Abstract
Saposin C is one of four homologous proteins derived from sequential cleavage of the saposin precursor protein, prosaposin. It is an essential activator for glucocerebrosidase, the enzyme deficient in Gaucher disease. Gaucher disease is a rare autosomal recessive lysosomal storage disorder caused by mutations in the GBA gene that exhibits vast phenotypic heterogeneity, despite its designation as a “simple” Mendelian disorder. The observed phenotypic variability has led to a search for disease modifiers that can alter the Gaucher phenotype. The PSAP gene encoding saposin C is a prime candidate modifier for Gaucher disease. In humans, saposin C deficiency due tomutations in PSAP results in a Gaucher-like phenotype, despite normal in vitro glucocerebrosidase activity. Saposin C deficiency has also been shown to modify phenotype in one mousemodel of Gaucher disease. The role of saposin C as an activator required for normal glucocerebrosidase function, and the consequences of saposin C deficiency are described, and are being explored as potential modifying factors in patients with Gaucher disease.
Keywords: Prosaposin, Lysosomal storage disorder, Glucocerebrosidase, Modifier, Activator
1. Introduction
1.1. The saposin proteins in lysosomal storage disorders
The saposins are a set of four small glycoproteins, referred to as saposin (Sap) A-D, that act as enzymatic activators in multiple stages of lysosomal sphingolipid degradation, as well as lysosomal membrane digestion [1–3]. These four homologous proteins are generated in the endosome via proteolysis of a 73 kDa precursor protein called prosaposin (pSap) [4,5]. The resulting Saps A-D each consist of approximately 80 amino acids, with six similarly-located cysteine residues that confer heat stability and a characteristic tertiary structure through the formation of three conserved disulfide bridges [3,6].
Mature Saps A-D assist lysosomal hydrolases in the degradation of sphingolipids. Deficiencies or dysfunctions of these activators can result in lysosomal storage disorders that mimic by deficiencies of the enzyme activated by that particular Sap [7–9]. Prosaposin deficiency results in the accumulation of sphingolipid substrates that are hydrolyzed by Sap-associated enzymes including sulfatides, glucosylceramide, lactosylceramide, galactosylceramide, digalactosylceramide, ceramide, and GM2 and GM3 gangliosides [7,10]. Patients with deficient pSap (OMIM ID: 611721), resemble type 2 (acute neuronopathic) Gaucher disease (GD; OMIM ID: 230900) [11]. Deficiencies of individual Saps also mimic specific disorders; Sap A deficiency resembles Krabbe disease [8], and inadequate Sap B resembles metachromatic leukodystrophy [12–14]. Six patients described with mutations in the Sap C domain of the PSAP gene (OMIM ID: 610539) have symptoms similar to either type 1 or type 3 (OMIM IDs: 230800, 231000) GD, despite normal glucocerebrosidase (GCase; EC 3.2.1.45) activity [9]. Sap C knockout mice also exhibit phenotypes most closely analogous to type 3 GD [15]. A specific deficiency of Sap D has not been reported in humans.
Saposin C has particular relevance for GD. It is a necessary activator for GCase, the enzyme deficient in this disorder due to mutations in the GBA gene (OMIM ID: 606463) [4,16,17]. GD is an autosomal recessive disorder, and the most common lysosomal storage disorder. Deficiency of GCase leads to accumulation of glucosylceramide in lysosomes, causing substrate storage in macrophages in the spleen, liver, bone marrow, and other organs. Patients with GD often exhibit hepatosplenomegaly, thrombocytopenia, bone lesions, and anemia [16]. In some cases, patients also develop neurological symptoms, including myoclonic epilepsy, ataxia, intellectual impairment, and abnormal horizontal saccadic eye movements [16,18]. Clinically, GD is classified into three types, based on whether the patient displays neurological symptoms, and the age at which these first manifest [17]. Type 1 (non-neuronopathic) GD, the most common form, does not involve the central nervous system, but the severity ranges from significant morbidity in childhood due to complications from cytopenia, liver dysfunction, failure to thrive, or skeletal involvement, to patients that remain asymptomatic or undiagnosed for much of their life [17,19].
Type 2 and type 3 GD, the acute and chronic neuronopathic forms, respectively, are characterized by neurological dysfunction [17]. Type 2 GD affects infants, who have a life expectancy of less than two or three years [17,20]. These patients exhibit rapid neurological decline, severe hepatosplenomegaly, failure to thrive, and ultimately opisthotonus. A subgroup dies from hydrops fetalis or congenital ichthyosis before or shortly after birth [21,22].
Type 3 GD results in a less severe phenotype than type 2 [17]. Neurological symptoms vary greatly, including myoclonus, seizures, ataxia, dementia, and slowed horizontal eye movements [16]. A subgroup of these patients display significant visceral involvement, including hepatosplenomegaly, and can have extensive bone disease.
Despite clinical categorization of GD into these three types, a wide spectrum of phenotypic heterogeneity is observed in this disorder. As an essential activator of GCase, Sap C is a potential disease modifier, and subtle changes in its expression may contribute to the array of phenotypes observed in GD.
1.2. History of saposin research
Sap C was discovered by Ho and O’Brien in 1971 [23]. It was extracted from spleen homogenate from a 12-year-old female with type 3 GD, following splenectomy. Further experiments showed that it was heat-stable and capable of restoring mutant GCase activity in vitro [23]. The common genetic origin of Sap C and Sap B, which was discovered in 1964, was confirmed in the late 1980s, when the cDNAs encoding each protein were cloned independently, and it was found that both derive from proteolytic processing of a 73 kDa precursor protein, later confirmed to be pSap [24–26]. In total, four homologous domains were found in the pSap protein. All were approximately 80 amino acid residues in length and had similarly located cysteine and proline residues, suggesting common secondary and tertiary structures. In addition, each domain had at least one glycosylation site. These results indicated the existence of two other mature Sap proteins, which correspond to Saps A and D [27–29].
The nomenclature for the Sap proteins has evolved over the years, and thus the literature is often confusing. The current term “saposin,” derived from “sphingolipid activator protein,” was coined by O’Brien and Kishimoto [28,29]. Sap C has taken many names, originally called factor P by its discoverers [23]. It was also referred to as heat stable factor, A1 activator, and co-β-glucosidase in the early literature. In 1984, the term sphingolipid activator protein (SAP) was applied to the two Saps known at the time. Sap B and Sap C were called SAP-1 and SAP-2, respectively [30]. The currently accepted saposin A-D nomenclature system arose from an improved understanding of the genetic basis for pSap and the mature Sap proteins. The Saps were named sequentially based on their position in the pSap amino acid sequence, starting from the N-terminus. This nomenclature identifies the individual Saps based on their amino acid sequences, rather than their activities, highlights their common origin from PSAP, and distinguishes them from other sphingolipid activator proteins [31].
2. Saposin C structure and function
2.1. The saposin precursor
Prosaposin is encoded by the ~17 kb PSAP gene, located on chromosome 10q21 [32]. The gene contains 15 exons and 14 introns. The four homologous gene domains, each encoding one Sap protein, suggest that the PSAP gene arose from multiple duplications of an ancient gene that encoded a single Sap. Three isoforms of pSap result from alternative splicing at exon 8, which consists of nine base pairs encoding three amino acids in Sap B. Human cDNA libraries contain cDNAs with all nine, the last six, or none of the base pairs in exon 8 [33]. Differential splicing of PSAP does not appear to have any phenotypic effects [34].
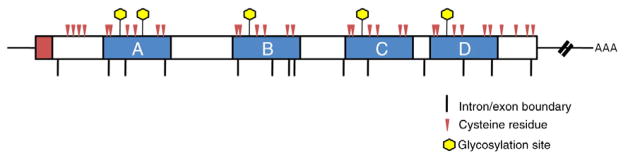
Prosaposin consists of 524 amino acids, including the four Sap domains, a 16-residue signal peptide, and five polypeptide sequences that separate the Sap domains. The protein contains five glycosylation sites, two in the Sap A domain, and one in each Sap B-D domains (Fig. 1) [2]. After synthesis, pSap, a 55 kDa, unglycosylated protein is transported through the endoplasmic reticulum to the Golgi apparatus, where sugar residues are added [35]. The extent to which pSap is glycosylated in the Golgi is believed to determine whether it is successfully reaches the lysosome via the sortilin receptor pathway [36,37]. Oligomerization of pSap may also prevent its entry to this pathway and proper trafficking to the lysosome [38]. In the lysosome, pSap is cleaved by cathepsin D in the interdomain sequences, yielding the four mature Sap proteins [5,38].
Fig. 1.
Prosaposin cDNA structure, with individual saposin domains noted. The 16-amino acid signal peptide is shaded in red, and functional saposin domains are shown in blue. Cysteine residues and glycosylation sites are indicated.
In addition to its role as the precursor of the Saps, pSap has intrinsic protein function. It is found uncleaved in brain, skeletal muscle, and cardiac muscle [39]. pSap is enriched in cerebrospinal fluid, bile, semen [40], and human milk [41], and is secreted by Sertoli cells to promote spermatogenesis [42]. It is a known neurotrophic factor [43] that can accelerate regeneration of ischemic hippocampal neurons [44], and may be involved in the early development of the brain [45].
2.2. The saposin C–GCase interaction
Saposin C is an 80 amino acid protein encoded by the third domain in PSAP (Fig. 2). It has a molecular weight of 9 kDa, although this can increase to 12 kDa after N-glycosylation at Asn 22 [46]. Six Cys residues form three disulfide linkages to shape the tertiary structure of Sap C. Pairings between Cys 5 and Cys 78, Cys 8 and Cys 72, and Cys 36 and Cys 47 stabilize a bundle of five α-helices, and also render Sap C heat stable at temperatures over 100 °C [23,47]. This arrangement of α-helices is sometimes referred to as the saposin fold. Sap C has an isoelectric point of pH 4.3–4.4 [4]. At pH 5.4 it undergoes a conformational change in which the α-helix bundle opens to a V-shaped, or “boomerang,” configuration. This exposes hydrophobic residues that were previously located inside the saposin fold, rendering Sap C capable of penetrating phospholipid membranes and binding anionic phospholipids [48,49].
Fig. 2.
Amino acid sequence of saposin C. The three conserved disulfide linkages are underlined and bolded, along with the N-glycosylation site (red).
Once the pH-dependent interaction between Sap C and lipid vesicles was known, the physical interaction between Sap C, lipids, and GCase was explored. To map the site of interaction with GCase, Weiler et al. measured GCase activities and kinetic constants of different synthetic Sap C sequences [50]. They showed that two polypeptide fragments of Sap C could each individually promote GCase activity to 90% of control levels. The fragments consisted of residues 27–34 of domain 1 of Sap C, and residues 41–49 of domain 2, respectively. These sites were proposed as activator regions for GCase. Residues 41–82, however, were proposed as the site of binding to GCase. Recently, Atrian et al. used a protein-protein rigid docking model to explore the interaction sites between GCase and Sap C [51]. Based on this study, residues 9, 20–25, and 56–74 of Sap C were suggested to be the sites of interaction with GCase, confirming the experimental data of Weiler et al. The computational docking model also revealed that specific GCase residues (314, 317, 318, 348, 358, 362, 365, 366, 369, 370, 372, 373, 441, 443–445, 463, 464, and 487; UniProt Knowledgebase, EBI-EMBL) can potentially interact with Sap C [51].
In an effort to characterize the Sap C-GCase complex, Ho et al. suggested a 2:1 stoichiometry between Sap C and GCase, forming fully-activated GCase [52]. The 1:1 stoichiometry showed only 28% of the activity of this complex. Using gel-permeation HPLC on human spleen extracts, Aerts et al. separated two forms of GCase: nonactivated (60 kDa) and fully activated (200 kDa) [53]. Earlier, Prence et al. had shown that incubation of rat liver with Sap C and phosphatidylserine shifts the size of GCase from 57 kDa to 188 kDa by sucrose density gradient ultracentrifugation [54]. Weiler et al. subsequently proposed a two-step mechanism of formation for the Sap C-GCase complex. A 2:1 Sap C-GCase intermediate complex forms first, followed by dimerization to a 4:2 complex with a molecular weight of 180–220 kDa [50]. Although both Sap C and GCase have been shown to be available as functional dimers [55,56], the 4:2 complex has not been crystallized [57].
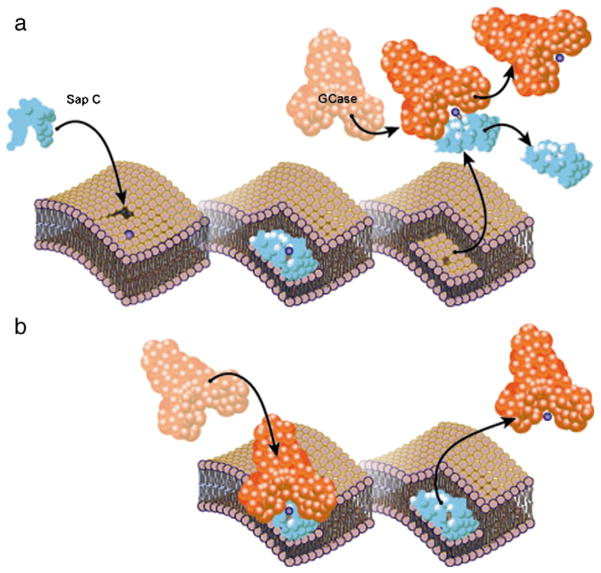
Saposin C is an established activator for the hydrolysis of glucosylceramide by GCase in lysosomes [9,23], but the mechanism by which Sap C promotes lysosomal hydrolysis is not fully known. Förster resonance energy transfer studies demonstrate associations between GCase and Sap C, GCase and membrane regions altered by Sap C, and GCase and hydrolysis products [58,59]. Sap C is believed to associate with both GCase and the phospholipid membrane, bringing them together so that GCase can hydrolyze endogenous glucosylceramide. Sap C may also help extract and solubilize the lipid substrate from the membrane, rendering it is accessible for hydrolysis [58] (Fig. 3).
Fig. 3.
Proposed mechanisms of GCase (orange) activation by Sap C (blue). Sap C embeds in preexisting defects in the bilayer membrane, resulting in a thinned membrane domain, and exposing the headgroup of glucocerebroside substrate (purple ball). Activated GCases is shown in red. a) Solubilizer model. Sap C dissociates from the membrane in a soluble complex with glucocerebroside, which is hydrolyzed by GCase in solution. b) Liftase model. Sap C stably associates with the membrane and exposes glucocerebroside for direct hydrolysis by GCase on the surface of thinned membrane domains.
To initiate activation of GCase by Sap C, a number of cationic lysine residues on two of the α-helices of Sap C may reversibly bind to the anionic membrane [47]. Once bound, a conformational change occurs in Sap C, exposing a hydrophobic region originally contained within the α-helix bundle [47,58]. Increased local concentration of anionic phospholipids within the bilayer augments the ability of Sap C to bind and destabilize the membrane [48]. The acidity of these phospholipids may create a pH gradient that helps to expose the hydrophobic domain of Sap C, or to encourage interaction with lysine residues. Sap C then localizes at preexisting membrane defects, where the lipids are less densely packed and the membrane is weakened [58]. From the edges of these defects, Sap C can further destabilize the membrane, restructuring it into patch-like domains of varying thickness [58,60–62]. The thinned regions are believed to result from the removal of the upper leaflet of the membrane, revealing the headgroups of internal lipids [59], or from the removal of endogenous lipids by Sap C for transfer to hydrolases [58].
It is currently unknown how the lipid is transferred from Sap C to GCase. One hypothesis, referred to as the solubilizer model (Fig. 3a), is that Sap C extracts the lipid from the bilayer in a soluble complex, and then dissociates from the membrane to form a complex with GCase, handing over the substrate for hydrolysis [58,59]. Another possibility, described by the liftase model [59], is that Sap C remains bound to the thinned regions of the membrane where glycosphingolipids are exposed. GCase can associate with this complex and cleave the exposed glucosylceramide headgroups that sit above the thinned portions of the membrane [47,58]. The liftase model (Fig. 3b) is currently the preferred theory to explain how Sap C stimulates GCase [47]. Studies of lipid antigen transfer to CD1 molecules by Sap C, where the protein is shown to be incapable of forming a soluble complex with lipid antigens, support this hypothesis [63]. Other Saps, such as Saps B and D, are best described as solubilizers [47,64].
In addition to its role as an activator for hydrolysis, Sap C also protects GCase from proteolytic degradation [65]. GCase activity is reduced in pSap knockout mice compared to wildtype mice, and GCase degradation is enhanced in both mouse and human pSap-deficient fibroblasts [65]. Treatment with the protease inhibitors leupeptin or pepstatin A restored GCase activity and protein levels in pSap-deficient fibroblasts. Finally, the addition of Sap C to pSap-deficient fibroblasts restores GCase activity in a manner similar to the protease inhibitors, suggesting that it also protects the enzyme from proteolytic degradation [65]. This protective effect was not observed for other Sap proteins tested.
2.3. Animal models of saposin C deficiency
A mouse model of specific Sap C deficiency was created by knock-in of a Cys to Pro mutation in the Sap C domain of the PSAP gene [15]. At 12 months, these mice developed weakness in the hind limbs and progressive ataxia. Foamy storage cells were observed in the spinal ganglion at around 25 weeks of age, and gradual loss of Purkinje cells began at 2 months, with most cerebellar Purkinje cells lost by 20 months. Inclusion bodies in neurons and axons in the spinal cord also developed around this time, along with pro-inflammatory activated astrocytes and microglia. GCase activity in brain, lung, liver, and spleen tissue samples was approximately 50% that of wildtype mice. A slight increase of glucosylceramide in the spinal cord was observed but, surprisingly, glycosphingolipid levels in the brain and visceral organs were not elevated.
In addition, Sun et al. generated a mouse model with both Sap C deficiency and GD [66]. In this model, Sap C-knockout mice were crossbred with knock-in GD mice homozygous for the gba mutation V394L. V394L homozygous mice are a viable model of chronic neuronopathic (type 3) GD [66–68]. They display neurological deficits, and liver and spleen GCase activity is reduced to 4–11% of wildtype levels. GCase activity in the brain is 25% that of wildtype levels [67]. Sap C deficiency caused by a point mutation in PSAP in these mice, resulted in a more severe phenotype. Sap C−/−/V394L homozygous mice showed even greater reduction in GCase activity, and increased storage of glucosylceramide and glucosylsphingosine. The elevated levels of glucosylsphingosine likely contributed to the CNS involvement, which was not seen in V394L homozygotes with wildtype Sap C. Sap C−/−/V394L mice developed hindlimb paresis and ataxia. They died, on average, at age 48 days from neurological complications, while V394L homozygotes remained viable. Sap C was confirmed as a disease modifier for GD in this model due to the increased severity of the phenotype in its absence.
3. The role of saposin C in the pathogenesis of Gaucher disease
Reduced GCase activity in GD is primarily caused by protein mis-folding and improper trafficking of the enzyme to the lysosome, or the total absence of the protein resulting from null alleles [69]. However, some mutations in GCase result in diminished residual enzyme activity in vitro by causing structural or biochemical changes that interfere with the mechanism of activation by Sap C [70]. The N370S mutation causes an amino acid substitution that limits the mutant enzyme’s capacity to interact with Sap C and to bind anionic phospholipid-containing membranes [71]. Computational models suggest that this mutation also decreases the protein’s flexibility around the active site, particularly in loop 1 (residues 311-319). This rigidity may reduce the conformations available to the enzyme, preventing it from properly establishing a binding interface for interaction with Sap C [72,73].
Salivioli et al. showed that Sap C is unable to reconstitute the hydrolytic activity of N370S GCase to the level of wildtype enzyme [71]. In the absence of Sap C, N370S GCase is only 10–15% as active as wildtype enzyme. Under optimal conditions [19,74] in patient fibroblasts, the addition of Sap C restores the activity of N370S GCase to 50–70% that of wildtype [71]. This increase is less pronounced when Sap C is added to other mutant forms of the enzyme [70]. The activating capability of Sap C toward N370S GCase depends primarily on the concentration of phospholipids present in the membrane [71]. Sap C can restore N370S GCase activity to 70% that of wildtype enzyme when there is a high (60–75%) membrane concentration of phosphatidylserine, but its activity was only 15% that of wildtype when the phosphatidylserine concentration was less than 20% of the total membrane composition. GCase activity correlated most strongly with the phosphatidylserine concentration of the membrane, and concentration-dependent increases in activity due to the addition of Sap C were smaller by comparison [71]. Compared to wildtype enzyme, N370S GCase had a decreased binding affinity for membranes with low phosphatidylserine concentrations [71].
The defective interaction between N370S GCase and Sap C may be improved by chaperone molecules, such as N-butyl-deoxynojirimycin (NB-DNJ) [73]. Chemical chaperones are small molecules that promote proper folding of a protein to accurately establish its tertiary structure [75]. NB-DNJ and isofagomine (IFG) act as chaperones for newly synthesized mutant GCase as it passes through the endoplasmic reticulum [73,76]. These molecules result in proper folding of the protein to ensure that it is transported to the lysosome, and not broken down by endoplasmic-reticulum-associated degradation (ERAD). Enhancement of mutant GCase activity by chaperone therapy is primarily due to improved trafficking to the lysosome. However, computational models suggest that once the mutant enzyme has reached the lysosome, NB-DNJ can also restore N370S GCase activity by promoting the interaction of the enzyme with Sap C and the anionic phospholipid membrane [73]. The N370S mutation appears to destabilize GCase within the Sap C binding interface, preventing its association with the enzyme [51,72,73]. NB-DNJ may stabilize the residues of loop 1, inducing a conformational change that improves their interaction with Sap C [73].
4. Clinical consequences of saposin C deficiency
Six patients with Sap C deficiency due to mutations in the PSAP gene have been reported in the literature, all of whom had symptoms resembling GD [9,77–83]. Two of these patients exhibited a type 1 GD phenotype, while two had neurological involvement resembling type 3 GD [9,77,80]. The two remaining patients, an adult brother and sister, were originally described as having type 1 GD, but a recent update reports that they have begun to display mild neurological deterioration [78,79]. Three of the patients have mutations causing non-synonymous substitution of a cysteine residue, breaking one of the three disulfide bridges in Sap C. Mutations in the start codon of pSap, resulting in a null allele, are present in three patients. Patient 6 is homozygous for a seven-amino acid deletion in Sap C, while the others are all compound heterozygotes for their respective mutations. Descriptions of the clinical findings in these patients are summarized in Table 1.
Table 1.
Summary of reported mutations in the Sap C domain of the PSAP gene. These result in Sap C deficiency and phenotypes resembling either type 1 or type 3 Gaucher disease.
| Patient | cDNA mutation (Allele 1/Allele 2) | Protein mutation (Allele 1/Allele 2) | GD Phenotype | Reference |
|---|---|---|---|---|
| 1 | c.1145 G>T/unknown | p.Cys382Phe | Type 3 | [77,82] |
| 2 | c.1144 T>G/c.1288 C>T | p.Cys382Gly/p.Gln430* | Type 3 | [80,81,83] |
| 3 | c.1A>T/c.1046 T>C | p.M1L/p.L349P | Type 1 | [78] |
| 4 | c.1A>T/c.1046 T>C | p.M1L/p.L349P | Type 1 | [78] |
| 5 | c.1A>G/c.943 T>A | p.M1V/p.Cys315Ser | Type 3 | [9] |
| 6 | c.1024_1044delTTTGACAAAATGTGCTCGAAG | p.Phe342_Lys348del/p.Phe342_Lys348del | Type 1 | [9] |
5. Conclusions
It is becoming increasingly apparent that GD encompasses a spectrum of phenotypes, and that there is great phenotypic heterogeneity associated with this single-gene disorder [84]. Over 250 mutations associated with GD have been identified in GBA, yet direct correlations between genotype and phenotype are limited [68]. Two notable exceptions are N370S, which is found exclusively in type 1 GD, and L444P, which when homozygous, is typically associated with neurological involvement [85]. However, general observations of patients with GD indicate that patients with the same genotype can often display drastically different clinical symptoms, while other patients may have similar phenotypes arising from different genotypes [16]. The lack of direct causal relationships between specific mutations and phenotypes suggests the involvement of disease modifiers that influence the constellation of symptoms in patients with GD [86].
Some patients with GD also develop Parkinson disease (PD), and mutations in GBA are the most common known genetic risk factor for the development of parkinsonism [87]. The odds ratio for a patient with PD to carry a GBA mutation, relative to controls, is 5.4 (95% CI, 3.89 to 7.57) [87]. Still, only a small fraction of patients with GD develop PD during their lifetime. Given that pSap is a neurotrophic factor, it is conceivable that it plays a role in determining which patients with GD develop PD. The neurotrophic region of pSap is mapped to domain 1 of the Sap C sequence, and residues 13–18 of Sap C showed neurotrophic activity in both in vitro [88] and in vivo experiments [44]. pSap expression is also increased in the hippocampal regions CA1, CA3, and the dentate gyrus during prenatal brain development in rats [45]. Considering that hippocampal regions CA2-4 are the main sites of involvement in GD [89] and are also involved in Lewy body disorders [89], it is possible that altered Sap C levels in patients with GD could play a role in the development of neurological manifestations or parkinsonism in some cases.
A modifier gene is any gene that affects the phenotypic expression caused by a disease allele, but may not itself be disease-causing when mutated [90]. Disease modifiers can be genes in other loci that interact with the disease-causing allele, or may result from additional mutated sites on the same allele [91,92]. While it is easy to recognize situations where modifier genes may be involved, it is far more difficult to identify specific genetic interactions that influence disease manifestations.
A number of potential modifiers for GD, both within the GBA locus and at unrelated loci have been proposed [91,93,94]. Recently, the SCARB2 gene (OMIM ID: 602257) was confirmed to be a disease modifier for GD in one unique family [95]. This gene encodes lysosomal integral membrane protein type 2 (LIMP-2), which is essential for accurate localization of GCase to the lysosome. Mutations in SCARB2 cause improper trafficking of GCase to the lysosome, reducing the enzyme’s overall activity toward glucosylceramide. However, there may be alternative lysosomal trafficking pathways for GCase, since mutations in SCARB2 alone do not result in a Gaucher phenotype.
Because of the essential role of Sap C as an activator of GCase, the PSAP gene is a logical candidate modifier for GD. It has long been known that Sap C and Sap A, both of which are capable of promoting GCase activity, are increased in the spleens of patients with GD [23,96] by approximately 17- and 60-fold, respectively, relative to controls [96]. These activators may be elevated in affected tissues in order to compensate for decreased GCase activity, boosting any residual enzyme available. Since Sap C deficiency produces clinical features similar to those seen in GD despite normal GCase levels, Sap C deficiency in conjunction with inherited GD mutations affecting GCase activity would likely compound the severity of the disease. While this scenario has been explored in the mouse model created by Sun et al. [66], variation in Sap C levels among GD patients with discordant phenotypes has yet to be studied.
Another reason to identify factors that can modify the disease course is to assess their therapeutic potential. It is interesting to speculate that modifying levels of Sap C in patients might enhance the activity of mutant GCase, resulting in clinical improvement. Determining whether discordant phenotypes in patients with GD patients may result from differences in Sap C expression remains an area for future investigation. Further studies of the molecular genetics of the PSAP gene and the processing of its products may reveal other factors that mediate genotype-phenotype correlation in GD. Sap C merits further investigation as a modifier for GD given its role as an essential activator of GCase, as well as the strong phenotypic similarity between patients with Sap C deficiency and GD. Moreover, as we better understand the impact of Sap C on GCase activity, therapies directed toward altering Sap C levels or function may play a role in patient management.
Acknowledgments
This work was supported by the Intramural Research Program of the National Human Genome Research Institute and National Institutes of Health. The authors thank Darryl Leja for his assistance with Fig. 3.
Abbreviations
- GCase
glucocerebrosidase
- GD
Gaucher disease
- PD
Parkinson disease
- Sap
saposin
- pSap
prosaposin
- FRET
Förster resonance energy transfer
- NB-DNJ
N-butyl-deoxynojirimycin
- IFG
isofagomine
- LIMP-2
lysosomal integral membrane protein type 2
Footnotes
Conflcts of interest
The authors declare no conflicts of interest.
Contributor Information
Rafael J. Tamargo, Email: tamargorj@mail.nih.gov.
Arash Velayati, Email: velayatia@mail.nih.gov.
Ehud Goldin, Email: goldine@mail.nih.gov.
Ellen Sidransky, Email: sidranse@mail.nih.gov.
References
- 1.Darmoise A, Maschmeyer P, Winau F. The immunological functions of saposins. Adv Immunol. 2010;105:25–62. doi: 10.1016/S0065-2776(10)05002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kishimoto Y, Hiraiwa M, O’Brien JS. Saposins: structure, function, distribution, and molecular genetics. J Lipid Res. 1992;33:1255–1267. [PubMed] [Google Scholar]
- 3.Vaccaro AM, Salvioli R, Tatti M, Ciaffoni F. Saposins and their interaction with lipids. Neurochem Res. 1999;24:307–314. doi: 10.1023/a:1022530508763. [DOI] [PubMed] [Google Scholar]
- 4.Futerman AH, Zimran A. Gaucher disease. CRC/Taylor & Francis; Boca Raton: 2006. [Google Scholar]
- 5.Hiraiwa M, Martin BM, Kishimoto Y, Conner GE, Tsuji S, O’Brien JS. Lysosomal proteolysis of prosaposin, the precursor of saposins (sphingolipid activator proteins): its mechanism and inhibition by ganglioside. Arch Biochem Biophys. 1997;341:17–24. doi: 10.1006/abbi.1997.9958. [DOI] [PubMed] [Google Scholar]
- 6.Vaccaro AM, Salvioli R, Barca A, Tatti M, Ciaffoni F, Maras B, Siciliano R, Zappacosta F, Amoresano A, Pucci P. Structural analysis of saposin C and B. Complete localization of disulfide bridges. J Biol Chem. 1995;270:9953–9960. doi: 10.1074/jbc.270.17.9953. [DOI] [PubMed] [Google Scholar]
- 7.Kuchař L, Ledvinová J, Hřebíček M, Myšková H, Dvořáková L, Berná L, Chrastina P, Asfaw B, Elleder M, Petermöller M, Mayrhofer H, Staudt M, Krägeloh-Mann I, Paton BC, Harzer K. Prosaposin deficiency and saposin B deficiency (activator-deficient metachromatic leukodystrophy): Report on two patients detected by analysis of urinary sphingolipids and carrying novel PSAP gene mutations. Am J Med Genet A. 2009;149A:613–621. doi: 10.1002/ajmg.a.32712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spiegel R, Bach G, Sury V, Mengistu G, Meidan B, Shalev S, Shneor Y, Mandel H, Zeigler M. A mutation in the saposin A coding region of the prosaposin gene in an infant presenting as Krabbe disease: first report of saposin A deficiency in humans. Mol Genet Metab. 2005;84:160–166. doi: 10.1016/j.ymgme.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Vaccaro AM, Motta M, Tatti M, Scarpa S, Masuelli L, Bhat M, Vanier MT, Tylki-Szymanska A, Salvioli R. Saposin C mutations in Gaucher disease patients resulting in lysosomal lipid accumulation, saposin C deficiency, but normal prosaposin processing and sorting. Hum Mol Genet. 2010;19:2987–2997. doi: 10.1093/hmg/ddq204. [DOI] [PubMed] [Google Scholar]
- 10.Bradova V, Smid F, Ulrich-Bott B, Roggendorf W, Paton BC, Harzer K. Prosaposin deficiency: further characterization of the sphingolipid activator protein-deficient sibs. Multiple glycolipid elevations (including lactosylceramidosis), partial enzyme deficiencies and ultrastructure of the skin in this generalized sphingolipid storage disease. Hum Genet. 1993;92:143–152. doi: 10.1007/BF00219682. [DOI] [PubMed] [Google Scholar]
- 11.Hulková H, Cervenková M, Ledvinová J, Tochácková M, Hrebícek M, Poupetová H, Befekadu A, Berná L, Paton BC, Harzer K, Böör A, Smíd F, Elleder M. A novel mutation in the coding region of the prosaposin gene leads to a complete deficiency of prosaposin and saposins, and is associated with a complex sphingolipidosis dominated by lactosylceramide accumulation. Hum Mol Genet. 2001;10:927–940. doi: 10.1093/hmg/10.9.927. [DOI] [PubMed] [Google Scholar]
- 12.Deconinck N, Messaaoui A, Ziereisen F, Kadhim H, Sznajer Y, Pelc K, Nassogne MC, Vanier MT, Dan B. Metachromatic leukodystrophy without arylsulfatase A deficiency: a new case of saposin-B deficiency. Eur J Pediatr Neurol. 2008;12:46–50. doi: 10.1016/j.ejpn.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Kretz KA, Carson GS, Morimoto S, Kishimoto Y, Fluharty AL, O’Brien JS. Characterization of a mutation in a family with saposin B deficiency: a glycosylation site defect. Proc Natl Acad Sci U S A. 1990;87:2541–2544. doi: 10.1073/pnas.87.7.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlote W, Harzer K, Christomanou H, Paton BC, Kustermann-Kuhn B, Schmid B, Seeger J, Beudt U, Schuster I, Langenbeck U. Sphingolipid activator protein 1 deficiency in metachromatic leucodystrophy with normal arylsulphatase A activity. A clinical, morphological, biochemical, and immunological study. Eur J Pediatr. 1991;150:584–591. doi: 10.1007/BF02072213. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Ran H, Zamzow M, Kitatani K, Skelton MR, Williams MT, Vorhees CV, Witte DP, Hannun YA, Grabowski GA. Specific saposin C deficiency: CNS impairment and acid beta-glucosidase effects in the mouse. Hum Mol Genet. 2010;19:634–647. doi: 10.1093/hmg/ddp531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidransky E. Gaucher disease: complexity in a “simple” disorder. Mol Genet Metab. 2004;83:6–15. doi: 10.1016/j.ymgme.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Beutler E, Grabowski GA. Gaucher Disease. In: Scriver C, Beaudet A, Sly W, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York, NY: 2001. pp. 3635–3688. [Google Scholar]
- 18.Steet RA, Chung S, Wustman B, Powe A, Do H, Kornfeld SA. The iminosugar isofagomine increases the activity of N370S mutant acid beta-glucosidase in Gaucher fibroblasts by several mechanisms. Proc Natl Acad Sci U S A. 2006;103:13813–13818. doi: 10.1073/pnas.0605928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aerts JM, Sa Miranda MC, Brouwer-Kelder EM, Van Weely S, Barranger JA, Tager JM. Conditions affecting the activity of glucocerebrosidase purified from spleens of control subjects and patients with type 1 Gaucher disease. Biochim Biophys Acta. 1990;1041:55–63. doi: 10.1016/0167-4838(90)90122-v. [DOI] [PubMed] [Google Scholar]
- 20.Prows CA, Sanchez N, Daugherty C, Grabowski GA. Gaucher disease: enzyme therapy in the acute neuronopathic variant. Am J Med Genet. 1997;71:16–21. [PubMed] [Google Scholar]
- 21.Eblan MJ, Goker-Alpan O, Sidransky E. Perinatal lethal Gaucher disease: a distinct phenotype along the neuronopathic continuum. Fetal Pediatr Pathol. 2005;24:205–222. doi: 10.1080/15227950500405296. [DOI] [PubMed] [Google Scholar]
- 22.Stone DL, Sidransky E. Hydrops fetalis: lysosomal storage disorders in extremis. Adv Pediatr. 1999;46:409–440. [PubMed] [Google Scholar]
- 23.Ho MW, O’Brien JS. Gaucher’s disease: deficiency of ‘acid’-glucosidase and reconstitution of enzyme activity in vitro. Proc Natl Acad Sci U S A. 1971;68:2810–2813. doi: 10.1073/pnas.68.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujibayashi S, Wenger DA. Synthesis and processing of sphingolipid activator protein-2 (SAP-2) in cultured human fibroblasts. J Biol Chem. 1986;261:15339–15343. [PubMed] [Google Scholar]
- 25.Fujibayashi S, Wenger DA. Biosynthesis of the sulfatide/GM1 activator protein (SAP-1) in control and mutant cultured skin fibroblasts. Biochim Biophys Acta. 1986;875:554–562. doi: 10.1016/0005-2760(86)90077-9. [DOI] [PubMed] [Google Scholar]
- 26.O’Brien JS, Kretz KA, Dewji N, Wenger DA, Esch F, Fluharty AL. Coding of two sphingolipid activator proteins (SAP-1 and SAP-2) by same genetic locus. Science. 1988;241:1098–1101. doi: 10.1126/science.2842863. [DOI] [PubMed] [Google Scholar]
- 27.Furst W, Machleidt W, Sandhoff K. The precursor of sulfatide activator protein is processed to three different proteins. Biol Chem Hoppe Seyler. 1988;369:317–328. doi: 10.1515/bchm3.1988.369.1.317. [DOI] [PubMed] [Google Scholar]
- 28.Morimoto S, Martin BM, Kishimoto Y, O’Brien JS. Saposin D: a sphingomyelinase activator. Biochem Biophys Res Commun. 1988;156:403–410. doi: 10.1016/s0006-291x(88)80855-6. [DOI] [PubMed] [Google Scholar]
- 29.Morimoto S, Martin BM, Yamamoto Y, Kretz KA, O’Brien JS, Kishimoto Y. Saposin A: second cerebrosidase activator protein. Proc Natl Acad Sci U S A. 1989;86:3389–3393. doi: 10.1073/pnas.86.9.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inui K, Wenger DA. Biochemical, immunological and structural studies on a sphingolipid activator protein (SAP-1) Arch Biochem Biophys. 1984;233:556–564. doi: 10.1016/0003-9861(84)90479-x. [DOI] [PubMed] [Google Scholar]
- 31.O’Brien JS, Kishimoto Y. Saposin proteins: structure, function and role in human lysosomal storage disorders. FASEB J. 1991;5:301–308. doi: 10.1096/fasebj.5.3.2001789. [DOI] [PubMed] [Google Scholar]
- 32.Furst W, Sandhoff K. Activator proteins and topology of lysosomal sphingolipid catabolism. Biochim Biophys Acta. 1992;1126:1–16. doi: 10.1016/0005-2760(92)90210-m. [DOI] [PubMed] [Google Scholar]
- 33.Nakano T, Sandhoff K, Stumper J, Christomanou H, Suzuki K. Structure of full-length cDNA coding for sulfatide activator, a Co-beta-glucosidase and two other homologous proteins: two alternate forms of the sulfatide activator. J Biochem. 1989;105:152–154. doi: 10.1093/oxfordjournals.jbchem.a122629. [DOI] [PubMed] [Google Scholar]
- 34.Cohen T, Ravid L, Altman N, Madar-Shapiro L, Fein A, Weil M, Horowitz M. Conservation of expression and alternative splicing in the prosaposin gene. Brain Res Mol Brain Res. 2004;129:8–19. doi: 10.1016/j.molbrainres.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 35.Igdoura SA, Morales CR. Role of sulfated glycoprotein-1 (SGP-1) in the disposal of residual bodies by Sertoli cells of the rat. Mol Reprod Dev. 1995;40:91–102. doi: 10.1002/mrd.1080400112. [DOI] [PubMed] [Google Scholar]
- 36.Lefrancois S, Zeng J, Hassan AJ, Canuel M, Morales CR. The lysosomal trafficking of sphingolipid activator proteins (SAPs) is mediated by sortilin. EMBO J. 2003;22:6430–6437. doi: 10.1093/emboj/cdg629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igdoura SA, Rasky A, Morales CR. Trafficking of sulfated glycoprotein-1 (prosaposin) to lysosomes or to the extracellular space in rat Sertoli cells. Cell Tissue Res. 1996;283:385–394. doi: 10.1007/s004410050549. [DOI] [PubMed] [Google Scholar]
- 38.Yuan L, Morales CR. Prosaposin sorting is mediated by oligomerization. Exp Cell Res. 2011;317:2456–2467. doi: 10.1016/j.yexcr.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 39.Sano A, Hineno T, Mizuno T, Kondoh K, Ueno S, Kakimoto Y, Inui K. Sphingolipid hydrolase activator proteins and their precursors. Biochem Biophys Res Commun. 1989;165:1191–1197. doi: 10.1016/0006-291x(89)92728-9. [DOI] [PubMed] [Google Scholar]
- 40.Hineno T, Sano A, Kondoh K, Ueno S, Kakimoto Y, Yoshida K. Secretion of sphingolipid hydrolase activator precursor, prosaposin. Biochem Biophys Res Commun. 1991;176:668–674. doi: 10.1016/s0006-291x(05)80236-0. [DOI] [PubMed] [Google Scholar]
- 41.Kondoh K, Hineno T, Sano A, Kakimoto Y. Isolation and characterization of prosaposin from human milk. Biochem Biophys Res Commun. 1991;181:286–292. doi: 10.1016/s0006-291x(05)81415-9. [DOI] [PubMed] [Google Scholar]
- 42.Morales CR, Zhao Q, Lefrancois S, Ham D. Role of prosaposin in the male reproductive system: effect of prosaposin inactivation on the testis, epididymis, prostate and seminal vesicles. Arch Androl. 2000;44:173–186. doi: 10.1080/014850100262146. [DOI] [PubMed] [Google Scholar]
- 43.O’Brien JS, Carson GS, Seo HC, Hiraiwa M, Kishimoto Y. Identification of prosaposin as a neurotrophic factor. Proc Natl Acad Sci U S A. 1994;91:9593–9596. doi: 10.1073/pnas.91.20.9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotani Y, Matsuda S, Wen TC, Sakanaka M, Tanaka J, Maeda N, Kondoh K, Ueno S, Sano A. A hydrophilic peptide comprising 18 amino acid residues of the prosaposin sequence has neurotrophic activity in vitro and in vivo. J Neurochem. 1996;66:2197–2200. doi: 10.1046/j.1471-4159.1996.66052197.x. [DOI] [PubMed] [Google Scholar]
- 45.Xue B, Chen J, Gao H, Saito S, Kobayashi N, Shimokawa T, Nabeka H, Sano A, Matsuda S. Chronological changes in prosaposin in the developing rat brain. Neurosci Res. 2011;71:22–34. doi: 10.1016/j.neures.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Kleinschmidt T, Christomanou H, Braunitzer G. Complete amino-acid sequence and carbohydrate content of the naturally occurring glucosylceramide activator protein (A1 activator) absent from a new human Gaucher disease variant. Biol Chem Hoppe Seyler. 1987;368:1571–1578. doi: 10.1515/bchm3.1987.368.2.1571. [DOI] [PubMed] [Google Scholar]
- 47.Rossmann M, Schultz-Heienbrok R, Behlke J, Remmel N, Alings C, Sandhoff K, Saenger W, Maier T. Crystal structures of human saposins C and D: implications for lipid recognition and membrane interactions. Structure. 2008;16:809–817. doi: 10.1016/j.str.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 48.Vaccaro AM, Ciaffoni F, Tatti M, Salvioli R, Barca A, Tognozzi D, Scerch C. pH-dependent conformational properties of saposins and their interactions with phospholipid membranes. J Biol Chem. 1995;270:30576–30580. doi: 10.1074/jbc.270.51.30576. [DOI] [PubMed] [Google Scholar]
- 49.de Alba E, Weiler S, Tjandra N. Solution structure of human saposin C: pH-dependent interaction with phospholipid vesicles. Biochemistry. 2003;42:14729–14740. doi: 10.1021/bi0301338. [DOI] [PubMed] [Google Scholar]
- 50.Weiler S, Kishimoto Y, O’Brien JS, Barranger JA, Tomich JM. Identification of the binding and activating sites of the sphingolipid activator protein, saposin C, with glucocerebrosidase. Protein Sci. 1995;4:756–764. doi: 10.1002/pro.5560040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atrian S, Lopez-Vinas E, Gomez-Puertas P, Chabas A, Vilageliu L, Grinberg D. An evolutionary and structure-based docking model for glucocerebrosidase-saposin C and glucocerebrosidase-substrate interactions - relevance for Gaucher disease. Proteins. 2008;70:882–891. doi: 10.1002/prot.21554. [DOI] [PubMed] [Google Scholar]
- 52.Ho MW, Rigby M. Glucocerebrosidase: stoichiometry of association between effector and catalytic proteins. Biochim Biophys Acta. 1975;397:267–273. doi: 10.1016/0005-2744(75)90199-0. [DOI] [PubMed] [Google Scholar]
- 53.Aerts JM, Donker-Koopman WE, van Laar C, Brul S, Murray GJ, Wenger DA, Barranger JA, Tager JM, Schram AW. Relationship between the two immunologically distinguishable forms of glucocerebrosidase in tissue extracts. Eur J Biochem. 1987;163:583–589. doi: 10.1111/j.1432-1033.1987.tb10907.x. [DOI] [PubMed] [Google Scholar]
- 54.Prence E, Chakravorti S, Basu A, Clark LS, Glew RH, Chambers JA. Further studies on the activation of glucocerebrosidase by a heat-stable factor from Gaucher spleen. Arch Biochem Biophys. 1985;236:98–109. doi: 10.1016/0003-9861(85)90609-5. [DOI] [PubMed] [Google Scholar]
- 55.John M, Wendeler M, Heller M, Sandhoff K, Kessler H. Characterization of human saposins by NMR spectroscopy. Biochemistry. 2006;45:5206–5216. doi: 10.1021/bi051944+. [DOI] [PubMed] [Google Scholar]
- 56.Choy FY, Woo M, Potier M. In situ radiation-inactivation size of fibroblast membrane-bound acid beta-glucosidase in Gaucher type 1, type 2 and type 3 disease. Biochim Biophys Acta. 1986;870:76–81. doi: 10.1016/0167-4838(86)90010-5. [DOI] [PubMed] [Google Scholar]
- 57.Lieberman RL. A Guided Tour of the Structural Biology of Gaucher Disease: Acid-beta-Glucosidase and Saposin C. Enzyme Res. 2011;2011:973231. doi: 10.4061/2011/973231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alattia JR, Shaw JE, Yip CM, Prive GG. Direct visualization of saposin remodelling of lipid bilayers. J Mol Biol. 2006;362:943–953. doi: 10.1016/j.jmb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Alattia JR, Shaw JE, Yip CM, Prive GG. Molecular imaging of membrane interfaces reveals mode of beta-glucosidase activation by saposin C. Proc Natl Acad Sci U S A. 2007;104:17394–17399. doi: 10.1073/pnas.0704998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.You HX, Qi X, Yu L. Direct AFM observation of saposin C-induced membrane domains in lipid bilayers: from simple to complex lipid mixtures. Chem Phys Lipids. 2004;132:15–22. doi: 10.1016/j.chemphyslip.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 61.Wilkening G, Linke T, Sandhoff K. Lysosomal degradation on vesicular membrane surfaces. Enhanced glucosylceramide degradation by lysosomal anionic lipids and activators. J Biol Chem. 1998;273:30271–30278. doi: 10.1074/jbc.273.46.30271. [DOI] [PubMed] [Google Scholar]
- 62.Vaccaro AM, Tatti M, Ciaffoni F, Salvioli R, Serafino A, Barca A. Saposin C induces pH-dependent destabilization and fusion of phosphatidylserine-containing vesicles. FEBS Lett. 1994;349:181–186. doi: 10.1016/0014-5793(94)00659-8. [DOI] [PubMed] [Google Scholar]
- 63.Leon L, Tatituri RV, Grenha R, Sun Y, Barral DC, Minnaard AJ, Bhowruth V, Veerapen N, Besra GS, Kasmar A, Peng W, Moody DB, Grabowski GA, Brenner MB. Saposins utilize two strategies for lipid transfer and CD1 antigen presentation. Proc Natl Acad Sci U S A. 2012;109:4357–4364. doi: 10.1073/pnas.1200764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahn VE, Faull KF, Whitelegge JP, Fluharty AL, Prive GG. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc Natl Acad Sci U S A. 2003;100:38–43. doi: 10.1073/pnas.0136947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun Y, Qi X, Grabowski GA. Saposin C is required for normal resistance of acid beta-glucosidase to proteolytic degradation. J Biol Chem. 2003;278:31918–31923. doi: 10.1074/jbc.M302752200. [DOI] [PubMed] [Google Scholar]
- 66.Sun Y, Liou B, Ran HM, Skelton MR, Williams MT, Vorhees CV, Kitatani K, Hannun YA, Witte DP, Xu YH, Grabowski GA. Neuronopathic Gaucher disease in the mouse: viable combined selective saposin C deficiency and mutant glucocerebrosidase (V394L) mice with glucosylsphingosine and glucosylceramide accumulation and progressive neurological deficits. Hum Mol Genet. 2010;19:1088–1097. doi: 10.1093/hmg/ddp580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu YH, Quinn B, Witte D, Grabowski GA. Viable mouse models of acid beta-glucosidase deficiency: the defect in Gaucher disease. Am J Pathol. 2003;163:2093–2101. doi: 10.1016/s0002-9440(10)63566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29:567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 69.Yu Z, Sawkar AR, Kelly JW. Pharmacologic chaperoning as a strategy to treat Gaucher disease. FEBS J. 2007;274:4944–4950. doi: 10.1111/j.1742-4658.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 70.Liou B, Kazimierczuk A, Zhang M, Scott CR, Hegde RS, Grabowski GA. Analyses of variant acid beta-glucosidases: effects of Gaucher disease mutations. J Biol Chem. 2006;281:4242–4253. doi: 10.1074/jbc.M511110200. [DOI] [PubMed] [Google Scholar]
- 71.Salvioli R, Tatti M, Scarpa S, Moavero SM, Ciaffoni F, Felicetti F, Kaneski CR, Brady RO, Vaccaro AM. The N370S (Asn370–>Ser) mutation affects the capacity of glucosylceramidase to interact with anionic phospholipid-containing membranes and saposin C. Biochem J. 2005;390:95–103. doi: 10.1042/BJ20050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei RR, Hughes H, Boucher S, Bird JJ, Guziewicz N, Van Patten SM, Qiu H, Pan CQ, Edmunds T. X-ray and biochemical analysis of N370S mutant human acid beta-glucosidase. J Biol Chem. 2011;286:299–308. doi: 10.1074/jbc.M110.150433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Offman MN, Krol M, Silman I, Sussman JL, Futerman AH. Molecular basis of reduced glucosylceramidase activity in the most common Gaucher disease mutant N370S. J Biol Chem. 2010;285:42105–42114. doi: 10.1074/jbc.M110.172098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Weely S, van den Berg M, Barranger JA, Sa Miranda MC, Tager JM, Aerts JM. Role of pH in determining the cell-type-specific residual activity of glucocerebrosidase in type 1 Gaucher disease. J Clin Invest. 1993;91:1167–1175. doi: 10.1172/JCI116276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 76.Chang HH, Asano N, Ishii S, Ichikawa Y, Fan JQ. Hydrophilic iminosugar active-site-specific chaperones increase residual glucocerebrosidase activity in fibroblasts from Gaucher patients. FEBS J. 2006;273:4082–4092. doi: 10.1111/j.1742-4658.2006.05410.x. [DOI] [PubMed] [Google Scholar]
- 77.Christomanou H, Aignesberger A, Linke RP. Immunochemical characterization of two activator proteins stimulating enzymic sphingomyelin degradation in vitro. Absence of one of them in a human Gaucher disease variant. Biol Chem Hoppe Seyler. 1986;367:879–890. doi: 10.1515/bchm3.1986.367.2.879. [DOI] [PubMed] [Google Scholar]
- 78.Tylki-Szymanska A, Czartoryska B, Vanier MT, Poorthuis BJ, Groener JA, Lugowska A, Millat G, Vaccaro AM, Jurkiewicz E. Non-neuronopathic Gaucher disease due to saposin C deficiency. Clin Genet. 2007;72:538–542. doi: 10.1111/j.1399-0004.2007.00899.x. [DOI] [PubMed] [Google Scholar]
- 79.Tylki-Szymanska A, Groener JE, Kaminski ML, Lugowska A, Jurkiewicz E, Czartoryska B. Gaucher disease due to saposin C deficiency, previously described as non-neuronopathic form - No positive effects after 2-years of miglustat therapy. Mol Genet Metab. 2011;104:627–630. doi: 10.1016/j.ymgme.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 80.Christomanou H, Chabas A, Pampols T, Guardiola A. Activator protein deficient Gaucher’s disease. A second patient with the newly identified lipid storage disorder. Klin Wochenschr. 1989;67:999–1003. doi: 10.1007/BF01716064. [DOI] [PubMed] [Google Scholar]
- 81.Rafi MA, de Gala G, Zhang XL, Wenger DA. Mutational analysis in a patient with a variant form of Gaucher disease caused by SAP-2 deficiency. Somat Cell Mol Genet. 1993;19:1–7. doi: 10.1007/BF01233949. [DOI] [PubMed] [Google Scholar]
- 82.Schnabel D, Schroder M, Sandhoff K. Mutation in the sphingolipid activator protein 2 in a patient with a variant of Gaucher disease. FEBS Lett. 1991;284:57–59. doi: 10.1016/0014-5793(91)80760-z. [DOI] [PubMed] [Google Scholar]
- 83.Diaz-Font A, Cormand B, Santamaria R, Vilageliu L, Grinberg D, Chabas A. A mutation within the saposin D domain in a Gaucher disease patient with normal glucocerebrosidase activity. Hum Genet. 2005;117:275–277. doi: 10.1007/s00439-005-1288-x. [DOI] [PubMed] [Google Scholar]
- 84.Goker-Alpan O, Schiffmann R, Park JK, Stubblefield BK, Tayebi N, Sidransky E. Phenotypic continuum in neuronopathic Gaucher disease: an intermediate phenotype between type 2 and type 3. J Pediatr. 2003;143:273–276. doi: 10.1067/S0022-3476(03)00302-0. [DOI] [PubMed] [Google Scholar]
- 85.Charrow J, Andersson HC, Kaplan P, Kolodny EH, Mistry P, Pastores G, Rosenbloom BE, Scott CR, Wappner RS, Weinreb NJ, Zimran A. The Gaucher registry: demographics and disease characteristics of 1698 patients with Gaucher disease. Arch Intern Med. 2000;160:2835–2843. doi: 10.1001/archinte.160.18.2835. [DOI] [PubMed] [Google Scholar]
- 86.Goker-Alpan O, Hruska KS, Orvisky E, Kishnani PS, Stubblefield BK, Schiffmann R, Sidransky E. Divergent phenotypes in Gaucher disease implicate the role of modifiers. J Med Genet. 2005;42:e37. doi: 10.1136/jmg.2004.028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, Chen CM, Clark LN, Condroyer C, De Marco EV, Durr A, Eblan MJ, Fahn S, Farrer MJ, Fung HC, Gan-Or Z, Gasser T, Gershoni-Baruch R, Giladi N, Griffith A, Gurevich T, Januario C, Kropp P, Lang AE, Lee-Chen GJ, Lesage S, Marder K, Mata IF, Mirelman A, Mitsui J, Mizuta I, Nicoletti G, Oliveira C, Ottman R, Orr-Urtreger A, Pereira LV, Quattrone A, Rogaeva E, Rolfs A, Rosenbaum H, Rozenberg R, Samii A, Samaddar T, Schulte C, Sharma M, Singleton A, Spitz M, Tan EK, Tayebi N, Toda T, Troiano AR, Tsuji S, Wittstock M, Wolfsberg TG, Wu YR, Zabetian CP, Zhao Y, Ziegler SG. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hiraiwa M, Campana WM, Mizisin AP, Mohiuddin L, O’Brien JS. Prosaposin: a myelinotrophic protein that promotes expression of myelin constituents and is secreted after nerve injury. Glia. 1999;26:353–360. [PubMed] [Google Scholar]
- 89.Wong K, Sidransky E, Verma A, Mixon T, Sandberg GD, Wakefield LK, Morrison A, Lwin A, Colegial C, Allman JM, Schiffmann R. Neuropathology provides clues to the pathophysiology of Gaucher disease. Mol Genet Metab. 2004;82:192–207. doi: 10.1016/j.ymgme.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 90.Romeo G, McKusick VA. Phenotypic diversity, allelic series and modifier genes. Nat Genet. 1994;7:451–453. doi: 10.1038/ng0894-451. [DOI] [PubMed] [Google Scholar]
- 91.Latham T, Grabowski GA, Theophilus BD, Smith FI. Complex alleles of the acid beta-glucosidase gene in Gaucher disease. Am J Hum Genet. 1990;47:79–86. [PMC free article] [PubMed] [Google Scholar]
- 92.Kiesewetter S, Macek M, Jr, Davis C, Curristin SM, Chu CS, Graham C, Shrimpton AE, Cashman SM, Tsui LC, Mickle J, et al. A mutation in CFTR produces different phenotypes depending on chromosomal background. Nat Genet. 1993;5:274–278. doi: 10.1038/ng1193-274. [DOI] [PubMed] [Google Scholar]
- 93.Montfort M, Chabas A, Vilageliu L, Grinberg D. Functional analysis of 13 GBA mutant alleles identified in Gaucher disease patients: Pathogenic changes and “modifier” polymorphisms. Hum Mutat. 2004;23:567–575. doi: 10.1002/humu.20043. [DOI] [PubMed] [Google Scholar]
- 94.Winfield SL, Tayebi N, Martin BM, Ginns EI, Sidransky E. Identification of three additional genes contiguous to the glucocerebrosidase locus on chromosome 1q21: implications for Gaucher disease. Genome Res. 1997;7:1020–1026. doi: 10.1101/gr.7.10.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Velayati A, Depaolo J, Gupta N, Choi JH, Moaven N, Westbroek W, Goker-Alpan O, Goldin E, Stubblefield BK, Kolodny E, Tayebi N, Sidransky E. A mutation in SCARB2 is a modifier in gaucher disease. Hum Mutat. 2011;32:1232–1238. doi: 10.1002/humu.21566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morimoto S, Yamamoto Y, O’Brien JS, Kishimoto Y. Distribution of saposin proteins (sphingolipid activator proteins) in lysosomal storage and other diseases. Proc Natl Acad Sci U S A. 1990;87:3493–3497. doi: 10.1073/pnas.87.9.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]