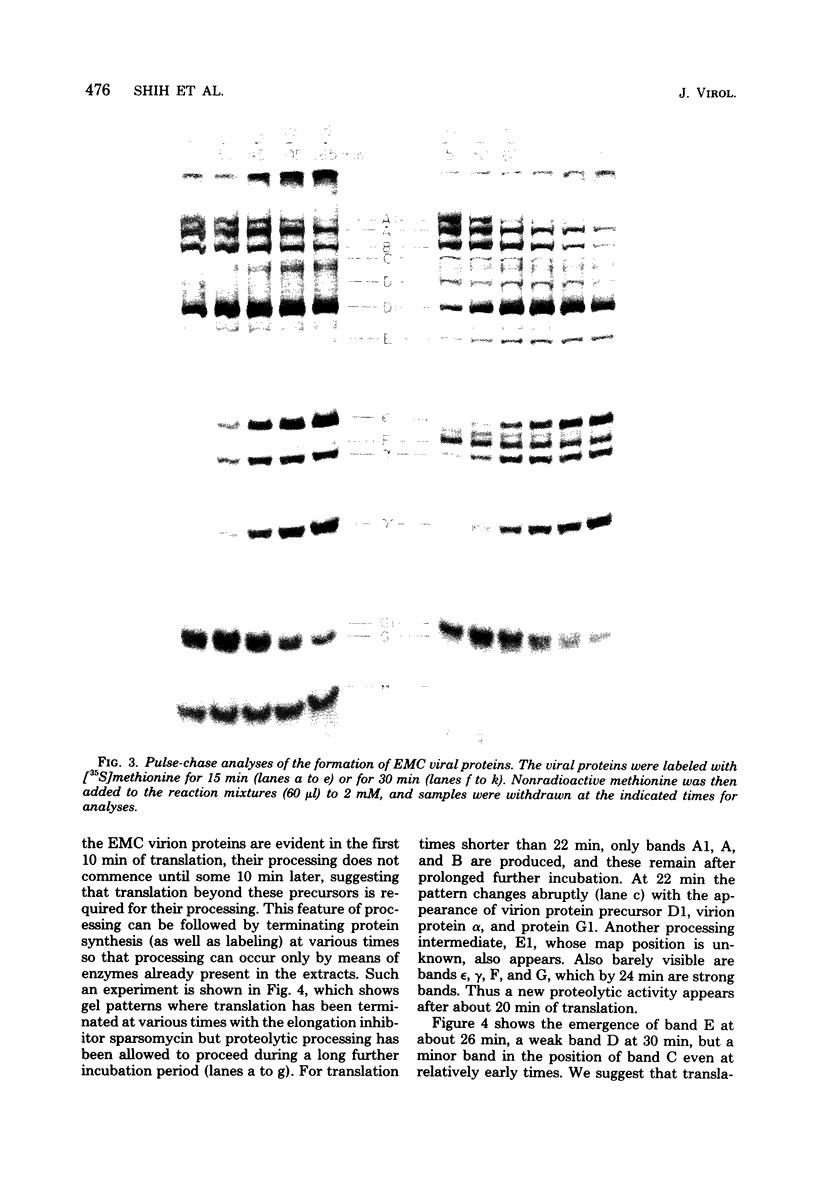
Abstract
In cell-free extracts derived from rabbit reticulocytes, encephalomyocarditis RNA can be translated completely, and the products can be processed extensively to give encephalomyocarditis virion proteins and several nonvirion proteins, including a genome-coded protein required for processing. The latter is probably a protease. Translation is very efficient. Under typical conditions, each EMC RNA is translated approximately eight times during a 3-h period. Kinetic analyses (time-course experiments, pulse-chase experiments, and pulse-stop experiments) have been used to determine the time of appearance of major products, and these times have been correlated with map positions. The gene for the putative protease is located near the middle of the genome downstream from the virion protein genes. Ribosomes can travel the length of encephalomycarditis RNA within 30 min, but there is a delay in their progress along the RNA at some point soon after they traverse the region coding for virion protein precursors. This delay results in the accumulation of precursors for a period of about 10 min before the putative protease is made and virion proteins (epsilon, alpha, and gamma) are released by proteolysis.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown F., Newman J., Stott J., Porter A., Frisby D., Newton C., Carey N., Fellner P. Poly(C) in animal viral RNAs. Nature. 1974 Sep 27;251(5473):342–344. doi: 10.1038/251342a0. [DOI] [PubMed] [Google Scholar]
- Butterworth B. E., Hall L., Stoltzfus C. M., Rueckert R. R. Virus-specific proteins synthesized in encephalomyocarditis virus-infected HeLa cells. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3083–3087. doi: 10.1073/pnas.68.12.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth B. E., Korant B. D. Characterization of the large picornaviral polypeptides produced in the presence of zinc ion. J Virol. 1974 Aug;14(2):282–291. doi: 10.1128/jvi.14.2.282-291.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth B. E., Rueckert R. R. Kinetics of synthesis and cleavage of encephalomyocarditis virus-specific proteins. Virology. 1972 Nov;50(2):535–549. doi: 10.1016/0042-6822(72)90405-9. [DOI] [PubMed] [Google Scholar]
- Chumakov K. M., Agol V. I. Poly(C) sequence is located near the 5'-end of encephalomyocarditis virus RNA. Biochem Biophys Res Commun. 1976 Jul 26;71(2):551–557. doi: 10.1016/0006-291x(76)90822-6. [DOI] [PubMed] [Google Scholar]
- Hall L., Rueckert R. R. Infection of mouse fibroblasts by cardioviruses: premature uncoating and its prevention by elevated pH and magnesium chloride. Virology. 1971 Jan;43(1):152–165. doi: 10.1016/0042-6822(71)90233-9. [DOI] [PubMed] [Google Scholar]
- Lawrence C., Thach R. E. Identification of a viral protein involved in post-translational maturation of the encephalomyocarditis virus capsid precursor. J Virol. 1975 Apr;15(4):918–928. doi: 10.1128/jvi.15.4.918-928.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Lenard J. Cleavage of mengovirus polyproteins in vivo. J Virol. 1974 Aug;14(2):261–269. doi: 10.1128/jvi.14.2.261-269.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor S. Evidence for the existence of protomers in the assembly of encephalomyocarditis virus. J Virol. 1975 May;15(5):1107–1120. doi: 10.1128/jvi.15.5.1107-1120.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paucha E., Seehafer J., Colter J. S. Synthesis of viral-specific polypeptides in Mengo virus-infected L cells: evidence for asymmetric translation of the viral genome. Virology. 1974 Oct;61(2):315–326. doi: 10.1016/0042-6822(74)90269-4. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. Translation of encephalomyocarditis virus RNA in vitro yields an active proteolytic processing enzyme. Eur J Biochem. 1978 Apr 17;85(2):457–462. doi: 10.1111/j.1432-1033.1978.tb12260.x. [DOI] [PubMed] [Google Scholar]
- Shih D. S., Shih C. T., Kew O., Pallansch M., Rueckert R., Kaesberg P. Cell-free synthesis and processing of the proteins of poliovirus. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5807–5811. doi: 10.1073/pnas.75.12.5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y. V., Agol V. I. Complete translation of encephalomyocarditis virus RNA and faithful cleavage of virus-specific proteins in a cell-free system from Krebs-2 cells. FEBS Lett. 1978 Mar 1;87(1):7–11. doi: 10.1016/0014-5793(78)80121-5. [DOI] [PubMed] [Google Scholar]
- Svitkin Y. V., Lyapustin V. N., Lashkevich V. A., Agol V. I. A comparative study on translation of flavivirus and picornavirus RNAs in vitro: apparently different modes of protein synthesis. FEBS Lett. 1978 Dec 1;96(1):211–215. doi: 10.1016/0014-5793(78)81096-5. [DOI] [PubMed] [Google Scholar]
- Villa-Komaroff L., Guttman N., Baltimore D., Lodishi H. F. Complete translation of poliovirus RNA in a eukaryotic cell-free system. Proc Natl Acad Sci U S A. 1975 Oct;72(10):4157–4161. doi: 10.1073/pnas.72.10.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmern D., Kaesberg P. 3'-terminal nucleotide sequence of encephalomyocarditis virus RNA determined by reverse transcriptase and chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4257–4261. doi: 10.1073/pnas.75.9.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]