Abstract
Stress induces long-lasting changes in neuronal gene expression and cholinergic neurotransmission, but the underlying mechanism(s) are incompletely understood. Here, we report that chromatin structure and histone modifications are causally involved in this transcriptional memory. Specifically, the AChE gene encoding the acetylcholine-hydrolyzing enzyme acetylcholinesterase is known to undergo long-lasting transcriptional and alternative splicing changes after stress. In mice subjected to stress, we identified two alternative 5′ exons that were down-regulated after stress in the hippocampus, accompanied by reduced acetylation and elevated trimethylation of H3K9 at the corresponding promoter. These effects were reversed completely by daily administration of the histone deacetylase (HDAC) inhibitor sodium butyrate for 1 wk after stress. H3K9 hypoacetylation was associated with a selective, sodium butyrate-reversible promoter accumulation of HDAC4. Hippocampal suppression of HDAC4 in vivo completely abolished the long-lasting AChE-related and behavioral stress effects. Our findings demonstrate long-lasting stress-inducible changes in AChE's promoter choices, identify the chromatin changes that support this long-term transcriptional memory, and reveal HDAC4 as a mediator of these effects in the hippocampus.
Keywords: HDAC inhibitors, ChIP, chromatin immunoprecipitation, histone methylation, histone acetylation
Stress induces long-lasting changes in neuronal gene expression and cholinergic neurotransmission (1, 2), but the underlying mechanism(s) are incompletely understood. Neuronal gene expression, like other transcriptional processes, is modulated by a dynamic process of chromatin modifications (3–5). According to the histone code hypothesis, different modifications of histones at a particular promoter region, alone or in combination, define a specific epigenetic state that balances between gene activation and gene silencing (6, 7). Although there are exceptions, histone hyperacetylation at promoters generally indicates an increase in gene activity, whereas hypoacetylation usually marks a decrease in activity (8). Histone methylation, on the contrary, correlates with either transcriptional activation (i.e., H3K4, H3K36) or repression (i.e., H3K9, H3K27, and H4K20). Histone methylation also may facilitate DNA methylation, which causes further repression of the affected genes (9, 10).
Chromatin modification is increasingly recognized as a crucial mechanism in several important phenomena in the brain, including neuronal differentiation, neurodegeneration, circadian rhythm, seizure, memory formation, and drug addiction (11, 12). Here, we studied the involvement of acetylcholinesterase (AChE) chromatin structure in the long-term neuroadaptations to stress in the hippocampus, which is one of the major brain areas involved in mammalian stress responses (13).
The AChE gene contains five short putative alternative first exons in mice (14). Each exon contains its own unique promoter region (14), which can modulate the expression of one splice variant over another. Defeat stress/seizures/posttraumatic stress can induce long-lasting changes in the promoters of several genes, including BDNF, GDNF, GRE, GLUR2, and RELN through chromatin-related modifications (12, 15–19). Here we studied how the stress-induced long-lasting changes in AChE expression in the brain (2, 14) are maintained and regulated at the chromatin level. We describe the long-lasting stress-induced expression changes of AChE 5′ splice variants, the related histone modifications at the corresponding promoters, reversal of these effects by histone deacetylase (HDAC) inhibitors, and a potential mediator, HDAC4, of stress-related transcriptional memory in the hippocampus.
Results
Regulation of AChE Expression by Forced-Swim Stress.
We previously showed long-lasting elevation of the 3′ “readthrough” variant of AChE, AChE-R, after 4 d of forced-swim stress (FSS) (2). We subsequently wished to test whether the AChE 5′ alternative transcripts (E1a, E1b, E1c, E1d1, and E1e; Fig. 1A) also display long-lasting changes in expression following the same repeated stress paradigm. To this end, we subjected mice to two FSS sessions a day, 4 min each, for 4 consecutive days (2). We killed the mice 2 wk later along with control mice that had undergone no stress and surgically dissected the hippocampus (Fig. 1B). To reveal the neuronal content in our hippocampal preparations, we dispersed the hippocampi to single cells using enzymatic digestion and sorted the cells by FACS using MAP2 antibodies. This sorting showed that neurons constituted >45% of our hippocampi preparations (Fig. S1). Therefore, for all subsequent experiments, we used hippocampal preparations without sorting. After extraction of hippocampal RNA, we analyzed transcript levels using real-time quantitative PCR (qPCR). Because transcript levels of several of the 5′ alternative exons were too low for proper quantification with qPCR, we also used semiquantitative conventional RT-PCR. We found a differential expression of some of these exons after stress: mE1b and mE1c transcripts were down-regulated, but all other 5′ transcripts remained essentially unchanged (P < 0.05; Fig. 1 C and D). To correlate the expression level of the 5′ transcripts to the 3′ transcripts, we analyzed expression levels of the two brain 3′ alternative transcripts, the soluble AChE-R and the synapse-associated AChE-S variant (Fig. 1A), using qPCR. Although, as expected, AChE-R mRNA was markedly elevated (P < 0.05; Fig. 1E), AChE-S mRNA was significantly reduced (Fig. 1F) in a manner that resembles the down-regulation of the 5′ transcript mE1c after stress (P < 0.05; Fig. 1D).
Fig. 1.
AChE expression after FSS. (A) The structure of the AChE gene. The mouse AChE gene contains five alternative exons, mE1a–mE1e, at the 5′ end of the gene. The AChE gene also contains unique promoter regions, mP1a, mP1c, mP1d, and mP1e, upstream of the various exons. For mRNA analysis of AChE mE1a–E1e, primers were designed to recognize each exon specifically. For ChIP analysis, primers were designed around the promoters, mP1a–mP1e. Alternative splicing of AChE at the 3′ end generates two isoforms: the main, synaptic form, AChE-S, and the stress-induced, soluble form, AChE-R (B) Schematic representation of the FSS paradigm. Mice were subjected to two FSS sessions a day, 4 min each, for 4 consecutive days, and were killed 2 wk later (n = 4). (C) Semiquantitative RT-PCR analysis of the AChE 5′ transcripts (mE1a–mE1e). mE1b and mE1c transcripts were down-regulated, whereas all other 5′ transcripts remained essentially unchanged. (D–F) Real-time PCR analysis before and after FSS of mE1c (D), AChE-R (E), and AChE-S (F). AChE-S mRNA is 80 times more abundant than AChE-R mRNA. AChE-R mRNA was elevated, whereas AChE-S and mE1c mRNAs were significantly reduced in the hippocampus of stressed mice. *P < 0.05, two-tailed u test.
Regulation of AChE Expression by FSS After Treatment with HDAC Inhibitors.
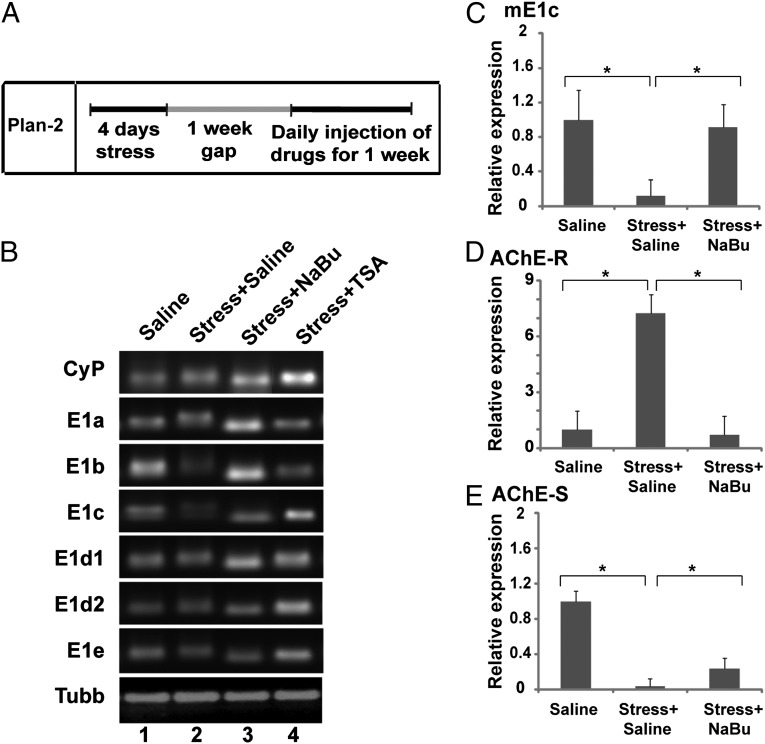
In light of the decreased expression of mE1b and mE1c after stress, we wished to test whether expression levels can be restored using histone deacetylase (HDAC) inhibitors (HDACi). HDACi were shown to improve symptoms of several brain-related pathologies; including stress-induced memory impairments (20, 21). To this end, we repeated the FSS experiment, but this time after the FSS protocol, the mice were placed back in their original cages for 1 wk and then injected with either sodium butyrate (NaBu) or trichostatin A (TSA) daily for 1 wk (Fig. 2A). Mice treated with saline injection after stress served as the control group; again, mice were killed 2 wk after the FSS. Remarkably, we found that NaBu completely restored the down-regulation of mE1b and mE1c exons at the 5′ end (Fig. 2B, lane 3). TSA, on the other hand, showed variable effects (Fig. 2B, lane 4). No changes were observed at any of the other exons (mE1a, mE1d1, mE1d2, or mE1e) with either NaBu or TSA (Fig. 2B). The data were supported further by qPCR confirming the down-regulation of mE1c after stress (P < 0.05; Fig. 2C) and its restoration after NaBu treatment (Fig. 2C, P < 0.05). We found similar restoration of AChE-R levels (P < 0.05; Fig. 2D) and partial restoration of AChE-S levels (P < 0.05; Fig. 2E) at the 3′ end, likely reflecting stress-induced changes in the AChE-S-selective microRNA-132 (22). These results indicate that the long-lasting stress-induced changes in AChE expression can be partly restored by TSA and fully rescued by NaBu.
Fig. 2.
Differential regulation of AChE exons after FSS and drug treatment. (A) Schematic representation of the course of the experiment. After the FSS protocol had been followed for 4 d, the mice were returned to their original cages for 1 wk and then were injected with NaBu, TSA, or saline daily for 1 wk and were killed 2 wk thereafter (n = 4). (B) Semiquantitative RT-PCR analysis of the AChE 5′ transcripts (mE1a–mE1e) in the hippocampus of control or stressed mice treated with saline or an HDACi (NaBu or TSA). Lane 1, saline; lane 2, stress + saline; lane 3, stress + NaBu; lane 4, stress + TSA. NaBu completely restored the down-regulation of mE1b and mE1c exons at the 5′ end. TSA, on the other hand, showed variable effects. (C–E) Real time PCR of mE1c (C), AChE-R (D), and AChE-S (E) in the hippocampus of control mice, stressed mice, and stressed mice treated with either saline or NaBu. At the 3' end, similar restoration was found in AChE-R mRNA levels, while AChE-S mRNA levels were partially restored with NaBu treatment. *P < 0.05, 2-tailed u test.
Chromatin Regulation of AChE After Forced Swim Stress.
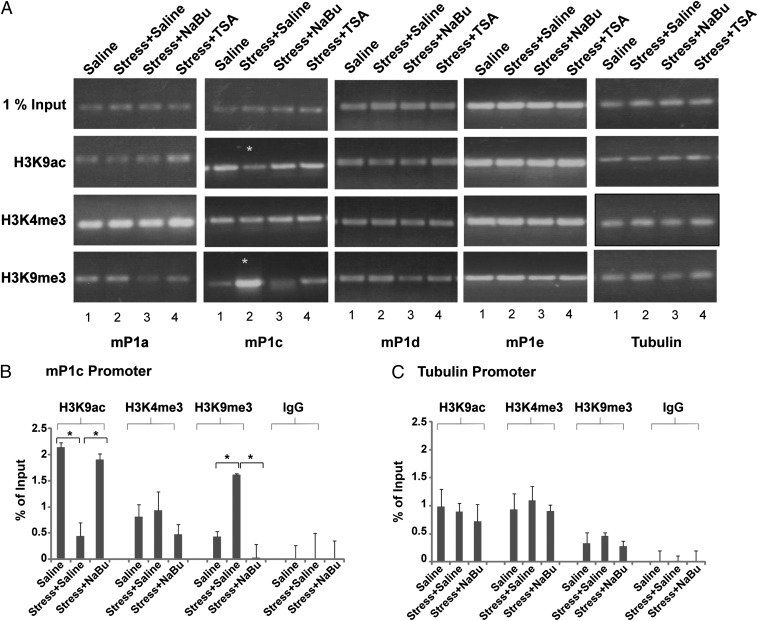
The long-lasting down-regulation of two of the 5′ transcripts mE1b and mE1c prompted us to investigate the mechanism by which the corresponding promoters elicit transcriptional memory and are differentially regulated by stress. We therefore analyzed the changes in several histone modifications of the corresponding promoters of the 5′ alternative transcripts after stress using ChIP assays. The ChIP technique was standardized on hippocampal tissues by optimizing cross-linking conditions, sonication, and antibody concentrations. For all assays, we used nonspecific IgG antibodies as negative controls to check the specificity of each antibody used. The β-tubulin III (Tubb3) gene, which was shown to be unaffected by stress or seizures in the hippocampus (16, 23), was used as control. Using ChIP, we tested the relative amounts of the active markers histone H3 lysine 9 acetylation (H3K9ac) and histone H3 lysine 4 trimethylation (H3K4me3) as well as the heterochromatin-associated marker H3 lysine 9 trimethylation (H3K9me3). Promoter regions mP1a, mP1d, and mP1e did not show any changes in the levels of the tested histone modifications after stress (Fig. 3A; lanes 1 and 2 in each promoter panel), whereas mP1c displayed a significant decrease in H3K9ac levels and a concomitant significant increase in H3K9me3 levels (Fig. 3A, mP1c, lanes 1 and 2) but no change in H3K4me3, indicating the transition from an active to a repressed chromatin state at this region. The hypoacetylation and hypermethylation of H3K9 at the mP1c promoter was confirmed further by ChIP-qPCR (Fig. 3 B and C). These results demonstrate long-lasting changes in histone modifications at the mP1c promoter after FSS that are correlated with and may explain the changes observed in mE1c expression.
Fig. 3.
Changes in active and repressive markers after stress and treatment with HDACi. (A) ChIP assays were performed to measure the levels of several histone modifications at the AChE promoters mP1a, mP1c, mP1d, and mP1e in the hippocampus after stress and treatment with HDACi (NaBu and TSA) using specific antibodies for each modification. H3K9ac was unaltered at the mP1a, mP1d, and mP1e promoters but showed pronounced decreased acetylation at the mP1c promoter after stress. Normal levels were restored when stressed mice were treated with NaBu or TSA. H3K4me3 was unaltered in all promoters. H3K9me3 was enriched after stress exclusively at the mP1c promoter. NaBu restored H3K9 methylation levels. TSA, in contrast, had little effect. (B–C) ChIP-qPCR for H3K9ac, H3K4me3, and H3K9me3 on the mP1c promoter (B) and the tubulin promoter (C). *P < 0.05, two-tailed u test; n = 4.
Regulation of AChE Promoter Structure After Treatment with HDACi.
To examine the effects of the two HDACi, TSA and NaBu, on the chromatin state of the AChE promoter region, we performed ChIP experiments for H3K9ac, H3K4me3, and H3K9me3 after HDAC inhibition. In line with the rescue of the expression level of AChE after HDAC inhibition, we found that the chromatin state of the mP1c promoter also was restored to its initial state: H3K9ac increased and H3K9me3 decreased (Fig. 3A, lane 3) back to their normal levels, whereas, once again, H3K4me3 did not change. The data were confirmed by qPCR on the AChE mP1c promoter (Fig. 3B). The Tubb3 promoter served as control (Fig. 3C). We note that although there was no change in the levels of H3K9me3 at the mP1a promoter after stress (Fig. 3A, lanes 1 and 2), this repressive mark showed decreased levels when stressed mice were treated with NaBu (Fig. 3A, lane 3). These results indicate that AChE 5′ alternative transcripts are regulated at the chromatin level by changes in acetylation and methylation of H3K9. They also show that HDACi rescue the stress-induced down-regulation of the AChE alternative exons mE1b and mE1c by modulating chromatin structure and causing the acetylation and methylation levels of H3K9 to revert to their normal levels.
Recently it was shown that during learning aged mice display a specific deregulation of histone H4 lysine 12 (H4K12) acetylation and fail to initiate a hippocampal gene-expression program associated with memory consolidation (24). It was shown further that histone H4 lysine 12 acetylation (H4K12ac) levels are altered in mouse neurons by a class of macrocyclic HDACi that shows mixed inhibition kinetics (25). To check whether H4K12ac may also be involved in the regulation of the long-lasting effect on AChE in our stress model, we performed ChIP assays for H4K12ac before and after stress, with and without HDACi treatment. We found a slight, NaBu-reversible increase in H4K12ac at the mP1a and mP1e promoters but no change in the levels of H4K12ac at the relevant mP1c promoter (Fig. S2), indicating that H4K12ac does not directly regulate the expression of mE1c or the acetylation state of the mP1c promoter.
DNA Methylation Remains Unaltered at the mP1c Promoter.
Several lines of evidence demonstrated that histone lysine methylation facilitates DNA methylation at specific promoter regions and that occasionally DNA methylation and histone methylation are linked functionally (10). To investigate whether the observed histone hypermethylation at the AChE mP1c promoter correlates with increased DNA methylation, we analyzed the DNA methylation status at CpG dinucleotides at the mP1c promoter region using bisulfite sequencing. We found that none of the CpG sites was methylated at the AChE mP1c promoter region before stress, and none became methylated after stress or after NaBu treatment (Fig. S3A). We supported these data further by performing ChIP with an antibody recognizing 5-methylcytidine (mDIP) on the mP1c promoter region in stressed mice and in stressed mice treated with NaBu. Once again we found that DNA methylation remained unchanged at the mP1c promoter (Fig. S3B). Mouse embryonic fibroblasts showing hypermethylation at the Oct4 promoter and R1 cells (mouse embryonic stem cells) showing hypomethylation at the Oct4 promoter region were used as controls (Fig. S3B). We therefore conclude that DNA methylation does not play a role in the long-term repression of the AChE mE1c promoter after stress.
HDAC4 Accumulates in Hippocampal Neurons After Stress.
To test the mechanism by which histone acetylation is reduced at the AChE mP1c promoter after stress, we examined the expression levels of several different HDACs before and after stress. Most HDACs were shown to be expressed predominantly in neurons in the adult brain (26). Therefore we selected HDACs that are inhibited by NaBu (i.e., HDAC1, 2, 4, 5, and 7) (27) and analyzed their expression level using qRT-PCR. We found a significant increase in the levels of HDACs 1, 2, 4, and 7 mRNAs 2 wk after repeated FSS and that HDAC4 displayed the most conspicuous rise in expression (∼20-fold increase) after stress (P < 10−5; Fig. 4). Remarkably, in all cases, the expression level reverted to normal when mice also were treated with NaBu (Fig. 4).
Fig. 4.
HDAC4 is elevated in the mouse hippocampus after stress. mRNA levels of Hdacs 1, 2, 4, 5, and 7 in the hippocampus were measured by qRT-PCR in nonstressed and stressed mice treated with either saline or NaBu. Hdac4 mRNA was significantly up-regulated (∼20-fold) in stressed mice as compared with controls. *P < 0.05; two-tailed u test; n = 4.
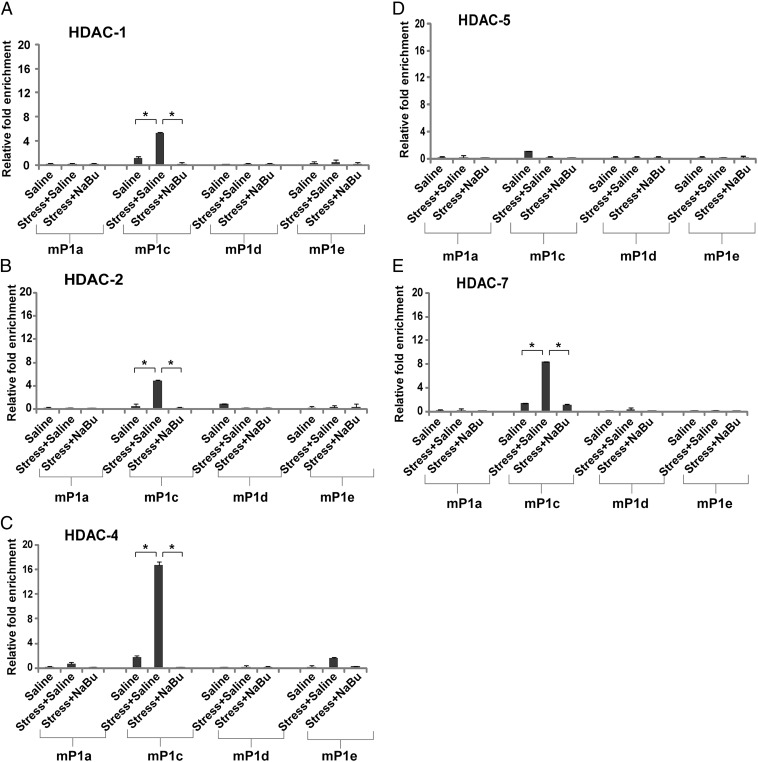
To test whether the increase in the expression level of the different HDACs resulted in the accumulation of their corresponding proteins on the AChE promoter, we performed further ChIP assays on the different HDACs, examining their association with the AChE mP1c promoter. Reassuringly, HDACs 1, 2, 4, and 7 displayed significant enrichment at the AChE mP1c promoter after stress (Fig. 5 A–C and E), and this enrichment also was restored completely when mice were treated with NaBu (Fig. 5 A–C and E). No changes were observed for HDAC5 (Fig. 5D). These results are in line with the expression results of the different HDACs (Fig. 4) and, once again, demonstrate the most pronounced enrichment of HDAC4. In silico analysis of the HDAC4 promoter revealed several potential glucocorticoid response elements, which likely respond to the increased corticosterone levels after stress, thereby increasing HDAC4 expression (Fig. S4). These data strengthen the notion that, of the different HDACs, HDAC4 is most likely to be involved directly in regulating the long-lasting effect of AChE after stress.
Fig. 5.
HDAC4 is enriched on the AChE mP1c promoter after stress. (A–E) ChIP was performed on different AChE promoters using antibodies for HDAC1 (A), HDAC2 (B), HDAC4 (C), HDAC5 (D), and HDAC7 (E) in the hippocampus of control, stressed, and NaBu-treated mice. HDAC4 showed almost exclusive enrichment (∼16-fold) at the AChE mP1c promoter. *P < 0.05; two-tailed u test; n = 4.
To test specifically the involvement of HDAC4 in AChE’s long-lasting stress-induced effects, we subjected mice to FSS for 4 d as in previous experiments, placed them back in their original cages for 1 wk, and injected lentiviral vectors expressing HDAC4 shRNAs or control shRNAs directly into the CA1 region of the hippocampus in the left hemisphere (Fig. 6A). Nonstressed mice injected with control shRNAs served as additional controls (Fig. 6A). We first confirmed that the virus silenced HDAC4 in vitro by immunoblots and by qRT-PCR (P < 0.05; Fig. S5 A and B) and in vivo by qRT-PCR (P < 0.05; Fig. 6B). To ensure selective knockdown, we further tested the expression levels of HDAC1, 2, and 7 after HDAC4 knockdown. We found that HDAC4 shRNAs do not affect other HDACs (1, 2, 7) and are selective for HDAC4 (Fig. S5C).
Fig. 6.
Regulation of AChE after HDAC4 silencing. (A) Schematic representation of the course of the experiment. After the FSS protocol was followed for 4 d, the mice were returned to their original cages for 1 wk and then were injected with either control shRNA (Ct-shRNA) virus or HDAC4 shRNA virus in the CA1 region of the hippocampus in the left hemisphere. Mice were killed 1 wk thereafter (n = 6). (B) In vivo validation of HDAC4 silencing. HDAC4 mRNA levels were increased significantly (approximately eightfold) after stress but were decreased by ∼80% when HDAC4 shRNA was administered. (C) Real-time RT-PCR analysis for mE1c in the hippocampus of unstressed or stressed mice treated with either control shRNA or HDAC4-shRNA. Lane 1, no stress/control shRNA; lane 2, stress/control shRNA; lane 3, stress/HDAC4 shRNA. HDAC4 silencing completely restored the levels of mE1c expression. (D) ChIP-qPCR for H3K9ac, H3K4me3, and H3K9me3 at the AChE promoter mP1c in the hippocampus after stress and HDAC4 silencing. (E) Time spent in the open arms in the elevated plus maze. Knockdown of HDAC4 restored the stress-induced anxiety-like behavior to normal behavior. *P < 0.05; two-tailed u test).
Depletion of HDAC4 in vivo completely restored the expression level of mE1c (P < 0.05; Fig. 6C) as well as the expression level of AChE- R and AChE-S (Fig. S6 A and B). We further checked whether HDAC4 silencing affects the structure of the AChE promoter. Using ChIP, we once again tested the active marks H3K9ac and H3K4me3 and the repressive mark H3K9me3. Although promoter regions mP1a, mP1d, and mP1e did not show any changes in the levels of the various histone modifications tested (Fig. S7), mP1c displayed full restoration of H3K9ac levels and a concomitant decrease of H3K9me3 to basal levels (P < 0.05; Fig. 6D), as was observed after NaBu administration (Fig. 3 B and C). H3K4me3 remained unaltered in all conditions (Fig. 6D). Finally, to test the functional significance of HDAC4 depletion in the hippocampus, we examined its effect on a well-documented stress-induced behavioral test, namely the elevated plus maze. Repeated stress normally causes anxiety-like behavior, demonstrated by a significant reduction in the time spent in the open arms of the maze (Fig. 6E) (28). Remarkably, hippocampal administration of HDAC4 shRNAs restored the time spent in the open arms nearly to the normal levels observed in the unstressed group (P < 0.05; Fig. 6E).
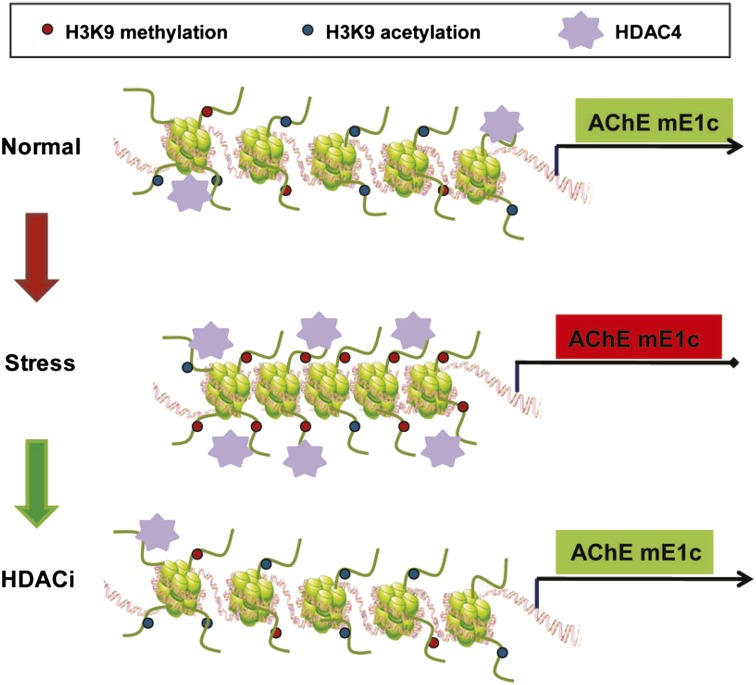
Taken together, our results depict a model in which stress induces long-lasting changes in AChE expression, modulated by hypoacetylation and hypermethylation of H3K9 at a specific promoter of AChE (mP1c). These results explain and suggest a mechanism for our previous observations showing long-lasting changes in AChE expression after stress (2). Intriguingly, restoration of the ground state of the AChE chromatin structure and the anxiety-like behavior after stress is mediated, at least partially, by HDAC4.
Discussion
A major challenge in neurocognition is identifying molecular mechanisms that underlie the enhanced formation of memory after exposure to stress. The hippocampus is a critical component of the neuroanatomical stress circuit. The FSS method is commonly used to examine stress-like behavior in mice. The reversal of stress by HDACi may offer an additional avenue to treat stress/depression in humans, and this model may be relevant to other psychiatric phenomena, such as fear, anxiety, social phobia, and posttraumatic stress disorder, as well. Using the FSS model, we found several lasting modifications in the hippocampus at the level of chromatin regulation at the AChE gene which precisely paralleled the changes in AChE gene expression.
Chromatin Regulation of the AChE Gene.
We view the AChE gene as a prototype case study of how the long-lasting effects of stress and HDAC inhibition presumably may affect changes in gene expression in the hippocampus and other brain regions at the level of chromatin regulation. Future genomewide experiments will be aimed at identifying these other genes and providing new leads toward understanding the pathophysiology and treatment of stress/depression. Although many questions remain regarding the mechanisms by which the long-lasting effects of stress and HDAC inhibition cause changes in chromatin architecture at the AChE gene and other genes, the results of the present study provide fundamental information concerning the detailed molecular mechanisms underlying the deleterious effects of stress on the brain and their potential reversal by treatment with HDACi. More generally, our results provide further support for the notion that chromatin regulation is an important mechanism controlling long-term adaptive changes in the brain associated with complex psychiatric conditions.
Changes in Histone Methylation at AChE Gene Promoters.
We found that H3K9me3 is enriched significantly after stress at one of the alternative 5′ AChE promoters and that the accumulation of this histone mark was reversed by treatment with NaBu. This modification is long-lasting, persisting at the AChE mP1c promoter 2 wk after the cessation of stress, suggesting a mechanism for long-term transcriptional memory. Indeed, treatment with HDACi significantly alters the hypermethylation level of H3K9me3 after stress. This finding could have profound implications for the treatment of anxiety-related disorders, and future work should focus on the timing and mode of administration. Current antidepressants are not very effective in curing stress/depression, and symptoms reappear in many patients after the discontinuation of treatment (29, 30). The concept that histone methylation regulates stress and memory also may suggest the inclusion of several histone demethylases (31, 32). Thus, the identification of these enzymes may lead to more selective therapeutic interventions that include the use of histone demethylase activators or inhibitors. This work underscores the importance of better understanding of the pathological effects of posttraumatic stress on the brain and the need for better therapeutic agents to reverse these effects. Our findings suggest that histone hypermethylation may represent a stable stress-induced molecular mechanism in the hippocampus, and perhaps elsewhere, and that the search for newer antidepressant agents will identify those that are more effective in demethylating histones at repressed genes.
Regulation of Chromatin by Acetylation and Deacetylation.
NaBu induced long-lasting H3 hyperacetylation at the AChE mP1c promoter in stressed mice. However, TSA showed variable effects. Our data suggest a model in which stress induces repression and HDAC inhibition induces derepression of the AChE mP1c promoter in the hippocampus. Specifically, FSS induces the specific trimethylation of histone H3K9, which persists long after the cessation of stress. This chromatin modification leads to a more “closed” chromatin state and thereby likely mediates the stable repression of two of the 5′ transcripts of the AChE gene (mE1b and mE1c). Hyperacetylation of the promoter, using HDACi, overcomes its methylation-induced repression and leads to a more “open” chromatin state at the AChE promoter, thus causing derepression of the AChE gene and possibly contributing to the antidepressant activity of HDACi. Because H3K9 can be either methylated (mono-, di-, or trimethylation) or acetylated, but not both, it is likely that reduced H3K9ac leads to H3K9 hypermethylation. This idea is supported by the observations that K9 methyltransferases are a part of the HDAC complexes (33, 34).
Chromatin regulation is a dynamic process in which several histones can be modified within close temporal and spatial proximity. As suggested by the histone code hypothesis, a combination of several histone modifications ultimately may determine the outcome of gene expression. Stress-induced changes in histone acetylation could be regulated via recruitment of specific HDACs or proteins that regulate these enzymes. It generally is believed that HDACs are controlled mainly at the level of their recruitment to target promoters (35). Previously, it was shown that HDAC2 has a negative function in regulating memory formation and that HDAC5 is associated with the down-regulation of certain brain-derived neurotrophic factor transcripts in a social defeat paradigm, a phenomenon that can be reversed by HDAC inhibition (12, 20).
A notable finding of the current study is that several HDACs (and especially HDAC4) are up-regulated after stress, that they (and especially HDAC4) accumulate at the AChE mP1c promoter, and that their overexpression and accumulation can be reversed by NaBu and by HDAC4 silencing. The fact that HDAC4 is accumulated at one specific promoter among the different promoters of AChE is intriguing and suggests local chromatin regulation, likely by accessibility. We postulate that we observe all changes in one particular promoter (mP1c) not only because it is the main promoter driving AChE expression but also because it is the only one that is open for manipulation in this poststress paradigm. The mechanism underlying this significant regulation effect could provide a unique experimental system for understanding this action. The role established for HDACs in long-term memory formation and long-lasting changes (12, 20) exemplifies how epigenetic mechanisms have the capacity to influence chromatin plasticity and stability in the nervous system. Additional characterization of these changes in HDAC4 will contribute to our understanding of the mechanisms of action of stress (potentially leading to treatment for severe stress/depression) and, more generally, shed light on the further molecular mechanisms governing gene regulation in the brain in vivo.
HDAC's Dual Regulation of Cognition and Inflammation?
That NaBu could restore the soluble AChE-R fully but restore the synaptic AChE-S only partially likely reflects stress-induced changes in the ability of acetylcholine to block inflammation (22) and thus also reduce anxiety (36). Given this specificity, our current findings suggest dual contributions of the long-lasting suppression of AChE-S after stress: (i) HDAC4-induced suppression of mP1c transcription that limits cholinergic neurotransmission and avoids stress-inducible anxiety and cognitive damages (37), and (ii) AChE-R–mediated destruction of extrasynaptic acetylcholine that reduces the cholinergic blockade of the production of proinflammatory cytokines (22, 38). The expression levels of the 5′ transcript mE1c and the 3′ transcripts AChE-R and AChE-S suggest that mE1c goes hand in hand with AChE-S. Unfortunately, we could not amplify the entire transcript from mE1c to AChE-S/AChE-R. It also should be noted that if the expression of one of the 3′ AChE variants goes up, the expression of other splice variant goes down, as we have seen in our analyses. Therefore, repressed AChE-R could result from overexpressed AChE-S and a shift of splicing rather than from mere repression. Additionally, other factors, such as the splicing factor SC35 (39), likely contribute to the AChE-S/AChE-R ratio, resulting in a complex regulatory network that operates in response to stress.
HDACi in Stress.
There are only a few examples of drug-induced chromatin regulation in the brain (40). The data show that changes induced in chromatin structure persist long after the treatment with HDACi and raise several questions regarding the underlying mechanisms involved. Such long-lived changes in chromatin regulation might be one of the crucial mechanisms for HDACi-induced neuroadaptations in the brain. HDACi cause increased acetylation of histones in intact cells and are being explored as treatment for several diseases, such as certain cancers (41), poly-glutamine-related (PolyQ) diseases (42, 43), schizophrenia (44), and spinal muscular atrophy (45). Our data presented here suggest that HDACi, and especially HDAC4, also may be used for treating stress-related neuropathologies. In addition, these findings complement an emerging literature suggesting an important role for epigenetic molecular mechanisms in memory formation. These data suggest that inhibition of HDACs might be a suitable therapeutic avenue for neurodegenerative diseases associated with learning and memory impairment and raise the possibility of recovery of long-term memories in patients with stress/depression.
Materials and Methods
Antibodies.
The following ChIP-grade antibodies were used: H3K9me3 (T. Jenuwein, Max Planck Institute, Freiburg, Germany); H3K4me3 (05–745; Millipore); H3K9ac (06–942; Millipore); HDAC1 (PA1-860; ABR); HDAC2 (05–814; Millipore); HDAC4 (H9411; Sigma); HDAC5 (H8163; Sigma), and HDAC7 (H6663; Sigma). 2 μg/mL of each antibody was used for all ChIP experiments.
Animals.
Adult (7- to 8-wk-old) male FVB/N mice were used in all experiments. The FSS protocol was as described (2). In all experiments, mice were singly housed and maintained on a 12-h light/dark cycle with access to food and water ad libitum. All animal procedures were carried out in accordance with Institutional Animal Care and Use guidelines. Mice were injected daily i.p. with sodium butyrate (100 mg/kg body weight), TSA (1 mg/kg), or saline after 1 wk of stress. Mice were killed 24 h after the last injection by quick anesthesia with isoflurane (<15 s) followed by cervical dislocation. Mice were dissected, and hippocampi were collected and snap-frozen in liquid nitrogen and stored at −80 °C.
mRNA Analysis.
RNA was purified from hippocampal tissue using the RNeasy kit (Qiagen) with DNase treatment (Promega). mRNA was reverse transcribed to cDNA using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Primer sequences are provided in Table S1. CyP and Tubb were used as controls. All PCR reactions were repeated at least three independent times.
ChIP Assays.
ChIP assays were performed as described (46). More details are provided in SI Materials and Methods.
Lentiviral Procedures.
GIPZ lentiviral shRNA microRNA (miR) clones for HDAC4 and Nonsilencing/Ct-GIPZ lentiviral shRNA miR control (Open Biosystems) were packaged by second-generation packaging viruses according to standard protocols as described (47). The packaged virus was collected at 24 and 48 h posttransfection and concentrated using ultracentrifugation at 25,000 × g for 90 min at 4 °C. The concentrated virus was diluted, and HEK-293T cells were infected with the diluted virus. The resulting titer (1× 109 infectious particles/mL) was assessed for shRNA viruses. Viral-mediated HDAC4-silencing levels were confirmed in vitro by qRT-PCR and Western blotting on primary cultured neurons, which were grown from embryonic day 14 mice embryos described (46).
Stereotactic Surgery.
Mice were anesthetized by i.p. injections of ketamine (50 mg/kg) (Fort Dodge) and Domitor (0.5 mg/kg) (Orion Pharma) mix and then were mounted on a stereotactic apparatus for intrahippocampal injections. Coordinates of the injection sites (in mm) relative to bregma were anteroposterior, 2.0; lateral, 1.8; and dorsoventral, −1.6. Injections of 0.5-μL lentiviral suspensions were conducted using a 10-μL Hamilton syringe on the CA1 region of the left hippocampus. After each injection, the needle was left in situ for 5 min before retraction to allow complete diffusion. All mice were fully awake and functional within 10 min after anesthesia was discontinued. Mouse behavior was tested 1 wk after viral injection.
Supplementary Material
Acknowledgments
We thank T. Jenuwein for H3K9me3 antibodies. This research was supported by grants from the Edmond J. Safra Foundation (to E.M.) and the Israel Institute for Psychobiology (to E.M.) and by the Gatsby Foundation and Grant 1799/10 from the Israel Science Foundation (to H.S.). E.M. is a Joseph H. and Belle R. Braun Senior Lecturer in Life Sciences. B.S.S. is a Safra Fellow, and G.Z. was awarded a predoctoral fellowship by the Safra Center. D.C-C. is supported by an Eshkol Fellowship from the Israel Ministry of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 21197 (volume 109, number 52).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209990110/-/DCSupplemental.
References
- 1.Kaufer D, Friedman A, Seidman S, Soreq H. Acute stress facilitates long-lasting changes in cholinergic gene expression. Nature. 1998;393(6683):373–377. doi: 10.1038/30741. [DOI] [PubMed] [Google Scholar]
- 2.Meshorer E, et al. Alternative splicing and neuritic mRNA translocation under long-term neuronal hypersensitivity. Science. 2002;295(5554):508–512. doi: 10.1126/science.1066752. [DOI] [PubMed] [Google Scholar]
- 3.Huang LT, et al. Maternal deprivation stress exacerbates cognitive deficits in immature rats with recurrent seizures. Epilepsia. 2002;43(10):1141–1148. doi: 10.1046/j.1528-1157.2002.14602.x. [DOI] [PubMed] [Google Scholar]
- 4.Crosio C, Heitz E, Allis CD, Borrelli E, Sassone-Corsi P. Chromatin remodeling and neuronal response: Multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J Cell Sci. 2003;116(Pt 24):4905–4914. doi: 10.1242/jcs.00804. [DOI] [PubMed] [Google Scholar]
- 5.Guan Z, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111(4):483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 6.Abad M, et al. Ing1 mediates p53 accumulation and chromatin modification in response to oncogenic stress. J Biol Chem. 2007;282(42):31060–31067. doi: 10.1074/jbc.M701639200. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 9.Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116(Pt 11):2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 10.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14(3):286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 11.Takizawa T, Meshorer E. Chromatin and nuclear architecture in the nervous system. Trends Neurosci. 2008;31(7):343–352. doi: 10.1016/j.tins.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Tsankova NM, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9(4):519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 13.McEwen BS. Plasticity of the hippocampus: Adaptation to chronic stress and allostatic load. Ann N Y Acad Sci. 2001;933:265–277. doi: 10.1111/j.1749-6632.2001.tb05830.x. [DOI] [PubMed] [Google Scholar]
- 14.Meshorer E, et al. Combinatorial complexity of 5′ alternative acetylcholinesterase transcripts and protein products. J Biol Chem. 2004;279(28):29740–29751. doi: 10.1074/jbc.M402752200. [DOI] [PubMed] [Google Scholar]
- 15.Meaney MJ, Szyf M. Environmental programming of stress responses through DNA methylation: Life at the interface between a dynamic environment and a fixed genome. Dialogues Clin Neurosci. 2005;7(2):103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsankova NM, Kumar A, Nestler EJ. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J Neurosci. 2004;24(24):5603–5610. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levenson JM, et al. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279(39):40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 18.Duman RS. Pathophysiology of depression: The concept of synaptic plasticity. Eur Psychiatry. 2002;17(Suppl 3):306–310. doi: 10.1016/s0924-9338(02)00654-5. [DOI] [PubMed] [Google Scholar]
- 19.Uchida S, et al. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69(2):359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459(7243):55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haggarty SJ, Tsai L-H. Probing the role of HDACs and mechanisms of chromatin-mediated neuroplasticity. Neurobiol Learn Mem. 2011;96(1):41–52. doi: 10.1016/j.nlm.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaked I, et al. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31(6):965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Newton SS, et al. Gene profile of electroconvulsive seizures: Induction of neurotrophic and angiogenic factors. J Neurosci. 2003;23(34):10841–10851. doi: 10.1523/JNEUROSCI.23-34-10841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 25.Marcaurelle LA, et al. An aldol-based build/couple/pair strategy for the synthesis of medium- and large-sized rings: Discovery of macrocyclic histone deacetylase inhibitors. J Am Chem Soc. 2010;132(47):16962–16976. doi: 10.1021/ja105119r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broide RS, et al. Distribution of histone deacetylases 1-11 in the rat brain. J Mol Neurosci. 2007;31(1):47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 27.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133(7, Suppl):2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 28.Walf AA, Frye CA. The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat Protoc. 2007;2(2):322–328. doi: 10.1038/nprot.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nestler EJ, et al. Neurobiology of depression. Neuron. 2002;34(1):13–25. doi: 10.1016/s0896-6273(02)00653-0. [DOI] [PubMed] [Google Scholar]
- 30.Manji HK, Drevets WC, Charney DS. The cellular neurobiology of depression. Nat Med. 2001;7(5):541–547. doi: 10.1038/87865. [DOI] [PubMed] [Google Scholar]
- 31.Gupta S, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30(10):3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA. 2009;106(49):20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292(5514):110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 34.Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293(5539):2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 35.West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3(12):921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- 36.Zimmerman G, et al. Post-traumatic anxiety associates with failure of the innate immune receptor TLR9 to evade the pro-inflammatory NFκB pathway. TranslPsychiatry. 2012;2:e78. doi: 10.1038/tp.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaltiel G, et al. Hippocampal microRNA-132 mediates stress-inducible cognitive deficits through its acetylcholinesterase target. Brain Struct Funct. 2012 doi: 10.1007/s00429-011-0376-z. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosas-Ballina M, et al. Acetylcholine-synthesizing T cells relay neural signals in a vagus nerve circuit. Science. 2011;334(6052):98–101. doi: 10.1126/science.1209985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meshorer E, et al. SC35 promotes sustainable stress-induced alternative splicing of neuronal acetylcholinesterase mRNA. Mol Psychiatry. 2005;10(11):985–997. doi: 10.1038/sj.mp.4001735. [DOI] [PubMed] [Google Scholar]
- 40.Covington HE, 3rd, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29(37):11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marks PA, et al. Histone deacetylases and cancer: Causes and therapies. Nat Rev Cancer. 2001;1(3):194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 42.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 43.Gray SG. Targeting histone deacetylases for the treatment of Huntington’s disease. CNS Neurosci Ther. 2010;16(6):348–361. doi: 10.1111/j.1755-5949.2010.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guidotti A, et al. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology. 2011;60(7-8):1007–1016. doi: 10.1016/j.neuropharm.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minamiyama M, et al. Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2004;13(11):1183–1192. doi: 10.1093/hmg/ddh131. [DOI] [PubMed] [Google Scholar]
- 46.Sailaja BS, Takizawa T, Meshorer E. Chromatin immunoprecipitation in mouse hippocampal cells and tissues. Methods Mol Biol. 2012;809:353–364. doi: 10.1007/978-1-61779-376-9_24. [DOI] [PubMed] [Google Scholar]
- 47.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1(1):241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]