Abstract
Mutations causing antibiotic resistance usually incur a fitness cost in the absence of antibiotics. The magnitude of such costs is known to vary with the environment. Little is known about the fitness effects of antibiotic resistance mutations when bacteria confront the host's immune system. Here, we study the fitness effects of mutations in the rpoB, rpsL, and gyrA genes, which confer resistance to rifampin, streptomycin, and nalidixic acid, respectively. These antibiotics are frequently used in the treatment of bacterial infections. We measured two important fitness traits—growth rate and survival ability—of 12 Escherichia coli K-12 strains, each carrying a single resistance mutation, in the presence of macrophages. Strikingly, we found that 67% of the mutants survived better than the susceptible bacteria in the intracellular niche of the phagocytic cells. In particular, all E. coli streptomycin-resistant mutants exhibited an intracellular advantage. On the other hand, 42% of the mutants incurred a high fitness cost when the bacteria were allowed to divide outside of macrophages. This study shows that single nonsynonymous changes affecting fundamental processes in the cell can contribute to prolonged survival of E. coli in the context of an infection.
INTRODUCTION
A major component of bacterial adaptation in the context of infectious diseases is their rapid evolution to tackle the immune system and antibiotics. Escherichia coli is a commensal and versatile pathogen that can cause death (1). Given these characteristics, it is an ideal organism for studying the transition of commensalism to pathogenicity. E. coli colonizes the infant gastrointestinal tract within hours after birth, and typically a mutualistic relation builds up. However, even the harmless E. coli can cause an infection when gastrointestinal barriers are broken (2) or in immunosuppressed hosts (3). Healthy hosts are also susceptible to highly adapted E. coli pathogenic clones, which can cause many different types of infections. There is evidence that some of the pathogenic strains evolved from the commensal E. coli through the acquisition of new genes and mutations (1). A fundamental part of the ecology of E. coli during the infection process is its interaction with the host immune system cells, in particular with macrophages (Mϕs). It is, however, not known whether E. coli harboring antibiotic resistance can have an advantage or disadvantage in the context of an interaction with the immune system. This knowledge is important given the high frequency of antibiotic resistance within commensal E. coli in healthy individuals (4, 5), which may lead to an increased risk of treatment failure during an infection process, because of limited therapeutic options.
Mutations that cause antibiotic resistance often produce associated fitness costs in bacteria (6, 7). When the environment contains an antibiotic, resistant bacteria exhibit an advantage. However, when the antibiotic is absent, resistant bacteria typically have reduced growth rates, although this depends on the genetic background (8, 9). This is not surprising, since mutations which cause antibiotic resistance often target physiologically important functions in the cell, such as transcription and protein synthesis, cell wall synthesis, or nucleic acid synthesis (6). Interestingly, the fitness effect of a resistance mutation can be detrimental in one environment and beneficial in another (10–14). For example, Trindade et al. (14) showed increased variation in fitness effects of resistant mutations in E. coli with increased environmental stress. Similarly, Hall et al. (11) demonstrated that the costs of 24 different rpoB mutations vary greatly among 41 environments with different carbon source. Having in mind that fitness effects of resistant mutations exhibit strong genotype-by-environment interactions, it is important to determine the effects of resistance in an environment imposed by the host. Despite its importance, to our knowledge there are only a few studies that explicitly address fitness effects of antibiotic resistant under conditions that are closer to the growth conditions in a host (15, 16). Furthermore, it has been shown that the fitness effects of antibiotic-resistant mutations vary substantially in different in vivo and in vitro models (17–20).
One important interaction that bacteria face in natural conditions is the interaction with cells from the immune system that are able to phagocytize them. There is little information available on fitness effects of antibiotic resistance in this important context. The aim of the research reported here is to determine whether or not single point mutations conferring rifampin (RIF), streptomycin (STR), and nalidixic acid (NAL) resistance can affect reproduction and survival of commensal E. coli in the face of professional phagocytes. We show that commensal bacteria carrying specific resistance mutations can survive better in the intracellular environment of professional phagocytes. This may have important consequences in designing therapeutic treatments and may be important for understanding the spread of drug resistance.
MATERIALS AND METHODS
Media and growth conditions.
The RAW 264.7 murine macrophage (Mϕ) cell line was maintained in an atmosphere containing 5% CO2 at 37°C in RPMI 1640 (Gibco) supplemented with 2 mM l-glutamine (Invitrogen), 1 mM sodium pyruvate (Invitrogen), 10 mM HEPES (Invitrogen), 100 U of penicillin-streptomycin (Gibco)/ml, 50 μM 2-mercaptoethanol solution (Gibco), 50 μg of gentamicin (Sigma)/ml, and 10% heat-inactivated fetal calf serum (standard RPMI complete medium). Before infection assays, the Mϕs were maintained in the same conditions, but in antibiotic-free RPMI medium (without penicillin-streptomycin and gentamicin). Bacterial strains were grown and competed in antibiotic-free RPMI medium in an atmosphere containing 5% CO2 at 37°C or in Luria-Bertani (LB) medium at 37°C, with aeration (Grant-Bio PHP-4 type Thermo-Shaker at 700 rpm).
Construction of strains.
Susceptible MG1655-YFP and MG1655-CFP strains (MG1655, galK::CFP/YFP, and ΔlacIZYA) containing yellow (YFP) and cyan (CFP) fluorescent proteins under constitutive expression were created by moving yfp or cfp chromosomal inserts by P1 transduction from previously described strains (MC4100, galK::CFP/YFP, ampR [pZ12], and strR [rpsL150]), that were kindly given by R. Kishony (21). To ensure the constitutive expression of YFP or CFP, the lac operon was deleted from an MG1655 background. Ampicillin resistance (pZ12) was removed from the yfp or cfp locus using the Wanner and Datsenko method (22). Mutations conferring resistance to RIF (in the rpoB gene), STR (in the rpsL gene), and NAL (in the gyrA gene) were previously constructed in E. coli K-12 MG1655 background (Table 1) (9). General transduction using P1 bacteriophage was performed as previously described (23) in order to place resistance mutations in the new E. coli K-12 MG1655-YFP and MG1655-CFP background. To confirm these mutations, each antibiotic resistance target gene was amplified and then sequenced. The primers used were as follows: to amplify part of the rpoB gene, 5′-CGTCGTATCCGTTCCGTTGG-3′ and 5′-TTCACCCGGATAACATCTCGTC-3′; to amplify the rpsL gene, 5′-ATGATGGCGGGATCGTTG-3′ and 5′-CTTCCAGTTCAGATTTACC-3′; and to amplify the gyrA gene, 5′-TACACCGGTCCACATTGAGG-3′ and 5′-TTAATGATTGCCGCCGTCGG-3′. Each resistant clone was grown from a single colony in LB medium supplemented with the respective antibiotic and stored in 15% glycerol at −80°C.
Table 1.
Genotypes of single-point mutations used in the study
| Gene | Genotype amino acid change | Nucleotide change | Antibiotic resistancea |
|---|---|---|---|
| rpsL | K43N | AAA to AAC | STR |
| rpsL | K43T | AAA to ACA | STR |
| rpsL | K43R | AAA to AGA | STR |
| rpsL | K88R | AAA to AGA | STR |
| rpoB | S531F | TCC to TTC | RIF |
| rpoB | H526Y | CAC to TAC | RIF |
| rpoB | I572F | ATC to TTC | RIF |
| rpoB | R529H | CGT to CAT | RIF |
| rpoB | S512F | TCT to TTT | RIF |
| rpoB | H526D | CAC to GAC | RIF |
| gyrA | S83L | TCG to TTG | NAL |
| gyrA | D87Y | GAC to TAC | NAL |
STR, streptomycin; RIF, rifampin; NAL, nalidixic acid.
Competitive fitness in conditions where bacteria can divide: test for effects on reproduction.
To estimate the fitness cost of resistance mutations, we performed competition assays (as are commonly done to estimate fitness effects of mutations [24]) in three different environments: LB medium, RPMI medium alone, and in RPMI medium with Mϕs. The resistant mutants constructed in the MG1655-CFP (or the MG1655-YFP) strain were competed against a susceptible MG1655-YFP (or susceptible MG1655-CFP) strain in an antibiotic-free environment at a ratio of 1:1. For competitions in LB medium, both resistant and susceptible strains were grown separately for 48 h for acclimatization (the bacteria were diluted at 1:103 after 24 h for passage) at 37°C with aeration and then mixed, and 10 μl of a 10−2 dilution was inoculated to a final volume of 150 μl of LB medium in 96-well microtiter plates (Costar, catalog no. 3595) for 24 h of competition. The plates were arranged in a checkerboard configuration wherein half of the wells were without cells to control for well-to-well and external contamination. For competitions in RPMI medium, resistant and susceptible strains were grown in antibiotic-free RPMI medium for 48 h (the bacteria were diluted at 1:10−3 after 24 h for acclimatization) at 37°C with 5% CO2. Competitions were performed in a 24-well cell culture tissue plates (containing 1 ml of culture medium in each well), by inoculating 10 μl of 10−1 dilution (approximately 5 × 104 bacteria). For competitions in the presence of the Mϕs, strains were competed in the same conditions used for competitions in the RPMI medium, except that Mϕs were present. In the infection with 106 E. coli with 106 Mϕs (RAW 264.7), after 3 h the number of CFU inside Mϕs is ∼104, and the CFU count for the outside area is 105. Mϕs were seeded in a 24-well tissue culture plate at approximately 2 × 105 to 3 × 105 cells per well and allowed to attach overnight. The cells were then washed, resuspended in fresh antibiotic-free RPMI medium, and activated with 2 μg of CpG-ODN 1826 (5′-TCCATGACGTTCCTGACGTT-3′ Σ)/ml for 24 h. After 24 h, the cells were washed from the remaining CpG-ODN, fresh antibiotic-free RPMI medium was added, and Mϕ were infected with bacteria as described above. The initial and final ratios of resistant and susceptible strains were determined by flow cytometry. The fitness cost of each of the resistance mutations was measured four times (twice in the YFP background and twice in the CFP background). The selection coefficient, a measure of competitive fitness, was estimated as: Scoeff = ln[(Nfb/Nfa)/(Nib/Nia)]/ln[Nfa/Nia] (25), where Scoeff is a selection coefficient of the resistant strain b against the susceptible strain a, Nfa and Nfb are the numbers of resistant (b) and susceptible (a) bacteria after competition, and Nia and Nib are the initial numbers of resistant (b) and susceptible (a) bacteria before the competition (Fig. 1B, D, and F).
Fig 1.
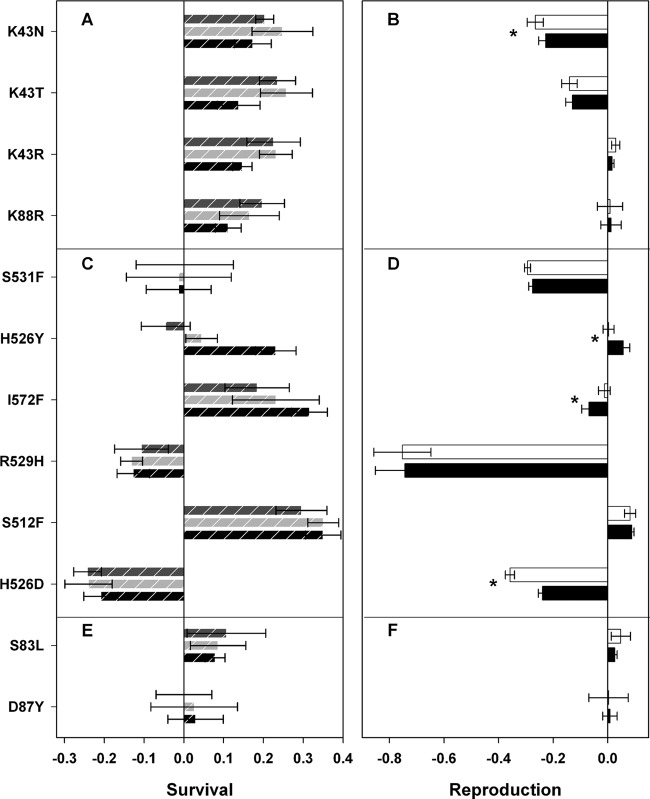
Effects of resistance on survival (left panel) and reproduction (right panel) of mutations in rpsL (A and B), rpoB (C and D), and gyrA (E and F) in E. coli. Panels A, C, and E show the fitness effects on survival inside Mϕs after 5 h (black dashed bars), 24 h (light gray dashed bars), or 48 h (dark gray dashed bars) postinfection. Panels B, D, and F show the effects of mutations when bacteria can reproduce in the presence (white bars) or absence (black bars) of Mϕs. All fitness effects were estimated using competition assays against a susceptible strain. The asterisk (*) represents significant differences (P < 0.05) determined using the Wilcoxon sum rank test.
Competitive fitness inside the Mϕs: test of the effect on survival.
Nonpathogenic E. coli does not replicate inside the Mϕs and thus, in this niche, survival is the most important fitness component (26). To estimate fitness the effect of the resistance mutations on survival inside phagocyte cells, Mϕs were prepared in the manner described above, infected with 5 × 106 bacteria (1:1, resistant versus susceptible strains), and centrifuged at 203 × g (1,000 rpm) for 5 min to enhance bacterial internalization. After 2 h of infection, the Mϕs were washed from the extracellular bacteria, and fresh cell culture medium containing 100 μg of gentamicin/ml was added to kill the remaining extracellular bacteria. After incubation for an additional hour, the medium was removed, monolayers of Mϕs were washed, and RPMI medium containing 20 μg of gentamicin/ml was added (0 h postinfection time point). To determine the number of intracellular bacteria after 5 and 24 h of incubation, infected Mϕs were washed three times with phosphate-buffered saline (PBS), and 0.1% Triton-X was added for 30 min at 37°C in order to lyse the Mϕs. The Mϕs were then centrifuged at 10,600 × g (10,000 rpm) for 5 min and washed in PBS, and the overall number of bacteria was counted by plating them on LB agar plates. To measure intracellular survival at 48 h postinfection, fresh culture medium containing gentamicin (20 μg/ml) was added 24 h postinfection to the infected cells.
Survival inside the Mϕs was estimated as the change in relative frequency (ΔX), calculated as follows: ΔX = Nfb/(Nfa + Nfb) − Nib/(Nia + Nib), where Nfa and Nfb are the numbers of resistant (b) and susceptible (a) bacteria after competition, and Nia and Nib are the initial numbers of resistant (b) and susceptible (a) bacteria before the competition (Fig. 1A, C, and E).
Survival of STR-resistant mutants in response to oxidative stress.
Given that all STR-resistant mutants showed a survival advantage inside Mϕs, we sought to determine whether the mutants would also show an advantage during nutrient limitation in the stationary growth phase and under oxidative stress, which are characteristics of the environment inside Mϕs.
To determine whether STR-resistant clones have differential fitness advantage in the exponential (4 h), early-stationary (24 h), and late-stationary (48 h) phases, competition assays between STR-resistant and -susceptible strains were performed. Briefly, STR-resistant and -susceptible strains were grown in antibiotic-free RPMI medium separately for 48 h at 37°C with 5% CO2 (the bacteria were diluted at 1:103 after 24 h for acclimatization) and then mixed at a ratio of 1:1 (1 resistant to 1 susceptible strain) plus 10 μl of a 10−1 dilution inoculated into 1 ml of culture medium. At 4, 24, and 48 h, samples of bacterial suspension were plated onto LB plates to estimate the ratios of STR-resistant to STR-susceptible strains at different growth phases (i.e., before exposure to H2O2 Σ; Fig. 2A).
Fig 2.
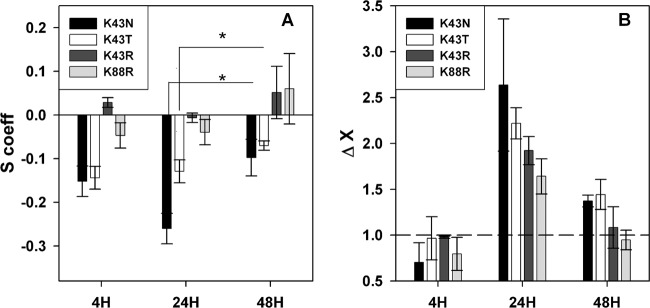
Starvation and oxidative stress diminish the fitness cost of STR-resistant mutations. (A) Effects on reproduction of STR-resistant mutations during 4-, 24-, and 48-h competition assays against a susceptible strain in RPMI medium. (B) Advantage of STR-resistant mutants against a susceptible strain after exposure to H2O2 at different phases of bacterial growth in RPMI medium. The bars above the dashed line represent an increased survival of the STR-resistant mutant against a susceptible strain. The asterisk (*) represents a statistical significant difference (P < 0.05) determined using the Wilcoxon sum rank test.
To determine whether STR-resistant clones would show an advantage for surviving oxidative stress during different growth phases, a mixture of STR-resistant and -susceptible strains (see the description above [before exposure to H2O2]) was treated with different concentrations of H2O2 (10 mM at 4 h, 20 mM at 24 h, and 40 mM at 48 h) for 30 min at 37°C. Appropriate dilutions were immediately plated onto LB medium to determine the relative numbers of STR-resistant to -susceptible strains after exposure to H2O2. Different concentrations of H2O2 were chosen because of the higher cell mortality at the exponential phase compared to the stationary phase in response to the same concentration of H2O2 (27). Four independent replicate experiments were performed for each strain (two in the YFP background and two in the CFP background). The survival of oxidative stress was calculated by dividing the relative frequencies of the STR-resistant mutant strains after and before exposure as follows: ΔX(H2O2) = [Nfb/(Nfa + Nfb)]/[Nib/(Nia + Nib)], where Nfa and Nfb are the numbers of resistant (b) and susceptible (a) bacteria after exposure to H2O2, and Nia and Nib are the numbers of resistant (b) and susceptible (a) bacteria before exposure (Fig. 2B).
Statistical analysis.
The Wilcoxon signed-rank test and Wilcoxon sum rank test with the Bonferroni correction (when multiple comparisons across mutants were made) were performed. The Kruskal-Wallis sum rank test was performed for comparisons across postinfection times. All statistical analysis was performed using R software (http://www.r-project.org/). Analysis of the linear regression between survival and reproduction of antibiotic-resistant mutants in the presence of the Mϕs (Fig. 3) was performed using SigmaPlot 9.0 software (Systat Software, Inc., Chicago, IL).
Fig 3.
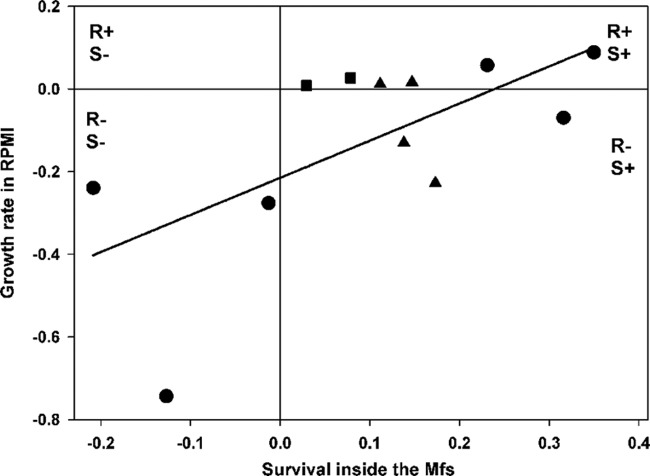
Test for correlation between survival inside Mϕs and growth rate. The intracellular survival of resistant mutants against a susceptible strain was measured at 5 h after bacterial internalization. The reproduction in RPMI medium without Mϕs was measured after a 24-h competition assay. The slope of the regression line (solid line) is 0.9 ± 0.34 (standard error) (P = 0.02), with R2 = 0.41. The graph is divided into quarters where R+ and R− (or S+ and S−) represent the advantage and disadvantage for reproduction (or survival), respectively. Symbols: ▲, STR-resistant mutants; ●, RIF-resistant mutants; ■, NAL-resistant mutants.
RESULTS
We studied 12 different antibiotic resistance mutations in rpsL, rpoB, and gyrA, conferring resistance to STR, RIF, and NAL antibiotics, respectively (Table 1). These mutations had been previously studied for fitness costs in LB medium when present in another genetic background (9). Because the fitness of antibiotic-resistant clones can depend on the genetic background (8), we measured the competitive fitness of these 12 mutants in LB medium and found that all showed a cost in LB medium. The costs of antibiotic-resistant mutations were not significantly different in the new genetic background in LB medium (Wilcoxon sum rank test with Bonferroni corrections; 4 of 12 mutations were significantly different without Bonferroni corrections [see Fig. S1 in the supplemental material]).
To determine the fitness effects of antibiotic-resistant clones in the presence of Mϕs, competition experiments between the susceptible and the resistant mutants were performed. Two main fitness traits are important during the infection process: reproduction, which occurs outside Mϕs, and survival, which is the main fitness component inside Mϕs. The effects on both traits—reproduction outside the Mϕs and survival inside the Mϕs—were measured (see Materials and Methods). In order to estimate the fitness effects of resistance in bacterial reproduction, competition assays in RPMI cell culture medium in the presence (+Mϕs) or absence (−Mϕs) of Mϕs were performed.
Global survival advantage of STR-resistant mutants inside the Mϕs.
Figure 1A shows the effects on survival of E. coli strains carrying mutations K43N, K43T, K43R, or K88R, which confer resistance to STR. Surprisingly, all STR-resistant mutants showed a survival advantage inside Mϕs at 5, 24, and 48 h postinfection. There was no significant differences in the survival effects of STR-resistant mutants between the postinfection periods (Kruskal-Wallis rank sum test, P > 0.05), except for the K43R mutant, which demonstrated increased survival inside Mϕs at later time points (Kruskal-Wallis rank sum test, P = 0.04). In contrast to the global fitness survival advantage inside Mϕs, two mutants showed a cost, and two other mutants were neutral when bacteria are allowed to reproduce, which was measured in competitive fitness assays against the susceptible strain in the presence or absence of the Mϕs (Fig. 1B). The cost of one mutation (K43N) differed significantly due to the presence of the Mϕs (Wilcoxon sum rank test, P = 0.04), whereas the costs of other three mutations were not different. In summary, single point mutations in the rpsL gene provided a survival fitness advantage in commensal E. coli in the intracellular niche of Mϕs, leading to an increased risk of treatment failure during an infection process.
Variable fitness effects in RIF-resistant mutants.
Half of the RIF mutants (S512F, I572F, and H526Y) showed a survival advantage inside the Mϕs (Fig. 1C). These were neutral or only slightly advantageous in competitive fitness assays where growth can occur outside Mϕs (Fig. 1D, white bars). Two RIF-resistant mutants showed impaired survival inside Mϕs (R529H and H526D) and also showed the highest fitness costs for reproduction (Fig. 1C and D). For the S531F mutation, no effect was detected on survival, but a deleterious effect was measured on reproduction. There was no overall difference for effects on survival of RIF-resistant mutants between different postinfection periods (Kruskal-Wallis rank sum test, P > 0.05), except for one mutant (H526Y), which ceased to be advantageous for survival inside the Mϕs at later time points (Kruskal-Wallis rank sum test, P = 0.01). The fitness effects on reproduction of three E. coli RIF-resistant mutants (Wilcoxon sum rank test, P = 0.01 for H526Y, P = 0.009 for I572F, and P = 0.005 for H526D) were significantly different due to the presence of the Mϕs, while effects for reproduction of other three mutants did not differ between presence or absence of Mϕs in the environment (Fig. 1D).
NAL-resistant mutants are advantageous or neutral.
The fitness of the S83L mutant was higher than susceptible in competitive fitness assays, for both reproduction in the culture media and survival inside Mϕs, whereas the fitness of D87Y mutant remained neutral (Fig. 1E and F). There was no difference in survival at the different postinfection time points (Kruskal-Wallis rank sum test, P > 0.05). We did not observe significant differences in fitness effects for the reproduction for the two studied NAL-resistant mutants (S83L and D87Y) due to the presence of the Mϕs (Wilcoxon sum rank test, P > 0.05 for both mutations) (Fig. 1F).
Advantage of STR-resistant mutants in response to oxidative stress in the stationary phase.
Given the striking survival advantage of all STR-resistant mutants, we tried to determine whether such results could be caused by the specific stress that bacteria face upon internalization, namely, nutrient starvation and/or oxidative stress. To test this hypothesis, competition assays were performed during the exponential phase, wherein bacteria are growing, and during the stationary phase, wherein growth is resumed. A possible advantage to oxidative stress was tested during these phases by adding H2O2. Although the fitness cost for reproduction was the highest after 24 h of bacterial growth, it was relieved after 48 h for the two most costly STR-resistant mutants, K43N and K43T (Wilcoxon sum rank test, P = 0.03 for K43N, and P = 0.03 for K43T [Fig. 2A]), indicating that STR resistance mutations could be advantageous during the stationary phase induced by nutrient limitation. Interestingly, all STR-resistant mutants displayed an increased survival in response to oxidative stress after 24 h but not during exponential growth phase (Fig. 2B). The results therefore indicate that nutrient deprivation and oxidative stress are key factors in the survival advantage that these mutants exhibit inside Mϕs.
Trade-off between survival and growth.
It has been proposed that resistance to stress is associated with reduced resource uptake (28). This trade-off between self-preservation and nutritional competence, the so-called SPANC balance, has been observed in several studies (28, 29). Recently, the SPANC trade-off has been directly linked to the growth rate, stress resistance, outer membrane permeability, morphotype characteristics, and virulence properties of antibiotic-resistant E. coli isolates from deep and visceral infections in humans (5). In the present study, we tested for a trade-off between survival inside the Mϕs and growth rate without Mϕs (Fig. 3). We did not find evidence of a trade-off but instead found that antibiotic-resistant clones that survived better inside Mϕs also had a better growth rate (Fig. 3).
DISCUSSION
Drug-resistant bacteria pose a significant threat to human health, and it is important to understand how the fitness of such bacteria can be impaired during infection. Here, we studied how antibiotic resistance affects two important fitness traits: the ability to survive and the ability to reproduce in the presence of Mϕs. It is known that during entry into Mϕs, bacteria experience a set of environmental stresses, such as host-induced nutrient limitation, acidification, toxic peptides, osmotic stress, and reactive oxygen species (ROS), the latter of which is believed to be the major cause of bacterial killing (30). To our knowledge, ours is the first study that measures the fitness effects for survival of several antibiotic-resistant mutants in the intracellular environment of the Mϕs. Surprisingly, we found that all STR-resistant mutants had increased survival inside Mϕs. RIF-resistant mutants were highly variable, and NAL-resistant mutants showed survival advantage of small effect. Importantly, STR resistance, although carrying substantial fitness costs for growth rate, shows a global advantage for survival.
The experience of the early single use of STR in 1946 for treating Mycobacterium tuberculosis infections indicated that resistance to this drug could be acquired very rapidly (31). At present, STR-resistant isolates have been identified in many other important pathogens, such as Shigella flexneri, Vibrio cholerae, Pseudomonas aeruginosa, and even in commensal E. coli sampled from healthy individuals (32–35). A high resistance incidence to this drug is frequently due to point mutations in rpsL gene, with the most common mutations occurring at the codons K43 and K88 (36) that were examined here. These mutations were shown here to be beneficial in the intracellular environment of Mϕs in E. coli. One possible explanation for the fitness advantage of STR-resistant mutations could be the ∼7-fold improvement in the accuracy of ribosomes in rpsL mutants (37). It was shown that STR resistance mutations in rpsL gene often lead to hyperaccurate, but slower ribosomes (38). Indeed, all STR resistance mutations that were tested in our study are responsible for the increased fidelity of ribosomes (39). Although fast ribosomes are required in actively dividing cells, hyperaccurate ribosomes are advantageous in nondividing cells during starvation because they exhibit attenuated protein oxidation during growth arrest (40), and oxidized proteins are known to be more susceptible to proteolytic degradation (41). This should be extremely relevant upon entry to the Mϕs, where E. coli not only undergoes growth arrest and nutrient starvation but also has to deal with ROS generated by the Mϕs (30). Consistent with this hypothesis, we found that STR-resistant mutants have reduced fitness costs when nutrients are deprived and survive better than susceptible strains under oxidative stress in the stationary phase (Fig. 2). Certainly, the finding that most commonly identified mutations, conferring resistance to STR, enhanced the survival capacity of E. coli inside the Mϕs suggests that an advantage could exist in other bacterial species, such as M. tuberculosis and other pathogenic bacteria.
Many bacterial pathogens (42–44) acquired resistance to RIF in the last decade. It is known that in 96% of RIF-resistant clinical isolates associated with tuberculosis, resistance is due to mutations in the rpoB gene, with the most common mutations at codons 531 and 526 in distinct geographical locations (45, 46). In the present study, in addition to prevalent mutations in codons 531 and 526 (S531F, H526Y, and H526D), other mutations in codons 512, 529, and 572 (S512F, R529H, and I572F) were also included. The fitness effects on survival of RIF-resistant mutants varied in our study. Interestingly, different base substitutions leading to different amino acids even at the same codon position (see Fig. 1C, H526D and H526Y) gave differential outcomes for E. coli survival inside Mϕs. The mutation at the codon 526 has been shown to be responsible for oxidative stress sensitivity in E. coli and Staphylococcus aureus. However, the molecular mechanism for this remains unknown (47). Several reports have suggested that single point mutations in the rpoB gene encoding the β subunit of the RNA polymerase can have an effect on RNA polymerase interaction with several promoters and transcriptional regulators, leading to different phenotypes (48–50). For example, in Bacillus subtilis, the RNA polymerase complex interacts with every promoter in bacterial genome, so the mutations in RNA polymerase lead to global changes in gene transcription and, hence, affect several physiological processes, such as growth and metabolism, chemotaxis, competence, spore resistance, and many others (48). Since RIF mutations have been found to affect physiological processes to different extents, it may not be surprising that we found a great variation in their fitness effects of RIF-resistant mutants inside Mϕs.
The emergence of NAL-resistant isolates during the treatment of Shigella, Campylobacter, or Salmonella infections has been of great concern (51–53). Single point mutations in the quinolone resistance-determining region of the DNA subunit gene gyrA at codons 83 (42% frequency) and 87 (35% frequency) have been attributed to the high levels of resistance to this antibiotic (54). Although the fitness costs of these mutations appear to be low in laboratory medium (9), it is not known how resistance to this drug may affect the survival and replication of these bacteria in the context of infection. In E. coli we found no fitness costs (for the D87Y mutation) or even slightly enhanced fitness (for the S83L mutation) of NAL-resistant clones for survival inside Mϕs, a finding compatible with previous reports showing that NAL resistance is usually associated with very small fitness costs (16).
It was previously demonstrated that fitness effects for the reproduction of antibiotic-resistant bacteria generally increase under stressful conditions (14, 55). The effects on the reproduction of more than half (58%) of the antibiotic-resistant mutants were either neutral or slightly advantageous in the presence of the Mϕs; however, this was mainly attributed to growth in the RPMI cell culture medium that we used for the maintenance of eukaryotic cells. Still, this is altogether relevant, because RPMI cell culture medium is supposed to mimic abiotic conditions in the human host. Moreover, fitness effects for reproduction differed in 33% of the antibiotic-resistant cases due to the presence of the Mϕs (compare the black and white bars in Fig. 1B, D, and F). This is, however, not surprising, given that Mϕs not only inflict several different stresses on bacteria but can also modify the composition of the extracellular medium. This is consistent with earlier findings suggesting that the fitness costs of antibiotic-resistant mutants may vary in different environmental conditions (11, 14).
These findings have several medically relevant implications. First, this work shows that the presence of Mϕs can have drastic consequences for the biological fitness of antibiotic-resistant E. coli. This conclusion points toward measuring fitness costs in such environments in other bacterial species as well as studying mutational targets of widely used antibiotics in clinics. Second, we identify single point mutations that are advantageous for bacterial survival in Mϕs because of the environmental stresses imposed by Mϕs, such as exposure to H2O2. Our main finding is that the stressful intracellular environment of Mϕs can select for antibiotic resistance has important consequences for predictions of the spread of drug resistance.
Supplementary Material
ACKNOWLEDGMENTS
The research leading to these results received funding from the European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement 260421-ECOADAPT. I.G. acknowledges the salary support of LAO/ITQB and FCT.
Footnotes
Published ahead of print 22 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01632-12.
REFERENCES
- 1. Denamur E, Picard B, Tenaillon O. 2010. Population genetics of pathogenic Escherichia coli, p 269–286 In Bacterial population genetics in infectious disease. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 2. Rolhion N, Darfeuille-Michaud A. 2007. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm. Bowel Dis. 13:1277–1283 [DOI] [PubMed] [Google Scholar]
- 3. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 4. Bailey JK, Pinyon JL, Anantham S, Hall RM. 2010. Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J. Med. Microbiol. 59:1331–1339 [DOI] [PubMed] [Google Scholar]
- 5. Levert M, Zamfir O, Clermont O, Bouvet O, Lespinats S, Hipeaux MC, Branger C, Picard B, Saint-Ruf C, Norel F, Balliau T, Zivy M, Le Nagard H, Cruveiller S, Chane-Woon-Ming B, Nilsson S, Gudelj I, Phan K, Ferenci T, Tenaillon O, Denamur E. 2010. Molecular and evolutionary bases of within-patient genotypic and phenotypic diversity in Escherichia coli extraintestinal infections. PLoS Pathog. 6:e1001125 doi:10.1371/journal.ppat.1001125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8:260–271 [DOI] [PubMed] [Google Scholar]
- 7. Maclean RC, Hall AR, Perron GG, Buckling A. 2010. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat. Rev. Genet. 11:405–414 [DOI] [PubMed] [Google Scholar]
- 8. Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJ. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944–1946 [DOI] [PubMed] [Google Scholar]
- 9. Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I. 2009. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 5:e1000578 doi:10.1371/journal.pgen.1000578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bataillon T, Zhang T, Kassen R. 2011. Cost of adaptation and fitness effects of beneficial mutations in Pseudomonas fluorescens. Genetics 189:939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hall AR, Iles JC, Maclean RC. 2011. The fitness cost of rifampicin resistance in Pseudomonas aeruginosa depends on demand for RNA polymerase. Genetics 187:817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macvanin M, Bjorkman J, Eriksson S, Rhen M, Andersson DI, Hughes D. 2003. Fusidic acid-resistant mutants of Salmonella enterica serovar Typhimurium with low fitness in vivo are defective in RpoS induction. Antimicrob. Agents Chemother. 47:3743–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paulander W, Maisnier-Patin S, Andersson DI. 2009. The fitness cost of streptomycin resistance depends on rpsL mutation, carbon source, and RpoS (σS). Genetics 183:539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trindade S, Sousa A, Gordo I. 2012. Antibiotic resistance and stress in the light of Fisher's model. Evolution doi:10.1111/j.1558-5646.2012.01722.x [DOI] [PubMed] [Google Scholar]
- 15. Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. 2011. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145:39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bjorkman J, Hughes D, Andersson DI. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 95:3949–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bjorkholm B, Sjolund M, Falk PG, Berg OG, Engstrand L, Andersson DI. 2001. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 98:14607–14612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Enne VI, Delsol AA, Roe JM, Bennett PM. 2004. Rifampicin resistance and its fitness cost in Enterococcus faecium. J. Antimicrob. Chemother. 53:203–207 [DOI] [PubMed] [Google Scholar]
- 19. Giraud E, Cloeckaert A, Baucheron S, Mouline C, Chaslus-Dancla E. 2003. Fitness cost of fluoroquinolone resistance in Salmonella enterica serovar Typhimurium. J. Med. Microbiol. 52:697–703 [DOI] [PubMed] [Google Scholar]
- 20. Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, Zhang Q. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. U. S. A. 102:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hegreness M, Shoresh N, Hartl D, Kishony R. 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311:1615–1617 [DOI] [PubMed] [Google Scholar]
- 22. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. Chapter 1:Unit 117 [DOI] [PubMed] [Google Scholar]
- 24. Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457–469 [DOI] [PubMed] [Google Scholar]
- 25. Maree AF, Keulen W, Boucher CA, De Boer RJ. 2000. Estimating relative fitness in viral competition experiments. J. Virol. 74:11067–11072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hamrick TS, Havell EA, Horton JR, Orndorff PE. 2000. Host and bacterial factors involved in the innate ability of mouse macrophages to eliminate internalized unopsonized Escherichia coli. Infect. Immun. 68:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lim AC, Mak KCN, Ng NUT, Ng TKK. 2007. Multiple modes of protection against hydrogen peroxide-induced oxidative damage in stationary and exponential phase Escherichia coli by DNA-binding protein (Dps). J. Exp. Microbiol. Immunol. 11:86–92 [Google Scholar]
- 28. Ferenci T. 2005. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 57:1–8 [DOI] [PubMed] [Google Scholar]
- 29. De Paepe M, Gaboriau-Routhiau V, Rainteau D, Rakotobe S, Taddei F, Cerf-Bensussan N. 2011. Trade-off between bile resistance and nutritional competence drives Escherichia coli diversification in the mouse gut. PLoS Genet. 7:e1002107 doi:10.1371/journal.pgen.1002107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schlosser-Silverman E, Elgrably-Weiss M, Rosenshine I, Kohen R, Altuvia S. 2000. Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J. Bacteriol. 182:5225–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crofton J, Mitchison DA. 1948. Streptomycin resistance in pulmonary tuberculosis. BMJ 2:1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barreto A, Guimaraes B, Radhouani H, Araujo C, Goncalves A, Gaspar E, Rodrigues J, Igrejas G, Poeta P. 2009. Detection of antibiotic resistant Escherichia coli and Enterococcus spp. in stool of healthy growing children in Portugal. J. Basic Microbiol. 49:503–512 [DOI] [PubMed] [Google Scholar]
- 33. Hong S, Choi YH, Choo YA, Choi Y, Choi SY, Kim DW, Lee BK, Park MS. 2010. Genetic characterization of atypical Shigella flexneri isolated in Korea. J. Microbiol. Biotechnol. 20:1457–1462 [DOI] [PubMed] [Google Scholar]
- 34. Rahmani F, Fooladi AA, Marashi SM, Nourani MR. 2012. Drug resistance in Vibrio cholerae strains isolated from clinical specimens. Acta Microbiol. Immunol. Hung. 59:77–84 [DOI] [PubMed] [Google Scholar]
- 35. Tseng JT, Bryan LE, Van Den Elzen HM. 1972. Mechanisms and spectrum of streptomycin resistance in a natural population of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2:136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sreevatsan S, Pan X, Stockbauer KE, Williams DL, Kreiswirth BN, Musser JM. 1996. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob. Agents Chemother. 40:1024–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zaher HS, Green R. 2010. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol. Cell 39:110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bilgin N, Claesens F, Pahverk H, Ehrenberg M. 1992. Kinetic properties of Escherichia coli ribosomes with altered forms of S12. J. Mol. Biol. 224:1011–1027 [DOI] [PubMed] [Google Scholar]
- 39. Okamoto-Hosoya Y, Hosaka T, Ochi K. 2003. An aberrant protein synthesis activity is linked with antibiotic overproduction in rpsL mutants of Streptomyces coelicolor A3(2). Microbiology 149:3299–3309 [DOI] [PubMed] [Google Scholar]
- 40. Ballesteros M, Fredriksson A, Henriksson J, Nystrom T. 2001. Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 20:5280–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3:3–8 [PubMed] [Google Scholar]
- 42. Liu G, Xu X, He L, Ding Z, Gu Y, Zhang J, Zhou L. 2011. Primary antibiotic resistance of Helicobacter pylori isolated from Beijing children. Helicobacter 16:356–362 [DOI] [PubMed] [Google Scholar]
- 43. Sekiguchi J, Miyoshi-Akiyama T, Augustynowicz-Kopec E, Zwolska Z, Kirikae F, Toyota E, Kobayashi I, Morita K, Kudo K, Kato S, Kuratsuji T, Mori T, Kirikae T. 2007. Detection of multidrug resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 45:179–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yesilyurt M, Kilic S, Celebi B, Celik M, Gul S, Erdogan F, Ozel G. 2011. Antimicrobial susceptibilities of Francisella tularensis subsp. holarctica strains isolated from humans in the Central Anatolia region of Turkey. J. Antimicrob. Chemother. 66:2588–2592 [DOI] [PubMed] [Google Scholar]
- 45. Cavusoglu C, Turhan A, Akinci P, Soyler I. 2006. Evaluation of the Genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 44:2338–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tan Y, Hu Z, Zhao Y, Cai X, Luo C, Zou C, Liu X. 2012. The beginning of the rpoB gene in addition to the rifampin resistance determination region might be needed for identifying rifampin/rifabutin cross-resistance in multidrug-resistant Mycobacterium tuberculosis isolates from Southern China. J. Clin. Microbiol. 50:81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kawamura N, Kurokawa K, Ito T, Hamamoto H, Koyama H, Kaito C, Sekimizu K. 2005. Participation of Rho-dependent transcription termination in oxidative stress sensitivity caused by an rpoB mutation. Genes Cells 10:477–487 [DOI] [PubMed] [Google Scholar]
- 48. Maughan H, Galeano B, Nicholson WL. 2004. Novel rpoB mutations conferring rifampin resistance on Bacillus subtilis: global effects on growth, competence, sporulation, and germination. J. Bacteriol. 186:2481–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perkins AE, Nicholson WL. 2008. Uncovering new metabolic capabilities of Bacillus subtilis using phenotype profiling of rifampin-resistant rpoB mutants. J. Bacteriol. 190:807–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou YN, Jin DJ. 1998. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:2908–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghosh AR, Thanasekaran K, Roy S. 2011. Nalidixic acid-resistant Shigella sonnei infection among dysenteric children in Port Blair. J. Microbiol. Antimicrob. 3:87–93 [Google Scholar]
- 52. Kumar Y, Sharma A, Mani KR. 2009. High level of resistance to nalidixic acid in Salmonella enterica serovar Typhi in Central India. J. Infect. Dev. Countries 3:467–469 [DOI] [PubMed] [Google Scholar]
- 53. Wu TL, Su LH, Chia JH, Kao TM, Chiu CH, Kuo AJ, Sun CF. 2002. Molecular epidemiology of nalidixic acid-resistant campylobacter isolates from humans and poultry by pulsed-field gel electrophoresis and flagellin gene analysis. Epidemiol. Infect. 129:227–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hopkins KL, Arnold C, Threlfall EJ. 2007. Rapid detection of gyrA and parC mutations in quinolone-resistant Salmonella enterica using pyrosequencing technology. J. Microbiol. Methods 68:163–171 [DOI] [PubMed] [Google Scholar]
- 55. Petersen A, Aarestrup FM, Olsen JE. 2009. The in vitro fitness cost of antimicrobial resistance in Escherichia coli varies with the growth conditions. FEMS Microbiol. Lett. 299:53–59 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.