Abstract
We genotyped a Chinese and an Indian-origin rhesus macaque using the Affymetrix Genome-Wide Human SNP Array 6.0 and catalogued 85,473 uniquely mapping heterospecific SNPs. These SNPs were assigned to rhesus chromosomes according to their probe sequence alignments as displayed in the human and rhesus reference sequences. The conserved gene order (synteny) revealed by heterospecific SNP maps is in concordance with that of the published human and rhesus macaque genomes.
Using these SNPs’ original human rs numbers, we identified 12,328 genes annotated in humans that are associated with these SNPs, 3,674 of which were found in at least one of the two rhesus macaques studied. Due to their density, the heterospecific SNPs allow fine-grained comparisons, including approximate boundaries of intra- and extra-chromosomal rearrangements involving gene orthologs, which can be used to distinguish rhesus macaque chromosomes from human chromosomes.
Keywords: Macaca mulatta, single nucleotide polymoprhisms (SNPs), Homo sapiens, heterospecific sequence maps
1. Introduction
The rhesus macaque (Macaca mulatta) is the most commonly used non-human primate model in biomedical research. A clear understanding of this species’ genome in relation to the human genome is vital to its application in biomedical studies of complex disease and other traits. Determining the location and nature of common features of human and macaque genetic variation necessitates dense genetic maps of orthologous regions in both taxa. As well as facilitating the discovery of highly conserved areas that presumably reflect important components of primate genomes, these comparative genomic techniques can identify genomic regions that evolved rapidly, reflecting adaptations that are either unique to one species or shared by both.
Despite their phylogenetic distance resulting from 25 million years of divergence, high levels of genomic orthology between humans and rhesus have been reported [1, 2]. Using their initial draft assembly of the rhesus macaque reference genome, Gibbs and colleagues [1] identified vast tracks within the rhesus macaque genome that exhibited DNA sequences associated with specific human diseases listed in the Human Gene Mutation database (http://www.hgmd.org). Yan et al. [3] demonstrated that specific macaque genes display a high degree of sequence similarity with human disease gene orthologs and genetic targets of drug development. It is likely that more such similarities are distributed throughout both the human and macaque genomes.
It is widely recognized that more information on rhesus macaques genomes will be valuable to biomedical research and, consequently, the identification of rhesus–specific SNPs has increased since the publication of the species’ reference genome in 2007 [1, 4–7]. While little is known about the intra-specific genome-wide variation among rhesus macaques, far less is known about the distribution of relevant human-rhesus macaque gene orthologs and the concordance of gene order (synteny) in the two taxa. Genomic rearrangements of orthologs between humans and rhesus macaques are still not well-defined but could compromise the translation of candidate gene approaches between humans and rhesus macaques.
We used the Affymetrix (Santa Clara, CA) Genome-Wide Human SNP Array 6.0 that features 1.8 million genetic markers, including more than 906,600 SNPs to identify and catalogue heterospecific SNPs (SNPs that are not species-specific) and associated orthologs between humans and rhesus macaques. Affymetrix assembled their SNP panels from the high-density SNP maps developed by the International HapMap (haplotype mapping) Consortium [8, 9] to identify common disease genes in humans [10]. Therefore, comparing the genome sequences of the human and the rhesus macaque will not only contribute to the assessment of evolutionary relationships between these species but also provide insights into their differentially conserved and rearranged orthologous regions that relate to similarities and differences in responses to treatment effects in biomedical research. Approximately one in 300 nucleotides in the human genome is polymorphic [11] and SNPs may be as abundant in the rhesus macaque genome [1]. Low coverage sequencing by the Indian rhesus macaque genome initiative revealed approximately 5.8 SNPs/kb of sequence [12] which is consistent with a similarly high SNP frequency in the rhesus macaque genome. Genotype data generated from the use of the SNP array 6.0 could provide among the highest-resolutions to date of patterns of variation and conservation of gene orthologs across large tracks of the human and rhesus macaque genomes.
Daly et al. [12] demonstrated that haplotype blocks that span up to 100kb may contain five or more common SNPs. One 84 kb block containing 8 SNPs in their study exhibited only two distinct haplotypes accounting for 95% of the chromosomes studied [12]. Consequently, most humans should exhibit four to six haplotypes per block and only a few SNPs will be needed to identify which of the 4–6 common alternative haplotypes is present [11]. This finding implies that specific chromosomal regions need not be saturated with markers to provide a cohesive map for genome-wide analysis. Therefore, instead of genotyping hundreds of SNPs across small chromosomal regions, we chose to employ heterospecific SNPs as tag-SNPs to detect and characterize orthologous human-rhesus macaque chromosomal segments. Since all the loci included in the SNP 6.0 array have been localized within genes in the human genome, it is highly probable that a large number of the heterospecific genic SNPs can be mapped to the rhesus macaque genome via direct genomic comparison.
The significant genetic disparity between Chinese-origin and Indian-derived rhesus macaques that stems from 160,000 years of genomic divergence [13] is well documented. Based on an examination of 150 kb of genic sequence data, Hernandez et al. [13] identified 1,476 SNPs, only 33% of which were found in both regional varieties with the remaining 67% being Indian-specific or Chinese-specific. In contrast, Satkoski Trask et al. [14] reported that only one-third of the randomly identified SNPs in gene deserts and 3’ coding regions are Chinese or Indian rhesus macaque-specific. Despite the different estimates of the prevalence of subspecies-specific SNPs in the two studies, this stark genetic differentiation between the two populations is in agreement with other reports that have relied on SNPs [4], STRs [15], and mtDNA [16]. The geographic regions from which the founders were derived also strongly influences the MHC haplotype composition of rhesus macaques [17]. Phenotypic differences between the two regional varieties of rhesus macaques, such as susceptibility to SIV [18], undoubtedly results in part from such underlying genetic differences. It is possible that differences in susceptibility of the two macaque varieties to other diseases will be discovered as the use of non-human primate models increases [19]. Because of the substantial variation between the Chinese and Indian rhesus macaques [4, 15, 16], we examined the conserved heterospecific SNPs in both varieties of rhesus macaques using the SNP 6.0 array to evaluate differences involving orthologous regions in these two macaque varieties.
We used these single-hit heterospecific SNPs to illustrate multiple regions of homology as well as significant chromosomal rearrangements involving regions containing putative gene orthologs in humans and macaques. The resulting orthology map is unique, as the orthologous SNPs (1) exceed the number of biomedically relevant loci currently available for rhesus macaques; (2) are localized in both the human and rhesus genomes; (3) encompass only shared human-rhesus polymorphisms, making genetic investigation in a rhesus macaque model more efficiently applied to human disease research; and (4) represent a minimal genome screening set of single-hit SNPs useful for genome-wide scans across large samples of animals.
2. Results
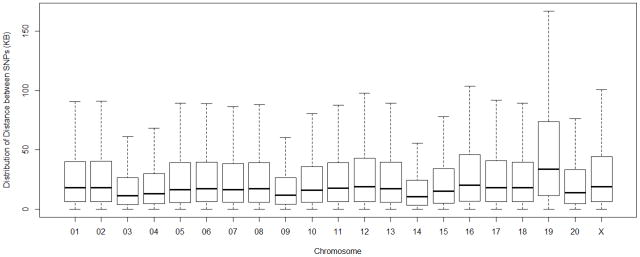
From an initial list of 906,000 SNPs on the array 6.0, more than half (553,000) satisfied the initial quality screening criteria and were regarded as potentially orthologous in the human and rhesus genomes, and only about 10% of the total number of SNPs on the array, i.e., 85,473 (81,805 in Chinese and 78,407 in Indian rhesus macaques), were scored as conserved SNPs. Although all adjacent SNPs were not perfectly equidistant from each other, the median inter-SNP distance among the 85,473 SNPs was approximately 18.5 kb with a range of 0.005 kb (Chromosome14) to 36,219.779 kb (Chromosome1), and the first and third quantiles were 6.5 kb and 42.5 kb, respectively. Figure 1 shows the distribution of single-hit heterospecific SNPs across each of the rhesus chromosomes (excluding the Y chromosome, which is not available in the rhesus draft sequence) and provides evidence of significant variability in heterospecific SNP density across human and rhesus chromosomes. The degree to which the single-hit heterospecific SNPs have been conserved in both human and rhesus macaque chromosomes is illustrated in Table 1.
Figure 1.
Density of single-hit heterospecific SNPs in rhesus macaque autosomes and the X chromosome (the same trend was observed when the Chinese and Indian individuals were analyzed separately). Black bars represent median values. Y chromosomes were not analyzed due to the absence of a Y-chromosome in the rhesus draft sequence.
Table 1.
Number of heterospecific SNPs conserved across the human and rhesus chromosomes. Many gene orthologs (and associated heterologous SNPs) have been translocated among non-homologous human and rhesus macaque chromosomes. Extensive heterologous SNP conservation is observed in chromosomes 1, 8, 18, and 19 (underlined). Rhesus X-linked SNPs are also observed in the human Y chromosome (bolded italics).
| Human chromosomes | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | ||
| Rhesus macaque chromosomes | 1 | 2,338 | 2255 | 0 | 0 | 63 | 1 | 0 | 9 | 0 | 2 | 0 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 |
| 2 | 5,526 | 1 | 0 | 5518 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | 7,696 | 0 | 2 | 1 | 0 | 1 | 0 | 6703 | 1 | 3 | 1 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 979 | 0 | 1 | 0 | |
| 4 | 6,658 | 1 | 0 | 1 | 1 | 2 | 6642 | 0 | 0 | 1 | 0 | 0 | 0 | 8 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 5 | 5,677 | 0 | 1 | 0 | 5667 | 0 | 1 | 4 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| 6 | 5,276 | 0 | 0 | 1 | 1 | 5266 | 1 | 3 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| 7 | 5,110 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 2596 | 2506 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| 8 | 4,317 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4313 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 9 | 5592 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 5582 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | |
| 10 | 2,971 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2129 | 0 | 834 | 0 | 0 | |
| 11 | 3,960 | 0 | 1 | 90 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3863 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | |
| 12 | 3,006 | 0 | 3006 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 13 | 3,866 | 0 | 3853 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | |
| 14 | 5,808 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 5804 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| 15 | 3,779 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 3770 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | |
| 16 | 1,807 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1799 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | |
| 17 | 2,782 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 2777 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 18 | 2,315 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2313 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 19 | 767 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 755 | 0 | 4 | 0 | 0 | 0 | |
| 20 | 2,414 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2409 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | |
| X | 3,808 | 0 | 0 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 3 | 0 | 0 | 0 | 3777 | 21 | |
| Total | 85473 | |||||||||||||||||||||||||
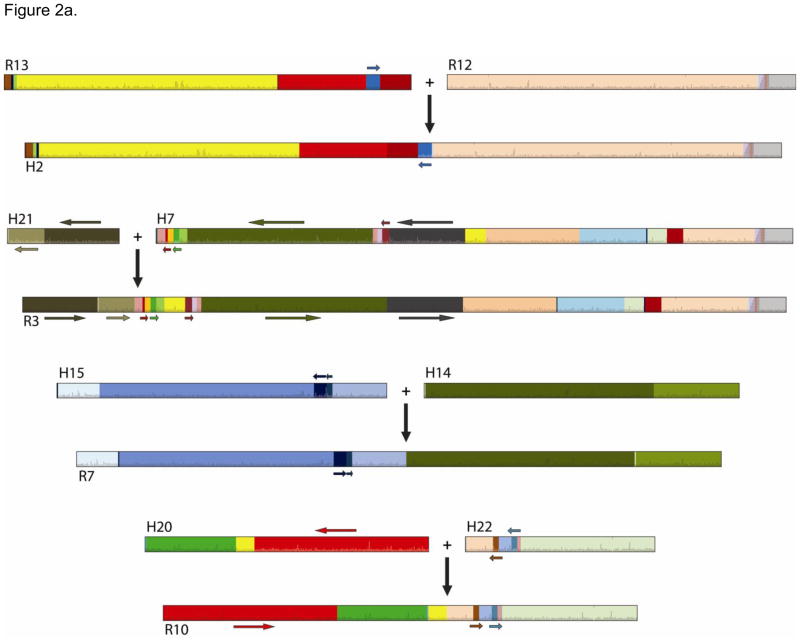
Synteny between rhesus and human chromosomes reflects large-scale rearrangements, including inversions and translocations that have occurred within and among most chromosomes. Chromosomes 1, 8, 18, 19 and X reflect strong conservation of synteny consistent with the human-rhesus short tandem repeat (STR) map of Rogers et al. [20]. Marked similarities in patterns of marker arrangement were observed between human chromosome 3 and rhesus chromosome 2, and between human chromosome 10 and rhesus chromosome 9. These findings are in agreement with previous reports of comparative homologies of human, baboon and rhesus macaque chromosomes [21, 22]. As illustrated in Figure 2a, rhesus chromosomes 12 and 13 corresponded to human chromosome 2, as previously observed in great apes [22] and baboons [23] based on G-banded karyotypes and in Japanese macaques [24] based on in situ suppression hybridization (reported using an older version of non-human primate chromosome nomenclature). This suggests that synteny among anthropoid chromosomes was maintained long before humans and chimpanzees diverged from their common ancestor. Because we relied on the macaque genome assembly to align our probes, our method detected most of the 23.6 Mb rhesus macaque genomic segment corresponding to the human chr2: 114,076,736-138 and 830,121 on rhesus chromosome 13, which represents a conflict between the rhesus genome assembly and cytogenetic inferences [25]. We also detected a much smaller span of human-rhesus homologous segment of approximately 0.1 Mb (i.e., from 1,298,361 –1,408,977) on rhesus chromosome 12, in agreement with Roberto et al.’s [25] cytogenetic results which map parts of the homologous section downstream of the human 2p/2q boundary mapping to both rhesus chromosomes 12 and 13.
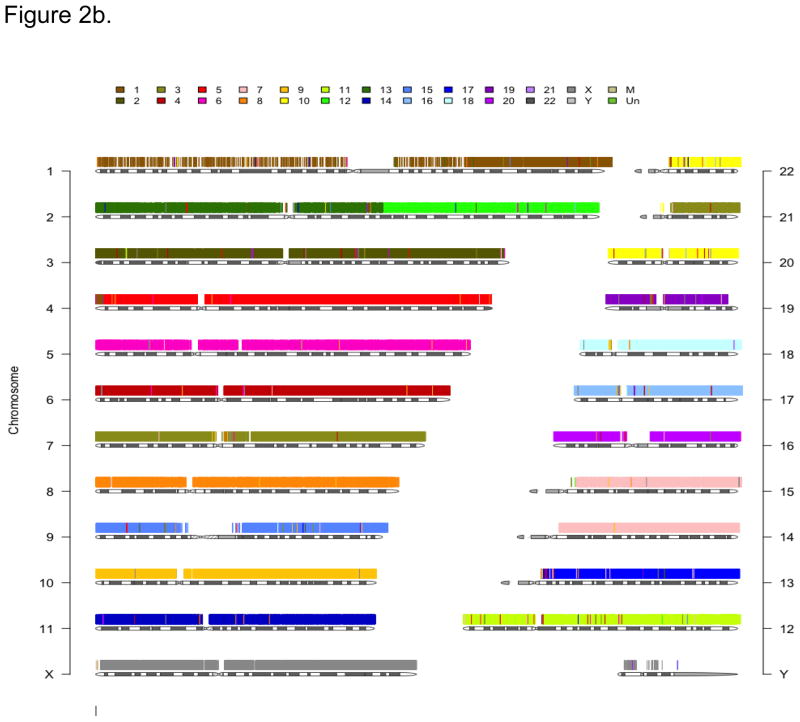
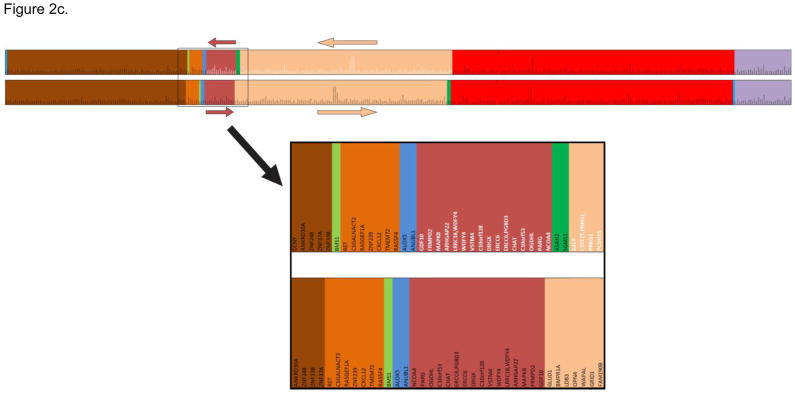
Figure 2.
Figure 2a. Associations among rhesus chromosomes and human chromosomes. Colors and horizontal arrows represent syntynic order and opposite directionality of heterospecific SNPs and vertical arrows denote chromosome fusion.
Figure 2b. A generalized image of conserved synteny of the rhesus macaque reference genome (RheMac2) chromosomal segments (colored) relative to the 23 human (HG19) chromosomes. Note that the absence of SNP probes in either species marks the approximate locations of centromeric regions. Colored vertical bars represent heterospecific SNPs within the conserved probes.
Figure 2c. Gene orthologs on rhesus macaque chromosome 9 and human chromosome 10 revealed by single-hit heterospecific SNPs. The bars are the conserved gene orthologs. The colored blocks define the conserved gene orders in both species; black font indicates human and rhesus macaque gene orders that are in the same direction, and white font highlights inverted gene orders (Top). An overlapping section of the human and rhesus chromosomes is magnified (Below).
In our study, human chromosomes 7 and 21, 14 and 15, and 20 and 22 closely resemble rhesus chromosomes 3, 7, and 10, respectively (Figure 2a), as previously reported by Rogers et al. based on STRs [20]. Reconstruction of ancestral karyotypes in anthropoids suggested that Papionini, including baboons, drills, and mangabeys, who share these correspondences to humans with macaques, display a derived fusion for homologs to human chromosomes 7 and 21, and 20 and 22 [26], resulting in the reduction of chromosome number from the ancestral catarrhine genome (2n=46) to that of macaques and Papionins (2n=42). The association between human chromosomes 14/15 and the rhesus chromosome 7 has also been reported by Pearson et al. [21], Wienberg et al. [24], Best et al. [22], and Murphy et al. [27]. According to Stanyon et al. [26], the syntenic association 14/15 predates placental mammals and fission of the 14/15 syntenic association occurred in the ancestral stock of the Hominoidea, causing the ancestral karyotype of these species to increase to 2n = 48 chromosomes.
Figure 2b illustrates chromosome segments with conserved SNP probe synteny in the rhesus macaque reference genome (RheMac2) that have been superimposed onto the 23 human (HG19) chromosomes; Figure 2c shows gene orthologs on rhesus macaque chromosome 9 in relation to human chromosome 10. These images reveal inter- and intra-chromosomal rearrangements that are fundamentally similar to those revealed by the human-on-rhesus macaque chromosome paints and rhesus macaque radiation hybrid (RH) maps [27–30].
The detail provided by our dense heterospecific SNP distribution is several magnitudes greater for effectively delineating gene order and small chromosomal rearrangements such as the rhesus X-chromosome SNPs transposed to the short arm of the human Y-chromosome in Table 1, which reflects a highly derived trait in humans [31]. However, specific assembly artifacts, including the microinversions of COMMD 8 and GABRB 1 markers reported by Karare et al. [32], were also reproduced in our data. Our heterospecific SNPs that were erroneously localized to rhesus macaque chromosome 5 reflected the same general gene order as observed by Karare et al. [32] and our SNPs detected twice as many gene orthologs compared to their study (see supplementary document).
While 96% of the heterospecific single-hit SNPs were shared between the Chinese and Indian rhesus macaques, 4,396 and 6,396 heterozygous SNPs were identified in the Chinese and Indian animals, respectively. These observed ratios of homozygous to heterozygous loci in the Chinese and Indian animals, respectively, are concordant with the higher estimates of SNP-based genetic diversity in Indian than in Chinese rhesus macaques [33].
A total of 79,367 human, 68,479 Chinese rhesus macaque and 65,801 Indian rhesus macaque heterospecific SNPs were successfully mapped to 12,328 human, 8,926 Chinese rhesus macaque and 8,798 Indian rhesus macaque gene orthologs, respectively (Table 2). Of these, 3,674 gene orthologs were found in the human and at least one of the two rhesus macaques, 3,601 and 3,563 of them in the Chinese and Indian rhesus macaque genomes, respectively, and 3,490 of these genes were shared among all three genomes. The low number of gene orthologs discovered in the rhesus genome is consistent with Fawcett et al.’s [34] observation that the rhesus genome has not yet been adequately annotated. On average each human gene contained between 1 and 271 SNPs with an average of 6.4, while a maximum of 226 (mean = 7.7) and 216 (mean = 7.5) SNPs per gene ortholog were identified in the Chinese and Indian rhesus macaques, respectively.
Table 2.
Number of single-hit heterospecific genic SNPs per gene ortholog in human and rhesus macaques. The list of gene orthologs has been provided as supplementary documentation. The numbers of Chinese and Indian gene orthologs also found in humans are parenthesized and the number of human gene orthologs found in at least one of the rhesus macaques is bracketed.
| Chinese rhesus macaque | Indian rhesus macaque | Human | |
|---|---|---|---|
| Number of gene orthologs detected in each group | 8,926 (3,601) | 8,798 (3,563) | 12,328 [3,674] |
| Number of genic SNPs in each group | 68,479 | 65,801 | 79,367 |
| Maximum number of SNPs per gene ortholog in each group | 226 | 216 | 271 |
| Average number of SNPs per gene ortholog in each group | 7.7 | 7.5 | 6.4 |
3. Discussion
The approximately 85,000 probes originally designed to interrogate genic regions in the human genome that also hybridized to regions of the rhesus genome imply that the rhesus genome contains a high degree of human sequence identity in genomic regions of potential biomedical interest. The discovery of a large number of heterospecific SNPs in only two rhesus macaques that uniquely map to both the human and rhesus genomes reflects the extensive amount of conserved variation in macaques that remains to be tapped.
While the usefulness of our mapping procedures is limited by the verity of published alignment frame and annotations [35], the concordance of our synteny maps with previously published work serves to validate our methods for localization of probes to published alignment frames. As alignment frames become more robustly validated and annotated, our methods for SNP discovery and ortholog localization will become powerful tools for assessing important genic diversity and genic linkages in human and Macaca genomes. This information will be valuable in determining the usefulness of macaques as research subjects for diseases of interest in humans.
Approximately 50% of the rhesus macaque genome is repetitive sequences, primarily transposable elements [1]. While the use of single-hit SNPs circumvented the problem of repetitive DNA, probes that mapped more than once to the rhesus genome may have aligned with authentic genes that occur in multiple copies in both humans [36, 37] and rhesus macaques [38, 39]. Analysis using the Galaxy program indicated that a significant number of potentially functional SNPs are shared between humans and rhesus macaques including many that are associated with known human traits and diseases. A total of 59,247 heterospecific SNPs were contained within all gene orthologs with an average of between 6 and 7 SNPs per gene in humans and approximately 8 SNPs per gene in rhesus macaques. Unlike Fawcett et al. [34] who observed SNP distributions to be random across the rhesus macaque chromosomes, the single-hit SNPs discovered in the present study are not evenly distributed, probably reflecting the non-random distributions of genic regions in the genome and the low frequency with which recombination events occur within conserved blocks.
Both Fawcett et al. [34] and Fang et al. [40] reported far more SNPs than the present study, i.e., between 3 and 5.5 million, respectively, of which only 4,472 and 18,324 were non-synonymous. The exclusive use of SNPs first identified in humans may have introduced an ascertainment bias that reduced the level of variability discovered in the rhesus macaque samples [41, 42]. However, unlike our SNPs, Fawcett et al. [34] and Fang et al.’s [40] SNPs have not been associated with pre-existing human rs numbers nor are they traceable in both genomes using related-tracks in the UCSC genome browser.
Were the rhesus gene annotation more complete and accurate [35], more rhesus macaque gene orthologs and other functional units would have been already identified, since many more genes with precise annotations have been identified in humans than in rhesus macaques. Over 3,674 human genes with corresponding SNP rs numbers are orthologous to annotated genes in the rhesus macaque draft sequence. While many rhesus genes with GenBank accession numbers and SNPs with rs numbers have been identified, many (2,939) currently do not have formal gene names but have been designated with “LOC” prefixes and gene ID number [LOC prefixes are used when a published symbol is not available and orthologs have not yet been determined (http://www.ncbi.nlm.nih.gov/books/NBK3840/#genefaq.Nomenclature)].
Comparison of human and rhesus chromosomes reveal that synteny is confined to conserved segments and inversions are the most common type of rearrangements. Many genes on a particular human chromosome have orthologs on a different rhesus chromosome. The overall data generated here reveals that genomic rearrangements, including inter-chromosomal translocations and intra-chromosomal inversions, have occurred frequently during primate genome evolution and could have contributed significantly to genetic diversity among primates. The extent to which single-copy gene orthologs are conserved in these human and rhesus chromosomes strongly implies that these chromosomes are likely to be functional equivalents in humans and rhesus macaques and conserved across evolutionary time.
The inclusion of a Chinese and an Indian animal in the present study allowed us to evaluate the intraspecific genetic differences involving orthologs that are specifically relevant to biomedical research. Fang et al. [40] calculated that 97% of the heterozygous positions in Indian rhesus macaques were also present in Chinese rhesus macaques. In the present study, Chinese rhesus macaques exhibited 96% of the human-Indian rhesus gene orthologs, but only 70% of the inferred heterozygous positions in the Indian rhesus macaque were also observed in the Chinese rhesus macaque. This is surprising since the single-hit SNPs were discovered in the present study without prior knowledge of their variability in either macaque variety. Furthermore, both these individuals exhibited comparable levels of heterozygosity across 2,808 rhesus-specific SNP loci [33]. Nevertheless, some of our 3,674 heterospecific SNPs that are annotated in the rhesus genome are probably non-synonymous and contribute to phenotypic variation within populations [43].
While conserved in humans and macaques, the gene orthologs can aid in phylogenetic studies by revealing highly derived differences involving large-scale genome rearrangements in both species. This unique resource will enable a SNP-based comparative mapping of human-macaque gene orthologs for describing gene order (synteny) and rearrangements across chromosomal segments in both taxa. Moreover, being amenable to large scale linkage analysis, this resource promises to be a useful tool for resolving assembly ambiguities in the rhesus draft genome. Additionally, it allows the addition of markers in a stepwise fashion to the growing framework map. By assaying an optimal set of heterospecific reference SNPs, our approach may provide a comprehensive framework for creating a map of biomedically relevant human gene orthologs in the rhesus genome. Framework maps for each individual rhesus chromosome will provide a backbone on which to build a series of ever more detailed physical maps. A detailed high resolution map would enhance our ability to characterize the genetic structure of captive rhesus macaque populations across orthologous regions of the genome. The combined ability to conduct interspecies and cross-population genome scans and comparisons with high density heterospecific SNPs may someday elucidate the complete genetic architecture of simple and complex disease susceptibility.
While studies that rely on genome-wide analyses of large cohorts of subjects can detect and map disease susceptibility genes [44], population stratification and relatedness can produce spurious associations if not properly addressed [45]. Significant genetic structure in captive populations of rhesus macaques has been reported [15, 17, 46] due to interspecies hybridization (e.g., between rhesus and cynomolgus macaques in Indochina), introgression between Chinese and Indian rhesus macaques, founder effects, and inter-generational genetic drift. Because our technique is applicable to large-scale tracking of genetic variation across large sample sets, it can help clarify the unknown genetic structures of orthologous regions that are conserved and/or rearranged in rhesus macaques. Moreover, our method facilitates detecting and tracing the transmission of heterospecific SNPs in members of multigenerational pedigrees to identify groups of linked markers.
4. Materials and Methods
4.1. SNP Genotyping
Genomic DNA (960 ng/ul) from one female Chinese and one female Indian rhesus macaque were obtained from the California National Primate Research Center. A human female DNA sample (1 ug/ul) was purchased from Zyagen (San Diego, CA). Each sample was hybridized to the Affymetrix Genome-Wide Human SNP Array 6.0, according to the manufacturer’s protocol. This array features 1.8 million genetic markers, including more than 906,600 single nucleotide polymorphisms (SNPs). The arrays were washed and stained on a Fluidics Station 450 and scanned on a GeneChip Scanner 3000. The Affymetrix GTC Console was used to assess the built-in quality control metrics using the Contrast Quality Control algorithm and SNP data that failed the quality control checks were eliminated from subsequent analyses. Subsequent to passing validation, SNP genotypes from both the human control sample and rhesus macaque test samples were processed with the CRLMM Bioconductor package [47]. More than 553,000 SNPs suspected to be orthologous in humans and rhesus macaques were identified at this stage.
4.2. Identification and validation of probes and SNP calls in orthologous gene regions
Since the SNP 6.0 array uses the hybridization of 25-mer probes to query both strands at multiple offsets with respect to each SNP [10], it was necessary to filter probes that occurred more than once in the human genome (i.e., probes that cannot be mapped to a unique position) in the rhesus macaque genome. SNP locations, genotypes, probe sequences, and annotation information was downloaded from the Affymetrix website (https://www.affymetrix.com). For each SNP, perfect match (PM) probes for both alleles were aligned to the UCSC rhesus macaque 2 genome (rheMac2) using BWA [48] and to expressed sequence tags (ESTs) from GenBank (also downloaded from the UCSC website: http://genome.ucsc.edu). An integrated analysis combining the rhesus genome and EST sequences increased the likelihood of detecting unreliable probe-template matches including probes that did not uniquely map to the rhesus genome. To further decrease the probability of spurious matches due to EST sequences that anneal to gene regions within the rhesus genome, procedures using the genome and EST sequences were performed separately. SNPs within probes that aligned to multiple locations to the rhesus genome or to multiple regions in the ESTs (i.e., multi-hit SNPs) were removed from the dataset, reducing the risk of querying paralogous sites. To prevent ambiguous hits and false matches, only one probe per SNP was required to perfectly align with the rhesus genome to ensure that at least one of the human SNPs matched at a specific SNP site in the rhesus genome.
The human and rhesus reference genomes were used as initial scaffolds to anchor the single-hit SNPs to chromosomal regions. Using the single-hit SNPs generated from the unique probe alignments in the rhesus genome, the aligned positions of these probes (in rheMac2) were compared to the rhesus chromosomal region shown as orthologous to that SNP’s position (in hg19) in the UCSC Genome Browser’s Primate Chain/Net Comparative Genomics track. This yielded a list of human SNPs that have probe sequences localized to areas of the rhesus genome that have already been determined to be orthologous to humans.
4.3. SNP-based comparative mapping
We used the alignment information of the entire set of single-hit probes in both rhesus macaques to locate each SNP on each rhesus chromosome and determine the distribution of single-hit SNPs across each of the rhesus autosomal chromosomes and the X chromosome. These SNPs were sorted according to their map locations on each chromosome to calculate inter-SNP distances. The grDevices package in R was utilized to calculate the quantile statistics of the distribution of SNPs across each chromosome, and these statistics were plotted using the Graphics package in R version 2.13.2 [49].
By explicitly mapping SNP alleles to both the human and rhesus genomes, we have increased the SNP 6.0 array’s sensitivity in detecting heterospecific SNPs. Using the Galaxy program [50], we mapped each of the single-hit rhesus SNPs to genes in NCBI’s annotated rhesus macaque genome (Build 1.2; ftp://ftp.ncbi.nih.gov/genomes/MapView/Macaca_mulatta/sequence/BUILD.1.2/initial_release/) and human genome (Build 37.3; ftp://ftp.ncbi.nih.gov/genomes/MapView/Homo_sapiens/sequence/BUILD.37.3/initial_release/). The genes for both species associated with specific SNP rs numbers were tallied before the gene information in human and rhesus macaques was compared.
Supplementary Material
Highlights.
Rhesus macaques are relevant animal models for post-genomic era biomedical research
Chromosomal rearrangements can confound candidate gene identification in macaques
Heterospecific SNPs allow fine-grained comparisons of human and macaque chromosomes
Synteny maps based on heterospecific SNPs can enable localization of gene orthologs
Acknowledgments
We thank Ryan Davis at the UC Davis MIND Institute for his helpful advice and contributions to this manuscript. This study was supported by the California National Primate Research Center base grant (No. RR000169-48), by an ARRA supplement awarded to SK and Nick Lerche, CNPRC (No. RR018144-07) and NIH grants RR005090 and RR025871 to DGS. Animals used in this research were managed in compliance with Institutional Animal Care and Use Committee (IACUC) regulations or in accordance with the National Institutes of Health guidelines or the US Department of Agriculture regulations prescribing the humane care and use of laboratory animals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu Y-s, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers Y-H, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang S-P, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csürös M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX-Z, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AFA, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han S-G, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu L-L, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O’Brien WE, Prüfer K, Stenson PD, Wallace JC, Ke H, Liu X-M, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS Rhesus Macaque Genome Sequencing Analysis Consortium. Evolutionary and Biomedical Insights from the Rhesus Macaque Genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 3.Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, Du H, Chen J, Chen R, Zhang P, Huang Z, Thompson JR, Meng Y, Bai Y, Wang J, Zhuo M, Wang T, Huang Y, Wei L, Li J, Wang Z, Hu H, Yang P, Le L, Stenson PD, Li B, Liu X, Ball EV, An N, Huang Q, Zhang Y, Fan W, Zhang X, Li Y, Wang W, Katze MG, Su B, Nielsen R, Yang H, Wang J, Wang X, Wang J. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotech. 2011;29:1019–1023. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson B, Street S, Wright H, Pearson C, Jia Y, Thompson S, Allibone P, Dubay C, Spindel E, Norgren R. Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta) BMC Genomics. 2007;8:43. doi: 10.1186/1471-2164-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malhi RS, Sickler B, Lin D, Satkoski J, Tito RY, George D, Kanthaswamy S, Smith DG. MamuSNP: A Resource for Rhesus Macaque (Macaca mulatta) Genomics. PLoS ONE. 2007;2:e438. doi: 10.1371/journal.pone.0000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Street S, Kyes R, Grant R, Ferguson B. Single nucleotide polymorphisms (SNPs) are highly conserved in rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques. BMC Genomics. 2007;8:480. doi: 10.1186/1471-2164-8-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satkoski J, Malhi R, Kanthaswamy S, Tito R, Malladi V, Smith D. Pyrosequencing as a method for SNP identification in the rhesus macaque (Macaca mulatta) BMC Genomics. 2008;9:256. doi: 10.1186/1471-2164-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PIW, Maller JB, Kirby A, Elliott AL, Parkin M, Hubbell E, Webster T, Mei R, Veitch J, Collins PJ, Handsaker R, Lincoln S, Nizzari M, Blume J, Jones KW, Rava R, Daly MJ, Gabriel SB, Altshuler D. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 11.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 12.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez RD, Hubisz MJ, Wheeler DA, Smith DG, Ferguson B, Rogers J, Nazareth L, Indap A, Bourquin T, McPherson J, Muzny D, Gibbs R, Nielsen R, Bustamante CD. Demographic Histories and Patterns of Linkage Disequilibrium in Chinese and Indian Rhesus Macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- 14.Trask J, Malhi R, Kanthaswamy S, Johnson J, Garnica W, Malladi V, Smith D. The effect of SNP discovery method and sample size on estimation of population genetic data for Chinese and Indian rhesus macaques (Macaca mulatta) Primates. 2011;52:129–138. doi: 10.1007/s10329-010-0232-4. [DOI] [PubMed] [Google Scholar]
- 15.Kanthaswamy S, Satkoski J, Kou A, Malladi V, Glenn Smith D. Detecting signatures of inter-regional and inter-specific hybridization among the Chinese rhesus macaque specific pathogen-free (SPF) population using single nucleotide polymorphic (SNP) markers. Journal of Medical Primatology. 2010;39:252–265. doi: 10.1111/j.1600-0684.2010.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanthaswamy S, Smith DG. Effects of geographic origin on captive Macaca mulatta mitochondrial DNA variation. Comp Med. 2004;54:193–201. [PubMed] [Google Scholar]
- 17.Kanthaswamy S, Kou A, Satkoski J, Penedo MCT, Ward T, Ng J, Gill L, Lerche NW, Erickson BJA, Smith DG. Genetic characterization of specific pathogen-free rhesus macaque (Macaca mulatta) populations at the California National Primate Research Center (CNPRC) American Journal of Primatology. 2010;72:587–599. doi: 10.1002/ajp.20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen J. Vaccine Studies Stymied by Shortage of Animals. Science. 2000;287:959–960. doi: 10.1126/science.287.5455.959. [DOI] [PubMed] [Google Scholar]
- 19.Hayden EC. US plans more primate research. Nature. 2008;453:439. doi: 10.1038/453439a. [DOI] [PubMed] [Google Scholar]
- 20.Rogers J, Garcia R, Shelledy W, Kaplan J, Arya A, Johnson Z, Bergstrom M, Novakowski L, Nair P, Vinson A, Newman D, Heckman G, Cameron J. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics. 2006;87:30–38. doi: 10.1016/j.ygeno.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Pearson PL, Roderick TH, Davisson MT, Garver JJ, Warburton D, Lalley PA, O’Brien SJ. Report of the committee on comparative mapping. Cytogenetic and Genome Research. 1979;25:82–95. doi: 10.1159/000131403. [DOI] [PubMed] [Google Scholar]
- 22.Best RG, Diamond D, Crawford E, Grass FS, Janish C, Lear TL, Soenksen D, Szalay AA, Moore CM. Baboon/human homologies examined by spectral karyotyping (SKY): a visual comparison. Cytogenetic and Genome Research. 1998;82:83–87. doi: 10.1159/000015070. [DOI] [PubMed] [Google Scholar]
- 23.Yunis J, Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- 24.Wienberg J, Stanyon R, Jauch A, Cremer T. Homologies in human and Macasa fuscata chromosomes revealed by in situ suppression hybridization with human chromosome specific DNA libraries. Chromosoma. 1992;101:265–270. doi: 10.1007/BF00346004. [DOI] [PubMed] [Google Scholar]
- 25.Roberto R, Misceo D, D’Addabbo P, Archidiacono N, Rocchi M. Refinement of macaque synteny arrangement with respect to the official rheMac2 macaque sequence assembly. Chromosome Research. 2008;16:977–985. doi: 10.1007/s10577-008-1255-1. [DOI] [PubMed] [Google Scholar]
- 26.Stanyon R, Rocchi M, Capozzi O, Roberto R, Misceo D, Ventura M, Cardone M, Bigoni F, Archidiacono N. Primate chromosome evolution: Ancestral karyotypes, marker order and neocentromeres. Chromosome Research. 2008;16:17–39. doi: 10.1007/s10577-007-1209-z. [DOI] [PubMed] [Google Scholar]
- 27.Murphy W, Stanyon R, O’Brien S. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biology. 2001;2:reviews0005.0001–reviews0005.0008. doi: 10.1186/gb-2001-2-6-reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy WJ, Menotti-Raymond M, Lyons LA, Thompson MA, O’Brien SJ. Development of a Feline Whole Genome Radiation Hybrid Panel and Comparative Mapping of Human Chromosome 12 and 22 Loci. Genomics. 1999;57:1–8. doi: 10.1006/geno.1998.5695. [DOI] [PubMed] [Google Scholar]
- 29.Murphy WJ, Page JE, Smith C, Desrosiers RC, O’Brien SJ. A Radiation Hybrid Mapping Panel for the Rhesus Macaque. Journal of Heredity. 2001;92:516–519. doi: 10.1093/jhered/92.6.516. [DOI] [PubMed] [Google Scholar]
- 30.Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 31.Page DC, Harper ME, Love J, Botstein D. Occurrence of a transposition from the X-chromosome long arm to the Y-chromosome short arm during human evolution. Nature. 1984;311:119–123. doi: 10.1038/311119a0. [DOI] [PubMed] [Google Scholar]
- 32.Karere GM, Froenicke L, Millon L, Womack JE, Lyons LA. A high-resolution radiation hybrid map of rhesus macaque chromosome 5 identifies rearrangements in the genome assembly. Genomics. 2008;92:210–218. doi: 10.1016/j.ygeno.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanthaswamy S, Trask JS, Ross CT, Kou A, Houghton P, Smith DG, Lerche N. A Large-Scale SNP-Based Genomic Admixture Analysis of the Captive Rhesus Macaque Colony at the California National Primate Research Center. American Journal of Primatology. 2012;74:747–757. doi: 10.1002/ajp.22025. [DOI] [PubMed] [Google Scholar]
- 34.Fawcett G, Raveendran M, Deiros D, Chen D, Yu F, Harris R, Ren Y, Muzny D, Reid J, Wheeler D, Worley K, Shelton S, Kalin N, Milosavljevic A, Gibbs R, Rogers J. Characterization of single-nucleotide variation in Indian-origin rhesus macaques (Macaca mulatta) BMC Genomics. 2011;12:311. doi: 10.1186/1471-2164-12-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Goodsell J, Norgren R. Limitations of the rhesus macaque draft genome assembly and annotation. BMC Genomics. 2012;13:206. doi: 10.1186/1471-2164-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O’Connell RJ, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK. The Influence of CCL3L1 Gene-Containing Segmental Duplications on HIV-1/AIDS Susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 37.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, Carter NP, Lee C, Stone AC. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Degenhardt JD, de Candia P, Chabot A, Schwartz S, Henderson L, Ling B, Hunter M, Jiang Z, Palermo RE, Katze M, Eichler EE, Ventura M, Rogers J, Marx P, Gilad Y, Bustamante CD. Copy number variation of CCL3-like genes affects rate of progression to simian-AIDS in Rhesus Macaques (Macaca mulatta) PLoS Genet. 2009;5:e1000346. doi: 10.1371/journal.pgen.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taormina P, Trask J, Houghton P, Smith D, Kanthaswamy S. CCL3L1 copy number variation (CNV) in rhesus macaques (Macaca mulatta) Comparative Medicine. (in press) [PMC free article] [PubMed] [Google Scholar]
- 40.Fang X, Zhang Y, Zhang R, Yang L, Li M, Ye K, Guo X, Wang J, Su B. Genome sequence and global sequence variation map with 5.5 million SNPs in Chinese rhesus macaque. Genome Biology. 2011;12:R63. doi: 10.1186/gb-2011-12-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen R. Population genetic analysis of ascertained SNP data. Human genomics. 2004;1:218–224. doi: 10.1186/1479-7364-1-3-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen R, Signorovitch J. Correcting for ascertainment biases when analyzing SNP data: applications to the estimation of linkage disequilibrium. Theoretical Population Biology. 2003;63:245–255. doi: 10.1016/s0040-5809(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 43.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Research. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanthaswamy S, Kou A, Smith DG. Population Genetic Statistics from Rhesus Macaques (Macaca mulatta) in Three Different Housing Configurations at the California National Primate Research Center. Journal of the American Association for Laboratory Animal Science. 2010;49:598–609. [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- 50.Blankenberg D, Kuster GV, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; 2001. Galaxy: A Web-Based Genome Analysis Tool for Experimentalists. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.