CLOVAR Differentiated/CLOVAR poor prognosis groups (Figure 2D and Supplemental Figure 4). Understanding the misclassification of these patients requires further investigation and will assist in more accurate prediction of PFS.
Discussion
Here, we presented a detailed analysis of the TCGA ovarian carcinoma expression data in terms of the 4 expression subtypes previously reported by the TCGA Research Network and others (20, 23). In the combination of data types analyzed here, we substantially expanded the description of the previously observed Mesenchymal, Differentiated, Proliferative, and Immunoreactive subtypes (20, 23, 24). Importantly, we showed that individual HGS-OvCa samples often expressed multiple subtype signatures, and classification into different mutually exclusive subtypes may therefore be less informative than in other cancers, such as breast cancer and glioblastoma (25, 33). Infiltrating cells had a profound effect on the expression patterns of HGS-OvCa, and analysis of the expression profiles of purified HGS-OvCa tumor cells may identify novel signatures that are currently dominated by the tumor microenvironment signals.
The most relevant result of this study was the development of the CLOVAR subtype and CLOVAR survival signatures and their significant ability to predict outcome to therapy in the validation set. Tumor samples not present in the current study may be classified using data available at TCGA ( https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/other/publications/ov_exp/). The classifier effectively subdivided HGS-OvCa patients in the large validation set into classes with different survival profiles, independent of and complemented by known risk-associated variables, such as size of residual tumor and age. Previous attempts to identify poor prognosis groups typically separated patient cohorts into 2 groups, in which median survival of the poor survival category ranged 33–41 months (9, 10, 29, 34). The large validation set allowed a detailed stratification resulting in identification of a subset of HGS-OvCa — comprising 23% of the validation set — with 23 months median survival and 63% platinum therapy resistance. Pinpointing a subset of very poor prognosis patients prompts consideration of additional treatment options that may improve survival for these patients. Studies of recurrence and platinum response are affected by the different definitions of tumor recurrence being used at different institutes (35). It is possible that greater prediction accuracy would be achieved when a standardized definition of progression is applied to all cases in the validation set. Investigation of incorrectly predicted cases, and factors such as BRCA pathway alterations and CCNE1 amplifications, may reveal additional predictive factors (34, 36). Importantly, the prognostic model presented here needs to be validated prospectively before it may be translated to clinical use. A prospective study would be most revealing when assessing the predictive capacities of the CLOVAR signatures in conjunction with other prognostic factors, such as BRCA mutation status (37, 38), age, grade, and residual disease. Concerns of prognosis are often among the first issues dealt with by ovarian cancer patients, and robust prognostic classification may aid in providing patients insight into this life-changing event.
The distinct biological characteristics of tumor samples associated with the each of the expression signatures suggest that the efficacy of experimental treatment modalities may be higher in some, but not all, prognostic groups. Clinical trials have been initiated in other cancers, such as glioma (trial ID RTOG 0825), that evaluate whether cases with high expression of Mesenchymal signatures are specifically sensitive to angiogenesis inhibitors. Similar efforts may be worthwhile in serous ovarian cancer, in which a subset of samples shows a mesenchymal phenotype. The effects of experimental therapies, such as dose-dense therapy or treatment with DNA damage repair inhibitors, may similarly be evaluated in the context of gene expression signatures (39–41). The CLOVAR signatures consist of 198 genes, making implementation using medium-throughput expression profiling platforms feasible. The association between the CLOVAR Mesenchymal/CLOVAR Immunoreactive signature and CLOVAR survival suggests an active role for the stromal tumor microenvironment in the pathogenesis of HGS-OvCa. Targeting of factors able to orchestrate immune response or stromal infiltration may therefore be used to design cancer therapies.
Studying HGS-OvCa in light of expression subtypes exposes features of ovarian carcinoma pathology that would otherwise have been neglected. The comprehensive CLOVAR framework presented here should provide a basis for improved understanding of ovarian carcinoma tumorigenesis that may ultimately lead to more effective treatments. Combining CLOVAR with clinically related features allows for robust survival classification. We expect that our findings will bring us closer to improving outcome in this devastating disease by providing an effective prognostic strategy combined with an approach to establishing new treatment modalities.
Methods
Patients and tumor samples.
HGS-OvCa samples were collected and processed through the TCGA Biospecimens Core Resource at the International Genomics Consortium; the same set of samples was used as in the prior TCGA ovarian carcinoma report (20). 489 ovarian tumors were selected according to the following criteria: (a) at least 70% tumor nuclei and less than 20% necrosis, (b) microarray quality controls within standards, and (c) high-quality data on each of the 3 gene expression platforms used. See Supplemental Table 1 for patient characteristics; all patient data have been deposited at the Data Coordinating Center of TGCA ( http://cancergenome.nih.gov).
Microarray experiments and data processing.
Gene expression profiles were established as described previously (20, 21, 25). In short, samples were assayed on 3 different microarray platforms: Affymetrix Human Exon 1.0 ST GeneChips, Affymetrix HT-HG-U133A GeneChips, and custom designed Agilent 244,000 feature gene expression microarrays. Microarray labeling and hybridization protocols and quality control measures for each platform were performed as described elsewhere. Probes on all 3 platforms were aligned to a transcript database consisting of RefSeq (36.1) and complete coding sequences from GenBank (version 161). Affymetrix HT-HGU133A and Exon platforms were normalized and summarized using robust multichip average (RMA). Agilent data were lowess normalized and log transformed, and the mean was used to calculate gene level summaries. Gene-centric expression values were generated for every gene with at least 5 (Affymetrix) or 3 (Agilent) perfect-match probes per gene, resulting in expression values for 12,042 (HT-HGU133A), 18,632 (Exon), and 18,623 (Agilent) genes, the intersect of the 3 platforms being 11,861 genes. All data are available at TCGA ( https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/other/publications/ov_exp/). The full data have been described in detail elsewhere (ref. 21 and TCGA Data Primer; https://wiki.nci.nih.gov/display/TCGA/TCGA+Data+Primer). Factor analysis was used to integrate these measurements together into a single estimate of the relative gene expression, as described previously (25, 42), and resulted in a unified gene estimate for each sample for 11,861 genes.
Subclass signature gene and gene set identification.
The significance analysis of microarrays (SAM) method was used to identify marker genes and gene sets of each subtype. Each class was compared with the other 3 classes combined (43). We have provided both rank order and test statistics for all these analyses to allow independent confirmation of our findings on future analyses and datasets. The 200 most representative genes per TCGA expression cluster were selected by ranking genes by the F score (the measure used by SAM to indicate the difference in gene expression between 2 groups) for a given gene (43). F scores that are further deviated from 0 indicate a larger difference in gene expression. Signature genes could be either down- or upregulated. We limited our analysis to patients whose expression profiles clustered with a positive silhouette width (n = 413), a measure of how well a given expression profile represents the cluster of which it is a member. For marker gene set identification, SAM was applied on gene set activation scores.
Validation datasets.
Class signatures were verified using expression profiles of 64 additional TCGA U133A expression profiles and publicly available stage II–III–IV ovarian serous carcinoma samples (GEO accession no. GSE9899; refs. 9, 10, 23, 29–31). Probes from Affymetrix HG-U133A or HG-U133plus2 GeneChip platforms were mapped to a transcript database and combined in 1 probe set per gene (44). Expression levels from the 4 Affymetrix datasets were individually established using RMA and quantile normalization (45). The Operon Human version 3 dataset of Crijns et al. (9) was downloaded from GEO (accession no. GSE13876). For each gene within each sample, expression values were averaged for the 2 dye swap profiles. Probes measuring the same gene were averaged to get 1 expression value per gene and sample.
We used ssGSEA (see below) to generate gene set activation scores for each of the 4 signatures, after dimension reduction of the 800 genes in the combined signatures to the 100 genes included in the CLOVAR subtype signatures to facilitate classification of new samples using medium-throughput expression technologies (Supplemental Table 7). We used a Correlation-based Feature Subset Selection algorithm available in Weka software, version 3.6.2 (46, 47). The search method was set to Best First search with default values. 10-fold cross validation was performed on the data. For each of the 10 runs, the algorithm output the best subset of genes it could find. Then, each of the genes was assigned a score based on how many subsets it was included in out of 10. We sorted the genes in descending order based on their scores: genes that appeared in all 10 subsets were at the top, followed by genes included in 9 subsets, and so on. To establish a 100-gene signature capable of predicting subtype, we selected the top 100 genes of this list that were additionally present in each of the 4 validation sets. The 100-gene set was used to train support vector machines for classification of samples in the validation set. Mean classification error when applying the 100-gene signature was 8% (CLOVAR Differentiated, 9%; CLOVAR Immunoreactive, 9%; CLOVAR Mesenchymal, 6%; CLOVAR Proliferative, 8%), which indicates that classification can be performed with small error rates using the 100-gene signature. Using the 100-gene signature and ssGSEA, we assigned 879 expression profiles in the validation set to 1 of the 4 classes.
Prediction of survival.
Using the 481 samples in the TCGA cohort for which survival annotation was supplied, each gene in the unified expression dataset (n = 11,861) was assigned a univariate Cox P value for correlation with survival. The 100 genes with lowest Cox P values were used to calculate a prognostic t score, defined as the 2-sided t statistic comparing, within each tumor profile, the average of the poor prognosis mRNAs with the average of the good prognosis mRNAs (i.e., the t score for a given tumor being high when both the poor prognosis mRNAs in the signature were high and the good prognosis mRNAs were low) (48).
ssGSEA scores and gene sets.
ssGSEA was used to generate gene set activation scores for the subtype, CLOVAR subtype, and CLOVAR survival expression signatures. For a given sample, gene expression values were rank normalized and rank ordered. The empirical cumulative distribution functions (ECDFs) of the genes in the signature and the remaining genes were calculated. A statistic was calculated by an integration of the difference between the ECDFs, which is similar to the one used in GSEA, but based on absolute expression rather than differential expression (25). The details of the procedure are as follows: for a given signature G of size NG and single sample S, of the dataset of N genes, the genes are replaced by their ranks according to their absolute expression L = {r1, r2, r3, . . . , rN} and rank ordered. An enrichment score ES(G,S) is obtained by a weighted sum (integration) of the difference between the ECDF of the genes in the signature PG and the ECDF of the remaining genes PNG.
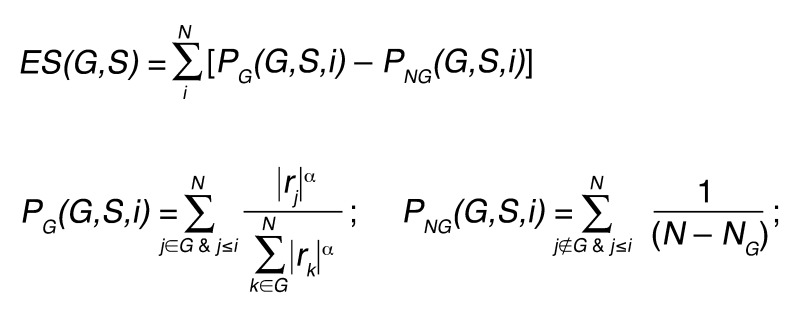
(Equation 1)
This calculation was repeated for each gene set in the database and each sample in the dataset. Notice that this quantity is signed, and that the exponent α = 3/4 adds a rank-associated weight.
The normalized ssGSEA scores are obtained in a similar manner as the normalized enrichment scores are obtained in standard GSEA (49). For a given sample, the ssGSEA normalized enrichment score is obtained by dividing the observed raw ssGSEA by the mean of the raw ssGSEA scores obtained for randomized versions of the gene set’s tags. This normalization uses the mean of the positive/negative permutation raw ssGSEA scores according to the sign (positive/negative) of the observed ssGSEA score. The normalization adjusts the ssGSEA scores to be in the same scale regardless of the size of the gene set and is motivated by the asymptotic scaling of the Kolmogorov-Smirnov statistic as a function of gene set size (see ref. 49 and Supplemental Methods).
Gene sets were downloaded from the Molecular Signatures Database (collection C2, version 3.0; http://www.broadinstitute.org/msigdb; ref. 28) and complemented by the Oncogene Pathway Activation database, a manually curated collection of 296 gene sets defined from GEO gene expression datasets and the biomedical literature as representing oncogene activation or tumor suppressor dysregulation (49). These 2 collections were augmented with additional “combined” signatures for the ones that have both upregulated (UP) and downregulated (DN) versions, producing a total of 7,553 gene sets. Subtypes were annotated using gene sets (see Supplemental Methods).
Subclass classification.
In order to establish the subtype of tumor samples in the validation set, CLOVAR subtype gene set scores were generated using ssGSEA. Scores were normalized to allow comparability between datasets from different expression platforms as described above. Of the TCGA samples originally clustering in the Immunoreactive subtype with a positive silhouette width, the lowest CLOVAR Immunoreactive signature normalized ssGSEA score was determined to be 0.63. Similarly, of the TCGA samples originally clustering in the Mesenchymal subtype, the lowest CLOVAR Mesenchymal signature ssGSEA score was determined to be 0.56. All samples in the validation set that expressed the CLOVAR Immunoreactive/CLOVAR Mesenchymal signature above these thresholds were assigned to the Immunoreactive (n = 280) and Mesenchymal (n = 282) groups per the higher of the 2 normalized ssGSEA scores. Samples in the validation set were assigned to 1 of the 4 classes per the highest of the 4 normalized ssGSEA scores. Normalized ssGSEA scores were additionally determined for the CLOVAR survival gene set, and cases were assigned to CLOVAR poor prognosis and CLOVAR good prognosis groups based on a score threshold of 0.5.
Associations of gene expression signatures with mutations and copy number alterations.
Segmented copy number profiles from the Agilent 244K platform were available for 489 ovarian carcinoma samples and matched normal controls. Preprocessing was performed using the GISTIC2.0 pipeline, as described previously (20, 50). All genes that were reported to be altered in at least 10 of 489 cases (1,437 deleted, 8,793 amplified) were categorized using sample-specific thresholds as previously described (21) into 1 of 3 levels: (a) deep deletion, (b) neutral copy number, and (c) high gain. Deletions and gains were tested independently of each other.
Nearly complete exon sequence data on 18,500 genes and 316 samples was used to report 19,356 somatic mutations in the previously reported integrated genomic analysis of ovarian carcinoma (20).
Association of copy number alterations and mutations with gene expression signatures was determined by comparing the normalized gene set scores of altered and nonaltered samples using a paired t test and using the Hochberg method implemented in p.adjust ( http://www.R-project.org) for controlling the family-wise error rate.
Expression profiling of normal fallopian tube.
Fresh Fallopian tube fimbria specimens were obtained from the Brigham and Women’s Hospital Department of Pathology with IRB approval. Only fimbria from nononcologic gynecological procedures were used. The tissue was incubated in a dissociation medium composed of MEM (Cellgro) supplemented with 3.5 mg/ml Pronase (Roche Diagnostics) and 0.25 mg/ml DNase (Sigma-Aldrich) for 48–72 hours in 4°C with constant mild agitation as previously described (51). The dissociated epithelial cells were harvested by centrifugation cultured in growth medium (DMEM/Ham’s F12, 1:1 [Cellgro], supplemented with 2% serum substitute Ultroser G [Pall Corp.] and 1% penicillin/streptomycin [PS; Invitrogen]) on culture dishes treated with human collagen IV (Sigma-Aldrich; 0.06 mg/ml in water). RNA from the cells was extracted using TRIzol reagent (Invitrogen) followed by RNeasy clean-up kit (Qiagen). Expression profiling was performed using Human Genome U133 Plus 2.0 microarray (Affymetrix).
Histopathological features.
For a subset of 98 TCGA ovarian carcinomas, a single slide stained for H&E was scanned at ×20 using Aperio XS slide scanner (Aperio). At low-power magnification, the extent of demoplastic stromal reaction was recorded as extensive if the tumor cell/stroma ratio was less than 2:1, i.e., the desmoplastic stroma accounted for at least one-third of the tumor area. Typically, tumor cells were seen in a nested pattern surrounded by abundant stroma. At high-power magnification, 10 fields were counted for the presence of intratumoral lymphocytes within the cellular borders of tumor cells. Prominent intratumoral lymphocytosis was defined as >10 lymphocytes per average high-power field.
Computation of risk scores.
To evaluate the CLOVAR survival signature for outcome prediction, 3 different models were considered. The first model included 2 covariates, CLOVAR survival score and CLOVAR subtype signatures (Figure 3A), grouping all samples into 3 categories: CLOVAR Immunoreactive, CLOVAR Mesenchymal, and others. In order to test whether including additional prognostic factors would improve the predictions, the first model was extended (Figure 3B) by adding the following 4 potential prognostic factors to the CLOVAR survival signature model: (a) stage, (b) grade, (c) age, and (d) residual disease. Next, the addition of “presence of BRCA1/BRCA2 germline mutations” to the second model was evaluated (Figure 3C). For this purpose, the third model included CLOVAR survival score, CLOVAR subtype signatures, stage, grade, age, residual disease, and presence of BRCA1/BRCA2 germline mutations as predictors of survival. To make a fair comparison among models, the same set of samples was used for all 3 models (TCGA training set, n = 273; validation set, n = 83), with nonmissing data on all variables. In each model, the risk score was computed for each patient, defined as a linear combination of predictor variables weighted by their respective Cox regression coefficients. Patients in the training set were then classified into good, intermediate, and poor outcome groups according to the risk scores, using tertile scores of the training set as cutoffs. Similarly, patients in the validation set were split into 3 risk groups using the same cutoffs as used for the training set. Differences in survival times were tested using the log-rank test.
Statistics.
Statistical analysis was performed using the R statistical computing environment. The SAM method was used to identify marker genes and gene sets of each subtype, ranking genes and gene sets by F test score and with a q value less than 0.05 considered significant. Log-rank likelihood models and Cox proportional hazards regression models were used to test for differences in survival, and a P value of less than 0.05 was considered significant. A paired 2-sided Student’s t test was used to test the significance of association between expression subtype and genomic alterations. P values were corrected for multiple testing using the Hochberg method for controlling the family-wise error rate. Corrected P values less than 0.05 were considered significant.
Study approval.
Ovarian cancer samples were collected and processed through the TCGA Biospecimens Core Resource at the International Genomics Consortium (Phoenix, Arizona, USA), as described elsewhere (20). All specimens were collected using IRB-approved protocols and were deidentified to ensure patient confidentiality. All data used in this study have been published previously, or through TCGA.
Supplementary Material
Acknowledgments
We thank the members of TCGA Research Network for their collaboration and comments. This work was supported by NIH grants U24CA143867 (to M. Meyerson) and U24CA143883 (to J.N. Weinstein); by the Chapman Foundation (to J.N. Weinstein); and by the National Health and Medical Research Council of Australia (to D.D. Bowtell and A. Defazio). No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest: Matthew Meyerson is an equity holder of and has received consulting fees from Foundation Medicine and has received consulting fees and research support from Novartis.
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Citation for this article: J Clin Invest. 2013;123(1):517–525. doi:10.1172/JCI65833.
References
- 1.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351(24):2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 2.Kobel M, et al. Differences in tumor type in low-stage versus high-stage ovarian carcinomas. Int J Gynecol Pathol. 2010;29(3):203–211. doi: 10.1097/PGP.0b013e3181c042b6. [DOI] [PubMed] [Google Scholar]
- 3.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9(6):415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markman M, et al. Phase III randomized trial of 12 versus 3 months of maintenance paclitaxel in patients with advanced ovarian cancer after complete response to platinum and paclitaxel-based chemotherapy: a Southwest Oncology Group and Gynecologic Oncology Group trial. J Clin Oncol. 2003;21(13):2460–2465. doi: 10.1200/JCO.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, Siegel R, Xu JQ, Ward E. Cancer Statistics, 2010. Ca-a Cancer Journal for Clinicians. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 6.Coleman MP, et al. Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and the UK, 1995-2007 (the International Cancer Benchmarking Partnership): an analysis of population-based cancer registry data. Lancet. 2011;377(9760):127–138. doi: 10.1016/S0140-6736(10)62231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7(8):573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 8.Spentzos D, et al. Gene expression signature with independent prognostic significance in epithelial ovarian cancer. J Clin Oncol. 2004;22(23):4700–4710. doi: 10.1200/JCO.2004.04.070. [DOI] [PubMed] [Google Scholar]
- 9.Crijns AP, et al. Survival-related profile, pathways, and transcription factors in ovarian cancer. PLoS Med. 2009;6(2):e24. doi: 10.1371/journal.pmed.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonome T, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68(13):5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jazaeri AA, et al. Gene expression profiles associated with response to chemotherapy in epithelial ovarian cancers. Clin Cancer Res. 2005;11(17):6300–6310. doi: 10.1158/1078-0432.CCR-04-2682. [DOI] [PubMed] [Google Scholar]
- 12.Hartmann LC, et al. Gene expression profiles predict early relapse in ovarian cancer after platinum-paclitaxel chemotherapy. Clin Cancer Res. 2005;11(6):2149–2155. doi: 10.1158/1078-0432.CCR-04-1673. [DOI] [PubMed] [Google Scholar]
- 13.Reed E. ERCC1 and clinical resistance to platinum-based therapy. Clin Cancer Res. 2005;11(17):6100–6102. doi: 10.1158/1078-0432.CCR-05-1083. [DOI] [PubMed] [Google Scholar]
- 14.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10(13):4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 15.Helleman J, et al. Mismatch repair and treatment resistance in ovarian cancer. BMC Cancer. 2006;6:201. doi: 10.1186/1471-2407-6-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 17.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 19.Lai A, et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol. 2011;29(2):142–148. doi: 10.1200/JCO.2010.30.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Network CGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Network T. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed AA, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221(1):49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tothill RW, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14(16):5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 24.Marquez RT, et al. Patterns of gene expression in different histotypes of epithelial ovarian cancer correlate with those in normal fallopian tube, endometrium, and colon. Clin Cancer Res. 2005;11(17):6116–6126. doi: 10.1158/1078-0432.CCR-04-2509. [DOI] [PubMed] [Google Scholar]
- 25.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JT, et al. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66(12):6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 27.Winslow MM, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473(7345):101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denkert C, et al. A prognostic gene expression index in ovarian cancer — validation across different independent data sets. J Pathol. 2009;218(2):273–280. doi: 10.1002/path.2547. [DOI] [PubMed] [Google Scholar]
- 30.Dressman HK, et al. An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer. J Clin Oncol. 2007;25(5):517–525. doi: 10.1200/JCO.2006.06.3743. [DOI] [PubMed] [Google Scholar]
- 31.Yoshihara K, et al. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PLoS One. 2010;5(3):e9615. doi: 10.1371/journal.pone.0009615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 33.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konstantinopoulos PA, et al. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol. 2010;28(22):3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang RS, et al. Platinum sensitivity-related germline polymorphism discovered via a cell-based approach and analysis of its association with outcome in ovarian cancer patients. Clin Cancer Res. 2011;17(16):5490–5500. doi: 10.1158/1078-0432.CCR-11-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Etemadmoghadam D, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15(4):1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolton KL, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307(4):382–390. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang D, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306(14):1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katsumata N, et al. Dose-dense paclitaxel once a week in combination with carboplatin every 3 weeks for advanced ovarian cancer: a phase 3, open-label, randomised controlled trial. Lancet. 2009;374(9698):1331–1338. doi: 10.1016/S0140-6736(09)61157-0. [DOI] [PubMed] [Google Scholar]
- 40.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 41.Ledermann J, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366(15):1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 42.Wang XV, Verhaak RG, Purdom E, Spellman PT, Speed TP. Unifying gene expression measures from multiple platforms using factor analysis. PLoS One. 2011;6(3):e17691. doi: 10.1371/journal.pone.0017691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, et al. AffyProbeMiner: a web resource for computing or retrieving accurately redefined Affymetrix probe sets. Bioinformatics. 2007;23(18):2385–2390. doi: 10.1093/bioinformatics/btm360. [DOI] [PubMed] [Google Scholar]
- 45.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 46.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor Newsl. 2009;11:10–18. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- 47. Hall MA.Correlation-Based Feature Selection For Machine Learning [PhD thesis}. Hamilton, New Zealand: Waikato University; 1999. [Google Scholar]
- 48.Creighton CJ, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26(25):4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levanon K, et al. Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene. 2010;29(8):1103–1113. doi: 10.1038/onc.2009.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.