Abstract
Few mycoplasmal polysaccharides have been described and little is known about their role in pathogenesis. The infection of mice with Mycoplasma pulmonis has been utilized in many in vivo and in vitro studies to gain a better understanding of host-pathogen interactions during chronic respiratory infection. Although alveolar macrophages have a primary role in host defense, M. pulmonis is killed inefficiently in vitro. One antiphagocytic factor produced by the mycoplasma is the family of phase- and size-variable Vsa lipoproteins. However, bacteria generally employ multiple strategies for combating host defenses, with capsular polysaccharide often having a key role. We show here that mutants lacking the EPS-I polysaccharide of M. pulmonis exhibit increased susceptibility to binding and subsequent killing by alveolar macrophages. These results give further insight into how mycoplasmas are able to avoid the host immune system and sustain a chronic infection.
Keywords: capsule, innate immunity, MH-S cells, yeast extract
Introduction
Mycoplasmas are species-specific pathogens of humans and animals. These obligate parasites commonly cause chronic infections of mucosal surfaces such as the respiratory and urogenital tracts and must be adept at avoiding host defenses, despite lacking a cell wall and having the smallest genome of any free-living bacterial species (Razin, et al., 1998). Mycoplasma pulmonis, the causative agent of murine respiratory mycoplasmosis, is used as a model for the study of mycoplasma-host interactions. Alveolar macrophages have a primary role in defense against mycoplasmal infection (Davis, et al., 1992; Hickman-Davis et al., 1997). The only proven antiphagocytic factor identified in mycoplasmas is the Vsa family of surface lipoproteins of M. pulmonis. The Vsa proteins are both phase- and size-variable. Phase variation results from site-specific DNA inversions that combine a previously silent vsa gene with the vsa expression site and plays a role in avoidance of adaptive immunity (Shen, et al., 2000; Chambaud, et al., 2001; Denison, et al., 2005). Size variation occurs in the tandem-repeat region of the protein and results from slipped-strand mispairing during DNA replication. Vsa size variation has a critical role in cytoadherence, biofilm formation, and the avoidance of killing from complement and alveolar macrophages (Simmons & Dybvig, 2003; Simmons, et al., 2004; Simmons & Dybvig, 2007; Shaw, et al., 2012). Not all species of mycoplasma produce proteins analogous to the Vsa proteins, and other types of antiphagocytic molecules should exist.
Capsular polysaccharide is a common antiphagocytic factor (Finlay & Falkow, 1989). Several Mycoplasma species including M. bovis, M. dispar, M. hyopneumoniae, M. mycoides subsp. mycoides, M. penetrans, M. pneumoniae, and M. pulmonis are thought to produce polysaccharides (Taylor-Robinson, et al., 1981; Tajima & Yagihashi, 1982; Tajima, et al., 1982; Almeida & Rosenbusch, 1991; Neyrolles, et al., 1998; Brooks, et al., 2004; Westberg, et al., 2004; Daubenspeck, et al., 2009). Essentially nothing was known about the functions of these polysaccharides until the discovery of the EPS-I polysaccharide of M. pulmonis and the isolation of EPS-I negative mutants (Daubenspeck, et al., 2009). It has recently been shown that EPS-I has roles in cytoadherence and in resistance to complement (Bolland & Dybvig, 2012; Bolland, et al., 2012). Here we report that the EPS-I polysaccharide reduces the binding of M. pulmonis to alveolar macrophages and protects the bacteria from killing.
Materials and methods
Mycoplasmas
The mycoplasmas used in this study are all derived from strain CT and have been described previously (Simmons & Dybvig, 2003; Simmons, et al., 2004). Strain CTG38 has a wild-type phenotype. Strain CTG1701 lacks the EPS-I polysaccharide due to the insertion of transposon Tn4001T in the gene MYPU_7410. The production of EPS-I was restored in CTG1701-C by complementing CTG1701 with the operon containing the genes MYPU_7410 and MYPU_7420. The exact location of Tn4001T in the genome of CTG1701 and details of the construction of the complemented strain have been described (Daubenspeck, et al., 2009). All of the mycoplasma strains used in this study produce a VsaG protein of identical size, containing 36 tandem repeats, and are thought to differ phenotypically only in the production of polysaccharide (Shaw, et al., 2012). Mycoplasmas were grown in mycoplasma broth, suspended in freezing medium, sonicated to break up aggregates and assayed for CFU on mycoplasma agar as described previously (Shaw, et al., 2012).
MH-S cells
The MH-S cells were derived from alveolar macrophages of a BALB/cJ mouse through bronchoalveolar lavage and transformation with simian virus 40 (Mbawuike & Herscowitz, 1989). The cell line consists of a heterogeneous population of complement receptor 3-negative and complement receptor 3-positive cells (Sankaran & Herscowitz, 1995). MH-S cells were grown in sodium bicarbonate-buffered Dulbecco's minimum essential medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum.
Interactions between mycoplasmas and MH-S cells
MH-S cells were prepared as described previously (Shaw, et al., 2012). The cells were activated with interferon gamma and lipopolysaccharide followed by three washes in assay buffer (Hank's Balanced Salt Solution buffered with 25 mM HEPES, pH 7.4, supplemented with 5% fetal bovine serum). After incubating for 1 hour, the number and viability of the washed cells was determined by using a hemacytometer and trypan blue exclusion. Only preparations of macrophages containing 95% or more viable cells were used.
Mycoplasmas were incubated with macrophages for up to 8 hours. Chloramphenicol was used to inhibit growth of the mycoplasma during this incubation period. This antibiotic was chosen because it is effective at inhibiting the growth of M. pulmonis and because studies had shown that chloramphenicol at concentrations up to 200 μg mL−1 does not diminish the ability of macrophages to phagocytose and kill bacteria (van den Broek, 1989). In our previous study (Shaw, et al., 2012), chloramphenicol was added to the reactions at a final concentration of 30 μg mL−1. This concentration of chloramphenicol inhibits the growth of mycoplasma strains CTG38 and CTG1701. However, strain CTG1701-C contains a chloramphenicol acetyltransferase gene, and 30 μg mL−1 of chloramphenicol was insufficient to inhibit its growth. It was determined that 90 μg mL−1 of chloramphenicol inhibited growth of CTG1701-C for up to 6 hours, but growth resumed after this time. Hence, for experiments with CTG1701-C co-incubated with MH-S cells for periods of time longer than 4 hours, the cells were initially incubated with 90 μg mL−1 of chloramphenicol and then an additional bolus of chloramphenicol (90 μg mL−1) was added at 4 hours. Using these conditions, chloramphenicol had no affect on the viability of CTG1701-C or MH-S cells and there was no detectable growth of CTG1701-C. However, CTG1701 and CTG38 lost viability when incubated with chloramphenicol at a final concentration of 90 μg mL−1. Hence, for experiments using these strains, the initial concentration of chloramphenicol was 30 μg mL−1 of assay buffer, and the bolus at 4 hours was also added to a final concentration of 30 μg mL−1 of assay buffer. The viability of these strains was unaffected at these concentrations of chloramphenicol.
The binding of mycoplasmas to MH-S cells and subsequent killing were examined as described (Shaw, et al., 2012). 1 × 106 MH-S cells were mixed with 1 × 108 CFU of the desired mycoplasma strain in a total volume of 1 ml of assay buffer containing either 90 or 30 μg/ml of chloramphenicol as indicated above. A sample was removed immediately for CFU determination. After incubation of the mixture for 40 minutes at 37°C with end-over-end rotation, the MH-S cells were harvested by centrifugation and washed 3 times with assay buffer to remove unbound mycoplasmas. The washed MH-S cells were suspended in assay buffer, gently sonicated to break up aggregates, and assayed for mycoplasma CFU. The number of recovered CFU after binding was divided by the number of CFU from the initial inoculation to determine the percentage of mycoplasmas bound. To examine killing, the MH-S cells with attached mycoplasmas were incubated at 37°C with samples taken at 4 and 8 hours. These samples were sonicated for 20 seconds to disrupt aggregates and assayed to determine the number of surviving mycoplasma CFU. The results were analyzed by ANOVA with multiple comparisons made by the Holm-Sidak method (SigmaPlot 11) with a P < 0.05 considered significant.
In some experiments, yeast extract was added to the assay buffer to examine its affect on the binding and killing of mycoplasmas. The results were analyzed by ANOVA as described above when comparing multiple strains of mycoplasma or the Student's T-test for comparison of a single strain with and without yeast extract added to the assay buffer.
Gas Chromatography/ Mass Spectrometry (GC/MS)
The EPS-I polysaccharide from the mycoplasmal strains was assessed by GC/MS using previously described methods (Daubenspeck, et al., 2009; Bolland, et al., 2012). Briefly, cells from stationary-phase cultures were harvested and washed three times by centrifugation and lysed by sonication. A 50-μl sample from each lysate was used to determine the amount of protein by using the BCA protein assay kit (Pierce). The remaining lysate was digested extensively with nucleases and proteinase K and dialyzed (2,000 molecular-weight cutoff) against water to remove small molecules, particularly monosaccharides. The dialyzed lysates were subjected to methanolysis and derivatized with the HMDS + TMCS + Pyridine, 3:1:9 (Sylon™ HTP) Kit (Sigma). Volumes equivalent to 100 μg of protein were analyzed by GC/MS with an Agilent Technologies 6890N Network GC System and a 5973 Network Mass Selective Detector with MSD Productivity Chemstation Software Rev. D.00.00. The amount of EPS-I was calculated from the area under the major galactose peak, with comparison to galactose standards of known amount.
Results
EPS-I protects mycoplasma from binding to and killing by MH-S cells
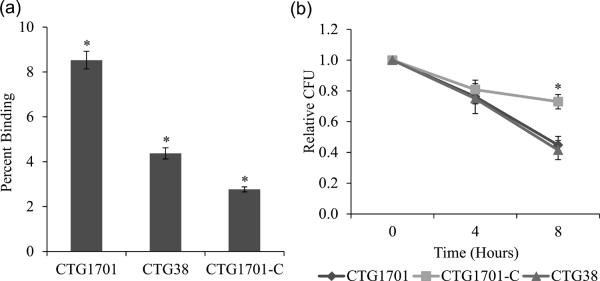
We investigated whether the production of EPS-I was involved with the ability of M. pulmonis to avoid binding to alveolar macrophages and to be killed (Fig. 1). Significantly more CTG1701, which lacks EPS-I, bound to macrophages than did CTG38 or the complemented CTG1701-C (Fig. 1a) (P < 0.001). By avoiding binding, and hence subsequent phagocytosis, EPS-I is antiphagocytic. Surprisingly, more CTG38 was bound by macrophages than was CTG1701-C (P < 0.001). Once the mycoplasmas were bound to the macrophages, there was no significant difference in the survival of bound CTG38 and CTG1701 at any time point (Fig. 1b). However, CTG1701-C survived significantly better at 8 hours than did CTG38 and CTG1701 (P < 0.006).
Fig. 1.
Binding and killing of M. pulmonis to MH-S cells. Panel (a) shows the binding of mycoplasmas strains to MH-S cells. Asterisks indicate significant differences in binding as compared to all other strains (P < 0.001). Error bars represent standard error of the mean. The results were replicated with n values of 6 for all strains. Panel (b) shows the survival of the mycoplasmas that are bound to MH-S cells. The only significant differences were at 8 hours of incubation with CTG1701-C surviving better than CTG38 (P = 0.003) or CTG1701 (P = 0.006). Error bars represent standard error of the mean. The results were replicated with n values of 6 (CTG1701 and CTG38) and 5 (CTG1701-C).
Relative amounts of EPS-I associated with mycoplasma strains
The difference between CTG38 and CTG1701-C in regard to binding to macrophages and subsequent killing was unexpected given that these two strains possess identical Vsa proteins and have no known differences other than the original mutation that disrupted MYPU_7410 and its complementation. We had noted in three separate experiments using three different media that the yield of EPS-I from CTG1701-C was relatively high. The amount of EPS-I associated with CTG38 and CTG1701-C was quantitated by GC/MS, by assaying equivalent amounts of lysate as determined by protein concentration. CTG38 and CTG1701-C had 24 ng and 123 ng EPS-I μg−1 protein, respectively, indicating a 5-fold difference in the amount of polysaccharide.
Enhanced killing with yeast extract added to the assay buffer
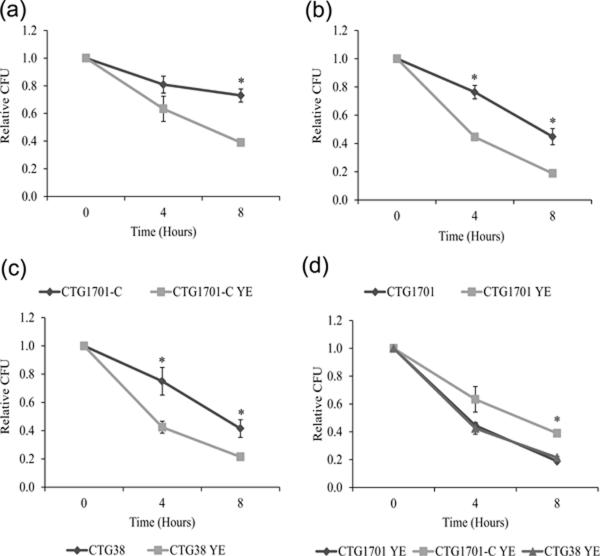
Depending on the medium and other culture conditions, there is a high degree of variability in the amount of mannose glycosides, detected by GC/MS, in M. pulmonis lysates. These results suggested that the mycoplasma is proficient at binding mannosylated molecules. Many such molecules would be encountered in the host and might affect phagocytosis. Although yeast extract is not present in the murine host, its mannosylated cell wall proteins might interact with the mycoplasma in a similar fashion as mannosylated host proteins. The addition of yeast extract to the assay buffer led to a significant increase in killing by alveolar macrophages for all three strains of mycoplasma, but CTG1701-C still survived significantly better at 8 hours than did CTG38 and CTG1701 (Fig. 2).
Fig. 2.
Effect of yeast extract on the survival of mycoplasmas that are bound to MH-S cells. Panels (a) to (c), survival of mycoplasma strains CTG1701-C, CTG1701, and CTG38, respectively, grown in assay buffer with and without yeast extract. Panel (d), comparative survival of CTG1701-C, CTG1701, and CTG38, bound to MH-S cells and incubated in assay buffer with yeast extract. In all panels, the abbreviation YE in the strain designations indicates that yeast extract was added to the assay buffer. Significant differences are indicated by an asterisk. Panel (a), the survival of CTG1701-C is significantly higher at 8 hours when yeast extract is absent (P < 0.001). Panel (b), the survival of CTG1701 is significantly higher at 4 and 8 hours (P < 0.001 and 0.002, respectively) when yeast extract is absent. Panel (c), the survival of CTG38 is significantly higher at 4 and 8 hours (P = 0.015 and 0.002, respectively) when yeast extract is absent. Panel (d), the survival of CTG1701-C was significantly higher than that of CTG1701 and CTG38 at 8 hours (P < 0.001 for both strains). Error bars represent standard error of the mean and in some cases are too small to visualize on the graphs. The results were replicated with n values of 6 for the data in all panels.
Discussion
Macrophages are thought to be a first line of defense against infection by mycoplasmas. We have shown previously that MH-S cells are not capable of killing M. pulmonis unless the mycoplasma is first bound by the macrophages. Killing is dependent on phagocytosis (Shaw, et al., 2012). Vsa proteins act as a shield that reduces binding to macrophages when the proteins are long with many tandem repeats. We show here that the EPS-I polysaccharide of M. pulmonis is a second shielding factor that inhibits binding to macrophages and hence is antiphagocytic. Both long Vsa protein and EPS-I have a role in protection from complement and inhibit biofilm formation (Bolland & Dybvig, 2012; Bolland, et al., 2012). The amount of EPS-I that is associated with the mycoplasma cell is about the same regardless of the length of the Vsa protein being produced (Bolland, et al., 2012). It is unknown whether the Vsa proteins and EPS-I interact directly, but it is apparent that full shielding from host defenses requires both a long Vsa protein and EPS-I. The shield primarily protects mycoplasmas from macrophages by inhibiting binding, but there are indications that a maximal shield may also inhibit phagocytosis of the bound mycoplasmas. Mycoplasmas bound to MH-S cells are not phagocytosed efficiently when they produce a very long VsaA protein (Shaw, et al., 2012). The relative resistance of CTG1701-C to killing even after being bound by the macrophages (Figs. 1b and 2d) suggests that the high level of EPS-I on CTG1701-C has the capability to inhibit phagocytosis.
There are several possible explanations for CTG1701-C producing as much as 5 times more EPS-I than CTG38. CTG1701 was complemented to generate CTG1701-C by inserting the 2-gene operon containing MYPU_7410 and MYPU_7420 along with its native promoter into the mycoplasmal genome by using transposon Tn4001C as the vector (Daubenspeck, et al., 2009). One possibility for increased production of EPS-I is that sequences upstream of the complementing operon in CTG1701-C have promoter activity that enhances transcription above that of the native promoter alone. Alternatively, the complementing operon might be missing regulatory sequences that are present in the native operon. Another possibility is the position of the complementing operon in the genome might enhance transcription, as has been shown for genes near the origin for DNA replication (Li, et al., 2003; Manna, et al., 2004).
Killing of M. pulmonis by MH-S cells was only modest. Host factors absent in the in vitro assays may be required for efficient phagocytosis. We show that yeast extract enhances killing. We view it likely that the mannosylated proteins from the yeast cell wall are responsible, possibly through interactions with complement receptor 3 or the mannose receptor on the macrophages. Complement receptor 3 contains a lectin domain that is believed to bind polysaccharide and increase killing of iC3b-oposonized microorganisms (Todd, 1996). The mannose receptor would also recognize and bind mannosylated proteins leading to internalization and subsequent killing (Underhill & Ozinsky, 2002). While mycoplasmas would not come into contact with mannosylated yeast cell wall proteins in the murine host, there are several mannosylated proteins produced in the mammalian lung. The mucins MUC5AC and MUC5B are mannosylated at sites containing the motif WXXW (Perez-Vilar, et al., 2004). These mucins bind to pathogens and are upregulated during bacterial infections including those of M. pneumoniae (Voynow, 2002; Kraft, et al., 2008; Voynow & Rubin, 2009).
The role of mycoplasmal capsule in host immune avoidance has for the most part not been previously studied, but Mycoplasma dispar is a possible exception. When co-cultured with lung fibroblasts, M. dispar became more resistant to killing by alveolar macrophages and had an increased amount of extracellular material on its surface that was observed by electron microscopy (Almeida, et al., 1992). Although the possibility that this material consists primarily of host molecules from the fibroblasts that adsorbed to the surface cannot be discounted, this material may be the result of increased capsule production induced by the fibroblasts. As with M. pulmonis, capsular polysaccharide in many species of mycoplasma might have prominent roles in resisting phagocytosis.
Acknowledgements
We thank P. Caldwell and P. Lao for technical assistance and D. Chaplin for providing the MH-S cell line. This work was supported by NIH grant AI64848.
References
- Almeida RA, Rosenbusch RF. Capsulelike surface material of Mycoplasma dispar induced by in vitro growth in culture with bovine cells is antigenically related to similar structures expressed in vivo. Infect. Immun. 1991;59:3119–3125. doi: 10.1128/iai.59.9.3119-3125.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RA, Wannemuehler MJ, Rosenbusch RF. Interaction of Mycoplasma dispar with bovine alveolar macrophages. Infect. Immun. 1992;60:2914–2919. doi: 10.1128/iai.60.7.2914-2919.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland JR, Dybvig K. Mycoplasma pulmonis Vsa proteins and polysaccharide modulate adherence to pulmonary epithelial cells. FEMS Microbiol. Lett. 2012;331:25–30. doi: 10.1111/j.1574-6968.2012.02551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland JR, Simmons WL, Daubenspeck JM, Dybvig K. Mycoplasma polysaccharide protects against complement. Microbiology. 2012;158:1867–1873. doi: 10.1099/mic.0.058222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BW, Lutze-Wallace CL, Lu P, Robertson RH. Identification and serological specificity of a polysaccharide component from Mycoplasma bovis. Vet. Res. Commun. 2004;28:197–208. doi: 10.1023/b:verc.0000017282.27591.92. [DOI] [PubMed] [Google Scholar]
- Chambaud I, Heilig R, Ferris S, et al. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 2001;29:2145–2153. doi: 10.1093/nar/29.10.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubenspeck JM, Bolland JR, Luo W, Simmons WL, Dybvig K. Identification of exopolysaccharide-deficient mutants of Mycoplasma pulmonis. Mol. Microbiol. 2009;72:1235–1245. doi: 10.1111/j.1365-2958.2009.06720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JK, Davidson MK, Schoeb TR, Lindsey JR. Decreased intrapulmonary killing of Mycoplasma pulmonis after short-term exposure to NO2 is associated with damaged alveolar macrophages. Amer. Rev. Resp. Dis. 1992;145:406–411. doi: 10.1164/ajrccm/145.2_Pt_1.406. [DOI] [PubMed] [Google Scholar]
- Denison AM, Clapper B, Dybvig K. Avoidance of the host immune system through phase variation in Mycoplasma pulmonis. Infect. Immun. 2005;73:2033–2039. doi: 10.1128/IAI.73.4.2033-2039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Falkow S. Common themes in microbial pathogenicity. Microbiol. Rev. 1989;53:210–230. doi: 10.1128/mr.53.2.210-230.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman-Davis J, Michalek SM, Gibbs-Erwin J, Lindsey JR. Depletion of alveolar macrophages exacerbates respiratory mycoplasmosis in mycoplasma-resistant C57BL mice but not mycoplasma-sensitive C3H mice. Infect. Immun. 1997;65:2278–2282. doi: 10.1128/iai.65.6.2278-2282.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft M, Adler KB, Ingram JL, Crews AL, Atkinson TP, Cairns CB, Krause DC, Chu HW. Mycoplasma pneumoniae induces airway epithelial cell expression of MUC5AC in asthma. Eur. Respir. J. 2008;31:43–46. doi: 10.1183/09031936.00103307. [DOI] [PubMed] [Google Scholar]
- Li Y, Youngren B, Sergueev K, Austin S. Segregation of the Escherichia coli chromosome terminus. Mol. Microbiol. 2003;50:825–834. doi: 10.1046/j.1365-2958.2003.03746.x. [DOI] [PubMed] [Google Scholar]
- Manna D, Breier AM, Higgins NP. Microarray analysis of transposition targets in Escherichia coli: The impact of transcription. Proc. Nat. Acad. Sci. USA. 2004:9780–9785. doi: 10.1073/pnas.0400745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbawuike IN, Herscowitz HB. MH-S, a murine alveolar macrophage cell line: morphological, cytochemical, and functional characteristics. J. Leukoc. Biol. 1989;46:119–127. doi: 10.1002/jlb.46.2.119. [DOI] [PubMed] [Google Scholar]
- Neyrolles O, Brenner C, Prevost MC, Fontaine T, Montagnier L, Blanchard A. Identification of two glycosylated components of Mycoplasma penetrans: a surface-exposed capsular polysaccharide and a glycolipid fraction. Microbiol. 1998;144:1247–1255. doi: 10.1099/00221287-144-5-1247. [DOI] [PubMed] [Google Scholar]
- Perez-Vilar J, Randell SH, Boucher RC. C-Mannosylation of MUC5AC and MUC5B Cys subdomains. Glycobiology. 2004;14:325–337. doi: 10.1093/glycob/cwh041. [DOI] [PubMed] [Google Scholar]
- Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran K, Herscowitz HB. Phenotypic and functional heterogeneity of the murine alveolar macrophage-derived cell line MH-S. J. Leukoc. Biol. 1995;57:562–568. doi: 10.1002/jlb.57.4.562. [DOI] [PubMed] [Google Scholar]
- Shaw BM, Simmons WL, Dybvig K. The Vsa shield of Mycoplasma pulmonis is antiphagocytic. Infect. Immun. 2012;80:704–709. doi: 10.1128/IAI.06009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Yu H, Gumulak J, French CT, Zou N, Dybvig K. Gene rearrangements in the vsa locus of Mycoplasma pulmonis. J. Bacteriol. 2000;182:2900–2908. doi: 10.1128/jb.182.10.2900-2908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons WL, Dybvig K. The Vsa proteins modulate susceptibility of Mycoplasma pulmonis to complement killing, hemadsorption, and adherence to polystyrene. Infect. Immun. 2003;71:5733–5738. doi: 10.1128/IAI.71.10.5733-5738.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons WL, Dybvig K. Biofilms protect Mycoplasma pulmonis cells from lytic effects of complement and gramicidin. Infect. Immun. 2007;75:3696–3699. doi: 10.1128/IAI.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons WL, Denison AM, Dybvig K. Resistance of Mycoplasma pulmonis to complement lysis is dependent on the number of Vsa tandem repeats: shield hypothesis. Infect. Immun. 2004;72:6846–6851. doi: 10.1128/IAI.72.12.6846-6851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima M, Yagihashi T. Interaction of Mycoplasma hyopneumoniae with the porcine respiratory epithelium as observed by electron microscopy. Infect. Immun. 1982;37:1162–1169. doi: 10.1128/iai.37.3.1162-1169.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima M, Yagihashi T, Miki Y. Capsular material of Mycoplasma gallisepticum and its possible relevance to the pathogenic process. Infect. Immun. 1982;36:830–833. doi: 10.1128/iai.36.2.830-833.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Robinson D, Furr PM, Davies HA, Manchee RJ, Mouches C, Bove JM. Mycoplasmal adherence with particular reference to the pathogenicity of Mycoplasma pulmonis. Isr. J. Med. Sci. 1981;17:599–603. [PubMed] [Google Scholar]
- Todd RF., III The continuing saga of complement receptor type 3 (CR3) J. Clin. Invest. 1996;98:1–2. doi: 10.1172/JCI118752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- van den Broek PJ. Antimicrobial drugs, microorganisms, and phagocytes. Rev. Infect. Dis. 1989;11:213–245. doi: 10.1093/clinids/11.2.213. [DOI] [PubMed] [Google Scholar]
- Voynow JA. What does mucin have to do with lung disease? Paediatr. Respir. Rev. 2002;3:98–103. doi: 10.1016/s1526-0550(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Voynow JA, Rubin BK. Mucins, mucus, and sputum. Chest. 2009;135:505–512. doi: 10.1378/chest.08-0412. [DOI] [PubMed] [Google Scholar]
- Westberg J, Persson A, Holmberg A, Goesmann A, Lundeberg J, Johansson KE, Pettersson B, Uhlen M. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP) Genome Res. 2004;14:221–227. doi: 10.1101/gr.1673304. [DOI] [PMC free article] [PubMed] [Google Scholar]