Abstract
We report the development of chemically-modified peptide nucleic acids (PNAs) as probes for qualitative and quantitative detection of DNA. The remarkable stability of PNAs toward enzymatic degradation makes this class of molecules ideal to develop as part of a diagnostic device that can be used outside of a laboratory setting. Using an enzyme-linked reporter assay, we demonstrate that excellent levels of detection and accuracy for anthrax DNA can be achieved using PNA probes with suitable chemical components designed into the probe. In addition, we report on DNA-templated crosslinking of PNA probes as a way to preserve genetic information for repetitive and subsequent analysis. This report is the first detailed examination of the qualitative and quantitative properties of chemically-modified PNA for nucleic acid detection and provides a platform for studying and optimizing PNA probes prior to incorporation into new technological platforms.
Introduction
Nucleic acid testing is highly specific and often provides definitive identification of a disease or pathogen. Methods to detect nucleic acid sequences are dominated by PCR,1 but applying PCR-based techniques outside of a modern laboratory is challenging. Samples collected in the field, for instance, typically contain inhibitors of the polymerases used in PCR amplification.2 These inhibitors can be naturally occurring (such as humic acids, urea, heme, Ca2+, proteinases, and/or polysaccharides) or come from items used to collect the samples (namely glove powder, NaCl, KCl, EDTA, SDS, and phenol).3 These inhibitors can be difficult to remove from samples. While there are other non-PCR-based detection platforms for nucleic acid analysis,1,4-7 they are similarly limited to a laboratory environment. To overcome the problems of stability to outside contamination, we are developing peptide nucleic acid (PNA) probes for use in a diagnostic system that does not rely on PCR.
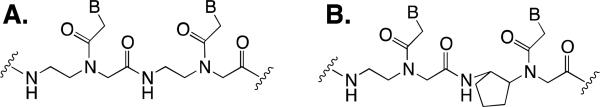
PNA binds to DNA using Watson-Crick basepairing, and it binds with greater stability and selectivity compared to a complementary DNA sequence (Figure 1A).8-10 In addition, PNAs are resistant to enzymatic proteolysis because of the aminoethylglycyl (aeg) backbone.11 While aegPNA is widely available and has been incorporated into some detection platforms, the development of chemically-modified PNA derivatives with enhanced binding properties12 provides many more opportunities for developing a diagnostic device. In theory, PNA-based nucleic acid probes can be coupled with emerging technological platforms for detection to afford a device that may be used outside of laboratory settings.1,13,14 The use of PNAs for nucleic acid testing should provide a high degree of stability to any diagnostic device and PNA probes could in principle be used to detect many different pathogenic agents and diseases (such as HIV, plague, influenza, or botulism). In our own work, we have found that incorporation of a cyclopentane ring into the backbone of aegPNA improves binding to complementary DNA and improves sequence specificity (Figure 1B).15 Furthermore, we have previously demonstrated that a single cyclopentane ring in a nucleic acid capture probe attached to a plastic surface can dramatically enhance detection. 16
Figure 1.
A. General structure of aegPNA showing two nucleobases (B = A, T, G, or C). This is a partial structure that may be extended in either direction to include a specific number of nucleobases. B. Structure of cyclopentane-modified PNA.
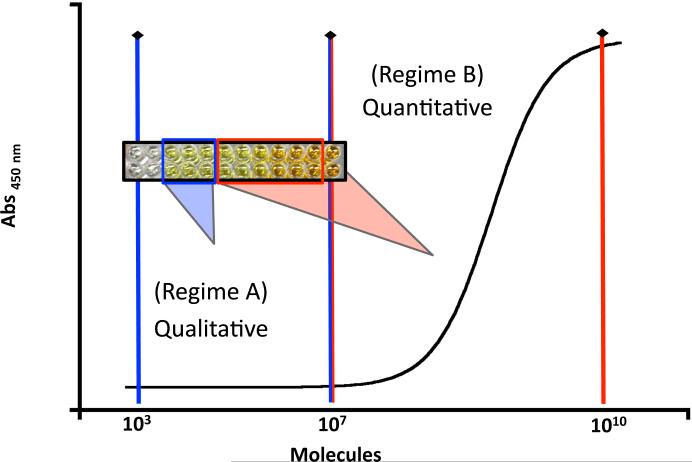
Other PNA analogs are similarly available, but there has been no rigorous evaluation of any modified PNAs to determine whether they can indeed be incorporated into a diagnostic device. The present report focuses on evaluating cyclopentane-PNA probes for detection of anthrax DNA, with a focus on evaluating the ability of these probes to provide both qualitative and quantitative detection of nucleic acids. In qualitative detection, the presence of an analyte can be confirmed but the amount remains unknown. In this case, there is a positive signal over background that corresponds to the presence of target DNA, but there is no variation in signal that correlates with the change in the amount of DNA. This situation is illustrated in Figure 2 (regime A) where the signal remains constant between 103 and 107 DNA molecules. Detection in regime A shows that the target DNA is present but the specific amount cannot be determined. In quantitative detection, both the presence and amount of an analyte can be determined. In this case, the signal varies proportionally with the amount of DNA so that the quantity can be determined. As shown in Figure 2 (regime B), the intensity of the readout signal correlates directly with the amount of DNA provided there are between 107 and 1010 molecules present. Determining both regimes is important for any diagnostic device and may need to be adjusted depending on the application.
Figure 2.
Quantitative vs. qualitative detection regimes for PNA probes. In regime A, only the presence of DNA, but not the amount can be detected. In regime B, the presence and amount of DNA can be detected.
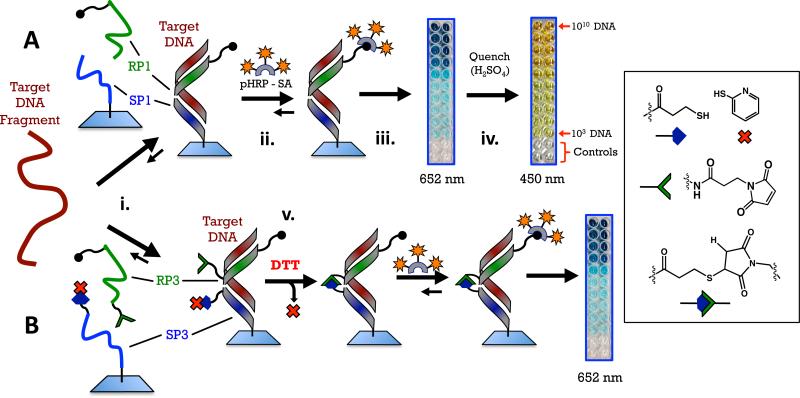
For the current study, we evaluate both qualitative and quantitative regions for nucleic acid detection and investigate two types of sandwich hybridization assays using PNA probes. The first type of sandwich hybridization uses two PNA probes (SP1 and RP1) that recognize adjacent portions of a target DNA sequence (Figure 3A). The surface probe (SP) is covalently attached via an amide bond to a plastic surface, while the reporter probe (RP) is free in solution. In addition, the RP has six biotin groups covalently attached to one end. If the target DNA is present, SP1 and RP1 both bind to DNA and a non-covalent complex that consists of SP1+RP1+DNA is formed on the plastic surface. Because RP1 contains several biotins, the presence of these groups can be determined using an enzyme that is covalently linked to streptavidin (for this study we use polyhorseradishperoxidasestreptavidin, pHRP-SA). The second type of sandwich hybridization uses PNA probes SP3 and RP3 that possess chemically reactive groups on the ends of the probes that will form a crosslink upon binding to the target DNA. These probes are otherwise identical to the SP1 and RP1 probes. The ability to covalently crosslink the SP3 and RP3 probes after recognition of target was developed to increase the stability of the complex with DNA on the surface of the plastic surface. Similar types of crosslinking strategies have been employed with PNA and DNA to enhance detection capabilities.17-20 Ultimately, this work establishes that PNA possesses properties suitable for use in detection platforms should incentivize incorporation of PNA into emerging technologies for detection.21
Figure 3.
Schemes for the two detection methods using PNA probes. A. Sandwich: PNA probes RP1 and SP1 recognize a 27-base sequence of anthrax DNA and form a three-component, non-covalent complex on a plastic surface (step i). Probe RP1 has six biotins on the C-terminal. If the target DNA is present and the complex forms, a color can be developed by introducing a polyhorseradish peroxidase+streptavidin conjugate followed by a solution of tetramethylbenzidine and peroxide (step ii). Enzymatic oxidation initially produces a blue color at 652 nm (step iii), quenching the enzyme with sulfuric acid produces a yellow color at 450 nm (step iv). B. Crosslinked sandwich: PNA probes RP3 and SP3 have reactive maleimide and thiol groups on their N- and C-terminal ends, respectively. A disulfide with 2-pyridylthiol is used as a protecting group for the pendant thiol on SP3. This group is reduced with dithiothreitol immediately following hybridization of the PNA probes with DNA to reveal a pendant thiol that reacts with maleamide on the adjacent PNA (step v).
Experimental
Nucleic Acid Detection Protocol
See the Supporting Information for details on the buffers.
Plate Preparation
Surface probe (SP) was dissolved in immobilization buffer (IB) [1.0 μM] with 100 μL added to each well of the plate. Some wells of the plate, designated as blanks, were left untouched throughout the process to serve as a blank against the rest of the plate. The plate was sealed with sealing tape for multiwell plates and stirred on a plate shaker (600-700 rpm) for 2 h at RT. Then 50 μL of capping solution (CAP) was added to the modified wells to give a final volume of 150 μL. This was again stirred at RT on a plate shaker for 30 min. Modified wells were aspirated and washed four times with 300 μL 1 × phoshphate buffer with Tween 20 (PBST) and four times with 1 × phosphate buffer (PBS). A final addition of 250 μL of 1 × PBS was used to store the wells until used in experiments. When the assay was ready to be performed, the 1 × PBS was aspirated and 200 μL blocking buffer with salmon sperm DNA (BLBs) was added to the wells. The plate was sealed and incubated with shaking for 30 min at 37 °C. The wells were immediately aspirated and used for sample detection.
DNA Sample Preparation at Different Concentrations for Detection
All samples were prepared in glass vials. The final volume for all samples used for DNA detection was 600 μL. Each of these samples had RP concentration fixed at 15 nM. The specific concentration of the target DNA for detection was prepared by a serial dilution of a stock 50 nM solution of the DNA target (TS) in 1 × PBS buffer prepared from the commercial DNA obtained from IDT. The initial (and highest concentration) DNA sample for detection was the first sample made, using the desired amount of DNA from the 50 nM stock solution and diluting with a solution of the reporter probe (RP, 15 nM) in 1 × PBS buffer. To dilute further from the initial DNA detection sample, a desired volume of DNA solution was removed and further diluted with the 15 nM reporter probe solution in 1 × PBS buffer. For example, to dilute an initial 450 pM DNA sample solution by three times, 200 μL of the initial DNA sample solution was removed and added to 400 μL of 15 nM reporter probe solution in 1 × PBS buffer to give a final DNA concentration of 150 pM. Once the final DNA concentration was obtained after dilution, the samples were heated to 95 °C in a sand bath for 5 min and then snap-cooled by placing the vials directly into an ice bath for 5 min. Then, the samples were directly used for DNA detection by removing 100 μL of each sample and individually adding the DNA solution to one well on the prepared 96-well plate (using a 200 μL pipettor). To construct a concentration dependent response curve, 10 different DNA concentrations were prepared and each concentration was prepared in duplicate. Two wells of each plate were dedicated to detection of one DNA concentration. After the samples were added to the wells, the plate was sealed and incubated with shaking at 37 °C for 3 h. Each well was aspirated following incubation, and then washed with 300 μL 1 × PBST for four times followed by 300 μL 1 × PBS for four times. If crosslinked PNA's were used, 50 μL of 3 mM dithiothreitol (DTT) (in 1 × PBS) was added to each well following the 3 h incubation to give a final volume of 150 μL and a final DTT concentration of 1 mM. The plate was incubated once again, with shaking again, at 37 °C for 30 min and then aspirated and washed as described above.
ELISA Protocol
The wells were incubated with 200 μL of blocking buffer (BLB). The plate was sealed and shaken at 37 °C for 30 min. Wells were then aspirated, followed by addition of 100 μL of 0.1 μg/mL pHRP-SA in BLB. The plate was sealed and incubated for 20 min at RT before aspirating. The wells were aspirated and followed a washing procedure that consisted of 300 μL four times of 1 × PBST and four times of 1 × PBS.
ELISA Readout
Following washes, 100 μL 1-Step™ Ultra tetramethylbenzidine (TMB) solution was added via a multichannel pipettor to facilitate nearly simultaneous initial starting points for all wells. The plate was immediately placed in a Molecular Devices (Sunnyvale, CA) SpectraMax M5 multi-mode microplate reader and monitored at 652 nm over the course of 30 min at RT. The plate was then removed, followed by addition of 50 μL of 2 M H2SO4 and mixed by hand for 10 s to quench the enzyme reaction. The plate was then returned to the plate reader and a final reading at 450 nm was performed. Plates were sealed with film following reading and placed in the refrigerator.
Plate Reruns
For the time-course assays performed after the initial assay, these plates were not quenched with H2SO4 after the initial assay. These plates were stored in a 4 °C refrigerator in the original unquenched TMB assay solution until assayed again. When the assay plate was rerun, the TMB solution was aspirated followed by 1 × PBST (4×) and 1 × PBS (4×) washes. The plate wells were blocked with 200 μL BLB for 30 min at 37 °C with shaking. 100 μL of 0.1 μg/mL pHRP-SA solution was added to the wells and incubated with shaking for 20 min at RT. The pHRP solution was aspirated, followed by 1 × PBST (4×) and 1 × PBS (4×) washes. Then 100 μL 1-Step™ Ultra TMB solution was added via a multichannel pipettor to all wells. The plate was immediately placed in a Molecular Devices (Sunnyvale, CA) SpectraMax M5 multi-mode microplate reader and monitored at 652 nm over the course of 30 min at RT.
Analysis of Results
Linear Regression Analysis of Absorbance (652 nm) Kinetics
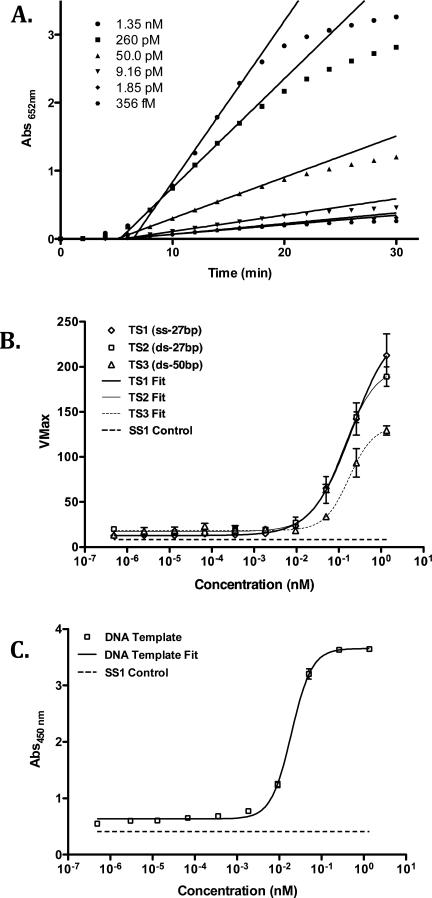
The absorbance was monitored at 2 min intervals over 30 min (16 pts) at each concentration of target nucleic acid. SoftMax Pro software was used to analyze the absorbance at 652 nm with respect to time (min). Linear regression analysis was applied to these results, ignoring any initial lag phase, using five points to return the steepest velocity (“VMax” - mAbs652 nm/min) over the course of the experiment. See Figure 4A & B for graphic examples.
Figure 4.
Detection of DNA using PNA probes SP1 and RP1. A. Absorbance data (652 nm) for detection of TS1 DNA over a range of concentrations and linear regression analyses of maximum velocities (VMax) for each concentration. B. Signal response plot (VMax vs. [DNA]) for each DNA in Table 2 and curve fits for each plot. C. Signal response plot (Absorbance at 450 nm vs. [TS1 DNA]) obtained after quenching of enzymatic oxidation with H2SO4.
Nonlinear Regression Analysis of Kinetic and Quenched Assay Results
The calculated VMax results were used as response values and plotted with respect to target nucleic acid concentration using Prism 5.0 software. These kinetic assay results were analyzed using 4-parameter logistic (4-PL) models. The resulting fits are graphically represented in Figure 4C and results are listed under Table S4. The protocol employing quenched TMB product used end point absorbance values at 450 nm plotted versus nucleic acid concentration and fit to a 4-PL model as for the kinetic data at 652 nm.
Results & Discussion
Detection of pag anthrax DNA via sandwich hybridization
Detection assays ideally confirm the presence of an agent and reveal the amount of the agent that is present. One way to identify the genetic material of biological agents uses Watson-Crick basepairing of a target DNA with two synthetic, nucleic acid probes to form a sandwich-hybridized complex that can be detected by one of several methods.22 In our work, the nucleic acid probes (Table 1) are peptide nucleic acids (PNAs), and one of the PNAs has a cyclopentane group added to one position in the middle of the sequence.23,24 Based on our prior work, PNA probes SP1 and RP1 have similar melting temperatures to their complimentary DNA sequences and afforded good initial detection results in the first iteration of our detection strategy.15
Table 1.
PNA Probes
| Probe | Sequence |
|---|---|
| SP1 | H2N-(mPEG)5-ATCCTTATcypCAATATT-CONH2 |
| SP2 | Ac-(mPEG)2-ATCCTTATCAATATT-Lys-(mPEG-Cys-NH2)-CONH2 |
| SP3 | H2N-(mPEG)2-ATCCTTATcypCAATATT-Lys(mPEG-SPDP)-CONH2 |
| RP1 | Ac-TAACAATAATCC-mPEG-{Lys[(mPEG-3)2-BT]}2-[Lys(mPEG-3-BT)]2-[(Lys(mPEG-BT)]2-Lys-CONH2 |
| RP2 | Mal-mPEG-TAACAATAATCC-Lys(mPEG-Ac)-CONH2 |
| RP3 | Mal-mPEG-TAACAATAATCC-mPEG-{Lys[(mPEG-3)2-BT]}2-[Lys(mPEG-3-BT)]2-[(Lys(mPEG-BT)]2-Lys-CONH2 |
Abbreviations: mPEG: 8-amino-3,6-dioxaoctanoic acid; mPEG-3: 11-amino-3,6,9-trioxaundecanoic acid; Mal: (N-maleimidopropionamido)-tetraeythyleneglycol; SPDP: 3-(2-pyridyldithio)-propionic acid; Tcyp: (S,S)-transcyclopentane modified thymine.
The PNA probes constitute two halves of a hybridization sandwich (Figure 3A). The N-terminus of the 15-bp surface probe (SP) is chemically immobilized via an amide bond to the wells of a “Nunc Immobilizer Amino” 96-well plate, while the 12-bp reporter probe (RP) has six biotins coupled to the C-terminus via a lysine reside with mPEG spacers. The two PNA probes were designed to hybridize to a 27-bp DNA sequence from the highly conserved pag-coding (PA) portion of the pX01 gene of B. anthracis (TS1, Table 2). In this study, SP2 and RP2 serve only as test sequences to examine the reactivity of the DNA-templated crosslink in solution (vide infra) and neither is used for DNA detection experiments (because these are test sequences the N-terminal of SP2 is capped with an acetyl group and contains no cyclopentane group since it is never attached to the surface of a 96-well plate, and RP2 has no biotins because it is not used in conjunction with enzymatic detection). The mPEG (8-amino-3,6-dioxaoctanoic acid) spacers in RP1 and RP3 were incorporated to provide sufficient spacing so that neighboring biotins would not interfere with binding to a poly-horseradish peroxidase-streptavidin conjugate (pHRP-SA). The six biotins provide multiple sites for pHRP-SA to bind to (i.e. label) the reporter probe to amplify signal resulting from DNA hybridization. The mPEG linkers in SP1 and SP3 serve to provide spacing between the PNA and the surface of the plate. Three fewer N-terminal mPEGs were used in SP3 than SP1 as the chemical yield was better with slightly fewer mPEG units. In this study, SP1 and RP1 form non-covalent complexes with the target DNA while the other probes in Table 1 were designed to form crosslinked dimers upon binding to the DNA target (Figure 3B, vide infra).
Table 2.
Nucleic Acid Targets
| Name | Nucleic Acid Type | Sequence |
|---|---|---|
| TS1 | DNA (27bp) | 5′ - GGA-TTA-TTG-TTA-AAT-ATT-GAT-AAG-GAT - 3′ |
| TS2 | DNA (27bp) | 5′ - GGA-TTA-TTG-TTA-AAT-ATT-GAT-AAG-GAT - 3′ |
| (ds) | 3′ - CCT-AAT-AAC-AAT-TTA-TAA-CTA-TTC-CTA - 5′ | |
| TS3 | DNA (50bp) (ds) | 5′ - GCTGAAATATAGGATTATTGTTAAATATTGATAAGGATGTAATGATAATA - 3′ |
| 3′ - CGACT TTATATCCTAATAACAATTTATAACTATTCCTACATTACT ATTAT - 5′ | ||
| TC mismatch | DNA (27bp) | 5′ - GGA-TTC-TTG-TTA-AAT-ATT-GAT-AAG-GAT - 3′ |
| TG mismatch | DNA (27bp) | 5′ - GGA-TTG-TTG-TTA-AAT-ATT-GAT-AAG-GAT - 3′ |
| TT mismatch | DNA (27bp) | 5′ - GGA-TTT-TTG-TTA-AAT-ATT-GAT-AAG-GAT - 3′ |
| SS1 | Control (24bp) | 5′ - TGC-AGT-CTG-TTA-CAA-TGA-CCT-ACT - 3′ |
Abbreviations: bp: base pairs; ds: double-stranded. Underlined portions of sequences indicate target regions of PNA probes.
After DNA hybridization with the PNA probes and careful washing to remove nonspecifically bound agents, any biotin retained on the surface as a result of both PNAs hybridizing to DNA can be detected via labeling the biotins with pHRP-SA followed by addition of a solution of tetramethylbenzidine (TMB), a substrate for HRP, and peroxide as the oxidant. When pHRP-SA is present, the enzyme uses peroxide to oxidize the TMB, which gives a blue color (652 nm). The enzymatic oxidation is quenched with the addition of H2SO4, resulting in a yellow color (450 nm) that can also be quantified.25 To evaluate the utility of this assay, we investigated the ranges of qualitative and quantitative detection by two methods: development of signal at 652 nm and signal values at 450 nm after quenching. In addition, we investigated the response of the assay to multiple forms of DNA targets, including single-stranded (ss) and double-stranded (ds) DNA.
Development of signal at 652 nm results from the buildup of oxidized TMB that is catalyzed by HRP. The rate of TMB oxidation is proportional to the amount of enzyme bound to the reporter PNA, which is itself proportional to the amount of target DNA. Therefore, each concentration of DNA results in a different sigmoidal curve that reflects the amount of the surface bound pHRP-SA after hybridization of the PNA probes to the target DNA.26 After PNA hybridization to target DNA and labeling with pHRP-SA, a solution of TMB and peroxide was added and development of signal at 652 nm was monitored for 30 minutes with absorbance values collected every 120 seconds. A linear best-fit line was applied to the rate of growth of absorption data to determine the maximal velocity VMax (as defined in the software package SpectaMax™). The initiation and leveling-off phases of the absorption data were excluded from the fit. The slope of the line fit was assigned to VMax, and this value was calculated for each concentration. Figure 4A shows the signal response detected by probes SP1 and RP1 for DNA TS1 over a concentration range of four-orders of magnitude. For data acquired at 652 nm, we consider a plot of VMax versus concentration of DNA to reflect the signal response of the assay (Figure 4B). Similar analyses for each nucleic acid in Table 2 were performed, and the results are plotted together on Figure 4B. The response curves were each fit to a 4-parameter logistic curve that reveals the qualitative and quantitative regions of the assay. In contrast to monitoring development of signal at 652 nm, the data obtained at 450 nm, which are obtained after quenching enzymatic oxidation with sulfuric acid, are single values of absorbance versus concentration of DNA. Figure 4C shows the signal response data at 450 nm. As expected, TS1 (which is ssDNA) provided the best signal across all methods, and the detection limit of the target DNA was ~6000 molecules (Table 3). Detection at this level could also be observed visually (Figure 3A) relative to the scrambled control SS1. Through the course of these studies, however, it became clear that detection between 6000 and 108 molecules of TS1 was only qualitative and could not be used to determine the precise quantity of DNA. Quantitative detection in this system begins at 108 molecules of TS1, and continues well into nM concentrations. Across the entire range of concentrations a scrambled control DNA sequence (SS1) gave no signal, demonstrating that the PNA probes are highly specific to the target DNA. Using more challenging DNA controls, a series of target DNAs in which only a single base mismatch was present could be clearly discerned in the same detection protocol, down to the picomolar concentration (see Supporting Information). In addition, signal resulting from PNA hybridization to target DNA was not affected if additional washes of the plate were performed, indicating that the PNA:DNA complex was tightly bound.
Table 3.
Detection of Nucleic Acids by PNAsa
| Nucleic Acid Type | Lower Limit of Qualititative Detection | Lower Limit of Quantitative Detectionb | |||
|---|---|---|---|---|---|
| Name | Template (SP1-RP1) | Crosslink (SP3-RP3) | Template (SP1-RP1) | Crosslink (SP3-RP3) | |
| TS1 | DNA (27bp) | 6.02 × 103 | 4.15 × 106 | 1.13 × 108 | 6.62 × 108 |
| (1.09 × 108)c | |||||
| TS2 | DNA (27bp) | 6.02 × 103 | 4.15 × 106 | 2.53 × 108 | 9.03 × 108 |
| (ds) | (2.19 × 107)c | ||||
| TS3 | DNA (50bp) (ds) | 2.14 × 107 | x | 4.49 × 109 | x |
| TC mismatch | DNA (27bp) | 6.02 × 103 | x | (1.86 × 108)c | x |
| TG mismatch | DNA (27bp) | 6.02 × 103 | x | (2.66 × 108)c | x |
| TT mismatch | DNA (27bp) | 6.02 × 103 | x | (1.60 × 108)c | x |
Unless otherwise noted, values were determined by curve fits to VMax (mAbs652 nm*min-1) vs. [DNA] data.
Values for quantitative detection are the number of molecules detected at 90% CI and 96% minimal distinguishable difference concentration.
Values determine from curve fit to absorbance at 450 nm vs. [DNA].
While detection of ssDNA is useful to explore the capabilities of PNA probes, a diagnostic device would have to bind the target sequence even if it is bound to its complementary sequence in a dsDNA complex. Commercial kits that are used to isolate DNA from samples for diagnostic analysis usually provide low molecular weight dsDNA fragments of highly variable sizes.27 Therefore, we investigated whether our assay could detect dsDNA. While PNA can invade dsDNA in some cases,28 we employed snap cooling29 to separate the complementary DNA strands and allow the PNA probes to compete for the target. Detection of the double stranded target (TS2, Tm = 55 °C) by following the development of signal at 652 nm (Figure 4B) showed detection similar to TS1 (qualitative detection at about 6000 molecules, quantitative detection starting around 108 molecules). Interestingly, the quantitative detection limit was lowered to 107 molecules when following the signal at 450 nm. The qualitative detection of a significantly longer dsDNA sequence (in this case a 50-bp fragment with the target sequence embedded within a central position in the DNA fragment (TS3, Tm = 65 °C)) rises to 107 molecules, and the quantitative detection limit increases to 109 molecules of dsDNA. This change is likely due to the increased stability of the DNA duplex that contains the target sequence.
pag anthrax DNA detection via DNA templated crosslinking of PNA probes
Rapid diagnosis can help to distinguish infected from non-infected patients, but re-testing of the initial result in a laboratory environment would provide a useful confirmation of the initial diagnosis. The main challenge for this type of follow-up testing is to preserve the integrity of samples collected in the field during the trip to a laboratory. Unfortunately, degradation of samples during transit frequently complicates analysis. We envisioned that a modified version of our current system could preserve nucleic acid information from an initial diagnosis so that transportation to a laboratory could be accomplished without fear of losing information. To accomplish this, the original PNA probes were re-designed with thiol and maleimide groups at each end so that a covalent thiomaleimide crosslink would form upon DNA binding. By forming a covalent bond between the surface and reporter PNA probes that is dependent on the presence of target DNA, the initial presence of the DNA target is preserved by the crosslinked PNAs. Once the PNAs are crosslinked, this complex should be stable to transport even if the original DNA is degraded.
To implement this strategy, a surface PNA probe (SP3) was synthesized with a protected thiol at the C-terminus (Figure 3B). The 2-pyridylthiol group protects the thiol until after DNA hybridization. This protecting group is removed by adding dithiothreitol (DTT) to the plate following hybridization. The reporter probe (RP3) contains a N-terminal maleimide, a Michael acceptor for nucleophilic attack by the thiol. Except for the DTT reduction step, the detection protocol remained the same. Before examining detection on a 96-well plate, we confirmed that covalent bond formation between PNAs SP2 and RP2 did occur in solution and was promoted by complementary DNA. For this experiment, PNA SP2 did not contain a cyclopentane group as we only wanted to confirm the reactivity of the DNA-templated crosslink. PNA SP2 was not used for any DNA detection experiments. Combination of just SP2 and RP2 in solution resulted in a very slow reaction, but in the presence of the complimentary DNA template the expected crosslink formed rapidly in solution and the cross-coupled product was confirmed by HPLC and mass spectrometry (see supporting information for details). Therefore, we felt that the same reaction could occur if RP3 was attached to the surface to the Nunc 96-well plate.
Using probes SP3 and RP3, DNA detection was investigated using the same methods previously described. For DNAs TS1 and TS2 (ss and dsDNA), quantitative detection starts around 108 DNA molecules, which is similar to the previous method with no crosslink (Table 3 and Figure S3 in Supporting Information). Unfortunately, qualitative detection is higher (around 106 molecules) and the probes do not detect the longer 50 bp DNA (TS3). To demonstrate the utility of the crosslink between PNAs SP3 and RP3, the 96-well plate was allowed to sit overnight in phosphate buffer after the initial detection protocol for TS1 was performed (see Figure S4 in Supporting Information). The next day, the plate was extensively washed and each well then relabeled with pHRP-SA. Remarkably, the average of all samples (which represents 10 different concentrations over the entire quantitative region of 7 orders of magnitude) was 59% of the detection signal compared to the previous day (Table 4 and Figure S5 in Supporting Information). For comparison, the same assay using PNAs without the crosslink capability displayed only 23% of the original signal. After another round of washing and relabeling with pHRP-SA, nearly all the signal from the assay using the non-crosslinked PNA probes was gone, but the crosslinked PNA probes still afforded 45% of the original signal. Similar results were obtained using TS2.
Table 4.
Comparison of Detection Signal Following Additional Labeling and Reanalysis.
| Template (SP1-RP1) | Crosslink (SP3-RP3) | |||||
|---|---|---|---|---|---|---|
| DNA | Rerun 1 | Rerun 2 | DNA | Rerun 1 | Rerun 2a | Rerun 3b |
| TS1 | 23% | 10% | TS1 | 59% | 45% | 30% |
| TS2 | 26% | 11% | TS2 | 51% | 35% | 31% |
Assay performed after a prior quench with H2SO4.
Assay performed 2 weeks after initial analysis.
Conclusion
The stability of PNA probes should translate to rugged devices for genetic detection. We have demonstrated in this manuscript that PNAs can be used as probes to signal the presence of DNA in both a qualitative and quantitative manner. Our method allowed us to identify double stranded DNA, which is important because DNA extracted from samples would contain fragments of double stranded DNA. The chemical crosslink between PNA probes provides a lasting link to the source of the initial sample so that additional assays no longer depend on the integrity of DNA collected at the sample site. Since introduction of the crosslink lowers sensitivity, it is likely that the reactive groups interfere with the enzymatic reactivity necessary for detection in the current format. Perhaps alternative crosslinking chemistries could avoid this issue.30 We have demonstrated the ability to quantitatively detect tens of millions (107) of dsDNA copies (Figure 2, regime B). Below this threshold, signal is qualitative and could be used to confirm the presence of DNA, but not the amount (Figure 2, regime A). This switch from quantitative to qualitative detection most likely results from insufficient enzyme labeling of the reporter PNA probe at low concentrations of target DNA. It is possible that increasing the number of biotins on the reporter probe could enhance the sensitivity. While the ELISA-based components of this detection system are not suitable for incorporation into a rugged diagnostic kit and the detection levels are not sufficient for use in the field, the well-established protocols associated with ELISA detection allow one to fully characterize the nucleic acid detection properties of different PNA probes prior to incorporation into a new technological platform.
We believe that current and future detection technologies for anthrax, as well as other pathogens, should consider using PNA as probes for nucleic acids. In particular, researchers developing new diagnostic platforms involving nanotechnology and microfluidics should consider incorporation of PNA probes into their design criteria to signal the presence of a pathogen. For this area, we feel it is important to have a general method to initially confirm the utility of any new PNA probe using standard procedures. This manuscript provides the basic method that can be used to validate sets of PNA probes before incorporation into a new type of technology for signal detection. In our lab, future versions of this diagnostic will focus on developing a tunable dynamic range of detection31 as well as other crosslinking technologies through the introduction of different chemical modifications.
Supplementary Material
Acknowledgement
We acknowledge support from the Intramural Research Program of NIDDK and the NIAID NIH/FDA Intramural Biodefense Research Program.
Footnotes
Supporting Information:
Synthesis and characterization data for all PNAs and all experimental procedures for detection and data analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ahmad F, Hashsham SA. Anal. Chim. Acta. 2012;733:1–15. doi: 10.1016/j.aca.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 2.Bessetti J. Profiles in DNA. 2007:9–10. [Google Scholar]
- 3.Radstrom P, Knutsson R, Wolffs P, Lovenklev M, Lofstrom C. Mol. Biotechnol. 2004;26:133–146. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 4.Inami H, Tsuge K, Matsuzawa M, Sasaki Y, Togashi S, Komano A, Seto Y. Biosensors & Bioelectronics. 2009;24:3299–3305. doi: 10.1016/j.bios.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 5.Ivnitski D, O'Neil DJ, Gattuso A, Schlicht R, Calidonna M, Fisher R. BioTechniques. 2003;35:862–869. doi: 10.2144/03354ss03. [DOI] [PubMed] [Google Scholar]
- 6.Lien KY, Lee GB. Analyst. 2010;135:1499–1518. doi: 10.1039/c000037j. [DOI] [PubMed] [Google Scholar]
- 7.Neuzil P, Novak L, Pipper J, Lee S, Ng LFP, Zhang CY. Lab on a Chip. 2010;10:2632–2634. doi: 10.1039/c004921b. [DOI] [PubMed] [Google Scholar]
- 8.Chakrabarti MC, Schwarz FP. Nucleic Acids Res. 1999;27:4801–4806. doi: 10.1093/nar/27.24.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen PE, Egholm M, Berg RH, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 11.Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchard O, Sonnichsen SH, Nielsen PE. Biochem. Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 12.Corradini R, Sforza S, Tedeschi T, Totsingan F, Marchelli R. Curr. Top. Med. Chem. 2007;7:681–694. doi: 10.2174/156802607780487759. [DOI] [PubMed] [Google Scholar]
- 13.Chin CD, et al. Nat. Med. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 14.de la Rica R, Stevens MM. Nat. Nanotechnol. 2012 doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- 15.Zhang N, Appella DH. J. Am. Chem. Soc. 2007;129:8424–8425. doi: 10.1021/ja072744j. [DOI] [PubMed] [Google Scholar]
- 16.Zhang N, Appella DH. J. Infect. Dis. 2010;201(Suppl 1):S42–5. doi: 10.1086/650389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Z, Taylor JS. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11159–11163. doi: 10.1073/pnas.97.21.11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Karalkar NB, Kool ET. Nat. Biotechnol. 2001;19:148–152. doi: 10.1038/84414. [DOI] [PubMed] [Google Scholar]
- 19.Sando S, Abe H, Kool ET. J. Am. Chem. Soc. 2004;126:1081–1087. doi: 10.1021/ja038665z. [DOI] [PubMed] [Google Scholar]
- 20.Cai J, Li X, Yue X, Taylor JS. J. Am. Chem. Soc. 2004;126:16324–16325. doi: 10.1021/ja0452626. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen PE. Molecular Biotechnology. 2004;26:233–248. doi: 10.1385/MB:26:3:233. [DOI] [PubMed] [Google Scholar]
- 22.Cosnier S, Mailley P. Analyst. 2008;133:984–991. doi: 10.1039/b803083a. [DOI] [PubMed] [Google Scholar]
- 23.Myers MC, Witschi MA, Larionova NV, Franck JM, Haynes RD, Hara T, Grajkowski A, Appella DH. Org. Lett. 2003;5:2695–2698. doi: 10.1021/ol0348811. [DOI] [PubMed] [Google Scholar]
- 24.Pokorski JK, Witschi MA, Purnell BL, Appella DH. J. Am. Chem. Soc. 2004;126:15067–15073. doi: 10.1021/ja046280q. [DOI] [PubMed] [Google Scholar]
- 25.Josephy PD, Eling T, Mason RP. J. Biol. Chem. 1982;257:3669–3675. [PubMed] [Google Scholar]
- 26.Wild D, editor. The Immunoassay Handbook. 3rd ed. Elsvier; 2005. [Google Scholar]
- 27.Levinthal C, Davison PF. J. Mol. Biol. 1961;3:674–&. doi: 10.1016/s0022-2836(61)80030-2. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen PE. Curr. Opin. Biotechnol. 2001;12:16–20. doi: 10.1016/s0958-1669(00)00170-1. [DOI] [PubMed] [Google Scholar]
- 29.Doty P, Marmur J, Eigner J, Schildkraut C. Proc. Natl. Acad. Sci. U. S. A. 1960;46:461–476. doi: 10.1073/pnas.46.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beal DM, Jones LH. Angew. Chem. Int. Ed. Engl. 2012;51:6320–6326. doi: 10.1002/anie.201200002. [DOI] [PubMed] [Google Scholar]
- 31.Liu JW, Lu Y. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.