Abstract
Viable mutants of polyoma with small deletions ranging in size from 2 to 75 base pairs were obtained by infecting 3T3 cells with polyoma DNA that had been cleaved once with HaeII endonuclease or with DNase-Mn2+ digestion. The HaeII endonuclease-cleaved DNA yielded mutants with deletions at map position 72--73, whereas the mutants generated by DNase I-Mn2+ digestion had deletions either at map position 72--73 or within the map coordinates 92 and 99. Both groups of mutants appeared to grow as well as wild-type virus in 3T3 cells. The deletions at map position 72--73 did not alter the virus's ability to transform rat cells. Hence, the region just to the early side of the origin of DNA replication is not essential for vegetative growth or transformation. But the mutants with deletions in the region between map coordinates 92 and 99, a segment thought to code for polyoma large and middle T antigens (Hutchinson et al., Cell 15:65--77, 1978; Smart and Ito, Cell 15:1427--1437, 1978; Soeda et al., Cell 17:357--370, 1979), transformed rat cells at 0.2 to 0.05 the efficiency of wild-type virus.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon J., Shenk T. E., Berg P. Biochemical procedure for production of small deletions in simian virus 40 DNA. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1392–1396. doi: 10.1073/pnas.72.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Landers T., Goff S. P., Manteuil-Brutlag S., Berg P. Physical and genetic characterization of deletion mutants of simian virus 40 constructed in vitro. J Virol. 1977 Oct;24(1):277–294. doi: 10.1128/jvi.24.1.277-294.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras R., Cole C., Berg P., Fiers W. Nucleotide sequence analysis of two simian virus 40 mutants with deletions in the late region of the genome. J Virol. 1979 Feb;29(2):789–793. doi: 10.1128/jvi.29.2.789-793.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W. Complementation between temperature-sensitive (ts) and host range nontransforming (hr-t) mutants of polyoma virus. Virology. 1977 Apr;77(2):589–597. doi: 10.1016/0042-6822(77)90484-6. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell A. J., Cowie A., Legon S., Kamen R. Multiple 5' terminal cap structures in late polyoma virus RNA. Cell. 1979 Feb;16(2):357–371. doi: 10.1016/0092-8674(79)90012-6. [DOI] [PubMed] [Google Scholar]
- Fluck M. M., Staneloni R. J., Benjamin T. L. Hr-t and ts-a: two early gene functions of polyoma virus. Virology. 1977 Apr;77(2):610–624. doi: 10.1016/0042-6822(77)90486-x. [DOI] [PubMed] [Google Scholar]
- Folk W. R., Wang H. C. Closed circular DNAs with tandem repeats of a sequence from polyoma virus. Virology. 1974 Sep;61(1):140–155. doi: 10.1016/0042-6822(74)90249-9. [DOI] [PubMed] [Google Scholar]
- Fried M., Griffin B. E. Organization of the genomes of polyoma virus and SV40. Adv Cancer Res. 1977;24:67–113. doi: 10.1016/s0065-230x(08)61013-1. [DOI] [PubMed] [Google Scholar]
- Friedmann T., LaPorte P., Esty A. Nucleotide sequence studies of polyoma DNA. The Hpa II 3/5 junction to the Hpa II 4/Hae III 18 junction, encoding the origin of DNA replication and the 5' end of the early region. J Biol Chem. 1978 Sep 25;253(18):6561–6567. [PubMed] [Google Scholar]
- Griffin B. E. Fine structure of polyoma virus DNA. J Mol Biol. 1977 Dec 5;117(2):447–471. doi: 10.1016/0022-2836(77)90137-1. [DOI] [PubMed] [Google Scholar]
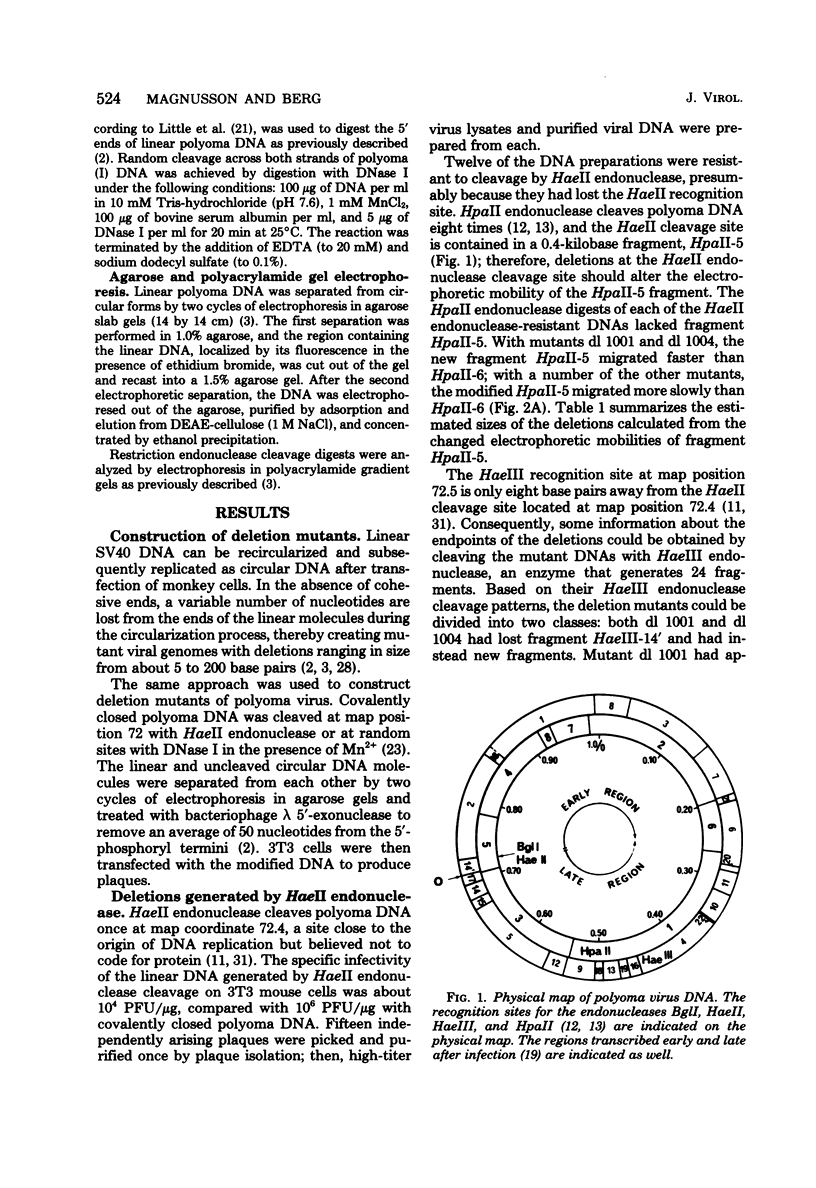
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
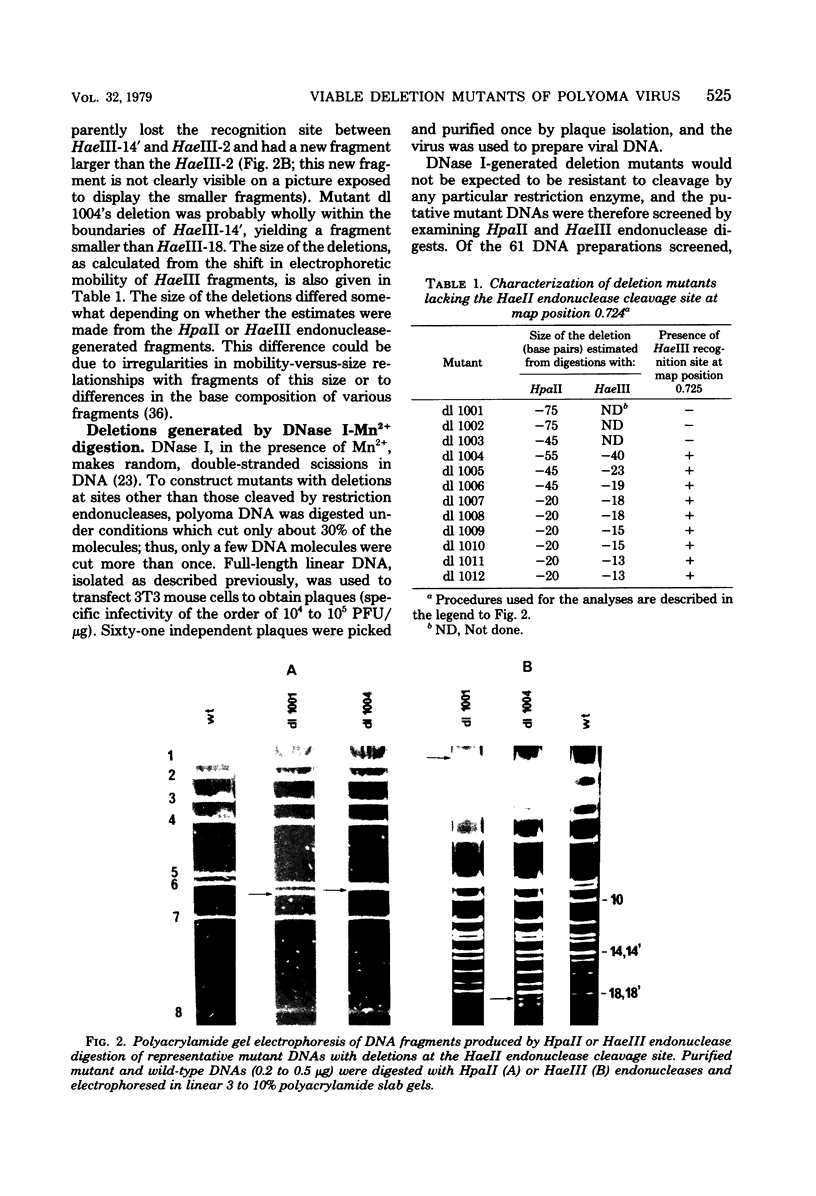
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hunter T., Gibson W. Characterization of the mRNA's for the polyoma virus capsid proteins VP1, VP2, and VP3. J Virol. 1978 Oct;28(1):240–253. doi: 10.1128/jvi.28.1.240-253.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Shure H. Topography of polyoma virus messenger RNA molecules. Cell. 1976 Mar;7(3):361–371. doi: 10.1016/0092-8674(76)90165-3. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. Deletion mutants of simian virus 40 generated by enzymatic excision of DNA segments from the viral genome. J Mol Biol. 1974 Oct 15;89(1):179–193. doi: 10.1016/0022-2836(74)90169-7. [DOI] [PubMed] [Google Scholar]
- Little J. W., Lehman I. R., Kaiser A. D. An exonuclease induced by bacteriophage lambda. I. Preparation of the crystalline enzyme. J Biol Chem. 1967 Feb 25;242(4):672–678. [PubMed] [Google Scholar]
- Magnusson G., Nilsson M. G. Multiple free viral DNA copies in polyoma virus-transformed mouse cells surviving productive infection. J Virol. 1977 Jun;22(3):646–653. doi: 10.1128/jvi.22.3.646-653.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melgar E., Goldthwait D. A. Deoxyribonucleic acid nucleases. II. The effects of metals on the mechanism of action of deoxyribonuclease I. J Biol Chem. 1968 Sep 10;243(17):4409–4416. [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Defective simian virus 40 genomes: isolation and growth of individual clones. Virology. 1974 Nov;62(1):112–124. doi: 10.1016/0042-6822(74)90307-9. [DOI] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of the genetic map of the polyoma genome. J Virol. 1976 Jun;18(3):824–832. doi: 10.1128/jvi.18.3.824-832.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morhenn V., Rabinowitz Z., Tomkins G. M. Effects of adrenal glucocorticoids on polyoma virus replication. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1088–1089. doi: 10.1073/pnas.70.4.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart J. E., Ito Y. Three species of polyoma virus tumor antigens share common peptides probably near the amino termini of the proteins. Cell. 1978 Dec;15(4):1427–1437. doi: 10.1016/0092-8674(78)90066-1. [DOI] [PubMed] [Google Scholar]
- Smith A. E., Kamen R., Mangel W. F., Shure H., Wheeler T. Location of the sequences coding for capsid proteins VP1 and VP2 on polyoma virus DNA. Cell. 1976 Nov;9(3):481–487. doi: 10.1016/0092-8674(76)90093-3. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Soeda E., Griffin B. E. Sequences from the genome of a non-transforming mutant of polyoma virus. Nature. 1978 Nov 16;276(5685):294–298. doi: 10.1038/276294a0. [DOI] [PubMed] [Google Scholar]
- Thimmappaya B., Shenk T. Nucleotide sequence analysis of viable deletion mutants lacking segments of the simian virus 40 genome coding for small t antigen. J Virol. 1979 Jun;30(3):668–673. doi: 10.1128/jvi.30.3.668-673.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn H., Cole C., Berg P., Fiers W. Nucleotide sequence analysis of two simian virus 40 mutants with deletions in the region coding for the carboxyl terminus of the T antigen. J Virol. 1979 Jun;30(3):936–941. doi: 10.1128/jvi.30.3.936-941.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert G., Feunteun J., Crawford L. V., Berg P., Fiers W. Nucleotide sequence deletions within the coding region for small-t antigen of simian virus 40. J Virol. 1979 Jun;30(3):674–682. doi: 10.1128/jvi.30.3.674-682.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger R. S., Salomon R., Dingman C. W., Peacock A. C. Role of base composition in the electrophoresis of microbial and crab DNA in polyacrylamide gels. Nat New Biol. 1972 Jul 19;238(81):65–69. doi: 10.1038/newbio238065a0. [DOI] [PubMed] [Google Scholar]