Abstract
The interaction of Candida albicans with phagocytes of the host's innate immune system is highly dynamic, and its outcome directly impacts the progression of infection. While the switch to hyphal growth within the macrophage is the most obvious physiological response, much of the genetic response reflects nutrient starvation: translational repression and induction of alternative carbon metabolism. Changes in amino acid metabolism are not seen, with the striking exception of arginine biosynthesis, which is upregulated in its entirety during coculture with macrophages. Using single-cell reporters, we showed here that arginine biosynthetic genes are induced specifically in phagocytosed cells. This induction is lower in magnitude than during arginine starvation in vitro and is driven not by an arginine deficiency within the phagocyte but instead by exposure to reactive oxygen species (ROS). Curiously, these genes are induced in a narrow window of sublethal ROS concentrations. C. albicans cells phagocytosed by primary macrophages deficient in the gp91phox subunit of the phagocyte oxidase do not express the ARG pathway, indicating that the induction is dependent on the phagocyte oxidative burst. C. albicans arg pathway mutants are retarded in germ tube and hypha formation within macrophages but are not notably more sensitive to ROS. We also find that the ARG pathway is regulated not by the general amino acid control response but by transcriptional regulators similar to the Saccharomyces cerevisiae ArgR complex. In summary, phagocytosis induces this single amino acid biosynthetic pathway in an ROS-dependent manner.
INTRODUCTION
Candida albicans is the most prominent fungal component of the human microbiome, residing within multiple body sites, including the skin, oral cavity, gastrointestinal tract, and vagina. C. albicans causes a spectrum of infections in otherwise healthy individuals with or without predisposing risk factors, such as vulvovaginal candidiasis, oropharyngeal thrush, and various cutaneous infections. In individuals with compromised immunity, disseminated candidiasis may result in severe disease with a mortality approaching 40%, a rate that has not changed in decades (1, 2). An increasing incidence of hospital-acquired disseminated candidiasis has been observed in recent years as the population of susceptible patients with impaired immune systems has risen. As a successful fungal pathogen, C. albicans possesses multiple virulence attributes, including filamentous growth, biofilm formation, numerous secreted hydrolases, and the ability to sense and adapt to numerous environmental changes within the host in order to facilitate establishment of an infection (reviewed in reference 3).
Invasive candidemia is prevented by the collective efforts of the innate immune system. Protective physical barriers such as epithelial integrity are the first line of defense against invading Candida. Macrophages and neutrophils, phagocytes that will internalize C. albicans upon recognition, are also key antifungal effectors. Following phagocytosis, a strong respiratory burst results in the production of antimicrobial reactive oxygen species (ROS) generated by the phagocyte NADPH oxidase and myeloperoxidase (MPO), nitric oxide (NO) generated by NOS2 (inducible nitric oxide synthase [iNOS]), and other reactive nitrogen species (RNS) (reviewed in reference 4). The contribution of this respiratory burst to the prevention of a C. albicans infection varies in different phagocytes and with different models of disease. Mice deficient in the gp91phox subunit of NADPH oxidase mimic the immune deficiencies observed in chronic granulomatous disease (CGD) and are more susceptible to C. albicans infections than normal mice (5, 6). However, phagocytes from gp91phox−/−/NOS2−/− mice were able to kill C. albicans just as effectively in vitro as cells from wild-type mice, even though these gp91phox−/−/NOS2 −/− mice are more susceptible to a variety of bacterial and fungal infections, including Candida guilliermondii and C. albicans (7, 8). Neutrophil MPO is critical in host defense against pulmonary and intraperitoneal C. albicans infections (9, 10). In a zebrafish larval model, loss of host NADPH oxidase activity results in enhanced filamentous growth of C. albicans, a critical virulence trait for this organism, and increased susceptibility to infection (11). It seems clear that phagocyte-derived ROS are an important, but by no means the only, component of antifungal defenses in whole animals. While hyphal growth is critical for the escape of C. albicans from phagocytic cells such as macrophages, numerous physiological changes are elicited upon recognition of phagocytes, and a striking diversity of means by which Candida modulates immune function is just beginning to be revealed (reviewed in reference 12).
Previously, transcript profiling experiments were performed to identify changes in C. albicans gene expression in response to macrophage phagocytosis (13). Upon internalization, C. albicans generates a rapid metabolic response by downregulating glycolysis genes while simultaneously upregulating genes involved in fatty acid utilization, the glyoxylate cycle, and gluconeogenesis. These alternative carbon utilization pathways are necessary for full virulence in the mouse tail vein injection model of disseminated candidiasis (14–16). While part of this reprogramming of transcription is broadly similar to changes in gene expression due to carbon starvation, hierarchal clustering identified a second set of genes whose regulation was independent of starvation. Genes belonging to this nonstarvation cluster included those involved in oxidative stress response pathways, DNA damage repair pathways, and uptake and utilization of peptides, metals, and other small molecules. The only metabolic gene pathway that did not partition with the starvation cluster but instead was part of the nonstarvation, stress response cluster was the arginine (ARG) biosynthetic pathway, where all but one gene was upregulated at least 3-fold (13). The ARG genes are also upregulated in cells phagocytosed by neutrophils, one of the few transcriptional responses shared by these two data sets (13, 17).
In this study, we demonstrated that the arginine biosynthetic pathway genes are induced specifically in phagocytosed cells. ARG pathway genes are also induced by multiple oxidative stress-inducing agents in vitro, and expression of ARG genes in phagocytosed cells is dependent on the macrophage oxidative burst, as this does not occur when the macrophages lack the NADPH oxidase. C. albicans strains containing a deletion of either ARG1 or ARG3 have no obvious sensitivity to ROS in vitro but possess hyphal morphogenesis delays within macrophages. We have identified homologs of the Saccharomyces cerevisiae arginine regulatory (ArgR) complex (Arg80p, Arg81p, Arg82p, and Mcm1p) and find that, while broadly similar, C. albicans lacks an ARG80 homolog but has duplicated ARG81. ArgR is primarily a repressor of gene expression, as in S. cerevisiae (18). The C. albicans ARG genes appear to be independent of the general amino acid control response, in which starvation for any one amino acid activates multiple biosynthetic pathways via the Gcn4p transcription factor in both yeast (reviewed in reference 19) and C. albicans (20). Gcn4p also targets ArgR to ARG promoters as both a positive and negative regulator in yeast (21), but we show that it serves mostly as a repressor of C. albicans ARG loci. Taken together, these data suggest that in response to the oxidative burst generated by macrophages following phagocytosis, C. albicans specifically induces arginine biosynthetic pathway genes to promote filamentous growth within macrophages.
MATERIALS AND METHODS
Strains and media.
C. albicans strains and plasmids are listed in Tables S1 and S2, respectively, in the supplemental material. Strains were propagated under standard conditions in YPD (1% yeast extract, 2% peptone, 2% glucose) or YNB (0.17% yeast nitrogen base without amino acids, 0.5% ammonium sulfate, 2% glucose) (22). Amino acids were added to YNB where indicated. Macrophages and cocultures were grown in RPMI with glutamine and HEPES.
Construction of C. albicans reporter and deletion strains.
To construct the reporter strains, the HIS1 gene from plasmid CIp20 (23) was incorporated into plasmids pGFP and pACT1-GFP (24) to generate pCJ1 and pCJ2, respectively. One thousand base pairs of the promoter region of ARG1 (pCJ5) and ARG3 (pCJ4), 674 bp of LYS1, and 500 bp of LEU2 were PCR amplified from genomic DNA and individually cloned into plasmid pCJ1 immediately 5′ of the translational start of green fluorescent protein (GFP). The resulting constructs as well as pCJ1 and pCJ2 plasmids were linearized with StuI and used to transform C. albicans RM1000 (ura3/ura3 his1/his1) by electroporation. Integration at the RPS10 locus was confirmed by PCR. Additionally, pCJ5 (ARG1-GFP) and pCJ4 (ARG3-GFP) were similarly integrated into gcn4Δ, arg81Δ, and arg83Δ C. albicans mutant strains derived from mutant libraries generated by Vandeputte et al. (25). The ADH1p-yCherry plasmid (11, 26) was linearized with SalI and transformed by electroporation into the ARG1, ARG3, ACT1, and promoterless GFP reporter strains. Integration at the ADH1 locus was confirmed by PCR. Strains and plasmids are described in Tables S1 and S2, respectively, in the supplemental material.
Mutations in ARG1 and ARG3 were generated using the SAT-flipper method (27) as described previously (28). Complementation of each mutant strain was achieved by cloning the open reading frames of ARG1 and ARG3 immediately adjacent to the first FLP recombination target (FRT) sequence within the FRT-SAT1-FLP-FRT cassette. This linearized construct was used to transform arg1Δ and arg3Δ mutants to nourseothricin resistance. Complementation was confirmed by both PCR and rescue of arginine auxotrophy (see Fig. S4 in the supplemental material). A SAT1-marked plasmid (pHZ130) encoding a constitutively expressed GFP was generated by replacing the URA3 gene in pACT1-GFP between the BamHI and SacI sites with a PCR fragment containing the nourseothricin resistance gene from pSFS1 (27). Plasmid pHZ130 was linearized with StuI and used to transform wild-type, arg1Δ, and arg3Δ cells to nourseothricin resistance, with integration at the RPS10 locus confirmed by PCR.
Fluorescence microscopy.
To test arginine-dependent expression of the ARG1-GFP and ARG3-GFP reporter constructs in vitro, overnight YPD cultures were diluted 1:100 in fresh YPD or YNB and grown for 4 h at 30°C. Five microliters of each culture was used for microscopy with an Olympus IX81-ZDC confocal inverted microscope and the SlideBook 5.0 digital microscopy software (Intelligent Imaging Innovations, Inc.). For the in vitro assays using YNB supplemented with different amino acids, YPD overnight cultures were diluted 1:100 in fresh YPD or YNB supplemented with 20, 100, and 200 μg/ml of l-arginine or 200 μg/ml of l-lysine or l-leucine and grown for 4 h at 30°C.
To normalize the fluorescent signal across different strains and experiments, we calculated the ratio between the background-subtracted fluorescence intensity of GFP and the constitutively expressed yCherry obtained using appropriate filter sets using SlideBook software, as described previously (11). A cell-free portion of each field was used as background, except in the bone marrow-derived macrophage (BMDM) experiments, in which the entire field was used as background to compensate for a low-level autofluorescence in the macrophages. At least 50 cells were counted for each reporter strain and each localization (intracellular versus extracellular, for instance).
For cocultures with J774.A cells, RAW264.7 cells, or bone marrow-derived macrophages, the cells were seeded onto coverslips in 12-well plates (5 × 106 cells/ml) at least 2 h prior to initiating the cocultures. C. albicans cells harboring the desired GFP reporter were grown overnight in YPD, diluted 1:100 in fresh YPD, and grown for 4 h at 30°C. Cells were then washed once with water, resuspended in phosphate-buffered saline (PBS), counted, and incubated with the macrophages at a 1:1 ratio. The cocultures were incubated at 37°C for 1 h and fixed with 4% paraformaldehyde. Fixed cells were stored at 4°C. Prior to imaging, cells were washed with PBS twice and stained with 35 μg/ml calcofluor white (CW) for 30 s prior to imaging.
Gene expression following exposure to ROS.
ARG3-GFP cells grown overnight in YPD were diluted 1:100 in fresh YPD and grown for 4 h at 30°C. Cells were washed once with water and resuspended in PBS. A total of 1 × 106 cells were inoculated onto coverslips on 12-well plates containing 2 ml of RPMI medium containing hydrogen peroxide (0 to 5.0 mM). Cells were incubated at 37°C for 1 h. Coverslips containing the attached cells were used for microscopy.
For Northern analysis, C. albicans cells grown overnight in YPD were diluted in fresh YPD to an optical density at 600 nm (OD600) of 0.25 and grown to an OD600 of 1.0. Cells were exposed to the indicated concentration of hydrogen peroxide, menadione, or tert-butyl hydroperoxide (t-BOOH), washed, and transferred to YNB for 15 min at 30°C. RNA was isolated from each sample using the hot acidic phenol method (29). A total of 10 ng of RNA was loaded and run on a 1% morpholinepropanesulfonic acid (MOPS)-formaldehyde agarose gel, transferred to nylon membranes, and hybridized with randomly labeled probes specific to each gene, using standard protocols (29). Blots were detected using a Storm 840 phosphorimager (Molecular Dynamics) and analyzed using the Image Quant 5.2 software.
Growth assays under oxidative stress conditions.
For assays on solid media, the indicated strains were grown overnight in YPD, washed once with sterile water, diluted to an OD600 of 0.1, and serially diluted 5-fold in 96-well plates. Cells were transferred to YPD plates containing the indicated concentrations of either H2O2 or menadione using a multipin replicating tool and incubated at 30°C for up to 3 days. To assess toxicity to acute exposures to H2O2, the indicated strains were grown overnight in YPD, washed once with sterile water, diluted to an OD600 of 0.1 in fresh YPD containing 0 to 20 mM H2O2, and cultured at 30°C for 1 h. Cells were diluted and plated onto YPD plates, and CFU were determined after 24 h of growth at 30°C.
Isolation of BMDMs.
Bone marrow-derived macrophages (BMDMs) were isolated as described previously (30). Briefly, leg bones from sacrificed ICR mice were collected and bone marrow flushed with Iscove's modified Dulbecco's medium (IMDM) plus 10% fetal bovine serum (FBS) plus penicillin-streptomycin (Pen-Strep) using a 27-gauge needle and syringe. The suspension was centrifuged at 1,000 rpm for 5 min, supernatant removed, and cell pellet resuspended in ammonium-chloride-potassium (ACK) lysis buffer (Lonza, Houston, TX) for 5 min at room temperature to lyse red blood cells. Following centrifugation and removal of the supernatant as described above, the cell pellet was resuspended in PBS and loaded into a 1-ml syringe containing a 27-gauge needle. These cells were flushed directly into 25 ml IMDM–10% FBS–Pen-Strep–10-ng/ml mouse granulocyte-macrophage colony-stimulating factor (GM-CSF), resuspended thoroughly, and distributed evenly across each well of a 6-well plate. Cells were incubated at 37°C with 5%CO2 for 7 days with supplementation of fresh medium containing 10 ng/ml GM-CSF every second day. Following the 7-day incubation, cells were harvested, counted, and replated in the absence of GM-CSF for 24 h before the coculture was initiated as described above.
RESULTS
Construction and validation of ARG1-GFP and ARG3-GFP reporter strains.
Microarray analysis of transcriptional changes following phagocytosis of C. albicans by mammalian macrophages indicated a unique regulation of genes of the arginine biosynthetic pathway: seven of the eight genes were induced, while no other amino acid biosynthetic genes were differentially regulated (the sole uninduced gene, ARG2, is regulated largely posttranslationally in S. cerevisiae [31]). Moreover, hierarchical clustering grouped the ARG pathway with stress response genes and not with the large set of genes responsive to carbon starvation (13). To confirm the induction of the ARG genes upon phagocytosis, transcriptional reporters in which the predicted 5′ promoter regions of ARG1 and ARG3 were fused to GFP were constructed. These two genes were chosen because they both showed the highest fold induction upon phagocytosis according to the microarray data (34.3- and 27.4-fold, respectively). These single-cell reporters were initially tested in vitro, and, as expected, ARG3-GFP cells grown in medium lacking amino acids (YNB) were strongly fluorescent, while the cells grown in complete medium (YPD) were not fluorescent (Fig. 1A). Using Northern blotting, we confirmed that GFP fluorescence from these reporters is representative of endogenous gene expression (data not shown).
Fig 1.
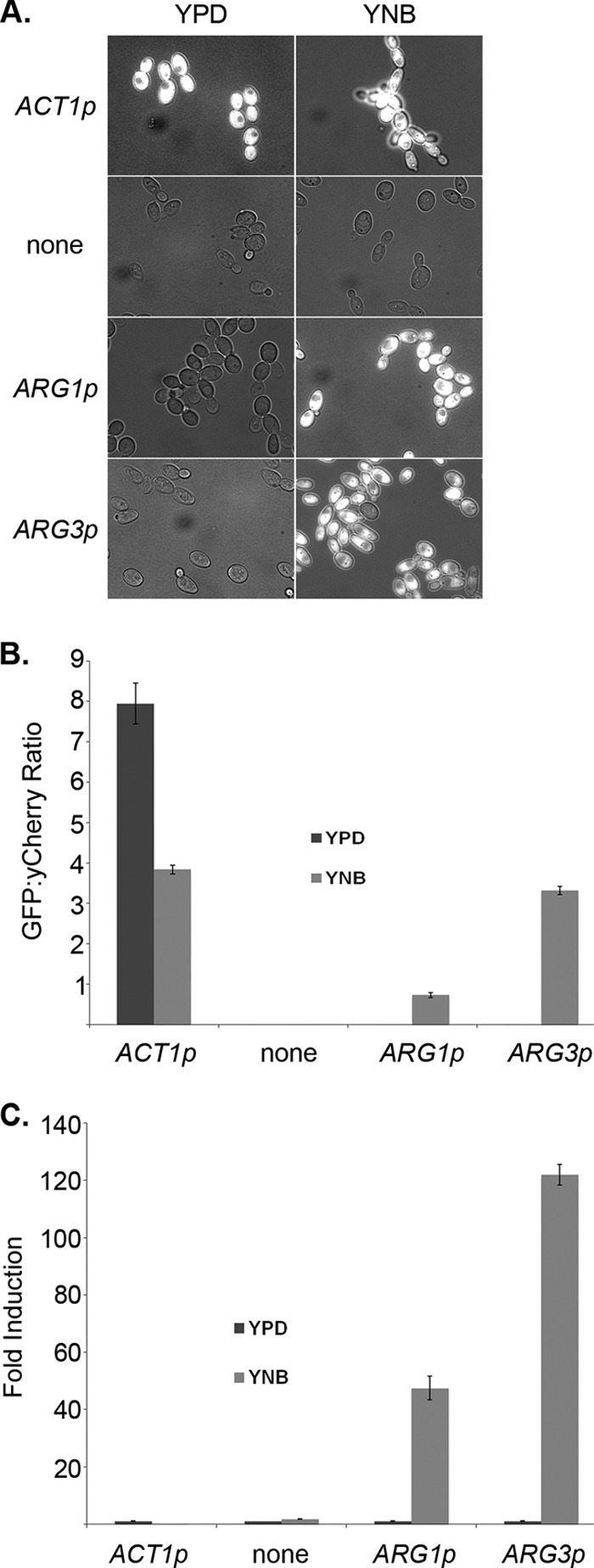
ARG1 and ARG3 promoters are induced in medium lacking arginine. (A) ACT1p-GFP (CJC26), promoterless GFP (CJC25), ARG1p-GFP (CJC28), and ARG3p-GFP (CJC28) strains were grown overnight in YPD and subcultured into fresh YPD or YNB for 4 h at 30°C. Images shown are overlaid GFP and DIC pictures. “None” indicates a GFP reporter construct with no promoter. (B) GFP fluorescence intensity was expressed as the average of the GFP/yCherry intensity ratios calculated from 50 cells per condition (YPD and YNB). Error bars represent the standard error. (C) Fold inductions were calculated, comparing each fluorescence intensity average to that shown by the same strain grown in YPD. Error bars represent the standard error.
These reporter strains were engineered to constitutively coexpress a second fluorescent marker, yCherry, integrated into the ADH1 locus to allow normalization and quantitation of the GFP signal. The background-subtracted fluorescence intensity was calculated for both reporters and converted into a ratio of green to red (11). As expected, we saw very strong fluorescence from the ACT1-GFP control and thus a high GFP/yCherry ratio (Fig. 1B). Expression of both the ARG1-GFP and ARG3-GFP constructs was dependent on the presence of arginine, with much higher ratios in arginine-deficient minimal YNB than in rich medium (Fig. 1B). The fluorescence ratios can be used to calculate fold induction, and the absence of arginine induces the ARG1 and ARG3 promoters 48- and 122-fold, respectively, while the controls show little difference between conditions.
Our initial ARG1-GFP reporter strain was not fluorescent under any conditions as a result of an apparent annotation error for this gene: the originally assigned ATG codon appends 77 amino acids to the amino terminus of the protein that are not homologous to the amino termini of Arg1p homologs of closely related species (see Fig. S1 in the supplemental material). An ARG1-GFP reporter constructed using a second in-frame ATG codon 231 nucleotides (nt) downstream was fluorescent in the absence of arginine (Fig. 1; see Fig. S1 in the supplemental material). Subsequent analysis by 5′ rapid amplification of cDNA ends (RACE) identified the primary transcriptional start site to be 187 bp 3′ to the annotated start codon (and 44 bp 5′ to the second ATG codon; see Fig. S1 in the supplemental material). Thus, translation of Arg1p likely begins at the downstream start codon.
To determine whether the promoters are induced by arginine deficiency specifically or by a more general amino acid starvation, reporter strains were grown in YNB supplemented with l-arginine (0 to 200 μg/ml), l-lysine, or l-leucine. Cells grown in YNB supplemented with >100 μg/ml arginine were not fluorescent (see Fig. S2 in the supplemental material). Conversely, ARG1 and ARG3 were highly expressed in cells grown in YNB supplemented with 200 μg/ml of either l-lysine or l-leucine, as assayed by both reporter fluorescence and Northern blotting. These results indicate that ARG1 and ARG3 respond specifically to arginine deprivation and that general starvation for other amino acids does not induce ARG gene expression. As discussed below, this implies that these ARG genes lie outside the general amino acid control response in C. albicans.
ARG genes are induced specifically in phagocytosed cells.
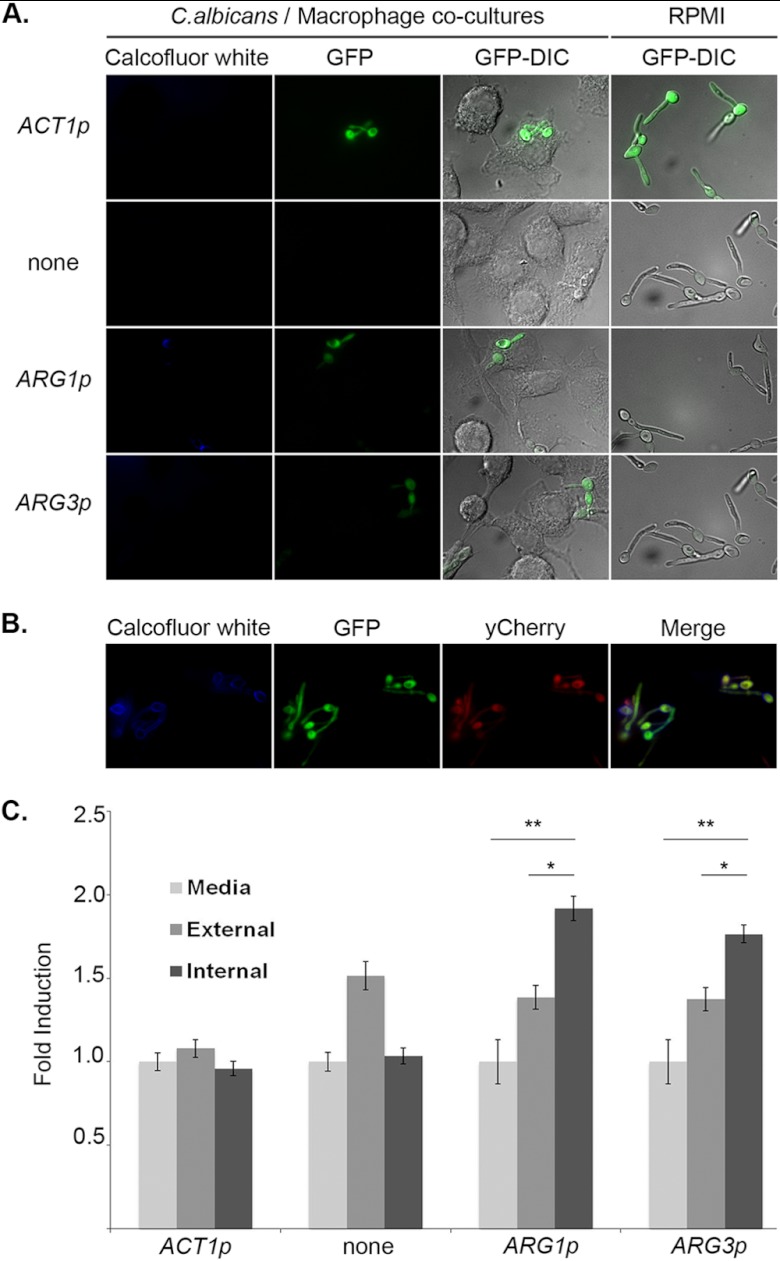
We next used our single-cell reporters to test whether induction of the ARG genes is specific to phagocytosed C. albicans cells. After coincubation of reporter strains with the macrophage cell line RAW264.7 for 1 h, cells were fixed and stained with calcofluor white (CW), a membrane-impermeant fluorescent compound that binds to chitin present in the cell wall of C. albicans, to discriminate between intracellular (CW-negative) and extracellular (CW-positive) fungal cells. As seen in Fig. 2A, phagocytosed ARG3-GFP reporter cells are substantially more likely to be fluorescent than extracellular ones, while the fluorescence of control reporter strains (ACT1-GFP, promoterless GFP) were unchanged based on localization. A representative image from all three fluorescent channels is shown in Fig. 2B. Specific induction of the ARG3-GFP reporter in phagocytosed cells can also be seen in time-lapse movies (see Movie S1 in the supplemental material).
Fig 2.
ARG1 and ARG3 promoters are induced specifically in phagocytosed cells. (A) ACT1p-GFP (CJC26), promoterless-GFP (CJC25; labeled “none”), ARG1p-GFP (CJC28), and ARG3p-GFP (CJC29) strains were cocultured for 1 h with murine macrophages or grown in RPMI alone. Cocultures were then fixed and stained with calcofluor white to distinguish intracellular cells before imaging. The images from left to right show calcofluor white, GFP fluorescence, and overlay of GFP and differential interference contrast (DIC) pictures for both cocultures and cells grown in RPMI medium alone. (B) Representative image of the ARG3-GFP reporter strain to show individually the calcofluor white, GFP, and yCherry fluorescence pictures as well as a merged image of the three fluorophores. (C) GFP fluorescence intensity was quantified as the average GFP/yCherry intensity ratio taken for 50 cells per condition (medium, external, and internal) and expressed as the fold induction relative to strains grown in RPMI medium. Error bars represent the standard error. *, P < 0.05; **, P < 0.001 (Student's t test).
Fluorescence intensity was quantified as described above for cells that were inside macrophages, outside, or grown in medium alone (Fig. 2C). These data show that the GFP fluorescence from the ARG1-GFP or ARG3-GFP reporter, while lower than that for the constitutive ACT1-GFP, is 1.8- to 2.0-fold higher in phagocytosed cells than in cells grown in medium alone. A small induction in extracellular C. albicans cells is seen; for reasons described below, we believe that this results from the oxidative burst generating extracellular ROS. Thus, we conclude that there is a modest but specific induction of ARG promoters following engulfment by macrophages.
To further examine ARG gene induction after phagocytosis, we turned to an in vivo model using zebrafish in which C. albicans cells can be visualized within macrophages in the context of a living animal (11). ARG3-GFP fluorescence intensity (measured as a ratio to a constitutively expressed dTomato) was similar for intracellular and extracellular C. albicans at 2 h postinfection (hpi), but GFP expression increased over time such that the ratio was significantly higher in intracellular cells at 4 and 24 hpi (see Fig. S3 in the supplemental material). Thus, phagocytosis by macrophages induced ARG3 expression in this live zebrafish model.
Regulation of the ARG pathway by Gcn4p and ArgR.
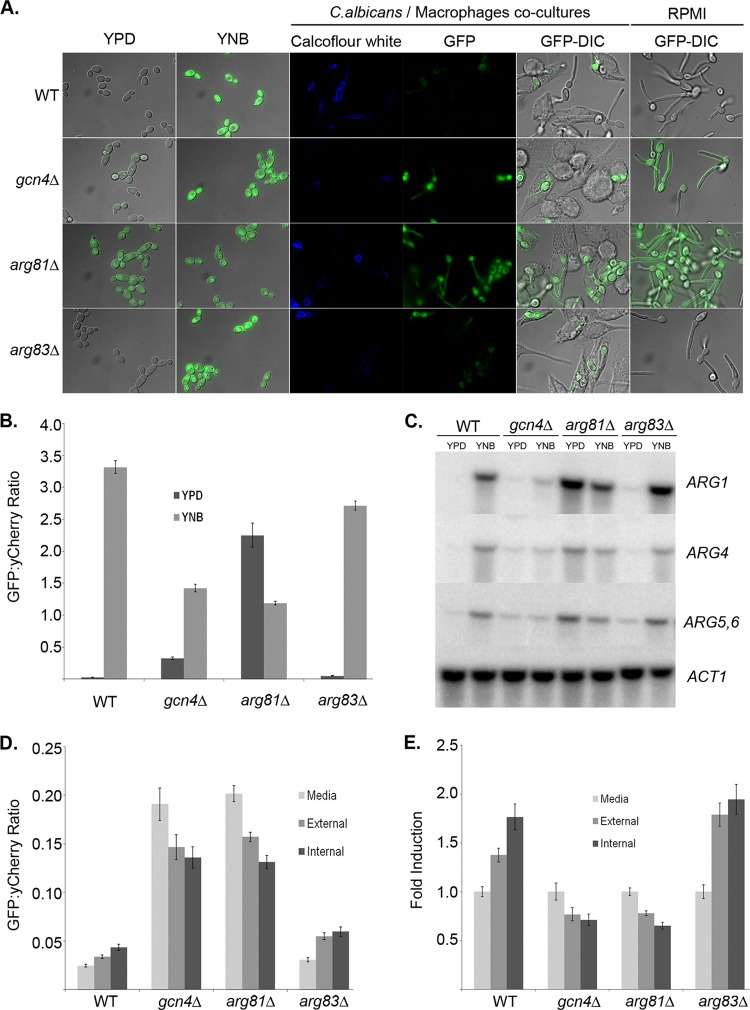
In S. cerevisiae, the ARG pathway is under complex and partially overlapping regulation by Gcn4p and the ArgR complex. Gcn4p is the central transcription factor of the general amino acid control response that, upon starvation for a single amino acid, upregulates a large number of genes for synthesis of multiple amino acids (reviewed in reference 19). A functional homolog of S. cerevisiae Gcn4p (ScGcn4p) has been identified in C. albicans (20). To test if this general amino acid control pathway regulates the ARG genes in vitro or in vivo, we integrated the ARG3-GFP and ADH1-yCherry reporters into a gcn4Δ strain obtained from a knockout library constructed by Vandeputte et al. (25). ARG3 was modestly derepressed under arginine-replete conditions (YPD) and further induced upon arginine starvation (YNB) (Fig. 3A, left panels), although quantitation of the fluorescence ratios demonstrated that the maximal expression in the gcn4Δ mutant was only 42% of that in the wild-type strain (Fig. 3B). Together, these two effects result in a 4.3-fold induction by arginine depletion in the gcn4Δ mutant versus 122-fold in the wild-type strain. Northern analysis confirmed the derepression in vitro, which was particularly notable for ARG4 and ARG5,6 (Fig. 3C). Derepression was more apparent in macrophage cocultures, in which strong fluorescence was observed in cells regardless of location (Fig. 3A and D). With the higher basal level of expression, the induction in phagocytosed cells is lost (Fig. 3E). Thus, Gcn4p appears to primarily repress ARG gene expression, perhaps by recruiting ArgR, as suggested for yeast (18).
Fig 3.
Gcn4p and the ArgR complex regulate the ARG genes. (A) Wild-type (WT) (CJC29), gcn4Δ (CJC35), arg81Δ (CJC32), and arg83Δ (CJC33) C. albicans cells transformed with both the ARG3p-GFP and ADH1p-yCherry reporter were grown in YPD or YNB for 4 h at 30°C, cocultured for 1 h with murine macrophages, or grown in RPMI medium alone. The two columns on the left show GFP-DIC overlaid pictures for the in vitro experiment (YPD versus YNB). For the cocultures, the images show calcofluor white fluorescence of extracellular C. albicans cells and GFP fluorescence individually. Overlays of GFP and DIC images are shown for both cocultures and cells grown in RPMI medium only. (B) GFP fluorescence intensity was expressed as the average of the GFP/yCherry intensity ratio taken from 50 cells per condition grown in vitro under arginine-replete (YPD) or -deficient (YNB) conditions. Error bars represent the standard error. (C) Wild-type (SC5314), gcn4Δ (DSY3233), arg81Δ (HZY28), and arg83Δ (DSY3426-2) C. albicans cells were grown in YPD to an OD600 of 1.0, washed, and grown in either YPD or YNB for 15 min. RNA was collected from each sample for Northern blot analysis with probes for the indicated genes. (D) The GFP/yCherry ratio was calculated from ARG3p-GFP-expressing cells grown in medium alone or in cells either extracellular or intracellular in macrophage cocultures. At least 50 cells were counted for each localization. (E) Fold induction, relative to medium alone for each strain, for the data in panel D.
The second transcriptional regulator, ArgR, is in yeast a complex of three DNA binding proteins, Mcm1p, Arg80p, and Arg81p, and an inositol phosphate kinase, Arg82p (32–35). C. albicans has close homologs of Mcm1p, Arg81p and Arg82p (called Ipk2p), but not Arg80p, which in yeast is both adjacent and highly homologous to Mcm1p, presumably the product of a tandem duplication. Instead, C. albicans has a second protein homologous to Arg81p, which is annotated as Arg83p. We identified arg81Δ and arg83Δ deletion strains in mutant libraries (25) and transformed both using the ARG3-GFP and ADH1-yCherry reporters. This reporter was strongly derepressed in the arg81Δ strain in the presence of arginine and was not further induced by arginine depletion or by phagocytosis (Fig. 3A to E). In contrast, the arg83Δ mutant showed normal repression of ARG3 in the presence of arginine and an induction in its absence both in vitro and in vivo.
ARG genes are induced by reactive oxygen species.
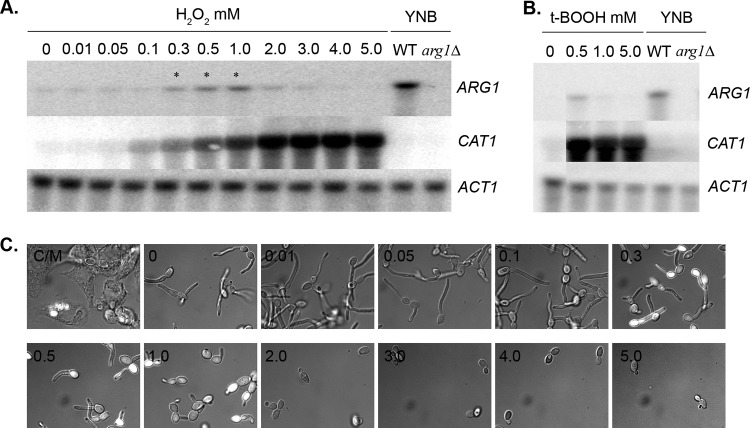
Why might phagocytosed cells specifically and uniquely upregulate the ARG pathway? We considered that the phagolysosome might be deficient in arginine; while it seemed unlikely that this environment would be devoid of one specific amino acid, arginine is the substrate for the inducible nitric oxide synthase (iNOS or NOS2), which generates NO within the phagolysosome. RPMI contains 300 μg/ml arginine, and supplementation up to 1 mg/ml did not affect ARG gene expression in phagocytosed cells (data not shown). Next, we considered that the ARG pathway is induced to generate polyamines, which have been shown to be required for the C. albicans yeast-to-hypha switch (36, 37), but the gene encoding the rate-limiting step of polyamine biosynthesis, ornithine decarboxylase (SPE1), is not induced by phagocytosis (13). An alternative possibility was suggested by a recent RNA-seq analysis of the C. albicans transcriptome in several host-relevant conditions by Bruno et al. (38). The ARG pathway is induced by moderate concentrations of hydrogen peroxide, which mimics the reactive burst of macrophages.
Northern analysis was initially used to assess whether the ARG pathway responds to ROS in vitro. The ARG1 message is expressed under arginine-replete conditions containing moderate concentrations (0.3 to 1.0 mM) of hydrogen peroxide, but expression is not detected at higher concentrations (>2 mM) (Fig. 4A); this is consistent with the RNA-seq analysis by Bruno et al. (38). ARG1 is also induced by moderate concentrations of tert-butyl hydroperoxide (t-BOOH) (Fig. 4B) and menadione (data not shown). In each case, the mRNA abundance for the gene encoding catalase (CAT1) continues to increase with the ROS concentration, indicating that the lack of ARG1 expression is not due to cell death from toxic ROS levels. These results were confirmed using the ARG3-GFP strain, which again showed induction only at moderate concentrations of ROS (Fig. 4C).
Fig 4.
ARG1 and ARG3 are induced by reactive oxygen species. (A) Wild-type (SC5314) C. albicans cells were grown in YPD to an OD600 of 1.0 and transferred to YNB or YPD supplemented with the indicated concentration of H2O2 for 15 min. RNA was collected from each sample, and Northern blots were probed for ARG1, CAT1, and ACT1. RNA from an arg1Δ strain grown in YNB was used as a control for the ARG1 probe specificity. (B) Wild-type C. albicans (SC5314) cells were grown in YPD to an OD600 of 1.0 and transferred to YNB or to YPD supplemented with the indicated concentration of tert-butyl hydroperoxide for 15 min, and Northern analysis was performed as for panel A. (C) ARG3p-GFP (CJC5) cells were grown in RPMI supplemented with different concentrations of H2O2 for 1 h. The images shown are GFP-DIC overlays. The numbers on each image represent the concentration of H2O2 in mM for each sample. C/M, 1-h coculture with macrophages as a comparison.
Because mRNA levels were increased for both ARG1 and ARG3 following H2O2 exposure, arginine biosynthesis might be necessary to counter the effects of prolonged exposures to oxidative stress. To test this possibility, we generated strains lacking either ARG1 or ARG3, along with complemented controls (28). As expected, these strains are arginine auxotrophs (see Fig. S4 in the supplemental material). We plated serial dilutions of arg1Δ and arg3Δ cells on plates containing increasing concentrations of either H2O2 or the superoxide anion (O2−)-generating compound menadione and allowed cells to grow for up to 3 days using the known ROS-sensitive hog1Δ mutant as a control (39) and found no alterations in growth relative to that of the wild-type control at any H2O2 concentration (see Fig S5 in the supplemental material). arg1Δ and arg3Δ mutants also did not increase the sensitivity to acute ROS toxicity as measured by the decrease in CFU after short-term exposure to higher concentrations of ROS (up to 20 mM) (data not shown). Thus, there are no obvious in vitro ROS-related phenotypes for disruption of the ARG pathway.
During coculture of C. albicans with macrophages, cells respond to one or more as-yet-unknown hypha-inducing signals to initiate germ tube formation, eventually rupturing the phagocyte. Arginine is catabolized into urea and ornithine by the cytosolic arginase Car1p, and urea is further degraded into CO2 and NH4+ by the urea amidolyase Dur1,2p. Others have proposed that this CO2 is a trigger for hyphal formation and have shown that both dur1,2Δ and arg4Δ mutations reduce hyphal growth in the macrophage (40). Similarly, the arg1Δ and arg3Δ mutants germinated more slowly than controls within the macrophage and generated shorter germ tubes. Cellular morphology of phagocytosed cells was quantified by counting, and nearly twice as many cells remained in the yeast form after 2 h (26% for the wild-type versus 48% for arg1Δ and 43% for arg3Δ) (Fig. 5). Hyphal elongation was also delayed, with more cells remaining in the germ tube phase in the mutants. Nevertheless, at longer time points there was no significant difference in fungus-induced macrophage damage, as assayed by lactate dehydrogenase release (data not shown). We tested the mutants in a mouse model of systemic candidiasis and found no alterations in virulence for either strain, as has been reported for arg4Δ mutants (41). Thus, despite the induction of these genes, disruption of the arginine biosynthetic pathway confers only minor alterations of the host-pathogen interaction.
Fig 5.
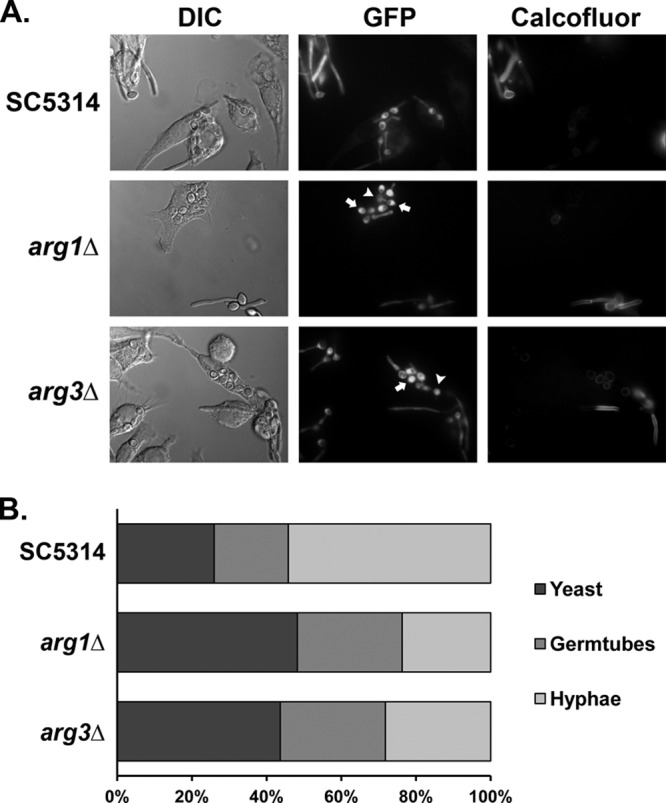
Mutants with mutations in ARG genes have abnormal germ tube dynamics following macrophage phagocytosis. (A) Wild-type (RGC1), arg1Δ (JRC47), and arg3Δ (JRC42) strains, each expressing an ACT1-GFP construct, were cocultured with RAW264.7 macrophages (1:1 Candida/macrophage ratio) for 2 h prior to chemical fixation with paraformaldehyde. To distinguish fungal cells that had been phagocytosed from extracellular fungi, fixed cocultures were stained with calcofluor white prior to imaging. Arrows highlight arg mutants that have not initiated germ tube formation. Arrowheads highlight arg mutants containing very short filaments. (B) Quantitation of cellular morphologies observed for phagocytosed GFP-SC5314, GFP-arg1Δ/Δ, and GFP-arg3Δ/Δ strains. For each strain, the morphologies of at least 150 phagocytosed cells were determined. Fisher's exact test (two tailed) was used to compare the distribution of yeast, germ tube, and hyphal morphologies between GFP-SC5314 and GFP-arg strains (P < 0.0001 for comparisons between SC5314 and each arg mutant).
ARG induction in macrophages is at least partly ROS dependent.
To test whether ARG induction is due to phagocyte-derived ROS, we turned to primary bone marrow-derived macrophages (BMDMs). Both ARG1-GFP and ARG3-GFP are induced in cells phagocytosed by BMDMs isolated from wild-type mice relative to medium-grown cells. In contrast to the cultured macrophages, however, there was a less significant difference between phagocytosed and nonphagocytosed cells. The magnitude of the ROS burst from these primary cells should be greater than that from cultured cells, so it is possible that even external cells are exposed to a greater oxidative stress than in our earlier experiments.
We next cocultured our C. albicans reporter strains with BMDMs isolated from mutant mice lacking the gp91phox subunit of the phagocyte oxidase responsible for generating ROS in the macrophage (42). Though the overall phagocytic activity of these cells was similar to that of wild-type macrophages, the ARG1-GFP and ARG3-GFP reporters were induced just slightly in cells phagocytosed by the gp91phox-deficient macrophages (Fig. 6 and 7). We noted a weak autofluorescence in some gp91phox-deficient macrophages that may account for some of this apparent induction. Thus, induction of the ARG genes results primarily from exposure of cells to ROS generated by the phagocyte's oxidative burst.
Fig 6.
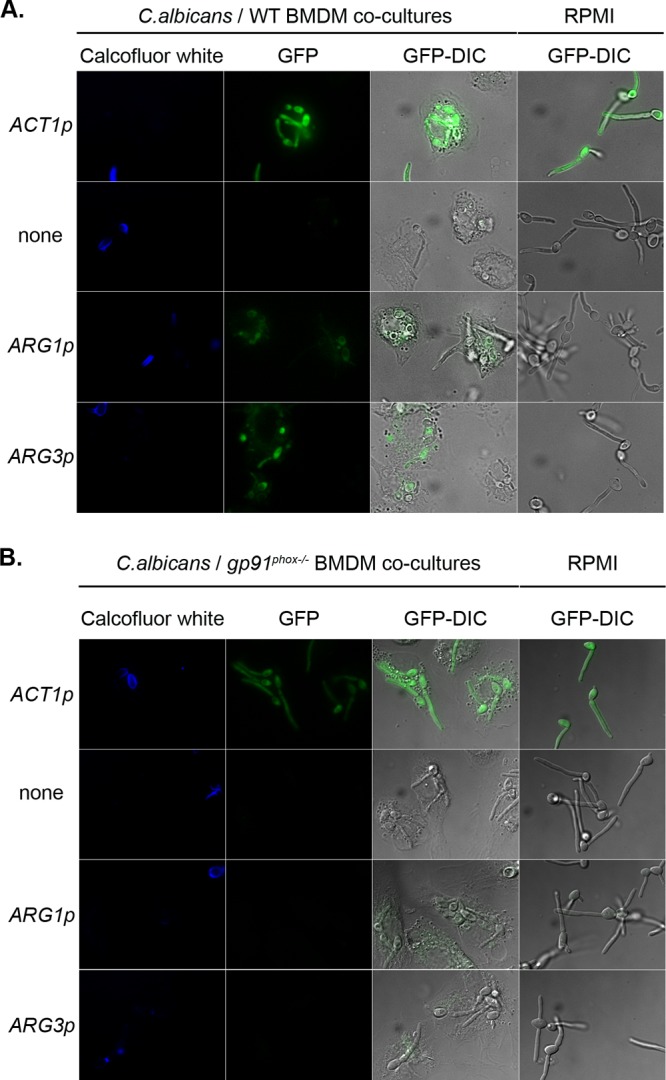
Induction of ARG genes is partially dependent on ROS. ACT1p-GFP (CJC26), promoterless GFP (CJC25), ARG1p-GFP (CJC28), and ARG3p-GFP (CJC29) cells were cocultured with BMDMs from wild-type mice (A) or gp91phox−/− mice (B) for 1 h. As a control, cells were also grown in RPMI medium alone. Images show calcofluor white, GFP, and GFP-DIC overlay images for both the cocultures and medium alone.
Fig 7.
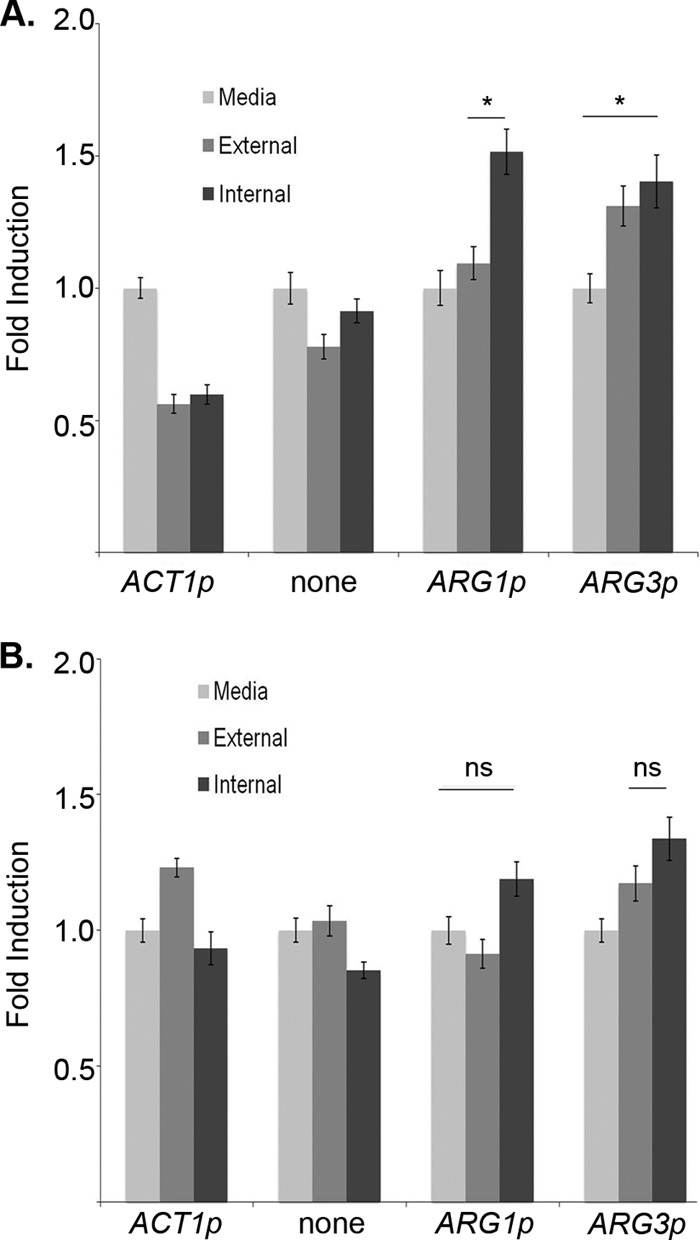
Quantification of ARG gene induction after phagocytosis by wild-type and gp91phox−/− BMDMs. GFP fluorescence intensity was calculated as the average of the GFP/yCherry intensity ratio taken from 50 cells per condition. The GFP fold induction was calculated based on the average GFP/yCherry ratio (n = 50 cells) for both intracellular and extracellular cells, relative to medium controls, in WT BMDMs (A) and gp91phox−/− BMDMs (B). Error bars represent the standard error. *, P < 0.01; ns, not significant (P > 0.05) (by Student's t test).
DISCUSSION
We demonstrate in this work that the genes encoding the arginine biosynthetic pathway are induced specifically in cells phagocytosed by either primary or cultured macrophages and that this induction results from exposure to moderate concentrations of phagocyte-derived ROS. Induction is dependent on the NADPH-dependent phagocyte oxidase, as macrophages lacking the gp91phox subunit do not activate the ARG1-GFP and ARG3-GFP reporter constructs. In vitro, the ARG genes are also induced by ROS at a narrow range of concentrations, which agrees with RNA-seq analysis (38). As expected, they are also strongly expressed when arginine is depleted in vitro.
An obvious alternative hypothesis to explain ARG gene induction in phagocytosed cells is that the phagolysosome is devoid of arginine. However, since arginine is unique among the amino acid synthesis pathways, this would imply that the phagolysosome was replete in at least most of the other 19 amino acids, and this seems unlikely, though it is possible that consumption of arginine as the substrate for iNOS (NOS2)-dependent generation of NO could lead to such an asymmetry. The data from the gp91phox macrophages (which still express a functional iNOS), in which the ARG1-GFP and ARG3-GFP reporters are not significantly induced, make this unlikely.
Mutation of ARG1 or ARG3 impairs hyphal morphogenesis in phagocytosed cells but does not obviously alter sensitivity to ROS in vitro and does not affect systemic virulence, as is also the case with an arg4Δ mutant (41). Why, then, does this pathway respond to ROS? One possibility is that increased arginine biosynthesis protects against ROS. In mammals arginine has been indirectly linked to increased resistance to ROS by enhancing generation of NO, which then induces antioxidant defenses. Fungi lack NOS-like enzymes, however, so this would not seem to be a likely mechanism. Nishimura et al. (43) identified a yeast N-acetyltransferase, Mpr1p, whose substrates are intermediates of the proline and arginine pathways and which contributes to resistance to endogenously generated ROS. Mpr1p, which is missing from the S288c genome but found in other strains of S. cerevisiae (e.g., Σ1278b) and in other fungi, including C. albicans, facilitates arginine production from proline intermediates, but the mechanism by which this is protective is unknown.
Alternatively, C. albicans cells may use moderate concentrations of ROS as a signal to activate antiphagocyte mechanisms, for instance, to promote hyphal growth inside the macrophage. Indeed, moderate ROS concentrations stimulate hyphal growth (44, 45). A role for arginine in hyphal induction was proposed by Ghosh et al. (40), who showed that C. albicans strains unable to synthesize (arg4Δ) or completely degrade (dur1,2Δ) arginine displayed fewer filaments within macrophages, findings consistent with the present study. Arginine is degraded by the arginase Car1p to generate ornithine and urea. Urea is subsequently degraded by the urea amidolyase Dur1,2p to generate ammonia and carbon dioxide, both of which are potent signals for hyphal morphogenesis (28, 46). Thus, both studies suggest that arginine synthesis within macrophages contributes to hyphal induction. The dur1,2Δ mutant was recently shown to be attenuated in a whole-animal model (47), in contrast to arg1Δ, arg3Δ, and arg4Δ mutants (this study and reference 41), which would indicate that arginine is not limiting in vivo but that its degradation does contribute to pathogenesis. In vitro, under macrophage-like conditions (amino acid rich and glucose poor), C. albicans cells rapidly neutralize acidic conditions. CAR1 is upregulated during this process, which is significantly slowed in a dur1,2Δ mutant (28), linking this phenomenon with arginine metabolism.
The pattern of expression in response to ROS is atypical: rather than increasing expression with increasing stress, the ARG genes are expressed only in a narrow range of sublethal concentrations, i.e., 0.3 to 1.0 mM H2O2. Inhibition of expression at higher concentrations cannot be from general toxicity of the ROS, because catalase (CAT1) expression remains very strong up to 5 mM.
In S. cerevisiae, the expression of at least some of the ARG genes is regulated by a complex called ArgR consisting of Mcm1p, Arg80p, Arg81p, and Arg82p (reviewed in reference 19). The first three of these are DNA binding proteins, with Arg80p a close homolog of the more general regulator Mcm1p. Gcn4p, which generally serves as an activator of transcription of other amino acid genes, recruits ArgR to ARG promoters, where it serves as a repressor under arginine-replete conditions (21). This appears to be conserved in C. albicans with one significant difference: the presence of arginine alone represses ARG gene transcription, even in the absence of other amino acids. Thus, despite the involvement of Gcn4p, the ARG pathway is largely outside the general control response system. C. albicans has a homolog of Mcm1p, which localizes to ARG promoters (48), though it lacks Arg80p, which appears to have been arisen in S. cerevisiae through a tandem duplication of Mcm1p. C. albicans has two Arg81p homologs, Arg81p, mutation of which derepresses expression in the presence of arginine, and Arg83p, for which we have not found a phenotype. Further study will be required to understand the function of the ArgR complex in response to different inputs in C. albicans.
Finally, the induction of the ARG genes may be the by-product of an expanded role for ArgR or Mcm1p. Recently, an arg81Δ mutant was shown to be among the most defective mutants tested in a model of biofilm attachment (49). As there is no obvious connection between arginine biosynthesis and biofilm formation, it is possible that ARG81 has acquired a new function in C. albicans. Rewiring transcription factor regulons has become a common theme in fungi, with proteins such as Gal4p, Rap1p, Tbf1p, and Mcm1p itself having different or additional functions in C. albicans than in S. cerevisiae (50–52). C. albicans Zap1p, a conserved regulator of zinc acquisition, has evolved a second function to control biofilm matrix production (53). A biofilm state is not conducive to escape from macrophages, so perhaps Arg81p is downregulated under such conditions, which would derepress ARG genes. Addressing this speculative proposal will require a more complete understanding of the regulons of the ArgR proteins.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dominique Sanglard, Aaron Mitchell, and Janet Quinn for strains and reagents. We thank other members of the Lorenz lab for fruitful discussions and Kevin Morano for assistance with the microscopy.
This work was supported by NIH awards R21AI071134 and R01AI075091 to M.C.L, R01AI081838 to R.A.C., and R15AI094406 to R.T.W.
Footnotes
Published ahead of print 9 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00290-12.
REFERENCES
- 1. Azie N, Neofytos D, Pfaller M, Meier-Kriesche HU, Quan SP, Horn D. 2012. The PATH (Prospective Antifungal Therapy) Alliance registry and invasive fungal infections: update 2012. Diagn. Microbiol. Infect. Dis. 73:293–300 [DOI] [PubMed] [Google Scholar]
- 2. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 3. Calderone RA, Clancy CJ. 2012. Candida and candidiasis. ASM Press, Washington, DC [Google Scholar]
- 4. Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. 2:820–832 [DOI] [PubMed] [Google Scholar]
- 5. Aratani Y, Kura F, Watanabe H, Akagawa H, Takano Y, Suzuki K, Dinauer MC, Maeda N, Koyama H. 2002. Critical role of myeloperoxidase and nicotinamide adenine dinucleotide phosphate-oxidase in high-burden systemic infection of mice with Candida albicans. J. Infect. Dis. 185:1833–1837 [DOI] [PubMed] [Google Scholar]
- 6. Aratani Y, Kura F, Watanabe H, Akagawa H, Takano Y, Suzuki K, Dinauer MC, Maeda N, Koyama H. 2002. Relative contributions of myeloperoxidase and NADPH-oxidase to the early host defense against pulmonary infections with Candida albicans and Aspergillus fumigatus. Med. Mycol. 40:557–563 [DOI] [PubMed] [Google Scholar]
- 7. Balish E, Warner TF, Nicholas PJ, Paulling EE, Westwater C, Schofield DA. 2005. Susceptibility of germfree phagocyte oxidase- and nitric oxide synthase 2-deficient mice, defective in the production of reactive metabolites of both oxygen and nitrogen, to mucosal and systemic candidiasis of endogenous origin. Infect. Immun. 73:1313–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shiloh MU, MacMicking JD, Nicholson S, Brause JE, Potter S, Marino M, Fang F, Dinauer M, Nathan C. 1999. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10:29–38 [DOI] [PubMed] [Google Scholar]
- 9. Aratani Y, Koyama H, Nyui S, Suzuki K, Kura F, Maeda N. 1999. Severe impairment in early host defense against Candida albicans in mice deficient in myeloperoxidase. Infect. Immun. 67:1828–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aratani Y, Miura N, Ohno N, Suzuki K. 2012. Role of neutrophil-derived reactive oxygen species for host defense and inflammation. Med. Mycol. J. 53:123–128 [DOI] [PubMed] [Google Scholar]
- 11. Brothers KM, Newman ZR, Wheeler RT. 2011. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryot. Cell 10:932–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collette JR, Lorenz MC. 2011. Mechanisms of immune evasion in fungal pathogens. Curr. Opin. Microbiol. 14:668–675 [DOI] [PubMed] [Google Scholar]
- 13. Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barelle CJ, Priest CL, Maccallum DM, Gow NA, Odds FC, Brown AJ. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lorenz MC, Fink GR. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83–86 [DOI] [PubMed] [Google Scholar]
- 16. Piekarska K, Mol E, van den Berg M, Hardy G, van den Burg J, van Roermund C, Maccallum D, Odds F, Distel B. 2006. Peroxisomal fatty acid β-oxidation is not essential for virulence of Candida albicans. Eukaryot. Cell 5:1847–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rubin-Bejerano I, Fraser I, Grisafi P, Fink GR. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 100:11007–11012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crabeel M, de Rijcke M, Seneca S, Heimberg H, Pfeiffer I, Matisova A. 1995. Further definition of the sequence and position requirements of the arginine control element that mediates repression and induction by arginine in Saccharomyces cerevisiae. Yeast 11:1367–1380 [DOI] [PubMed] [Google Scholar]
- 19. Hinnebusch AG. 2005. Translational regulation of gcn4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59:407–450 [DOI] [PubMed] [Google Scholar]
- 20. Tripathi G, Wiltshire C, Macaskill S, Tournu H, Budge S, Brown AJ. 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21:5448–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoon S, Govind CK, Qiu H, Kim SJ, Dong J, Hinnebusch AG. 2004. Recruitment of the ArgR/Mcm1p repressor is stimulated by the activator Gcn4p: a self-checking activation mechanism. Proc. Natl. Acad. Sci. U. S. A. 101:11713–11718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 [DOI] [PubMed] [Google Scholar]
- 23. Dennison PM, Ramsdale M, Manson CL, Brown AJ. 2005. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet. Biol. 42:737–748 [DOI] [PubMed] [Google Scholar]
- 24. Barelle CJ, Manson CL, MacCallum DM, Odds FC, Gow NA, Brown AJ. 2004. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast 21:333–340 [DOI] [PubMed] [Google Scholar]
- 25. Vandeputte P, Pradervand S, Ischer F, Coste AT, Ferrari S, Harshman K, Sanglard D. 2012. Identification and functional characterization of Rca1, a transcription factor involved in both antifungal susceptibility and host response in Candida albicans. Eukaryot. Cell 11:916–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keppler-Ross S, Noffz C, Dean N. 2008. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics 179:705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 28. Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. 2011. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio 2(3):e00055–11 doi:10.1128/mBio.00055-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ausubel FM, Brent B, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 2000. Current protocols in molecular biology. John Wiley & Sons, Edison, NJ [Google Scholar]
- 30. Saikolappan S, Estrella J, Sasindran SJ, Khan A, Armitige LY, Jagannath C, Dhandayuthapani S. 2012. The fbpA/sapM double knock out strain of Mycobacterium tuberculosis is highly attenuated and immunogenic in macrophages. PLoS One 7:e36198 doi:10.1371/journal.pone.0036198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wipe B, Leisinger T. 1979. Regulation of activity and synthesis of N-acetylglutamate synthase from Saccharomyces cerevisiae. J. Bacteriol. 140:874–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bechet J, Greenson M, Wiame JM. 1970. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur. J. Biochem. 12:31–39 [DOI] [PubMed] [Google Scholar]
- 33. Dubois E, Messenguy F. 1991. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol. Cell. Biol. 11:2162–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Messenguy F, Dubois E. 1993. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2586–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. 1999. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9:1323–1326 [DOI] [PubMed] [Google Scholar]
- 36. Herrero AB, Lopez MC, Garcia S, Schmidt A, Spaltmann F, Ruiz-Herrera J, Dominguez A. 1999. Control of filament formation in Candida albicans by polyamine levels. Infect. Immun. 67:4870–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ueno Y, Fukumatsu M, Ogasawara A, Watanabe T, Mikami T, Matsumoto T. 2004. Hyphae formation of Candida albicans is regulated by polyamines. Biol. Pharm. Bull. 27:890–892 [DOI] [PubMed] [Google Scholar]
- 38. Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. 2010. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 20:1451–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. 2004. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell 15:4179–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ghosh S, Navarathna DH, Roberts DD, Cooper JT, Atkin AL, Petro TM, Nickerson KW. 2009. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect. Immun. 77:1596–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pollock JD, Williams DA, Gifford MA, Li LL, Du X, Fisherman J, Orkin SH, Doerschuk CM, Dinauer MC. 1995. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9:202–209 [DOI] [PubMed] [Google Scholar]
- 43. Nishimura A, Kotani T, Sasano Y, Takagi H. 2010. An antioxidative mechanism mediated by the yeast N-acetyltransferase Mpr1: oxidative stress-induced arginine synthesis and its physiological role. FEMS Yeast Res. 10:687–698 [DOI] [PubMed] [Google Scholar]
- 44. DA Silva Dantas A, Patterson MJ, Smith DA, Maccallum DM, Erwig LP, Morgan BA, Quinn J. 2010. Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans. Mol. Cell. Biol. 30:4550–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nasution O, Srinivasa K, Kim M, Kim YJ, Kim W, Jeong W, Choi W. 2008. Hydrogen peroxide induces hyphal differentiation in Candida albicans. Eukaryot. Cell 7:2008–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schroppel K, Naglik JR, Eckert SE, Mogensen EG, Haynes K, Tuite MF, Levin LR, Buck J, Muhlschlegel FA. 2005. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 15:2021–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Navarathna DH, Lionakis MS, Lizak MJ, Munasinghe J, Nickerson KW, Roberts DD. 2012. Urea amidolyase (DUR1,2) contributes to virulence and kidney pathogenesis of Candida albicans. PLoS One 7:e48475 doi:10.1371/journal.pone.0048475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. 2008. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics 9:578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 8:e1002525 doi:10.1371/journal.ppat.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hogues H, Lavoie H, Sellam A, Mangos M, Roemer T, Purisima E, Nantel A, Whiteway M. 2008. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol. Cell 29:552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. 2007. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr. Biol. 17:1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38 doi:10.1371/journal.pbio.0060038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 7:e1000133 doi:10.1371/journal.pbio.1000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.