Abstract
Aeromonas hydrophila is a pathogenic bacterium that has been implicated in fish, animal, and human disease. Recently, a multidrug resistance (MDR) plasmid, pR148, was isolated from A. hydrophila obtained from a tilapia (Oreochromis niloticus) farm in Thailand. pR148 is a 165,906-bp circular plasmid containing 147 coding regions showing highest similarity to pNDM-1_Dok1, an MDR plasmid isolated from a human pathogen. pR148 was also very similar to other IncA/C plasmids isolated from humans, animals, food, and fish. pR148 contains a mercuric resistance operon and encodes the complete set of genes for the type 4 secretion system. pR148 encodes a Tn21 type transposon. This transposon contains the drug resistance genes qacH, blaOXA-10, aadA1, and sul1 in a class 1 integron; tetA and tetR in transposon Tn1721; and catA2 and a duplicate sul1 in a locus showing 100% similarity to IncU plasmids isolated from fish. The blaOXA-10 and aadA1 genes showed 100% similarity to those from the Acinetobacter baumannii AYE genome. The similarity of pR148 to a human pathogen-derived plasmid indicates that the plasmids were either transferred between different genera or that they are derived from a common origin. Previous studies have shown that IncA/C plasmids retain a conserved backbone, while the accessory region points to lateral gene transfer. These observations point out the dangers of indiscriminate use of antibiotics in humans and in animals and the necessity of understanding how drug resistance determinants are disseminated and transferred.
INTRODUCTION
The genes encoding microbial resistance are often carried on mobile elements called plasmids that have the ability to replicate and may have the potential of self transmission. Plasmids are a modular mosaic of mobile genetic elements (MGEs) which the plasmids can carry and transfer between individual bacteria of different species by lateral gene transfer (also called horizontal gene transfer) (1–3). In so doing, the plasmids can transfer these genes to the bacterial chromosome or can gain more drug resistance genes. This incessant transfer and collection of drug resistance genes has led to the rise of bacterial strains that are resistant to several antimicrobials, a condition called multidrug resistance.
The problem of antimicrobial resistance is of grave concern. The emergence of multidrug-resistant (MDR) strains and the possibility of transfer of this multidrug resistance to other bacteria has raised the grim specter of bacterial pathogens that cannot be treated by currently known antimicrobials and the reemergence of diseases that can cause large-scale global pandemics. The rise in incidence of MDR bacteria has been attributed to the indiscriminate use of antimicrobials in animal culture and in medicine.
Among the MDR plasmids, members of the IncA/C incompatibility group have been described from humans, animals, and fish and are regarded as of considerable public health risk due to their ability to spread across taxonomical barriers (4, 5). The IncA/C reference plasmid, pRA1, was first isolated in 1971 (6) from Aeromonas liquefaciens (later renamed Aeromonas hydrophila) as a transferable antimicrobial resistance plasmid conferring resistance to sulfonamides and tetracyclines. A. hydrophila is a pathogenic bacterium that is ubiquitous in most aquatic environments (7) and has been implicated in fish, amphibian, and human disease (8). In humans, A. hydrophila infections can be life threatening (9–11).
A. hydrophila, despite having a low conjugal transfer ability for some IncU plasmids (12), has been shown to harbor plasmids, including those of the IncA/C incompatibility group (5, 6, 13). We previously described an MDR plasmid (designated pR148) from A. hydrophila isolated from a tilapia farm in Thailand that had successively used several antimicrobials (13). Antimicrobial susceptibility testing revealed that this plasmid was shown to confer antimicrobial resistance against ampicillin, chloramphenicol, streptomycin, sulfamonomethoxine, and tetracycline, some of the same antimicrobials reportedly used on the farm. In the present study, we analyzed the whole sequence of this plasmid (pR148) and compared it to other closely related sequences. The definition of MDR has recently been standardized by a joint initiative of the European Centre for Disease Prevention and Control and the Centers for Disease Control and Prevention (14) as being nonsusceptible to ≥1 agent in ≥3 categories of antimicrobial agents, and pR148 possesses genes that confer resistance to 5 of these categories.
Since plasmids are a mosaic of sequences from divergent sources, it is important to devise a proper method of comparing them. Previous methods have focused on concatenating conserved genes to form a sequence-based phylogenetic analysis (5, 15). However, these approaches do not consider the relationships that can be inferred from genes that are shared only between some of the compared sequences. In our current approach, we determined the position of the plasmid in the network of mobile genetic elements and the phylogenetic relationship of all of the closely related sequences based on the full plasmid sequences.
MATERIALS AND METHODS
Bacterial source.
A. hydrophila containing plasmid pR148 was isolated from a tilapia (Oreochromis niloticus) farm in Thailand that recorded the consecutive use of multiple antimicrobials. The bacterial isolate was confirmed to be A. hydrophila using API20E and API20NE kits (bioMérieux, Marcy l'Etoile, France) in a previous study (13). The presence of a transferrable plasmid and antibiotic resistance were confirmed by conjugative transfer to Escherichia coli RC85 nal (resistant to nalidixic acid) and subsequently to E. coli HB101 str (resistant to streptomycin) using 1:1 broth cultures of the donor and host, incubated at 37°C overnight, and then plated on BTB-lactose-LB agar, which is composed of 0.0045% bromothymol blue (BTB), 1% lactose, 50 mg/ml nalidixic acid or 1,000 g/ml streptomycin (depending on the host E. coli strain), and one of the drugs to which A. hydrophila is resistant. This plasmid was shown to confer antimicrobial resistance against ampicillin, chloramphenicol, streptomycin, sulfamonomethoxine, and tetracycline using the standard agar dilution method by following Clinical and Laboratory Standards Institute guidelines (Japanese Society of Antimicrobials for Animals 2004).
Purification of plasmid for sequencing.
Plasmid DNA from E. coli HB101 was extracted using a GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO) as described previously (13) and sequenced by TaKaRa Bio, Inc. (Otsu, Japan), using a 454 GS-FLX system (Roche, Basel, Switzerland). The generated reads were assembled using Salmonella enterica pSN254(CP000604), A. hydrophila pRA1(FJ705807), and E. coli pAPEC-O2-R(AY214164) as reference plasmids. Remaining gaps were closed by standard PCR using manually designed primers.
Coding sequences (CDS) were detected using CRITICA version 1.05b (16) and Glimmer2 version 2.10 (17), as well as manually as described previously (18). The presence of genes coding for tRNA and rRNA was checked with tRNAScan-SE version 1.23 (http://lowelab.ucsc.edu/tRNAscan-SE/) or BLASTN (http://blast.ncbi.nlm.nih.gov/Blast.cgi), respectively. Function was predicted using COGs (clusters of orthologous groups; http://www.ncbi.nlm.nih.gov/COG/), and genes were initially annotated using IMC (In Silico Molecular Cloning, Genomics Edition, version 4.1.21D; In Silico Biology Inc., Yokohama, Japan).
The circular representation of pR148 was generated with IMC and edited to differentiate CDS based on COG functions and to add annotations. Linear comparative representations were based on results from DNAPlotter version 1.4 (Wellcome Trust Sanger Institute, United Kingdom).
Comparative analysis.
The nr/nt database was searched for closest matches to genes in pR148 with BLASTN in February 2012. Search parameters are given in the supplemental material. Sequences showing ≥50% query coverage were chosen for initial comparative analysis.
The backbone was determined by identifying the accessory region based on annotated integrase genes. The identity to other sequences in the nr/nt database was determined using BLASTN. Insertion/deletion (indel) regions were determined from the BLASTN results, and detailed analysis was conducted using Align Plus 4.10 in the Clone Manager Professional Suite (Scientific and Educational Software, Cary, NC).
Multiple plasmid sequence alignment analysis.
The MAUVE program filters and sorts internally identified matches into local colinear blocks (LCBs) (19). Each LCB represents a region of homologous sequence without any rearrangement of the input genomes. In our study, we used MAUVE to visualize the conserved (homologous) and indel regions among the compared sequences. To generate the progressive alignment of multiple plasmids by MAUVE 2.0 (http://asap.ahabs.wisc.edu/) (20), the start nucleotides (nucleotide number 1) of pNDM_Dok01, pNDM102337, and pSN254 were shifted to the repA gene. The sequences were aligned using the default seed weight, allowing MAUVE to determine LCBs with the default minimum weight and allowing for full alignment, iterative refinement, and sum-of-pairs LCB scoring with the default matrix (HOXD70), a gap opening score of −400, and a gap extension score of −30.
Protein family network of pR148.
To represent the evolutionary relationship of pP9014 with other plasmids as a network, we generated pairwise comparisons against other mobile genetic elements (MGEs) in the ACLAME (for a classification of mobile genetic elements, version 0.4) (21) database using the methodology modified from Lima-Melendez et al. (22). Briefly, a pairwise similarity comparison of each pR148-encoded protein (n = 147) to the total ACLAME database was obtained using BLAST (23) with an E value threshold of 0.0001 and refined using an E value of 1e−20 and a minimum identity of 20%.
The resulting output was parsed in the form of a matrix in which rows represent related MGEs and columns represent protein families. We then determined the similarity of pR148 to the related MGEs by obtaining a P value, which is the probability of finding a common number of protein families between each pair of MGE vehicles based on the hypergeometric formula
| (1) |
where c is the number of protein families in common, a is the number of protein families of MGEs in ACLAME hits, b is the number of protein families identified in plasmid pR148, p indicates the probability, i indicates the integer value for the number of protein families in common between two phages, and n is total number of protein families in the ACLAME database. Afterwards, the significance value (Sig) was obtained by using the formula
| (2) |
where the P value was obtained from equation 1 and T = (2,704 × 2,703)/2.
A new network of pR148 and the MGEs with a Sig value of >0 was constructed and merged with the existing ACLAME network in Cytoscape (http://www.cytoscape.org/) using an edge-weighted spring embedded model, wherein MGEs sharing more protein families (higher Sig value) appear closer in the network.
Phylogenetic analysis.
The alignments of the full genomic nucleotide sequences of pR148 and the plasmids chosen for comparative analysis were determined using the ClustalW implementation in MEGA5.05. To allow for longer gaps in the alignment, a gap-opening penalty of 10 and a gap extension penalty of 2 was allowed for both pairwise and multiple alignments with a transition weight of 0.2. Since ClustalW aligns linear sequences, the full nucleic acid sequences were parallelized by making sure all sequences are in the same orientation and choosing the start of the repA gene of all sequences as the zero point.
To generate the neighbor joining tree, a bootstrap analysis with 1,000 replications was conducted with substitutions for transitions and transversions of nucleotides using the maximum composite likelihood model, with gamma-distributed rates among sites, allowing for a heterogeneous pattern of substitution among lineages, and with a partial deletion of gaps for an 85% site coverage cutoff score.
Accession numbers.
The sequence of pR148 was deposited in GenBank (http://www.ncbi.nlm.nih.gov/GenBank) under the accession number JX141473. The accession numbers of the other sequences used in the study are the following (in parentheses): E. coli pNDM-1_Dok01 (AP012208), E. coli pNDM102337 (JF714412), E. coli pNDM10505 (JF503991), Klebsiella pneumoniae pNDM-KN (JN157804), Salmonella enterica serovar Typhimurium GI-VII-6 (AB571791), E. coli pUMNK88 (HQ023862), E. coli pAR060302 (FJ621588), Salmonella enterica serovar Newport pSN254 (CP000604), Salmonella enterica serovar Dublin pSD_174 (JF267651), E. coli pAPEC1990_61 (HQ023863), Providencia stuartii pMR0211 (JN687470), Salmonella enterica pAM04528 (FJ621587), Yersinia pestis serovar Orientalis pIP1202 (CP000603), Photobacterium damselae subsp. piscicida pP91278 (AB277724), P. damselae subsp. piscicida pP99-018 (AB277723), E. coli pPG010208 (HQ023861), Y. ruckeri pYR1 (CP000602), E. coli peH4H (FJ621586), A. hydrophila pRA1 (FJ705807), and Xenorhabdus nematophila XNC1_p (FN667743).
RESULTS AND DISCUSSION
Backbone of pR148.
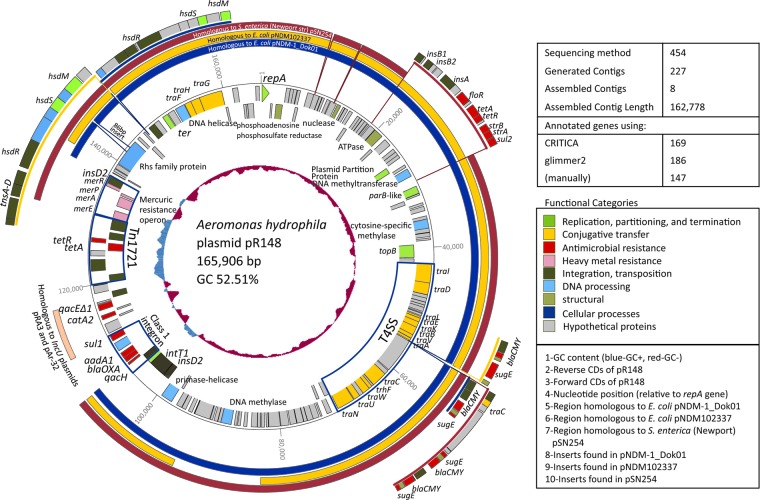
pR148 is a circular plasmid that is 165,906 bp long (Fig. 1). It contains 147 putative CDS as annotated manually. One hundred seventeen are on the same (forward) strand as the repA gene, and 30 are on the opposite (reverse) strand. Compared to other sequences that showed greater than 50% query coverage for the whole genome sequence (Fig. 2), pR148 has a highly conserved plasmid backbone, a feature of IncA/C plasmids that has been reported in previous studies (4, 5). The core backbone was determined from the full genomic sequence by identifying the accessory region flanked by two insD2 transposase proteins. The size of the core backbone was 128,699 bp, had 123 genes, and showed high synteny and very high identity (98 to 100%), high query coverage (>62%), and very high scores (>1 × 105) with the other compared sequences.
Fig 1.
Circular representation of pR148. The assembly and annotation information are shown. Circles from inside to outside: GC content (blue, GC+; red, GC−), reverse CDS, forward CDS, nucleotide position (relative to the repA gene, which is represented as an arrowhead), region in pR148 homologous to E. coli pNDM-1_Dok01, region homologous to E. coli pNDM-1_Dok01, region homologous to S. enterica (Newport) pSN254, inserts found in pNDM-1_Dok01, inserts found in pNDM102337, and inserts found in pSN254. Genes are color coded according to function. The locus for the type 4 secretion system (T4SS), the mercuric resistance operon, Tn1721, and the class 1 integron are enclosed in blue boxes.
Fig 2.
BLAST homology search results. The homology of the whole nucleotide sequence of pR148 was generated against the nr/nt database using BLASTN with a 2 to 3 match/mismatch scores, gap scores of 5 for existence and 2 for extension, and an expected threshold of 1,000 on 17 February 2012. Sequences showing ≥50% query coverage were chosen for initial comparative analysis. The black lines transverse to the alignment indicate regions of least similarity, while gaps indicate deletion regions (regions that contain nucleotide sequences in pR148 but not in the subject sequence).
repA, traD, traB, and traF.
The nucleotide sequences of the repA gene and the translated nucleotide sequences of repA, traD, traB, and traF genes were compared to those of other plasmids chosen for comparative analysis using BLASTN or TBLASTN. The repA gene showed 100% identity and 100% query coverage with the repA genes of pNDM-1_Dok01, pNDM102337, pNDM10505, and pNDM-KN and 99% identity and 100% query coverage for the rest of the plasmids in both nucleic acid and amino acid queries, with the exception of the BLASTN result for pRA1, which only showed 93% identity.
TraD, TraB, and TraF are important components of the T4SS (type 4 secretion system) and thus are important for conjugative transfer. The use of concatenated sequences of these proteins for comparative analysis of IncA/C plasmids have been performed already (5); however, in our study, TraD, TraB, and TraF showed very similar results except for peH4H, which does not contain TraD, pAPEC1990_61, which does not have TraB, and XNC1_p which does not contain TraD or TraF. GI-VII-6, which is a genomic island identified in S. Typhimurium and contains antimicrobial resistance genes (24), is not a plasmid and does not contain a repA gene. We considered that this approach was not sufficient to be able to establish the relationships of the plasmids, so we used different techniques that use more comprehensive methods.
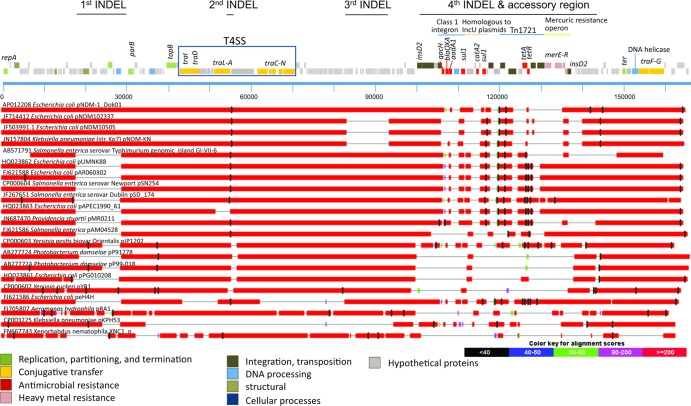
Multiple plasmid alignment constructed from MAUVE.
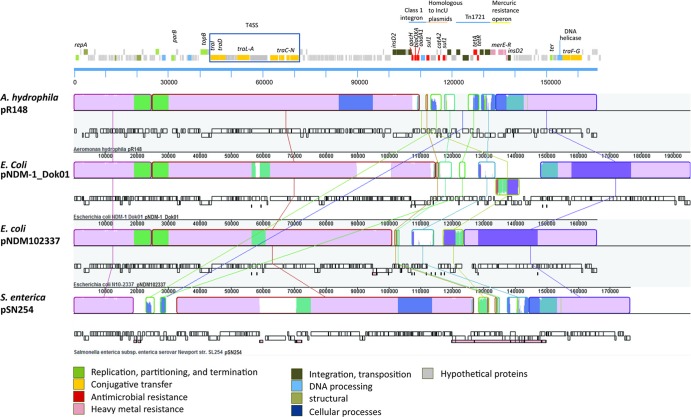
Based on the BLASTN homology result, pR148 showed very high similarity to several plasmids from the IncA/C family with indels in certain regions (Fig. 2). The compared sequences form similarity groups. One group is the NDM-1 group, containing sequences which include pNDM102337, pNDM10505, and pNDM-KN. These sequences have an indel region in the T4SS (at around 55,000 bp) and show a gap at around 90,000 bp. Another group contains pUMNK88, pAR060302, pSN245, pSD_174, pAPEC1990_61, pMR0211, and pAM04528 (Fig. 2). This group of plasmids has a gap at around 20,000 bp and is similar to the S. Typhimurium GI-VII-6. For graphical representations of the indel regions and for a more detailed comparative analysis using MAUVE, we selected three representative plasmids: pNDM-1_Dok01, the plasmid most similar to pR148, pNDM102337, a representative of the NDM-1 carrying plasmids, and pSN254, a well-studied plasmid and representative of the second group.
The MAUVE result (Fig. 3) revealed several regions of homology, called local colinear blocks (LCBs). Four indel regions were found at (i) ca. 25,000 bp, where pSN254 shows a major difference from the other sequences, (ii) ca. 55,000 bp in the T4SS, (iii) ca. 90,000 bp, which is a gap in pNDM102337, and (iv) ca. 140,000 bp, a region containing the RHS (retrotransposon hot spot) family protein in pR148. A comparative analysis using the full sequences of the plasmids revealed several important differences that can be visualized but cannot be quantified using MAUVE.
Fig 3.
MAUVE alignment of local colinear blocks (LCBs) of pR148, pNDM-1_Dok01, pNDM102337, and pSN254. The topmost panel is a linear representation of pR148 for reference, arranged from top to bottom representing reverse CDS, forward CDS, and nucleotide position relative to the repA gene. Genes are color coded according to function. The representation of each sequence from MAUVE contains, from top to bottom, the LCBs, forward CDs, reverse CDs, and annotated features. Lines are drawn to connect the similar LCBs, and a colored similarity plot is shown for each genome, the height of which is proportional to the level of sequence identity in that region. Mauve-colored segments are conserved among all sequences, while other colors indicate segments which are conserved only among some of the sequences.
First indel.
In the region equivalent to 17,944 to 28,929 bp of pR148, the plasmids pUMNK88, pAR060302, pSN254, pSD_174, pAPEC1990_61, pMR0211, pAM04528, pPG010208, peH4H, and the genomic island GI-VII-6 have an accessory element coding for antimicrobial resistance (Table 1). This antimicrobial resistance insert carries floR, tetA and tetR, strA and strB, and sul2 (see Fig. S1A in the supplemental material). A transcriptome analysis revealed that this region in pAR060302 is transcribed at lower levels compared to the backbone and other accessory elements but can be induced by antibiotic treatment (2). In pR148, pNDM102337, pNDM10505, pNDM-1_Dok01, and pYR1, this region is occupied by a highly conserved set of genes, including an AAA family ATPase which showed 100% identity and 100% query coverage with the AAA ATPase of P. damselae subsp. piscicida (YP_908585). This AAA ATPase has the walker A and B motifs and an arginine finger, a motif that senses ATP binding and hydrolysis and transmits conformational changes (25). Downstream of the AAA ATPase are genes for a DNA-binding plasmid partition protein and a DNA methylase. Further downstream at 73,743 to 74,711bp is a second walker-type AAA ATPase which is conserved in most other analyzed sequences except peH4H. This secondary AAA ATPase contains a CbbQ/NirQ/NorQ C terminal which may be involved in chaperone activity (25).
Table 1.
Regions of differentiation among the compared sequences
| Species | Element found in: |
Presence of: |
||||
|---|---|---|---|---|---|---|
| 1st indel (bp 17,944–28,929) | 2nd indel (bp 42,644–70,720) (T4SS) | 3rd indel (bp 82,806–93,637) | 4th indel (bp 139,675–143,928) (RHS) | Class 1 integron | Mer operon | |
| A. hydrophila pR148 | AAA ATPasea | T4SS | DNA methylase dcmd | RHS | Present | Present |
| E. coli pNDM-1_Dok01 | AAA ATPase | blaCMYc | DNA methylase dcm | HsdR | Present | |
| E. coli pNDM102337 | AAA ATPase | blaCMY | HsdR | Present | ||
| E. coli pNDM10505 | AAA ATPase | blaCMY | HsdR | Present | ||
| K. pneumoniae pNDM-KN | blaCMY | HsdR | Present | |||
| S. enterica (Typhimurium) GI-VII-6 | floRb | blaCMY | DNA methylase dcm | RHS | Present | |
| E. coli pUMNK88 | floR | blaCMY | DNA methylase dcm | RHS | Present | Present |
| E. coli pAR060302 | floR | blaCMY | DNA methylase dcm | RHS | Present | Present |
| S. enterica (Newport) pSN254 | floR | blaCMY (2×) | DNA methylase dcm | RHS | Present | Present |
| S. enterica (Dublin) pSD_174 | floR | blaCMY (2×) | DNA methylase dcm | RHS | Present | |
| E. coli pAPEC1990_61 | floR | blaCMY | DNA methylase dcm | RHS | Present | Present |
| P. stuartii pMR0211 | floR | blaCMY | DNA methylase dcm | qnrA1, sul1; No RHS | Present | |
| S. enterica pAM04528 | floR | blaCMY (2X) | DNA methylase dcm | RHS | Present | |
| Y. pestis (Orientalis) pIP1202 | T4SS | DNA methylase dcm | Inverted Tn21 | Present | ||
| P. damselae subsp. piscicida pP91278 | T4SS | DNA methylase dcm | RHS | |||
| P. damselae subsp. piscicida pP99-018 | T4SS | DNA methylase dcm | RHS | |||
| E. coli pPG010208 | floR | T4SS | DNA methylase dcm | RHS | ||
| Y. ruckeri pYR1 | AAA ATPase | T4SS | DNA methylase dcm | tetA, tetD, tetC | ||
| E. coli peH4H | floR | blaCMY | DNA methylase dcm | RHS | Present | Present |
| A. hydrophila pRA1 | T4SS | DNA methylase dcm | tetA, sul2; no RHS | |||
| X. nematophila XNC1_p | T4SS | DNA methylase dcm | RHS | |||
This region encodes an AAA ATPase, a DNA-binding plasmid partition protein, and a DNA methyltransferase.
This antimicrobial resistance insert encodes FloR, TetA/R, StrA/B, and Sul2 instead of the AAA ATPase found in pR148.
This antimicrobial resistance insert encodes BlaCMY, Blc, and SugE transecting the T4SS.
This region encodes the DNA methylase Dcm.
Second indel (T4SS).
From 42,644 to 70,720 bp, pR148 contains genes encoding the T4SS. The T4SS forms a channel composed of three core proteins. This channel is double walled and spans the bacterial membrane (26). T4SS are versatile systems that are used by bacteria to inject effectors to target eukaryotic cells and also for conjugative transfer to spread plasmids (27, 28).
Some plasmids have an insert within the T4SS between traA and a 1,840-aa (amino acid) hypothetical protein at 55,198 bp (Table 1). This insertion carries blaCMY, blc, and sugE (see Fig. S1B in the supplemental material). pSN254, pSD_174, and pAM04528 carry a duplicate of this region with the hypothetical proteins, a truncated traC and sugE, blc, and blaCMY genes in the opposite orientation and bordered by insC transposases.
Among the compared sequences, the indels in this region were more commonly detected than the first indel, but most sequences that contain the floR insert also contain this accessory element. This region has also been shown to be prone to excision over time (15), and the presence of β-lactamases has been shown to confer a significant fitness cost: a biological disadvantage under normal conditions that is inherent with the different metabolic demands involved in the expression of these genes, thus bacteria that do not carry these genes can outcompete bacteria that do carry them, and this can result in reduced dissemination of the plasmid. This has been shown with blaCMY-7 (29), blaSME-1 (30), and blaCMY-2 (31). The presence of this insert has also been shown to significantly reduce the conjugation frequency (32) and has been shown in the transcriptome analysis of pAR060302 (2) to be unresponsive to antibiotic treatment, but it is constantly transcribed more strongly than the floR region.
Third indel.
From 82,806 to 93,637 bp, there is a region that contains genes encoding the cytosine-specific DNA methylase dcm, which is a phosphoadenosine phosphosulfate reductase family protein and participates in repair of mismatches at 5-methylcytosine sites (33).
This indel region contains other genes of mostly unknown function. This region is absent from the NDM-containing plasmids pNDM102337, pNDM10505, and pNDM-KN (Table 1) but is highly conserved among the other compared plasmids (see Fig. S1C in the supplemental material). The absence of the dcm gene can cause a deficiency in the methylation of cytosine (34), making the plasmids more sensitive to EcoRII (CCATGG) restriction.
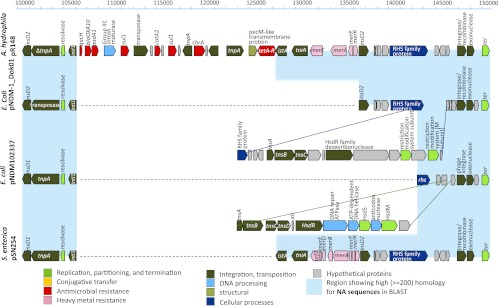
Fourth indel.
From 139,675 to 143,928 bp, pR148 codes for an RHS family protein. In pNDM-1_Dok01, pNDM102337, pNDM10505, and pNDM-KN, this region has an insert downstream of the RHS encoding HsdR family DNase protein set carried by a Tn7-like transposon (Fig. 4 and Table 1). In pYR1, there is an insert downstream of the RHS containing the tetracycline resistance genes rerA, tetD, tetC, and gltS. In pRA1, there is no RHS protein detected but there is an insert encoding tetA and sul2. RHS protein is also absent from pMR0211, but in its stead there is an insert encoding quinolone resistance (qnrA1), aminoglycoside resistance (aminoglycoside 6′-N-acetyltransferase), and sulfonamide resistance (sul1).
Fig 4.
Representation of the accessory and 4th indel and accessory region of pR148 compared to the flanking backbone sequences from the representative plasmids pNDM-1_Dok01, pNDM102337, and pSN254. The light blue background represents regions showing alignment scores of ≥200 in BLASTN. The scale on the top represents nucleotide positions in pR148. Genes are color coded according to function.
Members of the RHS family are known as hotspots for integration (35); however, among the compared sequences, only 5 plasmids show insertions, 2 plasmids do not have RHS family proteins, and the other sequences have the RHS family protein but no insertions.
Similarity to S. Typhimurium genomic island GI-VII-6.
pR148 shares a very high similarity to the genomic island GI-VII-6 (99% maximum identity and 73% query coverage of the total nucleotide sequence) derived from the chromosome of S. Typhimurium L-3553 isolated from cattle in Japan in 2004 (24). GI-VII-6 was reported to confer antimicrobial resistance to extended-spectrum cephalosporins in S. Typhimurium. GI-VII-6 is similar to pR148 regions from 149,813 to 161,083 bp and 9,119 to 106,217 bp. This region includes most of the backbone except for the region surrounding the repA gene and extends to the class 1 integron in the accessory region. GI-VII-6 is mostly similar to the plasmids pUMNK88, pAR060302, pAPEC1990_61, pMR0211, and peH4H containing both the inserted accessory element in the first indel and a single copy of the insert in the T4SS. GI-VII-6 does not contain the ter gene (encoding DNA replication terminus site-binding protein) and is flanked by IS26 transposases. As mentioned in a previous report, this genomic island may be derived from the transposition and/or homologous recombination of plasmid DNA (24).
Accessory region.
The core backbone does not contain any drug resistance genes, unlike some other IncA/C plasmids. All of the drug resistance genes are in an accessory region bordered by two insD2 sequences; the first is aligned on the negative strand from 101,147 to 100,143 bp, and the second is aligned on the positive strand from 136,345 to 137,349 bp. This accessory region contains 27 genes, is 37,207 bp long, and provides resistance against β-lactams (blaOXA-10), chloramphenicols (catA2), aminoglycosides (aadA1), sulfonamides (sul1), and tetracyclines (tetA/R) (Fig. 4). Similar drug resistance genes were detected in pR148 in our previous study using a microarray and PCR (13).
This accessory element shows the characteristics of a Tn21 transposon (34, 36), although tnpA on the 5′ end (ORF101) is truncated and is repeated downstream in the reverse orientation (ORF117). At the 5′ end, there is a class 1 integron with its variable region containing blaOXA-10 and aadA1. These genes have highest identity (≥99% query coverage and identity) to genes in a multidrug-resistant human pathogen (Acinetobacter baumannii AYE, NC_010410) that caused a nationwide outbreak in France in 2001 (37). blaOXA-10 is a class D extended-spectrum β-lactamase that had previously been detected in human pathogens. The presence of this gene in a fish pathogen may mean that there has been lateral gene transfer of the MGE carrying this gene. Whether this transfer happened from the fish to the human pathogen or vice versa or even through another intermediate carrier cannot be determined.
This variable region also encodes a sequence with 100% similarity and 97% sequence coverage with the group II intron maturase (JF681371) from Enterobacter cloacae strain GOCER. Group II introns are highly structured ribozymes found in bacteria and organellar genomes (38), and they are thought to be ancestors of nuclear pre-mRNA introns (39). Group II introns work on both RNA and DNA to promote splicing and mobility (40, 41). Class 1 integrons have been found in several Gram-negative bacteria (42) and in several of the compared sequences (Table 1). A notable difference is pIP1202, which contains a class 1 integron in an inverted Tn21 that has been inserted into the RHS.
Downstream of the class 1 integron there is a Tn1721-like transposon that provides tetracycline resistance through the tetA/R genes. pR148 also has a region between the class 1 integron and the Tn1721 transposon that most closely matches (96% query coverage and 100% maximum identity) some regions in IncU plasmids from A. hydrophila pRA3 (DQ401103), Aeromonas salmonicida pAr-32 (AJ517791), and E. coli pSa (L06822). This region also carries a catA2 gene and two sul1 genes. This region is followed by a PecM/EamA-like efflux protein (previously known as DUF6 and a member of the 10-TMS [transmembrane α helical spanners] DME [drug metabolite exporter] family [43]) and the chromate ion transporter chrA gene before Tn1721. Further downstream, there are genes encoding a heavy metal resistance operon. This region shares 99% maximum identity and 95% query coverage with similar regions in pAPEC1990_61, pUMNK88, pAR060302, pAM04528, peH4H, and pSN254, but it is not present in the other analyzed sequences. However, we could not detect significant resistance of pR148 against HgCl2 (MIC of 8 μM/ml versus 4 μM/ml for empty HB101) using the agar dilution method.
Comparative analysis of whole plasmid sequences.
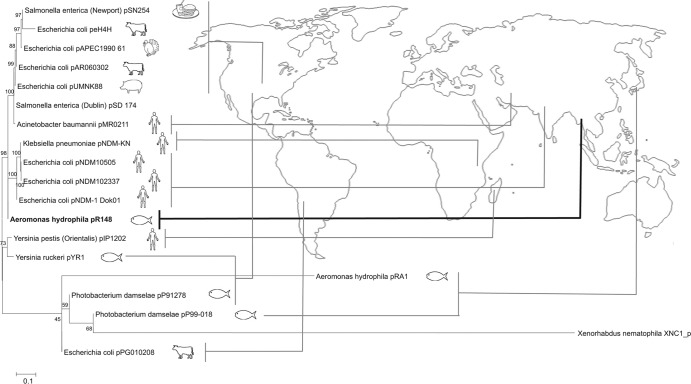
A phylogenetic tree based on whole plasmid sequences (Fig. 5) revealed that pR148 is most closely related to three E. coli-derived plasmids that contain blaNDM-1 (New Delhi metallo-β-lactamase): pNDM-1_Dok01, pNDM102337, and pNDM10505. A bacterium containing NDM-1 (FN396876) was first isolated in Sweden from a patient who had been hospitalized in India (44). NDM-1-containing bacteria have recently become a grave concern because of their increasing prevalence (45). pNDM-1_Dok01 was isolated from a Japanese traveler who had just returned from India in 2009 (46) and showed severe toxic symptoms. pNDM10505 was isolated from a patient with a travel history to India in 2010, and pNDM102337 was isolated in 2008 from a patient who had been hospitalized in Pakistan (47). It has been suggested that the blaNDM-1 gene is disseminated by plant pathogens or that plant pathogens serve as a reservoir of this gene (46, 48). The accessory region of pNDM-1_Dok01 bears a strong similarity to that of pNDM-HK(HQ451074), which was derived from an MDR E. coli strain in Hong Kong. The backbone of pNDM-HK in turn shows extensive similarity with plasmids from plant pathogens (48). pR148 may have been derived from pNDM-1_Dok01 with its accessory element replaced with other MGEs, or the two plasmids may have been derived from a common ancestral plasmid with one obtaining elements from plant pathogens (pNDM-1_Dok01) and the other from human and fish elements (pR148).
Fig 5.
Phylogenetic alignment and source map of closely related plasmids. The phylogenetic tree of parallelized sequences was aligned using ClustalW and was generated using the neighbor-joining method with 1,000 bootstrap replications.
The other fish-derived plasmids (pYR1, pP91278, and pP99018) form a cluster with human-derived pIP1202 and cattle-derived pPG010208. The fish-derived IncA/C reference strain pRA1 forms an outgroup (Fig. 5). It is interesting that all of the closely related plasmids were isolated from enterobacteria which all reside in the gut of the host. This may provide a clue into how these plasmids were transferred.
Since genetic information is exchanged in plasmids laterally, the relatedness of plasmids can best be visualized in a network, where MGE carriers (plasmids, phages, and genomic islands) can be connected when they share a homologous gene fragment above a certain threshold (49). This gene-sharing network can show the relationship of each individual protein of a given plasmid to those from other MGEs. ACLAME is a web-based database that provides a network classification of MGEs of various well-known plasmids, phages, and transposons in terms of their gene/protein contents (21, 50). Pairwise analysis in ACLAME assigns all annotated CDS to a protein family and compares the conservation of these protein families against other MGE vehicles (plasmids, phages, and genomic islands). It can take into account similarities that may be found in only a few of the compared sequences. One hundred forty-seven pR148-encoded proteins were analyzed against a total of 32,919 protein families from 2,704 plasmids, viruses, and prophages (as of March 2012). In this way, we were able to retrieve 134 predicted protein sequences from pR148 associated with ACLAME protein families encoded by various MGEs.
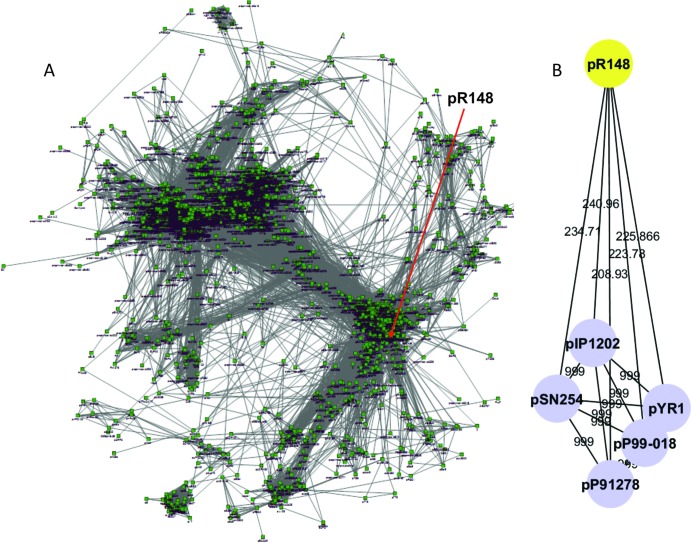
MGEs with a Sig value of more than 0 are shown in a matrix (see Fig. S2 in the supplemental material) and represented in a network (Fig. 6A). In this network, each node represents a genome, and nodes are connected if they are homologous to each other. The edges are weighted, meaning genomes sharing more families are closer to each other on the display. pR148 can be found clustered with other plasmids and MGE vehicles. Among the sequences chosen for comparison, the ACLAME database contained data for pSN254, pYR1, pIP1202, pP91278, and pP990-018. The network representation (Fig. 6B) shows that all of these MGEs are closely related to one another but pR148 has fewer shared elements.
Fig 6.
Network representation of the closely related plasmids. A network representation for pR148 was produced using Cytoscape by merging the available network for mobile genetic elements (MGEs) generated from the ACLAME database. Each node represents a genome, and nodes are connected if they share homologous DNA. The edges are weighted, meaning genomes sharing more families have a higher significance value and are closer on the display. (A) pR148 resides in a cluster of several MGEs that are closely related to one another. (B) The MGEs extracted from the ACLAME database that were also chosen for comparative analysis show that all of these MGEs are closely related to one another but pR148 has fewer shared elements. Circles, plasmids; lines, edges representing the significance value.
We determined the CLUSTALW alignment of all of the closely related sequences based on the BLASTN result of the full plasmid sequence from which we chose the closest relatives based on an artificial cutoff score (in the current case, ≥50% query coverage). We relaxed the gap-opening penalties in order to allow for alignments with larger gaps. This approach is simple; it relies on nucleic acid sequences, since MGEs are exchanged as nucleic acids, not as proteins, and it could discriminate between MGEs from the same protein families that have been acquired in different events and are in different positions in the genome. We believe this is an efficient and viable comparative method to determine the relatedness of closely related sequences.
This is the first report of this type of plasmid in A. hydrophila. The similarity of the accessory region of pR148 to elements from such diverse sources as human- and fish-derived plasmids is indicative of lateral gene transfer involving multiple modules. The high similarity of the backbone to human-derived plasmids indicates either that there was transfer between their hosts or that they are derived from a common ancestor. All of this is a serious cause for concern, since the judicious use of antimicrobials in animal and especially fish culture is not as rigorously enforced as it is in humans. This and other multidrug-resistant plasmids are a threat to the health not only of fish but also of humans.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the assistance of Gyemin Lee (Department of Information and Statistics, Gyeongsang National University) and Gipsi Lima-Mendez (Laboratoire de Bioinformatique des Génomes et des Réseaux, Université Libre de Bruxelles) for their assistance in the reticulate analysis.
This work was supported by the Japanese Society for the Promotion of Science (JSPS) Asian CORE Program. This work was also partially supported by a grant from the World Class University Program (R32-10253) funded by the Ministry of Education, Science and Technology of South Korea.
Footnotes
Published ahead of print 15 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01239-12.
REFERENCES
- 1. Aminov RI. 2011. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lang KS, Danzeisen JL, Xu W, Johnson TJ. 2012. Transcriptome mapping of pAR060302, a blaCMY-2-positive broad-host-range IncA/C plasmid. Appl. Environ. Microbiol. 78:3379–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Norman A, Hansen LH, Sorensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:2275–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P. 2012. Evolution of IncA/C blaCMY-2-carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 56:783–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, White DG, Cebula TA, Ravel J. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191:4750–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aoki T, Egusa S, Ogata Y, Watanabe T. 1971. Detection of resistance factors in fish pathogen Aeromonas liquefaciens. J. Gen. Microbiol. 65:343–349 [DOI] [PubMed] [Google Scholar]
- 7. Esteve C, Alcaide E, Blasco MD. 2012. Aeromonas hydrophila subsp. dhakensis isolated from feces, water and fish in Mediterranean Spain. Microbes Environ. [Epub ahead of print.dx.doi.org/10.1264/jsme2.ME12009 [DOI] [PMC free article] [PubMed]
- 8. Janda JM, Abbott SL. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23:35–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan FK, Ching JY, Ling TK, Chung SC, Sung JJ. 2000. Aeromonas infection in acute suppurative cholangitis: review of 30 cases. J. Infect. 40:69–73 [DOI] [PubMed] [Google Scholar]
- 10. Chim H, Song C. 2007. Aeromonas infection in critically ill burn patients. Burns 33:756–759 [DOI] [PubMed] [Google Scholar]
- 11. Okumura K, Shoji F, Yoshida M, Mizuta A, Makino I, Higashi H. 2011. Severe sepsis caused by Aeromonas hydrophila in a patient using tocilizumab: a case report. J. Med. Case Rep. 5:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bello-Lopez JM, Vazquez-Ocampo NJ, Fernandez-Rendon E, Curiel-Quesada E. 2012. Inability of some Aeromonas hydrophila strains to act as recipients of plasmid pRAS1 in conjugal transfer experiments. Curr. Microbiol. 64:332–337 [DOI] [PubMed] [Google Scholar]
- 13. Tipmongkolsilp N, del Castillo CS, Hikima J, Jung T, Kondo H, Hirono I, Aoki T. 2012. Multiple drug-resistant strains of Aeromonas hydrophila isolated from Tilapia farms in Thailand. Fish Pathol. 47:56–63 [Google Scholar]
- 14. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18:268–281 [DOI] [PubMed] [Google Scholar]
- 15. Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Badger JH, Olsen GJ. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512–524 [DOI] [PubMed] [Google Scholar]
- 17. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nho SW, Hikima J, Cha IS, Park SB, Jang HB, del Castillo CS, Kondo H, Hirono I, Aoki T, Jung TS. 2011. Complete genome sequence and immunoproteomic analyses of the bacterial fish pathogen Streptococcus parauberis. J. Bacteriol. 193:3356–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. 2009. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25:2071–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Darling AE, Mau B, Perna NT. 2010. Progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147 doi:10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leplae R, Hebrant A, Wodak SJ, Toussaint A. 2004. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 32:D45–D49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. 2008. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 25:762–777 [DOI] [PubMed] [Google Scholar]
- 23. Leplae R, Lima-Mendez G, Toussaint A. 2010. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 38:D57–D61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shahada F, Sekizuka T, Kuroda M, Kusumoto M, Ohishi D, Matsumoto A, Okazaki H, Tanaka K, Uchida I, Izumiya H, Watanabe H, Tamamura Y, Iwata T, Akiba M. 2011. Characterization of Salmonella enterica serovar Typhimurium isolates harboring a chromosomally encoded CMY-2 β-lactamase gene located on a multidrug resistance genomic island. Antimicrob. Agents Chemother. 55:4114–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Snider J, Houry WA. 2008. AAA+ proteins: diversity in function, similarity in structure. Biochem. Soc. Trans. 36:72–77 [DOI] [PubMed] [Google Scholar]
- 26. Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. 2009. Structure of a type IV secretion system core complex. Science 323:266–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7:703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hossain A, Reisbig MD, Hanson ND. 2004. Plasmid-encoded functions compensate for the biological cost of ampC overexpression in a clinical isolate of Salmonella Typhimurium. J. Antimicrob. Chemother. 53:964–970 [DOI] [PubMed] [Google Scholar]
- 30. Marciano DC, Karkouti OY, Palzkill T. 2007. A fitness cost associated with the antibiotic resistance enzyme SME-1 β-lactamase. Genetics 176:2381–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Subbiah M, Top EM, Shah DH, Call DR. 2011. Selection pressure required for long-term persistence of blaCMY-2-positive IncA/C plasmids. Appl. Environ. Microbiol. 77:4486–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poole TL, Edrington TS, Brichta-Harhay DM, Carattoli A, Anderson RC, Nisbet DJ. 2009. Conjugative transferability of the A/C plasmids from Salmonella enterica isolates that possess or lack blaCMY in the A/C plasmid backbone. Foodborne Pathog. Dis. 6:1185–1194 [DOI] [PubMed] [Google Scholar]
- 33. Lieb M. 1987. Bacterial genes mutL, mutS, and dcm participate in repair of mismatches at 5-methylcytosine sites. J. Bacteriol. 169:5241–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Korba BE, Hays JB. 1982. Partially deficient methylation of cytosine in DNA at CCATGG sites stimulates genetic recombination of bacteriophage λ. Cell 28:531–541 [DOI] [PubMed] [Google Scholar]
- 35. Wang YD, Zhao S, Hill CW. 1998. Rhs elements comprise three subfamilies which diverged prior to acquisition by Escherichia coli. J. Bacteriol. 180:4102–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JM, Raoult D, Medigue C, Weissenbach J, Cruveiller S. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805 doi:10.1371/journal.pone.0001805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zoschke R, Nakamura M, Liere K, Sugiura M, Borner T, Schmitz-Linneweber C. 2010. An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. U. S. A. 107:3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsuura M, Noah JW, Lambowitz AM. 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J. 20:7259–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G, Cavanagh S, Dunny GM, Belfort M, Lambowitz AM. 1997. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 11:2910–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rambo RP, Doudna JA. 2004. Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry 43:6486–6497 [DOI] [PubMed] [Google Scholar]
- 42. Fluit AC, Schmitz FJ. 2004. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 10:272–288 [DOI] [PubMed] [Google Scholar]
- 43. Jack DL, Yang NM, Saier MH., Jr 2001. The drug/metabolite transporter superfamily. Eur. J. Biochem. 268:3620–3639 [DOI] [PubMed] [Google Scholar]
- 44. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Babic M, Hujer AM, Bonomo RA. 2006. What's new in antibiotic resistance? Focus on β-lactamases. Drug Resist. Updates 9:142–156 [DOI] [PubMed] [Google Scholar]
- 46. Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, Hishinuma A, Arakawa Y, Kuroda M. 2011. Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS One 6:e25334 doi:10.1371/journal.pone.0025334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boyd D, Mataseje L, Graham M, Grant J, Mabon P, Moore D, Roscoe D, Tyler S, Van Domselaar G, Mulvey M. 2011. Complete sequences of 3 IncA/C plasmids harboring the NDM-1 metallo-β-lactamase. Abstr. 51st Intersci. Conf. Antimicrob. Agents Chemother, abstr. C1-525 [Google Scholar]
- 48. Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989 doi:10.1371/journal.pone.0017989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. 2010. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl. Acad. Sci. U. S. A. 107:127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leplae R, Lima-Mendez G, Toussaint A. 2006. A first global analysis of plasmid encoded proteins in the ACLAME database. FEMS Microbiol. Rev. 30:980–994 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.