Abstract
Interleukin-1 receptor-associated kinase 4 (IRAK4) is critical for MyD88-dependent Toll-like receptor (TLR) signaling, and patients with Irak4 mutations are extremely susceptible to recurrent bacterial infections. In these studies, mice homozygous for a mutant IRAK4 that lacks kinase activity (IRAK4KDKI) were used to address the role of IRAK4 in response to TLR agonists or bacterial infection. IRAK4KDKI macrophages exhibited diminished responsiveness to the TLR4 agonist LPS and little to no response to the TLR2 agonist Pam3Cys when compared to wild-type macrophages as measured by cytokine mRNA, cytokine protein expression and MAPK activation. Importantly, we identified two kinases downstream of the MAPKs, MNK1 and MSK1, whose phosphorylation is deficient in IRAK4KDKI macrophages stimulated through either TLR2 or TLR4, suggesting that IRAK4 contributes to TLR signaling beyond initial phosphorylation of MAPKs. Additionally, IRAK4KDKI macrophages produced minimal cytokine mRNA expression in response to the Gram-positive bacteria Streptococcus pneumoniae and Staphylococcus aureus compared to WT cells, and IRAK4KDKI mice exhibited increased susceptibility and decreased cytokine production in vivo upon S. pneumoniae infection. Treatment of infected mice with Poly IC:LC (Hiltonol®), a potent TLR3 agonist, significantly improved survival of both WT and IRAK4KDKI mice thereby providing a potential treatment strategy in both normal and immunocompromised patients.
Introduction
A major component of the mammalian immune system relies on the recognition of certain microbial components that are absent in host cells called pathogen-associated molecular patterns (PAMPs) (1, 2). Host cells recognize PAMPs through pattern recognition receptors (PRRs) that initiate a signal cascade that results in the upregulation of proinflammatory cytokines and the clearance of invading microbes. Of the PRRs, Toll-like receptors (TLRs) play a critical role in controlling microbial infection in both mice and humans. Mutations that occur in the TLR signaling pathway result in increased susceptibility to viral, bacterial and/or fungal infections depending on the specific signaling component affected. In the past decade, an increasing number of innate immune deficiencies have been identified in genes that encode TLR pathway components (3–7).
In humans, 10 TLRs have been identified. TLR2 heterodimerizes with TLR1 or TLR6, leading to the recognition of Gram positive bacterial components such as lipoproteins (8). TLR4, the first human TLR cloned (9, 10), is engaged by LPS found on Gram negative bacteria, while TLRs 3, 5, 7, 8, 9 and 11 recognize other bacterial and/or viral components such as double-stranded RNA (TLR3), flagellin (TLR5), single-stranded RNA (TLRs 7 and 8), CpG DNA (TLR9) and profilin (TLR11) (1). All TLRs activate NF-κB and MAPKs, but the intermediate signaling molecules used by a particular TLR can vary. While some proteins are common to all TLR signaling pathways, others are unique to a particular TLR or shared among only a subset of the TLRs. For example, cells deficient in the adapter protein MyD88 are completely refractory to signaling through nearly all TLRs with the notable exceptions of TLR3 and TLR4 (11, 12). TLR3 is completely independent of MyD88, while TLR4 has two signaling pathways, one of which is MyD88-dependent and the other that is MyD88-independent. Upon TLR engagement, MyD88 is recruited to the cytosolic TIR domain of a TLR and facilitates recruitment of the IRAK kinases, IRAK4, IRAK1 and IRAK2. Each of these proteins contributes to the assembly of a multi-protein structure, called the “myddosome,” that is critical for TLR-mediated signal transduction (13). IRAK1 and IRAK2 are thought to have redundant roles in myddosome formation and signal transduction, but the unique contributions, if any, of IRAK2 have not yet been thoroughly addressed (14). Once IRAK4 complexes with MyD88, it phosphorylates IRAK1, allowing IRAK1 to autophosphorylate and recruit the ubiquitin ligase TRAF6. TRAF6 ubiquinates both itself and IRAK1, enabling interaction of the complex with TAK1, the kinase responsible for IκB phosphorylation and degradation, leading to translocation of activated NF-κB to the nucleus. Another adapter protein, TIRAP (also called Mal), facilitates MyD88 recruitment to TLR2. Like TLR2, TLR4 uses TIRAP/MyD88 to initiate signaling through the MyD88-dependent pathway; however, TLR4 also utilizes another “bridging adapter,” TRAM, to recruit TRIF to the receptor complex and initiate TRIF-dependent signaling. TLR3 signals exclusively through the adapter TRIF.
Due to its role in propagating MyD88-dependent signaling, IRAK4 is presumably a critical component to most TLR signaling pathways, with the exception of TLR3 (15–18). Mice deficient in IRAK4 (IRAK4−/−) show characteristics reminiscent of MyD88−/− mice in that they are resistant to doses of certain TLR agonists, such as the LPS or CpG, that are lethal to wild-type (WT) mice (19, 20). However, there are discrepancies in the literature as to the whether the kinase activity of IRAK4 is required for TLR signaling or not (19–23). Interestingly, cytokine expression induced by LPS in macrophages expressing a mutant IRAK4 that lacks kinase activity was diminished, but not to the same extent as IRAK4−/− cells (20). These data suggest IRAK4 plays a role in propagating TLR signaling independent of its ability to phosphorylate IRAK1. In contrast, a separate study demonstrated diminished gene expression in both IRAK4−/− and IRAK4 kinase inactive cells, thus demonstrating a need for further exploration into the necessity of IRAK4 kinase activity in the propagation of TLR signaling (19).
Patients with mutations in IRAK4 present with recurrent bacterial infections but show no impaired defense against viral infections (presumably due to their retained ability to signal through TLR3 and other non-TLR viral receptors). S. pneumoniae is the most common infection found in these patients, followed by S. aureus and P. aeruginosa (4, 6). While others have observed impaired gene expression in IRAK4 deficient cells stimulated with synthetic TLR agonists, there is a paucity of data to address how this deficiency fares against infection with whole bacteria. In the studies presented in this report, we sought to clarify the literature regarding impaired TLR signaling in IRAK4KDKI macrophages and to develop a mouse model of infection to address the importance of IRAK4 kinase activity in controlling bacterial infection. We found that TLR4 signaling is partially deficient in macrophages derived from IRAK4KDKI mice, while TLR2 signaling appears to be fully dependent upon IRAK4 kinase activity for signaling. Macrophage responses were more impaired when exposed to bacteria known to be potent TLR2 simulators, while bacteria such as P. aeruginosa behaved more like a TLR4 agonist in that signaling was only partially impaired. Finally, differential susceptibility to S. pneumoniae was observed in WT vs. IRAK4KDKI mice, indicating the importance of IRAK4 during infection. Treatment of infected mice with Hiltonol®, a stabilized formulation of Poly IC:LC and potent TLR3 agonist, significantly improved the survival rate of both WT and IRAK4KDKI mice. These data represent a potential treatment strategy for both normal and immunocompromised patients.
Materials and Methods
Mice, macrophage isolation and reagents
IRAK4KDKI mice, generously provided by Dr. Kirk A Staschke and Dr. Raymond Gilmour (Lilly Research Laboratories, Indianapolis, IN), were derived on a C57BL/6J background and were bred homozygously at University of Maryland, School of Medicine. Age- and sex-matched C57BL/6J mice were purchased from Jackson Laboratories (Bar Harbor, ME). Peritoneal macrophages were obtained by performing a peritoneal lavage 5 days post-i.p administration of sterile thioglycollate (Remel, Lanexa, KS). For bone marrow-derived macrophages, bone marrow was cultured in standard medium supplemented with 20–25% LADMAC cell-conditioned medium (24). After 6 days, non-adherent cells were removed. Adherent cells were harvested using 0.25 % Trypsin-EDTA (Gibco, Grand Island, NY) after 7–14 days in culture and replated for experimental use in the absence of a CSF-1 source. All animal experiments were conducted under the guidelines and approval of the Institutional Animal care and Use Committee. All cellular experiments were conducted using DMEM (Gibco, Grand Island, NY) supplemented with 5% FCS, 2 mM L-glutamine and 1 % penicillin-streptomycin (P/S) (P/S was omitted in experiments using live or heat-killed bacteria) at 37° C and 5% CO2.
Agonists for TLR2, N-palmitoyl-S-[2,3-bis(palmitoyloxy)-(2RS)-propyl]-[R]-cysteinyl-[S]-seryl-[S]-lysyl-[S]-lysyl-[S]-lysyl-[S]-lysine (Pam3Cys), and TLR3, polyinosinic-polycytidylic acid (Poly I:C), were purchased (Invivogen, San Diego, CA). Poly IC:LC (Hiltonol) was generously provided by Dr. Andres Salazar (Oncovir, Inc., Washington, DC). Protein-free, phenol-water-extracted Escherichia coli K235 LPS was prepared as described elsewhere (25).
Quantitative real-time PCR (qPCR)
Macrophages were cultured in 12-well plates at a density of 2 × 106 per well. After stimulation, RNA was extracted using Tripure reagent (Roche, Indianopolis, IN) and 1 μg of total RNA was reversed transcribed (RT-PCR) using iScript (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. The resulting cDNA template was used for real-time PCR analysis with Power SYBR Green and the AB 7900HT Fast Time fluorescence detection system. The following primers were used: IL-1β sense, 5′-aaatacctgtggccttgggc and antisense, 5′-cttgggatccacactctccag; IFN-β sense, 5′-ttctccaccacagccctctc and antisense, 5′-ctttccattcagctgctccag; IL-12p40 sense, 5′-tctttgttcgaatccagcgc and antisense, 5′-ggaacgcacctttctggttaca; HPRT sense, 5′-gctgacctgctgattacattaa and antisense, 5′-tgatcattacagtagctcttcagtctga; KC sense, 5′-ccatggctgggattcacc and antisense, 5′-gaccattcttgagtgtggctatgac; IL-10 sense, 5′-atttgaattccctgggtgagaag and antisense, 5′-cacaggggagaaatcgatgaca; COX2 sense, 5′-actgggccatggagtggac and antisense, aatgacctgatatttcaattttccatc; PAI2 sense, 5′-tacaggcacaagcaggagataaaa and antisense, 5′-agctgagagaggagaaggctga; IL-6 sense, 5′-tgtctataccacttcacaagtcggag and antisense, 5′-gcacaactcttttctcatttccac; IP10 sense, 5′-gtgttgagatcattgccacga and antisense, 5′-tttttggctaaacgctttcattaa; S. pneumoniae 16S sense, 5′-gagtcgcaagccggtgacgg and antisense, 5′-cgtattcaccgcggcgtgct. Relative gene expression was calculated using the ΔCt calculation method in which all samples were normalized to the housekeeping gene HPRT (26).
Cytokine protein measurements
Macrophages were cultured in 12-well plates at a density of 2 × 106 per well. Six h after stimulation, supernatants were collected and sent to the University of Maryland Cytokine Core Laboratory (Baltimore, MD) for cytokine analysis using the Luminex Multianalyte System (R&D Systems, Minneapolis, MN). For in vivo measurements, serum samples were collected from mice 48 h post infection and processed as stated above.
Cell lysate preparation and Western blot analysis
Macrophages (3 × 106) were plated in 6-well plates and incubated with the indicated concentrations of TLR agonists or bacteria. Cells were washed with ice-cold PBS and resuspended in lysis buffer (50 mM Tris (pH 7.5), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 2 mM EDTA with a protease inhibitor mixture (Sigma-Aldrich, St. Louis, MO; P8340) and a phosphatase inhibitor cocktail (PhosStop, Roche, Indianopolis, IN)). Protein concentrations were determined (Bio-Rad, Hercules, CA) and lysates were boiled for 5 min in reducing sample buffer (Thermo Scientific, Pittsburgh, PA). Equal quantities of total protein were electrophoresed on SDS polyacrylamide gels and transferred onto PVDF membranes. Membranes were washed in TBS with 0.1% Tween 20 (TBST), incubated for 1 h at room temperature in 5% BSA or 5% nonfat milk in TBST, incubated overnight at 4° C with primary Ab, washed, incubated for 1 h at room temperature with HRP-labeled anti-rabbit or anti-mouse secondary Ab (Amersham Biosciences, Piscataway, NJ), and developed with the ECL detection kit (Thermo Scientific, Pittsburgh, PA). Anti-p-JNK, anti-p-ERK, anti-p-p38, anti-p38 total, anti-MSK1 and anti-MNK1 were purchased from Cell Signaling Technology (Danvers, MA); anti-H3ser10 and anti-H3 from Abcam (Cambridge, MA); anti-actin from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-eIF4e and anti-p-eIF4e Abs were generously provided by Dr. Nahum Sonenberg (McGill University, Montreal, Quebec, Canada).
Nuclear extract preparation and oligodeoxynucleotide (ODN) co-precipitation
Macrophages (8 × 106) were plated in 10 cm dishes and treated with TLR agonists. The cells were washed one time with PBS, pelleted by centrifugation and nuclear extracts were prepared using the Nuclear Extract Kit (Active Motif, Carlsbad, CA) according to the manufacturer’s protocol. A biotinylated ODN containing the NF-κB consensus sequence (AGTTGAGGGGACTTTCCCAGGC) was annealed to the complementary strand for 1 h at room temperature. This procedure was also performed with a negative control ODN with a mutated sequence (AGTTGAGGCGACATTCCCAGGC). Nuclear extract sample prepared from cells were diluted to 900 μl with co-precipitation buffer (0.1% Triton X-100, 10 mM HEPES (pH 7.3), 2 mM EDTA, 1 mM EGTA, 10% glycerol, supplemented with a protease inhibitor mixture (Sigma-Aldrich, St. Louis, MO; P8340)) and precleared with 60 μl of salmon sperm-agarose beads (Millipore, Billerica, MA) for 30 min at 4° C. Beads were removed by centrifugation and the supernatant incubated overnight at 4° C with 30 nM NF-κB-ODN. Streptavidin-agarose beads (Sigma, St. Louis, MO) were added for 1 h and ODN-protein complexes were then pelleted, washed three times and diluted in SDS-PAGE sample buffer. Boiled samples were centrifuged to remove beads, and the entire supernatant was loaded onto a 12% polyacrylamide gel. Western blots were performed as described above.
Mouse infections and bacteria
S. pneumoniae (6303, ATCC) was grown to log phase in BHI nutrient broth (Teknova, Hollister, CA), supplemented with 10% sterile glycerol and stocks were stored at −80°C. Mice were lightly anesthetized using isoflurane (VetOne) and bacteria were administered intranasally (i.n.) at the appropriate concentration in 30 uL saline total. For studies using Poly IC:LC, bacteria were administered intranasally in 50 μl of saline or Poly IC:LC solution (100 μg/mouse). Bacterial concentration was verified for each experiment by performing serial dilutions of the inoculum plated on Mueller-Hinton Agar supplemented with 5% sheep’s blood (Teknova, Hollister, CA). Mice were monitored twice a day for survival studies. For mRNA analysis of cytokine production, mice were sacrificed 24 or 48 h post-infection, bacterial load was assessed using serial plate dilutions and a portion of the left lobe of lung was excised and homogenized in Tripure RNA extraction reagent (Roche, Indianopolis, IN). RT- and qPCR was then performed as described above.
Results
IRAK4 kinase activity is required for optimal gene induction in response to TLR2 and TLR4, but not TLR3, agonists
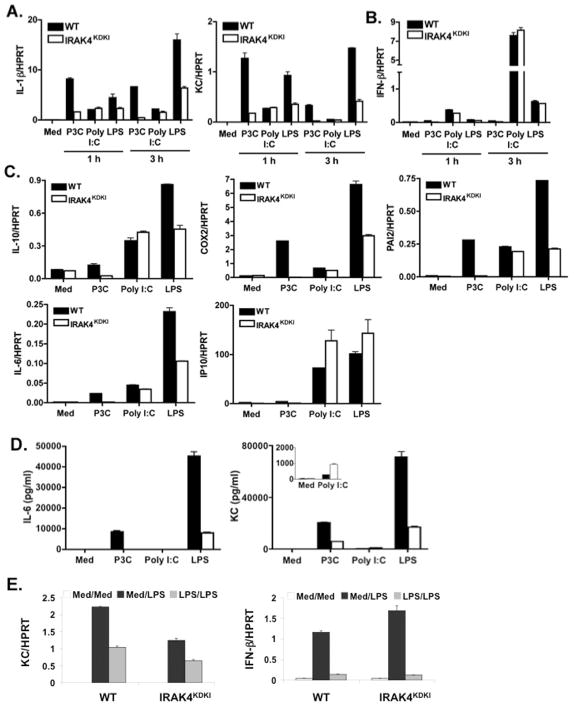
IRAK4 has been reported to be a critical kinase for propagating TLR-mediated signaling events leading to proinflammatory gene induction (14). To assess the requirement for IRAK4 kinase activity in MyD88-dependent TLR signaling, macrophages from WT and IRAK4KDKI mice were isolated and treated with the TLR2 agonist Pam3Cys (MyD88-dependent) or the TLR4 agonist LPS (MyD88- and TRIF-dependent). mRNA expression for MyD88-dependent and MyD88-independent genes was measured 1 or 3 h post-stimulation by qPCR. Induction of IL-1β and KC (Figure 1A) mRNA in Pam3Cys-treated IRAK4KDKI macrophages was minimal compared to the robust induction induced in WT cells. In contrast, LPS upregulated MyD88-dependent gene expression in IRAK4KDKI macrophages, although the level of IL-1β and KC mRNA was significantly less when compared to the full expression observed in WT cells. As the TLR3 signaling pathway is MyD88- and IRAK4-independent, we also stimulated macrophages with the TLR3 agonist Poly I:C to ensure that our IRAK4KDKI macrophages were capable of normal gene expression in response to an IRAK4- and MyD88-independent stimulus. Very little difference in mRNA induction in response to Poly I:C was observed in WT versus IRAK4KDKI macrophages, although the magnitude of gene expression was less than that induced by TLR2 or TLR4 signaling (Figure 1A)., IFN-β mRNA expression, which is induced by both LPS and Poly I:C in a MyD88-independent, TRIF-dependent fashion, was not inhibited in IRAK4KDKI macrophages stimulated with LPS or Poly I:C (Figure 1B). To examine whether these observations extend to additional genes, we performed additional qPCR analysis for the MyD88-dependent genes that encode IL-10, COX-2, PAI-2, IL-6, and the MyD88-independent gene IP-10 (Figure 1C). These data further support the observation that TLR2-induced gene expression is completely dependent on the kinase activity of IRAK4, while TLR4-induced gene expression is only partially dependent on the presence of this functional kinase. In contrast, LPS induction of IFN-β and IP-10 (Figure 1B, C) are independent of IRAK4 kinase activity. Additionally, we measured protein levels in the supernatants of both WT and IRAK4KDKI macrophages treated with Pam3Cys, Poly I:C or LPS (Figure 1D). In agreement with the mRNA data, Pam3Cys-treated IRAK4KDKI macrophages secreted very little IL-6 or KC while LPS induced some IL-6 and KC secretion in these cells, but not to the same extent as WT. While Poly I:C was not a potent inducer of either protein, it is interesting to note that KC protein expression was enhanced in IRAK4KDKI macrophages compared to WT macrophages (Figure 1D, insert). This confirms, at both the mRNA and protein levels, that the IRAK4KDKI macrophages are capable of responding to some TLR agonists (i.e., TLR3, TLR4) but not to others.
Figure 1. Cytokine gene expression in WT versus IRAK4KDKI macrophages.
WT and IRAK4KDKI macrophages were treated with medium only (Med) or stimulated with Pam3Cys (P3C; 1 μg/ml), Poly I:C (40 μg/ml), or LPS (100 ng/ml) for 1 or 3 h and analyzed for MyD88-dependent (A) and MyD88-independent (B) gene expression by qPCR. (C) Macrophages were treated as in (A) and (B) for 3 h and additional gene expression was analyzed by qPCR. (D) Macrophages were treated as in (A–C) for 6 h and supernatants were collected for cytokine measurement. (E) Macrophages isolated from WT and IRAK4KDKI mice were treated with medium only (Med/Med and Med/LPS) or LPS (100 ng/ml; LPS/LPS) for 18 h, washed 3 times then re-stimulated with medium (Med/Med) or LPS (100 ng/ml; Med/LPS and LPS/LPS) for 5 h. MyD88-dependent (KC) and MyD88-independent (IFN-β) gene expression was analyzed by qPC R. All samples were normalized to HPRT. Data are representative of at least 3 separate experiments.
We also sought to determine whether IRAK4 kinase activity is required to establish endotoxin tolerance in macrophages. As expected from previous studies (27), LPS-treated macrophages from WT mice that were previously exposed to LPS demonstrated diminished induction of both MyD88-dependent (i.e., KC, Figure 1E; LPS/LPS) and MyD88-independent (i.e., IFN-β, Figure 1E; LPS/LPS) genes when compared to macrophages not previously exposed to LPS (Figure 1E; Med/LPS). Consistent with the data in Figure 1A and B, stimulation of medium-pretreated IRAK4KDKI macrophages with LPS resulted in reduced KC but normal IFN-β mRNA levels (Figure 1E; Med/LPS). However, like WT macrophages, IRAK4KDKI macrophages that were previously exposed to LPS exhibited reduced expression of both KC and IFN-β mRNA compared to cells that were not previously exposed to LPS. We conclude that while IRAK4 kinase activity contributes to the MyD88-dependent pathway, it is not required to establish endotoxin tolerance in macrophages. Collectively, the data in Figure 1 suggest that IRAK4 kinase activity is critical for the activation of MyD88-dependent gene expression in response to both TLR2 and TLR4 agonists but is not required for all macrophage responses as endotoxin tolerance and the response to TLR3 remain intact in the absence of kinase active IRAK4.
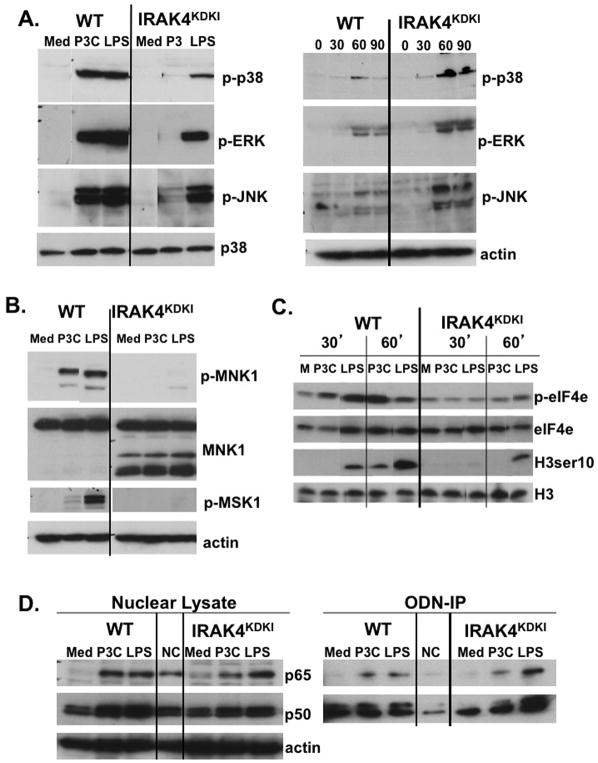
MAPK activation is severely deficient in TLR2-stimulated IRAK4KDKI macrophages, while only partially deficient in response to TLR4 stimulation
TLR signaling mediates activation of the MAPKs p38, ERK1/2 and JNK that ultimately promote proinflammatory gene expression. Therefore, we assessed whether IRAK4 kinase activity is required for activation of MAPKs. Macrophages from WT and IRAK4KDKI mice were treated with Pam3Cys to activate TLR2 or with LPS to activate TLR4, and the cells were lysed after 30 minutes. Activated MAPK proteins were measured by Western blot analysis using antibodies against phosphorylated (activated) proteins p38, ERK1/2 and JNK. While LPS-treated IRAK4KDKI macrophages showed diminished activation of p38 and ERK1/2 compared to WT macrophages, activation of JNK was minimally affected (Figure 2A). In contrast, Pam3Cys treatment of IRAK4KDKI macrophages yielded essentially no activation of any of the three MAPKs when compared to WT cells (Figure 2A). p38, ERK and JNK were all activated in response to the TLR3 agonist Poly I:C in both WT and IRAK4KDKI macrophages, although to a lower extent than was detected with Pam3Cys or LPS (Figure 2A, right panel). Interestingly, p38 and JNK activation were slightly augmented in IRAK4KDKI cells compared to WT. Membranes were re-probed with total p38 or actin to ensure equal loading.
Figure 2. Activation of MAPKs is diminished in IRAK4KDKI macrophages in response to Pam3Cys and LPS.
(A) WT and IRAK4KDKI macrophages were treated with Pam3Cys (P3C; 1 μg/ml) or LPS (100 ng/ml) for 30 min (left panel) or Poly I:C (50 μg/ml) for 30, 60 or 90 min and cell lysates were analyzed by Western blot. Membranes were probed with anti-phospho-p38, anti-phospho-ERK1/2, and anti-phospho-JNK antibodies. Membranes were stripped and re-probed using anti-total p38 (left panel) or anti-actin (right panel) to ensure equal loading. (B) WT and IRAK4KDKI macrophages were stimulated with Pam3Cys (P3C; 1 μg/ml) or LPS (100 ng/ml) for 30 min and cell lysates were analyzed by Western blot using anti-phospho-MNK1, anti-total MNK1 and anti-phospho-MSK1 antibodies. Membranes were re-probed with anti-actin to ensure equal loading. (C) WT and IRAK4KDKI macrophages were treated Pam3Cys (P3C, 1 μg/ml) or LPS (100 ng/ml) for 30 and 60 min and analyzed by Western blot using anti-phospho-eIF4e antibody and re-probed using anti-total eIF4e antibody to ensure equal loading or anti-phospho-histone H3 ser 10 and re-probed using anti-total histone H3 to ensure equal loading. (D) WT and IRAK4KDKI macrophages were treated as in (A) for 1 h. Nuclear extracts were prepared and analyzed by Western blot using anti-p65, anti-p50 and anti-actin (loading control) antibodies (left panel). The same lysates were also incubated overnight with a synthetic biotinylated ODN containing the NF-κB consensus sequence or with a mutated sequence as a negative control (‘NC’). Biotinylated-ODNs were precipitated using streptavidin-conjugated agarose beads, protein was eluted with reducing sample buffer and analyzed by Western blot using anti-p65 or anti-p50 antibodies (right panel). All results are representative of at least three independent experiments.
MNK1 is a kinase that is activated by both p38 and ERK (28, 29). Nearly complete inhibition of MNK1 phosphorylation was observed in IRAK4KDKI macrophages (Figure 2B), suggesting that the TLR4 signaling cascade is defective despite detectable, albeit reduced, upstream activation of p38 and ERK1/2. As expected, due to the lack of p38 and ERK activation observed in Figure 2A, Pam3Cys stimulation in IRAK4KDKI macrophages also failed to activate MNK1 in IRAK4KDKI but not in WT cells (Figure 2B). Membranes were re-probed with anti-MNK1 to ensure the lack of activation is not attributable to a loss of protein expression in IRAK4KDKI cells. Interestingly, MNK1 appears to be differentially expressed in IRAK4KDKI macrophages as demonstrated by the presence of lower molecular weight bands. These bands may represent degradation products of the protein as treatment with the proteasome inhibitor MG-132 reduced their expression (data not shown).
We also sought to determine whether our TLR agonists activate MSK1, a protein kinase that is similarly activated by ERK1/2 (30). As demonstrated by Western blot analysis, MSK1 is slightly activated by the TLR2 agonist Pam3Cys and robustly activated by LPS in WT macrophages, but phosphorylation of MSK1 was not detectable in IRAK4KDKI macrophages (Figure 2B). Together, these data suggest that despite the ability of IRAK4KDKI macrophages to retain the capacity to elicit some early MAPK activation, downstream activation is significantly impaired in response to both TLR2 and TLR4 stimuli.
eIF4e, a protein involved in translational regulation (31), is a known substrate for MNK1 (32). Phospho-eIF4e was increased in response to Pam3Cys and LPS in WT macrophages, albeit with different kinetics, but neither Pam3Cys nor LPS induced significant activation of eIF4e in the IRAK4KDKI cells (Figure 2C). We therefore conclude that a lack of MNK1 activation disrupts downstream signaling events that may ultimately contribute to reduced gene expression in IRAK4KDKI macrophages.
MSK1 phosphorylation of histone H3 at serine 10 leads to upregulation of immediate-early genes that promote cytokine transcriptional activation (i.e, c-jun, c-fos) (33–35). We therefore tested whether a lack of MSK1 activation in IRAK4KDKI macrophages led to a decrease in H3ser10 phosphorylation. Indeed, H3ser10 phosphorylation was decreased in IRAK4KDKI macrophages compared to WT in response to both Pam3Cys and LPS (Figure 2C).
Previous reports demonstrated IRAK4-independent NF-κB activation in response to TLR2 and TLR4 agonists in mice (19, 20), whereas other reports have shown reduced NF-κB activity in response to LPS or IL-1β stimulation in a human patient with IRAK4 deficiency (36). We therefore used both TLR2 and TLR4 agonists to explore the role of IRAK4 kinase activity for NF-κB activation in murine macrophages. As activation of NF-κB prompts its translocation into the nucleus, nuclear lysates from macrophages treated with Pam3Cys or LPS for 1 h were analyzed by Western blot using anti-p65 and anti-p50 antibodies. We found no deficiency of p65 in the nucleus of IRAK4KDKI in response to LPS. In contrast, there was a slight p65 deficiency in the nucleus of IRAK4KDKI macrophages compared to WT in response to Pam3Cys (Figure 2D, left panel). NF-κB p50 expression in the nucleus was diminished in IRAK4KDKI macrophages compared to WT in response to both Pam3Cys and LPS (Figure 2D, left panel).
To determine whether NF-κB binding to its cognate DNA sequence was impaired in IRAK4KDKI macrophages, we performed a co-precipitation assay using a biotinylated oligodeoxynucleotide (ODN) containing the NF-κB consensus binding sequence. The same nuclear extract preparations that were analyzed by Western blot in Figure 2D were incubated with biotinylated NF-κB-specific ODN or a mutated sequence that served as a negative control (‘NC’, LPS-stimulated nuclear extracts were incubated with the mutated sequence to control for non-specific binding). Streptavidin-conjugated beads were used to precipitate the biotinylated ODNs, co-precipitated proteins were resolved by Western blot and probed with anti-p65 and anti-p50 antibodies. There was no decrease in p65 binding in IRAK4KDKI macrophages in response to Pam3Cys or LPS (Figure 2D, right). Similarly, binding of NF-κB p50 was not impaired in Pam3Cys- or LPS-treated IRAK4KDKI cells despite the diminished quantity of p50 in the lysates used. Interestingly, binding of both p65 and p50 were slightly augmented in IRAK4KDKI macrophages in response to LPS when compared to WT cells. This may reflect augmented activation of p65 and p50 (independent of its translocation) and/or the activation of other transcription factors that enhance their DNA binding in IRAK4KDKI cells. Importantly, these data demonstrate that, unlike the complete attenuation of MAPK signaling in response to the MyD88-dependent TLR2 stimulus, Pam3Cys, NF-κB binding remains intact in IRAK4KDKI macrophages. Altogether, these data suggest that while TLR2-induced MAPK activation is completely dependent on IRAK4 kinase activity, TLR2-induced NF-κB activation is only partially dependent on the presence of a functional IRAK4 kinase (as demonstrated by decreased translocation of p65). In contrast, TLR4 activation of MAPKs is only partially dependent on IRAK4 kinase activity, yet the activation of NF-κB is independent of a functional IRAK4 kinase. To examine whether the NF-κB activation in IRAK4KDKI macrophages is a result of activation of an alternative pathway such as activation phosphoinositide 3-kinase (PI3K), we repeated our ODN precipitation in the presence of the PI3K inhibitor LY294002. We found the inhibitor did not decrease NF-κB activation at a dose that inhibited activation of PI3K (data not shown). Thus, the mechanism by which NF-κB achieves full activation despite the lack of IRAK4 kinase activity remains unclear but is independent of PI3K.
IRAK4 kinase activity is required for complete gene induction in macrophages in response to heat-killed bacteria
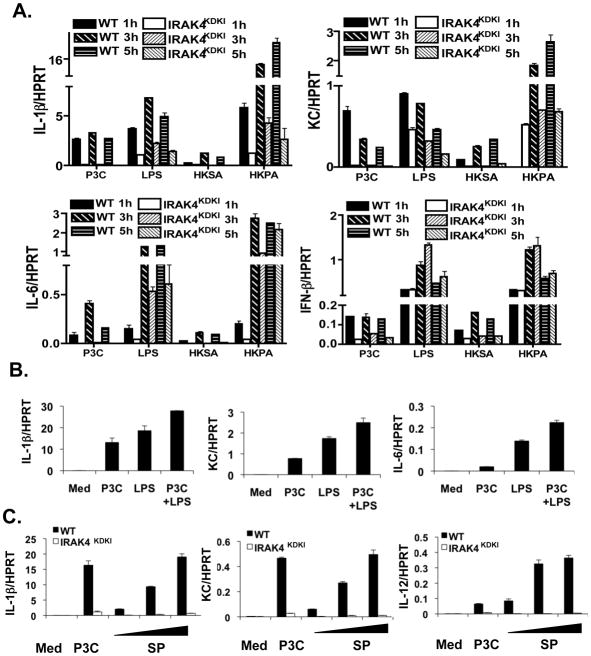
While previous work has largely examined the role of IRAK4 kinase activity in response to purified TLR agonists, few studies have addressed the importance of this kinase in response to whole bacteria. We therefore cultured macrophages from WT and IRAK4KDKI mice and stimulated them with heat-killed S. aureus (HKSA, a Gram positive bacterium) or P. aeruginosa (HKPA, a Gram negative bacterium) for 1, 3 or 5 h. Gene expression was measured by qPCR. As positive controls, we also treated these cells with the purified TLR agonists, Pam3Cys or LPS. We observed differences in the macrophage response to bacteria that were species-dependent. Figure 3A shows that IL-1β, KC and IL-6 mRNA expression in response to HKSA was completely dependent on IRAK4 kinase activity (similar to Pam3Cys) and was weaker than responses induced by LPS or HKPA. The responses to HKPA were much more robust than even that induced by LPS and only partially dependent on the presence of a functional IRAK4 kinase (Figure 3A). To determine whether Pam3Cys and LPS co-treatment elicits a more robust response than either agonist alone, we treated WT macrophages with Pam3Cys, LPS or a combination of the two for 3 h and measured gene expression by qPCR (Figure 3B). Macrophages treated with both Pam3Cys and LPS exhibited slightly higher levels of gene expression but did not synergize. Therefore, the extent to which responses to whole heat-killed Gram positive and Gram negative bacteria are IRAK4-dependent appear to reflect the predominant TLR agonist in the bacteria itself as HKPA expresses both TLR2 and TLR4 PAMPs, while HKSA expresses predominantly TLR2 agonists. The relative amounts of these agonists are likely to play a role in the magnitude of response.
Figure 3. Bacteria-induced cytokine gene expression is impaired in IRAK4KDKI macrophages.
(A) WT and IRAK4KDKI macrophages were treated with Pam3Cys (P3C, 1 μg/ml), LPS (100 ng/ml), heat-killed S. aureus (HKSA, 5 × 107 bacteria) or heat-killed P. aeruginosa (HKPA, 5 × 107 bacteria) for 1, 3 and 5 h. Samples were then analyzed for gene expression by qPCR. All samples were normalized to the housekeeping gene HPRT. Samples were tested in duplicate and each graph represents 3 separate experiments. (B) WT and IRAK4KDKI macrophages were treated with Pam3Cys (P3C, 1 μg/ml), LPS (100 ng/ml), or both for 3 h. Samples were then analyzed for gene expression by qPCR. (C) WT and IRAK4KDKI macrophages were treated with Pam3Cys (P3C, 1 μg/ml) or various concentrations of live S. pneumoniae (SP, MOI 1:1, 0.2:1 and 0.04:1). Samples were then analyzed for gene expression by qPCR. Each sample was normalized to HPRT and each graph represents 2 separate experiments.
S. pneumoniae is the leading killer of patients with deficiencies in IRAK4 (4, 6). We therefore sought to determine what response, if any, was induced in IRAK4KDKI macrophages infected with increasing numbers of S. pneumoniae (SP). Strikingly, there was virtually no mRNA induction of the proinflammatory cytokines IL1-β, KC or IL-12 p40 in IRAK4KDKI macrophages in response to infection with live bacteria despite robust responses observed in WT cells (Figure 3C).
IRAK4KDKI mice are more susceptible to S. pneumoniae infection than WT mice
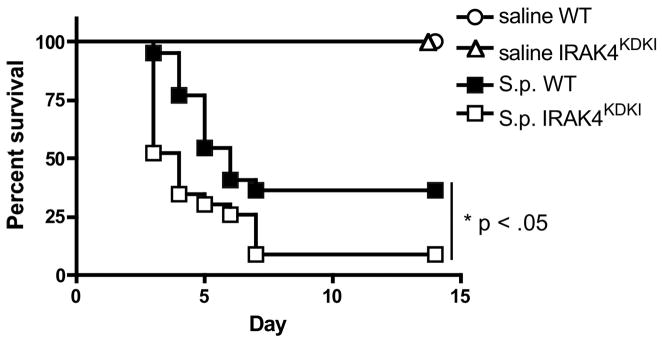
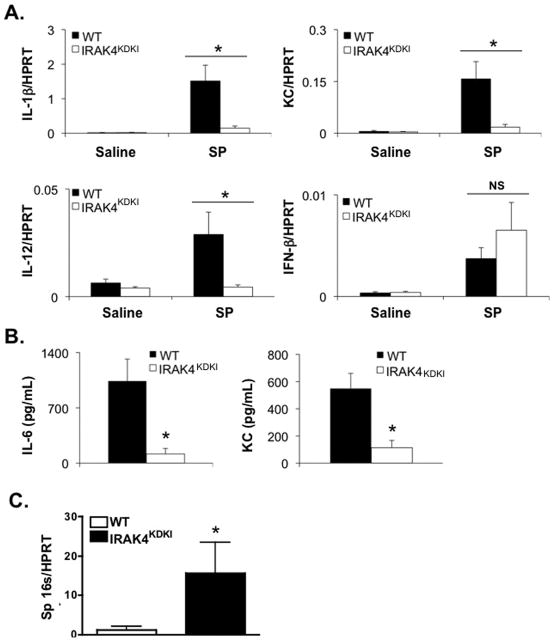
To assess whether IRAK4 kinase activity plays a role in controlling S. pneumoniae infection, WT and IRAK4KDKI mice were infected i.n. with S. pneumoniae and survival was assessed over time. While the majority of both WT and IRAK4KDKI succumbed to S. pneumoniae infection within 7 days, there was a significant increase in the rate of death in IRAK4KDKI mice compared to their WT counterparts (Figure 4; Log-rank test, p = 0.022). To determine whether this increased susceptibility was due to a deficiency in cytokine production, lungs were isolated from S. pneumoniae-infected mice 48 h post-infection, RNA was extracted and qPCR was used to quantify gene induction. As observed in vitro using isolated macrophages, there was a significant decrease in cytokine mRNA production in the IRAK4KDKI lungs when compared to WT in response to S. pneumoniae infection (Figure 5A). Interestingly, in contrast to stimulation of macrophages with Pam3Cys that does not induce IFN-β, infection with S. pneumoniae in vivo induced IFN-β in both WT and IRAK4KDKI mice, suggesting there are non-TLR2, IRAK4-independent ligands present on S. pneumoniae that can induce IFN-β mRNA, an observation that is consistent with other recent studies (37, 38). To characterize further differences in the course of infection between WT and IRAK4KDKI mice, we also measured serum cytokine levels (Figure 5B) and bacterial burden (Figure 5C) 48 h post-infection. IRAK4KDKI mice produced consistently lower levels of IL-6 and KC protein while demonstrating significantly higher bacterial burdens in their lung tissue. Interestingly, we did not observe any striking differences in tissue histology between WT and IRAK4KDKI mice when sacrificed at 48 or 72 h post-infection (data not shown). Both groups of mice showed low levels of inflammatory infiltration at both 48 h and 72 h (a time point when 40% of the IRAK4KDKI mice had already expired due to infection). Together, these data suggest that the absence of a functional IRAK4 kinase decreases proinflammatory cytokine production in response to S. pneumoniae infection while facilitating bacterial replication and thereby increasing the susceptibility of IRAK4KDKI mice to pneumococcal infection.
Figure 4. IRAK4KDKI mice are more susceptible than WT mice to S. pneumoniae infection.
WT or IRAK4KDKI mice were infected i.n. with 1 × 105 S. pneumoniae (S.p.) in a total volume of 30 μl or mock-infected with an equal volume of saline. Survival was assessed over the course of 14 days. Graph is the combined data of 4 separate experiments with a total of 19 mice per mouse strain (S.p. infected) or 4 mice per mouse strain (mock infected) in each group. (*) indicates log-rank test p = 0.022.
Figure 5. Gene induction is impaired in the lungs of S. pneumoniae-infected IRAK4KDKI mice.
(A) WT or IRAK4KDKI mice were infected i.n. with 5 × 105 S. pneumoniae (SP) in a total volume of 30 μl or mock-infected with an equal volume of saline. After 48 h, mice were sacrificed, lungs were harvested, RNA was extracted and gene expression was measured by qPCR. Each sample was normalized to HPRT. Data shown is combined data from at least 3 separate experiments using a total of 4 mice (saline) or at least 9 mice (SP infected) per group. (*) indicates Student’s t test p ≤ .05; (NS) indicates no statistical difference (p > .05). (B, C) WT or IRAK4KDKI mice were infected as in (A). After 48 h, serum cytokines (B) or bacterial rRNA (C) was measured. Data is the combined data of 2 separate experiments using a total of 9 (B) or 6 (C) mice per group. (**) indicates Student’s t test p < .01.
Poly IC:LC (Hiltonol™) improves survival of both WT and IRAK4KDKI infected with S. pneumoniae
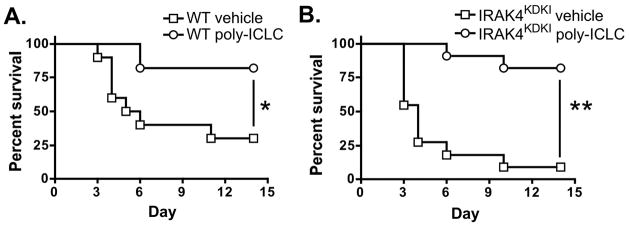
Our in vitro data demonstrates that TLR3 signaling remains intact in IRAK4KDKI macrophages. We therefore sought to determine whether administration of the TLR3 agonist Poly IC:LC (Hiltonol™), a stabilized Poly I:C, could improve survival in IRAK4KDKI infected with a lethal dose of S. pneumoniae. In these studies, vehicle only or 100 μg of Poly IC:LC was administered intranasally at the time of infection in both WT and IRAK4KDKI mice. There was a significant increase in survival of infected mice treated with Poly IC:LC compared to those treated with vehicle only (Figure 6; WT, log-rank test, p = 0.0097; IRAK4KDKI, log-rank test, p = 0.0001). Thus stimulation of TLR3, a MyD88- and IRAK4-independent innate immune pathway, provides a possible therapeutic strategy to boost immunity against bacterial pathogens in patients lacking key signaling components of MyD88- and/or IRAK4-dependent signaling pathways.
Figure 6. Poly IC:LC (Hiltonol™) treatment improves survival of mice infected with S. pneumoniae.
WT (A) or IRAK4KDKI (B) mice were infected i.n. with 1 × 105 S. pneumoniae in a total volume of 50 μl of Poly IC:LC (100 μg/mouse, open circle) or saline (vehicle, open square). Survival was assessed over the course of 14 days. The data represent the combined data of 3 separate experiments with a total of 11 mice per group (10 mice for WT vehicle group). (*) indicates log-rank test, p = 0.0097; (**) indicates log-rank test, p = 0.0001.
Discussion
TLR signaling is a critical activator of immune defense during infection. In this study, we have clarified the importance of IRAK4 kinase activity in generating full responses to Pam3Cys and LPS, agonists for TLR2 and TLR4, respectively. Importantly, cytokine production in IRAK4KDKI macrophages was also impaired in response to both heat-killed and live bacteria. We observed the most severe deficiency in IRAK4KDKI macrophages when these cells were infected with S. pneumoniae, reminiscent of the phenotype observed in patients with IRAK4 mutations (4, 6, 36). While early signaling events such as phosphorylation of p38, ERK1/2 and JNK were completely abrogated in response to Pam3Cys, IRAK4KDKI macrophages retained some ability to activate these proteins in response to LPS. However, activation of MNK1 and MSK1 in response to LPS was diminished in IRAK4KDKI macrophages suggesting that IRAK4 also plays a role downstream of p38, ERK1/2 and JNK activation in response to TLR4 agonists. Proinflammatory cytokine mRNA and protein expression remained partially intact in IRAK4KDKI macrophages in response to LPS, suggesting that early MAPK signaling is sufficient for upregulation of gene expression despite diminished downstream activity. However, our data also indicate that LPS activation of MAPKs is at least partially IRAK4-dependent as we observed diminished activation of their downstream substrates (such as MNK1 and MSK1, Figure 2B).
Our data support the hypothesis that IRAK4 kinase activity is most essential for MyD88-dependent TLR signaling. TLR2 is entirely MyD88-dependent and we show nearly a complete loss of signaling in response to the TLR2 agonist Pam3Cys as demonstrated by lowered cytokine mRNA expression (Figures 1 and 3) and loss of MAPK activation (Figure 2). Similar results were reported by Kawagoe et al. (19) using the TLR2 agonist MALP2, a TLR2/6 agonist (Pam3Cys is a TLR1/2 agonist). However, NF-κB activation in response to a TLR2 agonist was not diminished in IRAK4KDKI macrophages compared to WT (Figure 2D), suggesting that IRAK4 kinase-dependent activation of MAPKs is essential for upregulation of proinflammatory cytokine gene expression. We also report that TLR3 signaling, which is completely independent of MyD88, is not affected by the loss of IRAK4 kinase activity as demonstrated by equal cytokine mRNA expression in both WT and IRAK4KDKI macrophages in response to the TLR3 agonist Poly I:C, a result that agrees with previous studies (19, 20). However, currently published data does not agree on the role of IRAK4 kinase activity in response to the TLR4 agonist LPS. As LPS can propagate both MyD88-dependent and MyD88-independent (TRIF-dependent) signaling pathways, it is of critical importance that induction of genes that are pathway-dependent be considered separately. Our data confirm that LPS signaling is dependent on IRAK4 kinase activity as IRAK4KDKI macrophages exhibit diminished MyD88-dependent cytokine production in response to LPS (Figure 1). However, MyD88-indpendent genes, such as IFN-β and IP-10, are not affected by the loss of IRAK4 kinase activity. Furthermore, the loss in gene expression cannot be attributed to a loss of early signaling events such as activation of p38, ERK1/2 or JNK, as is the case for TLR2 signaling. Our results show LPS induced activation of MAPK in the absence of IRAK4 kinase activity, a result that agrees with previous studies showing MAPK activation in both IRAK4KDKI and IRAK4−/− macrophages (20, 43–44). Together, these data support a model in which LPS can induce MAPK activation completely independent of IRAK4 protein. This may occur via the LPS-induced MyD88-independent/TRIF-dependent pathway as MAPK activation by Poly I:C, a TLR3 agonist that also signals in a MyD88-independent/TRIF-dependent fashion, is not affected in cells lacking IRAK4 kinase activity (Figure 3A). Indeed, even MyD88−/− macrophages demonstrate MAPK activation in response to LPS (43, 45–46). Importantly, both MNK1 and MSK1, kinases that are activated downstream of activated MAPKs, were deficient in IRAK4KDKI macrophages in response to LPS. To confirm a functional consequence to this loss of kinase activation, we demonstrated diminished activation of their downstream substrates as well, i.e., eIF4e for MNK1 and histone H3 serine 10 for MSK1. Of particular note, the observation that LPS-induced tolerance was not affected IRAK4KDKI macrophages, despite the observed decrease in MyD88-dependent signaling, suggests either that the partial response to LPS observed in such macrophages achieves a threshold sufficient to elicit the chromatin remodeling suggested to underlie tolerance (39) or that tolerance induction is independent of IRAK4 activity. Studies are ongoing to determine the extent to which IRAK4 contributes to endotoxin tolerance.
Kim et al. suggest that while LPS-induced mRNA expression was not greatly affected in IRAKKDKI macrophages, protein levels were severely deficient due to a loss of mRNA stability and thus a reduction of translation of these cytokines (20). While our data clearly demonstrate diminished mRNA expression, our observation that eIF4e phosphorylation is also severely diminished in response to LPS (Figure 2C) suggests there may also be a secondary loss of translational activity for LPS-induced, MyD88-dependent genes. eIF4e plays a role in maintaining mRNA stability leading to efficient mRNA translation (31). If eIF4e activation were lost, as we found in response to both TLR2 and TLR4 agonists (Figure 2C), then it would be expected that translational activity would also be reduced. Interestingly, a similar pattern of MAPK activation (i.e., no defect in activation of p38 or ERK with downstream impairment of MNK1 and eIF4e activation) has been demonstrated in IRAK2-deficient macrophages in response to LPS (40). This suggests that activation of MNK1 and its downstream target, eIF4e, requires both the presence of IRAK2 as well as the kinase activity of IRAK4 (potentially to phosphorylate/activate IRAK2). In addition, we demonstrate little activation of MSK1 in response to LPS in IRAKKDKI macrophages. MSK1 phosphorylates histone H3 at serine 10 leading to increased gene expression, most notably of early immediate genes (33–35). Here we show MSK1 activation and subsequent phosphorylation of histone H3 serine 10 are both diminished in LPS-stimulated IRAK4KDKI macrophages, thus providing evidence for its potential role in the partial defect in the MAPK signaling cascade seen in response to LPS.
Patients with IRAK4 deficiency are most susceptible to S. pneumoniae infection. Similarly, we observed increased susceptibility to S. pneumonia infection in IRAK4KDKI mice. These mice died faster than WT mice, accompanied by decreased cytokine gene expression, decreased protein levels and increased bacterial burden in their lungs compared to WT mice (Figures 5 and 6). Combined with our in vitro data, this presents a feasible experimental model with which to explore therapeutic interventions that may compensate for the loss of IRAK4 signaling. To this end, our data show the IRAK4-independent, TLR3 agonist Poly IC:LC significantly increased the survival of S. pneumoniae-infected mice. This data is in contrast with Nakamura et al who found increased S. pneumoniae colonization after administration of repeated doses of Poly IC:LC (41). However, that report did not determine the survival rate of mice receiving Poly IC:LC treatment and there was no significant increase in colonization when Poly IC:LC was administered less frequently. Interestingly, the increased S. pneumoniae colonization was dependent on Poly IC:LC induction of IFN-β. As Poly IC:LC is a potent inducer of IFN-β in vivo (42), we are currently investigating the requirement of IFN-β to confer Poly IC:LC protection in our model. However, our data showing that IFN-β production in response to S. pneumoniae infection is not inhibited in IRAK4KDKI mice (Figure 5) suggests the mechanism may be independent of IFN-β induction. With further studies, such intervention could prove useful in patients with mutations in the MyD88/IRAK4-dependent pathway as our in vitro data suggest the activation of MAPKs with stimuli that are TLR/MyD88/IRAK4-independent could potentially compensate for deficiencies in early signaling proteins of the TLR cascade. With NF-κB signaling relatively intact in TLR2- or TLR4-stimulated IRAK4KDKI macrophages, our data would support the concept of increasing activation of the MAPK pathway and targeting its complete restoration in the absence or mitigation of IRAK4 kinase activity.
Acknowledgments
This work was supported by NIH grant AI18797 to S.N.V.
References
- 1.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 3.Ku CL, Picard C, Erdos M, Jeurissen A, Bustamante J, Puel A, von Bernuth H, Filipe-Santos O, Chang HH, Lawrence T, Raes M, Marodi L, Bossuyt X, Casanova JL. IRAK4 and NEMO mutations in otherwise healthy children with recurrent invasive pneumococcal disease. J Med Genet. 2007;44:16–23. doi: 10.1136/jmg.2006.044446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ku CL, von Bernuth H, Picard C, Zhang SY, Chang HH, Yang K, Chrabieh M, Issekutz AC, Cunningham CK, Gallin J, Holland SM, Roifman C, Ehl S, Smart J, Tang M, Barrat FJ, Levy O, McDonald D, Day-Good NK, Miller R, Takada H, Hara T, Al-Hajjar S, Al-Ghonaium A, Speert D, Sanlaville D, Li X, Geissmann F, Vivier E, Marodi L, Garty BZ, Chapel H, Rodriguez-Gallego C, Bossuyt X, Abel L, Puel A, Casanova JL. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 2007;204:2407–2422. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picard C, Casanova JL, Puel A. Infectious diseases in patients with IRAK-4, MyD88, NEMO, or IkappaBalpha deficiency. Clin Microbiol Rev. 2011;24:490–497. doi: 10.1128/CMR.00001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picard C, von Bernuth H, Ghandil P, Chrabieh M, Levy O, Arkwright PD, McDonald D, Geha RS, Takada H, Krause JC, Creech CB, Ku CL, Ehl S, Marodi L, Al-Muhsen S, Al-Hajjar S, Al-Ghonaium A, Day-Good NK, Holland SM, Gallin JI, Chapel H, Speert DP, Rodriguez-Gallego C, Colino E, Garty BZ, Roifman C, Hara T, Yoshikawa H, Nonoyama S, Domachowske J, Issekutz AC, Tang M, Smart J, Zitnik SE, Hoarau C, Kumararatne DS, Thrasher AJ, Davies EG, Bethune C, Sirvent N, de Ricaud D, Camcioglu Y, Vasconcelos J, Guedes M, Vitor AB, Rodrigo C, Almazan F, Mendez M, Arostegui JI, Alsina L, Fortuny C, Reichenbach J, Verbsky JW, Bossuyt X, Doffinger R, Abel L, Puel A, Casanova JL. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore) 2010;89:403–425. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suhir H, Etzioni A. The role of Toll-like receptor signaling in human immunodeficiencies. Clin Rev Allergy Immunol. 2010;38:11–19. doi: 10.1007/s12016-009-8135-0. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira-Nascimento L, Massari P, Wetzler LM. The Role of TLR2 in Infection and Immunity. Frontiers in Immunology. 2012;3:79. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 10.Poltorak A. Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 Gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Z, Zamanian-Daryoush M, Nie H, Silva AM, Williams BR, Li X. Poly(I-C)-induced Toll-like receptor 3 (TLR3)-mediated activation of NFkappa B and MAP kinase is through an interleukin-1 receptor-associated kinase (IRAK)-independent pathway employing the signaling components TLR3-TRAF6-TAK1-TAB2-PKR. J Biol Chem. 2003;278:16713–16719. doi: 10.1074/jbc.M300562200. [DOI] [PubMed] [Google Scholar]
- 12.Watters TM, Kenny EF, O’Neill LA. Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins. Immunol Cell Biol. 2007;85:411–419. doi: 10.1038/sj.icb.7100095. [DOI] [PubMed] [Google Scholar]
- 13.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4- IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flannery S, Bowie AG. The interleukin-1 receptor-associated kinases: critical regulators of innate immune signalling. Biochem Pharmacol. 2010;80:1981–1991. doi: 10.1016/j.bcp.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med. 2003;197:263–268. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki N, Saito T. IRAK-4--a shared NF-kappaB activator in innate and acquired immunity. Trends Immunol. 2006;27:566–572. doi: 10.1016/j.it.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki N, Suzuki S, Duncan GS, Millar DG, Wada T, Mirtsos C, Takada H, Wakeham A, Itie A, Li S, Penninger JM, Wesche H, Ohashi PS, Mak TW, Yeh WC. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature. 2002;416:750–756. doi: 10.1038/nature736. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki N, Suzuki S, Yeh WC. IRAK-4 as the central TIR signaling mediator in innate immunity. Trends Immunol. 2002;23:503–506. doi: 10.1016/s1471-4906(02)02298-6. [DOI] [PubMed] [Google Scholar]
- 19.Kawagoe T, Sato S, Jung A, Yamamoto M, Matsui K, Kato H, Uematsu S, Takeuchi O, Akira S. Essential role of IRAK-4 protein and its kinase activity in Toll-like receptor-mediated immune responses but not in TCR signaling. J Exp Med. 2007;204:1013–1024. doi: 10.1084/jem.20061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim TW, Staschke K, Bulek K, Yao J, Peters K, Oh KH, Vandenburg Y, Xiao H, Qian W, Hamilton T, Min B, Sen G, Gilmour R, Li X. A critical role for IRAK4 kinase activity in Toll-like receptor-mediated innate immunity. J Exp Med. 2007;204:1025–1036. doi: 10.1084/jem.20061825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraczek J, Kim TW, Xiao H, Yao J, Wen Q, Li Y, Casanova JL, Pryjma J, Li X. The kinase activity of IL-1 receptor-associated kinase 4 is required for interleukin-1 receptor/toll-like receptor-induced TAK1-dependent NFkappaB activation. J Biol Chem. 2008;283:31697–31705. doi: 10.1074/jbc.M804779200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lye E, Dhanji S, Calzascia T, Elford AR, Ohashi PS. IRAK-4 kinase activity is required for IRAK-4-dependent innate and adaptive immune responses. Eur J Immunol. 2008;38:870–876. doi: 10.1002/eji.200737429. [DOI] [PubMed] [Google Scholar]
- 23.Song KW, Talamas FX, Suttmann RT, Olson PS, Barnett JW, Lee SW, Thompson KD, Jin S, Hekmat-Nejad M, Cai TZ, Manning AM, Hill RJ, Wong BR. The kinase activities of interleukin-1 receptor associated kinase (IRAK)-1 and 4 are redundant in the control of inflammatory cytokine expression in human cells. Mol Immunol. 2009;46:1458–1466. doi: 10.1016/j.molimm.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Sklar MD, Tereba A, Chen BD, Walker WS. Transformation of mouse bone marrow cells by transfection with a human oncogene related to c-myc is associated with the endogenous production of macrophage colony stimulating factor 1. J Cell Physiol. 1985;125:403–412. doi: 10.1002/jcp.1041250307. [DOI] [PubMed] [Google Scholar]
- 25.McIntire FC, Sievert HW, Barlow GH, Finley RA, Lee AY. Chemical, physical, biological properties of a lipopolysaccharide from Escherichia coli K-235. Biochemistry. 1967;6:2363–2372. doi: 10.1021/bi00860a011. [DOI] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 27.Dobrovolskaia MA, Vogel SN. Toll receptors, CD14, and macrophage activation and deactivation by LPS. Microb Infect. 2002;4:903–914. doi: 10.1016/s1286-4579(02)01613-1. [DOI] [PubMed] [Google Scholar]
- 28.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waskiewicz AJ. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy CE, Campbell DG, Deak M, Bloomberg GB, Arthur JSC. Biochem J. 2005;387:507–517. doi: 10.1042/BJ20041501. C.E. McCoy and others - Regulatory phosphorylation of MSK1 2012: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhoads RE. eIF4E: new family members, new binding partners, new roles. J Biol Chem. 2009;284:16711–16715. doi: 10.1074/jbc.R900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the Cap-Binding Protein Eukaryotic Translation Initiation Factor 4E by Protein Kinase Mnk1 In Vivo. 2012 doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clayton AL, Rose S, Barratt MJ, Mahadevan LC. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 2000;19:3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lau PN, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci U S A. 2011;108:2801–2806. doi: 10.1073/pnas.1012798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 1999;18:4779–4793. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medvedev AE, Lentschat A, Kuhns DB, Blanco JC, Salkowski C, Zhang S, Arditi M, Gallin JI, Vogel SN. Distinct mutations in IRAK-4 confer hyporesponsiveness to lipopolysaccharide and interleukin-1 in a patient with recurrent bacterial infections. J Exp Med. 2003;198:521–531. doi: 10.1084/jem.20030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koppe U, Hogner K, Doehn JM, Muller HC, Witzenrath M, Gutbier B, Bauer S, Pribyl T, Hammerschmidt S, Lohmeyer J, Suttorp N, Herold S, Opitz B. Streptococcus pneumoniae stimulates a STING- and IFN regulatory factor 3-dependent type I IFN production in macrophages, which regulates RANTES production in macrophages, cocultured alveolar epithelial cells, and mouse lungs. J Immunol. 2012;188:811–817. doi: 10.4049/jimmunol.1004143. [DOI] [PubMed] [Google Scholar]
- 38.Parker D, Martin FJ, Soong G, Harfenist BS, Aguilar JL, Ratner AJ, Fitzgerald KA, Schindler C, Prince A. Streptococcus pneumoniae DNA initiates type I interferon signaling in the respiratory tract. MBio. 2011;2:e00016–11. doi: 10.1128/mBio.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 40.Wan Y, Xiao H, Affolter J, Kim TW, Bulek K, Chaudhuri S, Carlson D, Hamilton T, Mazumder B, Stark GR, Thomas J, Li X. Interleukin-1 receptor-associated kinase 2 is critical for lipopolysaccharide-mediated post-transcriptional control. J Biol Chem. 2009;284:10367–10375. doi: 10.1074/jbc.M807822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura S, Davis KM, Weiser JN. Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J Clin Invest. 2011;121:3657–3665. doi: 10.1172/JCI57762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caskey M, Lefebvre F, Filali-Mouhim A, Cameron MJ, Goulet JP, Haddad EK, Breton G, Trumpfheller C, Pollak S, Shimeliovich I, Duque-Alarcon A, Pan L, Nelkenbaum A, Salazar AM, Schlesinger SJ, Steinman RM, Sekaly RP. Synthetic double-stranded RNA induces innate immune responses similar to a live viral vaccine in humans. J Exp Med. 2011;208:2357–2366. doi: 10.1084/jem.20111171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliveira FS, Carvalho NB, Brandao AP, Gomes MT, de Almeida LA, Oliveira SC. Interleukin-1 receptor-associated kinase 4 is essential for initial host control of Brucella abortus infection. Infect Immun. 2011;79:4688–4695. doi: 10.1128/IAI.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koziczak-Holbro M, Glück A, Tschopp C, Mathison JC, Gram H. IRAK-4 kinase activity-dependent and -independent regulation of lipopolysaccharide-inducible genes. Eur J Immunol. 2008;38:788–796. doi: 10.1002/eji.200737886. [DOI] [PubMed] [Google Scholar]
- 45.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 46.Kim L, Butcher BA, Lee CW, Uematsu S, Akira S, Denkers EY. Toxoplasma gondii genotype determines MyD88-dependent signaling in infected macrophages. J Immunol. 2006;177:2584–2591. doi: 10.4049/jimmunol.177.4.2584. [DOI] [PubMed] [Google Scholar]