Abstract
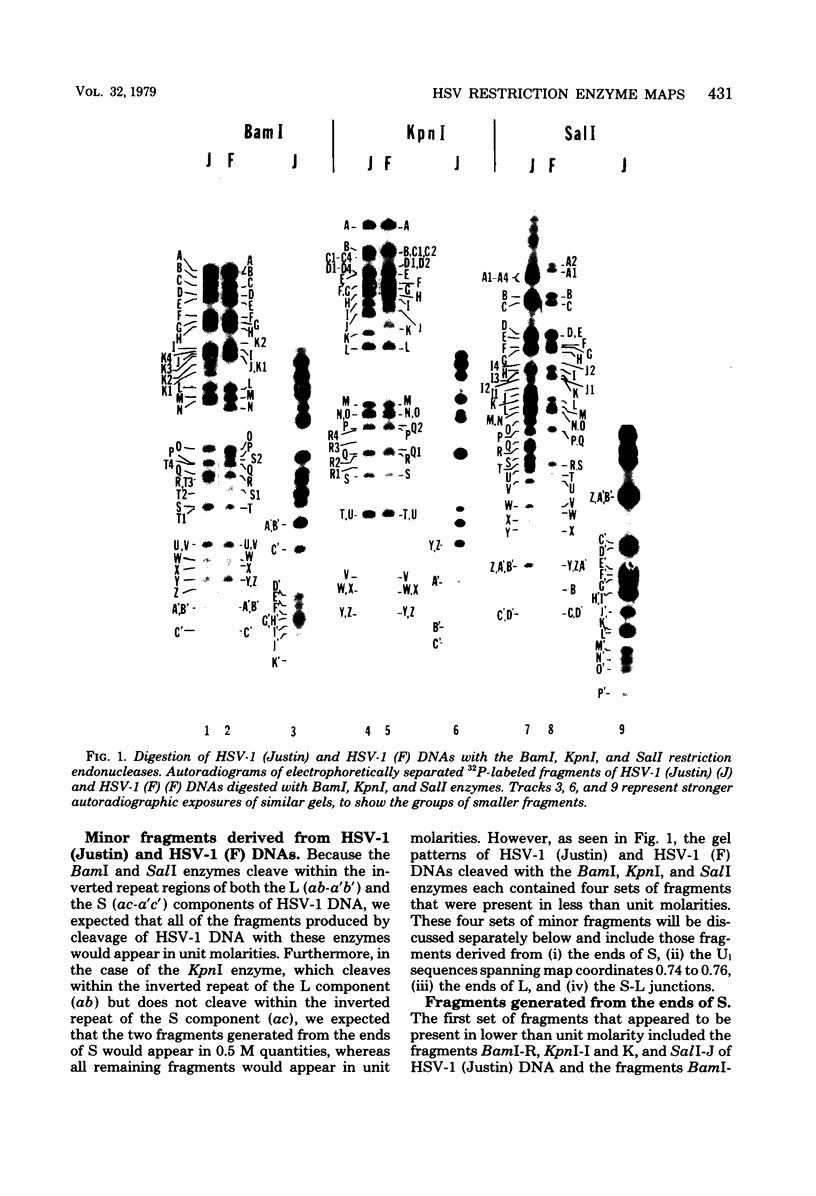
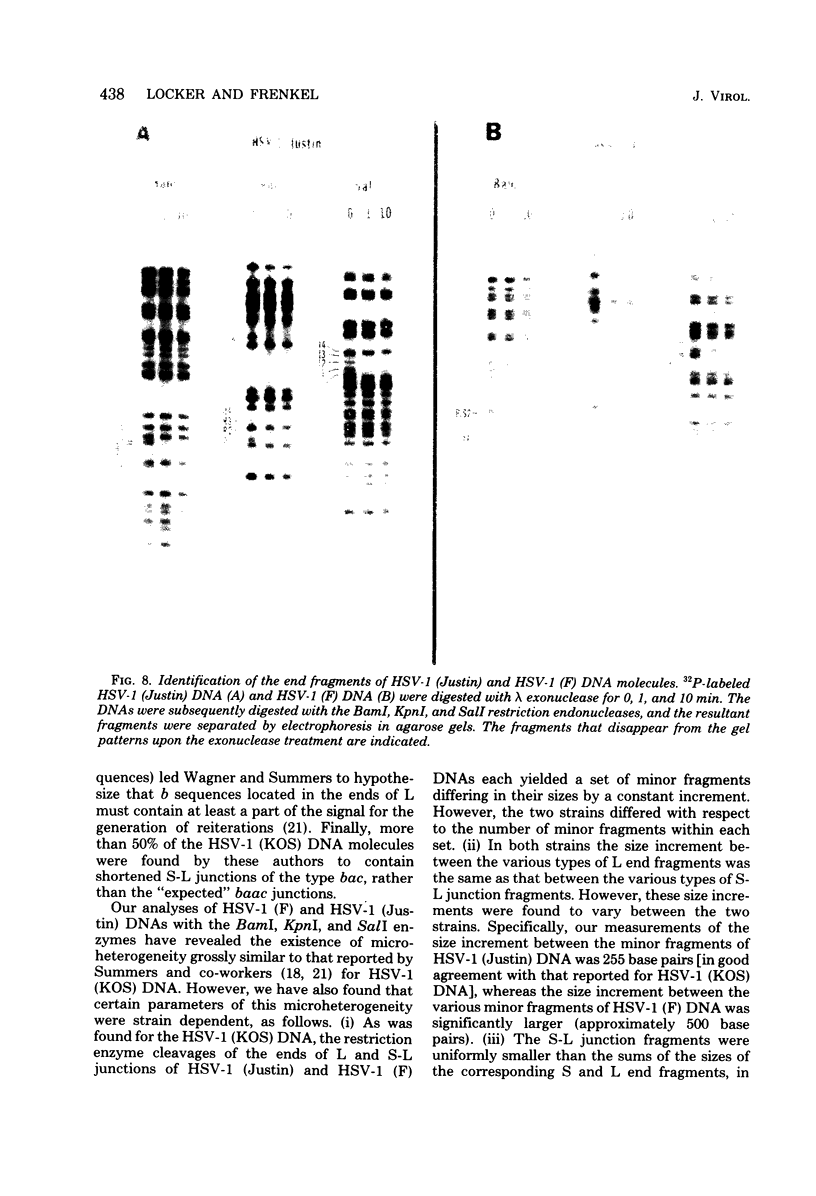
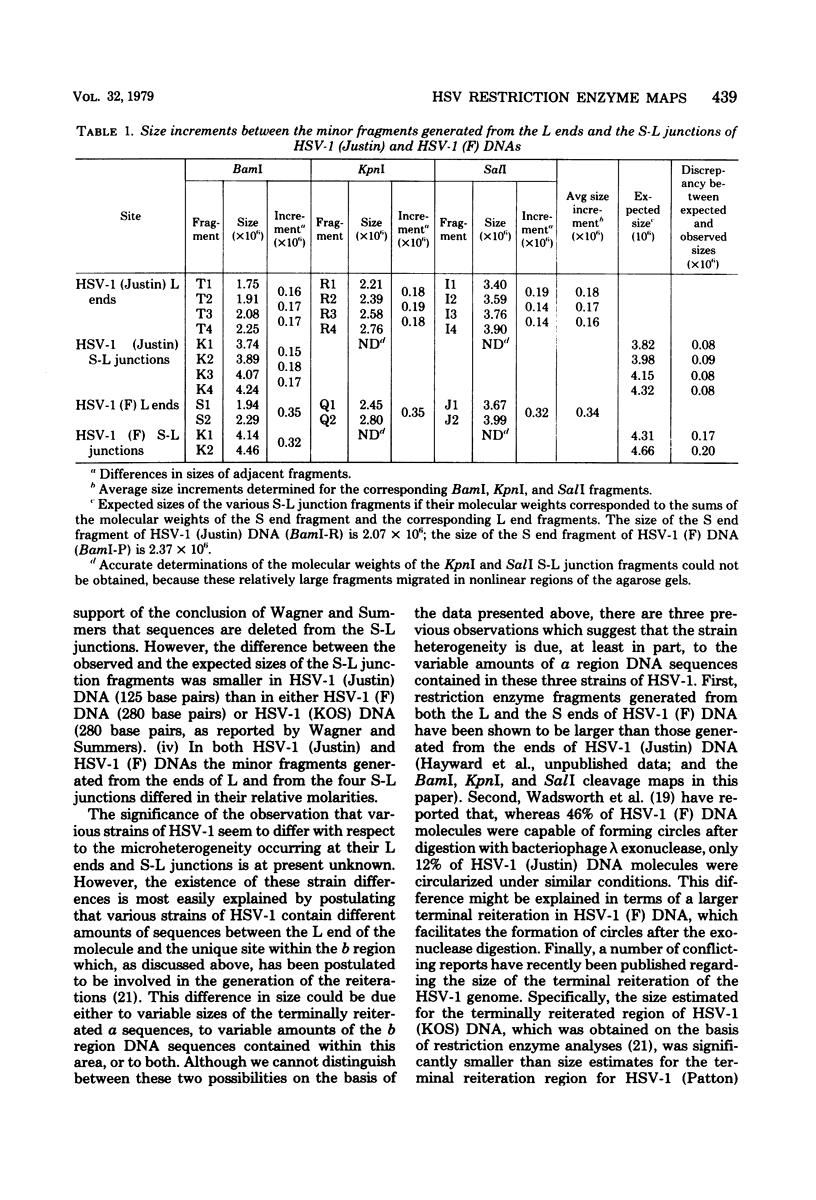
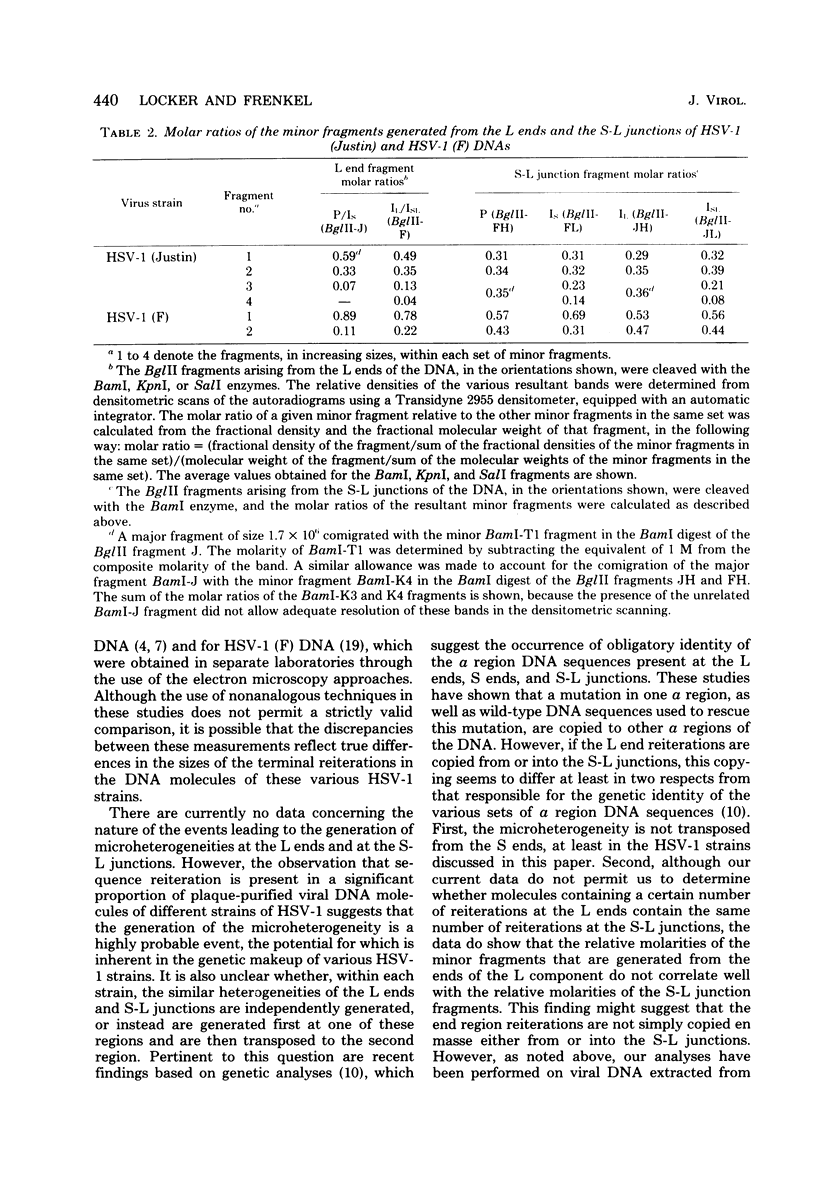
We present the locations of the cleavage sites for the BamI, KpnI, and SalI restriction endonucleases within the DNA molecules of herpes simplex virus type 1 (HSV-1) strains Justin and F. These restriction enzymes cleave the HSV-1 DNA at many sites, producing relatively small fragments which should prove useful in future studies of HSV-1 gene structure and function. The mapping data revealed the occurrence of heterogeneity within three regions of the viral genome including (i) the region spanning map coordinates 0.74--0.76, (ii) the ends of the large (L) DNA component, and (iii) the junction between the large (L) and the small (S) components. The heterogeneity in the ends of L and the S-L junctions of HSV-1 (Justin) and HSV-1 (F) DNAs was grossly similar to that previously reported to occur in the ends of L and the S-L junctions of the HSV-1 (KOS) DNA (M. J. Wagner and W. C. Summers, J. Virol. 27:374--387, 1978). Thus, cleavage of these regions with restriction endonucleases yielded sets of minor fragments differing in size by constant increments. However, the various strains of HSV-1 differed with respect to the numbers, size increments, and relative molarities of the various minor fragments, suggesting that the parameters of the heterogeneity are inherited in the structural makeup of the HSV-1 genome. The strain dependence of the pattern of heterogeneity can be most easily explained in terms of variable sizes of the terminally reiterated a sequence, contained in the DNA molecules of these three strains of HSV-1.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker Y., Dym H., Sarov I. Herpes simplex virus DNA. Virology. 1968 Oct;36(2):184–192. doi: 10.1016/0042-6822(68)90135-9. [DOI] [PubMed] [Google Scholar]
- Clements J. B., Cortini R., Wilkie N. M. Analysis of herpesvirus DNA substructure by means of restriction endonucleases. J Gen Virol. 1976 Feb;30(2):243–256. doi: 10.1099/0022-1317-30-2-243. [DOI] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W., Hyman R. W. The terminal repetition of herpes simplex virus DNA. Virology. 1975 Sep;67(1):144–157. doi: 10.1016/0042-6822(75)90412-2. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman R. W., Burke S., Kudler L. A nearby inverted repeat of the terminal sequence of herpes simplex virus DNA. Biochem Biophys Res Commun. 1976 Jan 26;68(2):609–615. doi: 10.1016/0006-291x(76)91189-x. [DOI] [PubMed] [Google Scholar]
- Jacob R. J., Morse L. S., Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979 Feb;29(2):448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W. An exonuclease induced by bacteriophage lambda. II. Nature of the enzymatic reaction. J Biol Chem. 1967 Feb 25;242(4):679–686. [PubMed] [Google Scholar]
- Locker H., Frenkel N. Structure and origin of defective genomes contained in serially passaged herpes simplex virus type 1 (Justin). J Virol. 1979 Mar;29(3):1065–1077. doi: 10.1128/jvi.29.3.1065-1077.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse L. S., Buchman T. G., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus DNA. IX. Apparent exclusion of some parental DNA arrangements in the generation of intertypic (HSV-1 X HSV-2) recombinants. J Virol. 1977 Oct;24(1):231–248. doi: 10.1128/jvi.24.1.231-248.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston V. G., Davison A. J., Marsden H. S., Timbury M. C., Subak-Sharpe J. H., Wilkie N. M. Recombinants between herpes simplex virus types 1 and 2: analyses of genome structures and expression of immediate early polypeptides. J Virol. 1978 Nov;28(2):499–517. doi: 10.1128/jvi.28.2.499-517.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. V. Terminally repetitive sequences. J Virol. 1976 Feb;17(2):503–512. doi: 10.1128/jvi.17.2.503-512.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. J., Summers W. C. Structure of the joint region and the termini of the DNA of herpes simplex virus type 1. J Virol. 1978 Aug;27(2):374–387. doi: 10.1128/jvi.27.2.374-387.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R. Sequence arrangement in herpes simplex virus type 1 DNA: identification of terminal fragments in restriction endonuclease digests and evidence for inversions in redundant and unique sequences. J Virol. 1976 Oct;20(1):211–221. doi: 10.1128/jvi.20.1.211-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]







