Abstract
Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae have emerged as major nosocomial pathogens. blaKPC, commonly located on Tn4401, is found in Gram-negative bacterial strains, with the two most common variants, blaKPC-2 and blaKPC-3, identified in plasmids with diverse genetic backgrounds. In this study, we examined blaKPC-4- and blaKPC-5-bearing plasmids recovered from two K. pneumoniae strains, which were isolated from a single New Jersey hospital in 2005 and 2006, respectively. IncN plasmid pBK31551 is 84 kb in length and harbors blaKPC-4, blaTEM-1, qnrB2, aac(3)-Ib, aph(3′)-I, qacF, qacEΔ1, sul1, and dfrA14, which confer resistance to β-lactams, quinolones, aminoglycosides, quaternary ammonium compounds, and co-trimoxazole. The conserved regions within pBK31551 are similar to those of other IncN plasmids. Surprisingly, analysis of the Tn4401 sequence revealed a large IS110- and Tn6901-carrying element (8.3 kb) inserted into the istA gene, encoding glyoxalase/bleomycin resistance, alcohol dehydrogenase, and S-formylglutathione hydrolase. Plasmid pBK31567 is 47 kb in length and harbors blaKPC-5, dfrA5, qacEΔ1, and sul1. pBK31567 belongs to a novel IncX subgroup (IncX5) and possesses a highly syntenic plasmid backbone like other IncX plasmids; however, sequence similarity at the nucleotide level is divergent. The blaKPC-5 gene is carried on a Tn4401 element and differs from the genetic environment of blaKPC-5 described in Pseudomonas aeruginosa strain P28 from Puerto Rico. This study underscores the genetic diversity of multidrug-resistant plasmids involved in the spread of blaKPC genes and highlights the mobility and plasticity of Tn4401. Comparative genomic analysis provides new insights into the evolution and dissemination of KPC plasmids belonging to different incompatibility groups.
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) have emerged as a major cause of nosocomial infections worldwide. The therapeutic challenges associated with these highly resistant strains correlate with higher morbidity and mortality, increased length of hospitalization, and an overall increase in health care costs (1). In the past decade, the spread of Klebsiella pneumoniae carbapenemase (KPC), a class A serine β-lactamase, has led to a rapid rise in prevalence of CRE infections in the United States and other global regions (2). Currently, 12 KPC variants (KPC-2 to KPC-13) have been identified since the initial report, with KPC-2 and KPC-3 identified as the most frequent types and K. pneumoniae as the most common host species (2, 3). Genotypic studies of emerging KPC-bearing K. pneumoniae strains from international sources indicate that the predominant strains are typically multilocus sequence type 258 (ST258), which is suggestive of pandemic spread by a single clone (2, 4–7).
KPC is encoded by the blaKPC gene, which is carried by Tn4401 or Tn4401-like genetic elements, and has been reported in a variety of transferable plasmids (8–14). Tn4401 is approximately 10 kb in size, is delimited by two 39-bp imperfect inverted repeats, and harbors insertion sequences ISKpn6 and ISKpn7 in addition to transposase and resolvase genes (15). Tn4401 and blaKPC genes have been reported in plasmids of various sizes, which differ in their ability to transfer within and between species, harbor different antibiotic resistance elements, and possess different incompatibility (Inc) groups, including IncFII, IncL/M, IncN, IncR, and ColE1 (4, 6, 10, 16).
Whole-plasmid sequencing and comparative genomic analyses have identified common functional modules and unique acquired genes that provide important molecular information for developing epidemiological tools to identify and track the spread of resistance plasmids within and among Enterobacteriaceae species (17–20). Various blaKPC-2- and blaKPC-3-harboring plasmids belonging to different Inc groups have been successfully sequenced, including IncN plasmids p9 and p12 (10), IncFII plasmids pKPHS2 (21), pKP048 (19), pKpQIL (20), pKpQIL-IT (18), and pSLMT (HQ589350), and ColE1 plasmid p15S (10). However, the complete sequence analysis of carbapenem resistance plasmids belonging to other Inc groups, or harboring other blaKPC variants, has not been described. Here we report the complete sequences of two plasmid genomes harboring blaKPC-4 and blaKPC-5, recovered from two K. pneumoniae isolates cultured at the same New Jersey hospital. Our analysis shows the genetic diversity of multidrug-resistant plasmids involved in the spread of the blaKPC genes and highlights the mobility and plasticity of Tn4401.
(This study was partially presented at the 52nd Annual Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC], 2012.)
MATERIALS AND METHODS
Retrospective analysis of carbapenem-resistant K. pneumoniae.
A total of 27 carbapenem-resistant K. pneumoniae strains isolated from 2005 to 2010 at a single New Jersey hospital were genetically characterized. Detection of blaKPC genes and identification of the epidemic K. pneumoniae ST258 clone were performed using multiplex real-time PCR methods described previously (22, 23), and full-length blaKPC genes were characterized by conventional PCR and DNA sequencing (24). Two KPC-4-harboring strains were detected: BK31551 (CK7) and BK31572 (CK180) were isolated from blood culture samples from two patients with bacteremia in September 2005 and November 2006, respectively. KPC-5-bearing strain BK31567 (CK102) was isolated from a blood culture sample from a patient with urospesis in April 2006.
Species were identified using the Vitek 2 system (bioMérieux), and MICs were determined by broth microdilution in cation-adjusted Mueller-Hinton broth (MHB) according to Clinical and Laboratory Standards Institute methods (25), using Sensititre GNX2F trays (Thermo Fisher Scientific, Waltham, MA). Carbapenem resistance was defined by MICs of higher than the 2012 CLSI breakpoints for one or more of the following carbapenem agents: imipenem, ≥4 μg/ml; meropenem, ≥4 μg/ml; ertapenem, ≥2 μg/ml; and doripenem, ≥4 μg/ml (25).
Characterization of blaKPC-4- and blaKPC-5-harboring plasmids and strains.
Plasmid DNA was extracted using a Qiagen plasmid Maxikit (Qiagen, Valencia, CA), followed by electroporation into Escherichia coli DH10B (Invitrogen) using a Gene Pulser II instrument (Bio-Rad Laboratories). E. coli DH10B transformants were selected on Luria-Bertani (LB) agar plates containing 100 μg/ml ampicillin or 1 μg/ml imipenem and then screened by multiplex real-time PCR for the presence of blaKPC genes (23). Plasmid size was estimated by S1 nuclease digestion of plasmid DNA, followed by pulsed-field gel electrophoresis (PFGE) using a Bio-Rad CHEF-DR III variable-angle system (26). Transformants with a single plasmid were then selected and subjected to susceptibility testing as described above. Plasmids isolated from E. coli DH10B transformants were digested with restriction endonuclease EcoRV (New England BioLabs, Boston, MA), and their restriction patterns were compared. Transferability of blaKPC-4- and blaKPC-5-bearing plasmids was examined by conjugation experiments using three clinical isolates (BK31551, BK31572, and BK31567) as donors and E. coli J53Azr as the recipient as described previously (27). E. coli J53 transconjugants with KPC-encoding plasmids were selected on LB plates containing 50 μg/ml sodium azide and 100 μg/ml ampicillin. The presence of the blaKPC-4 or blaKPC-5 gene in E. coli J53 transconjugants was confirmed by the aforementioned multiplex real-time PCR (23).
Plasmid incompatibility groups were determined using the multiplex PCR method described by Carattoli et al. (28). β-Lactamase genes, including blaCTX-M, blaSHV, blaTEM, blaGES, blaNDM, blaVIM, blaIMP, blaOXA-48, blaACT-1, blaACC, blaBIL-1, blaCMY, blaDHA, blaFOX, blaLAT, blaMIR-1, and blaMOX, were investigated by PCR using methods described elsewhere (29, 30). Multilocus sequence typing (MLST) of the parental strains (31) and plasmid MLST for IncN group plasmids (32) were performed using previously described methods. OmpK35 and OmpK36 porins were analyzed by SDS-PAGE, and their genes were amplified by PCR followed by DNA sequencing (33).
Plasmid sequencing and bioinformatics.
Plasmid DNA from E. coli DH10B transformants of BK31551 (KPC-4) and BK31567 (KPC-5) were extracted as described above using a Qiagen plasmid Maxikit (Qiagen, Valencia, CA). The plasmid DNA was fragmented, and genomic libraries were prepared and sequenced using a Roche 454 GS-FLX system. Sequencing reads were assembled into consensus de novo assembly contigs using the Roche genome sequencer FLX software GSA assembler, version 2.5.3. Gaps between contigs were closed by PCR with standard Sanger sequencing.
Open reading frames (ORFs) were predicted and annotated using the RAST (rast.nmpdr.org) server (34), followed by manual comparative curation and sequence similarity searches directed against the NCBI (www.ncbi.nlm.nih.gov/BLAST) and IS Finder (www-is.biotoul.fr) databases. MEGA 5.01 was used for sequences comparison and alignment (35). Mauve 2.3.1 was used to perform comparative genome alignment for different plasmids (36).
Nucleotide sequence accession numbers.
The complete nucleotide sequences of pBK31551 and pBK31567 have been deposited in GenBank under accession numbers JX193301 and JX193302, respectively.
RESULTS
Characteristics of clinical isolates.
Genotyping of the 27 carbapenem-resistant isolates indicated that they all possessed the blaKPC genes, including blaKPC-3 (n = 22; 81.5%) and blaKPC-2 (n = 2; 7.4%), while three strains were unexpectedly identified with blaKPC-4 (n = 2) and blaKPC-5 (n = 1). With the exception of one isolate, all blaKPC-2- and blaKPC-3-harboring strains belonged to the epidemic K. pneumoniae ST258 clone (22); however, neither the blaKPC-4- nor the blaKPC-5-carrying strains were ST258. Multilocus sequence typing (31) indicated that the three non-ST258 isolates belonged to uncommon sequence types (STs): ST834 (2-1-2-1-7-1-25), strain BK31551; ST964 (41-1-1-1-7-4-87), strain BK31572; and ST429 (2-1-2-1-9-1-116), strain BK31567. The differing PFGE profiles of these three strains also indicated they were not genetically related (data not shown). S1-PFGE on different E. coli DH10B transformants revealed that each strain carried a single plasmid ranging in size from 49 to 84 kb (49 kb for pBK31567, 75 kb for pBK31572, and 84 kb for pBK31551). Conjugation experiments showed that three blaKPC-4- and blaKPC-5-harboring plasmids could be successfully transferred to E. coli J53 recipients. In addition, SDS-PAGE and DNA sequence analysis showed no evidence of porin loss of OmpK35 and OmpK36 in three parental strains (data not shown).
A list of antimicrobial agents and their MICs against KPC-4- or -5-producing clinical isolates (and their transformants) is shown in Table 1. The parental strains were resistant to aztreonam, cefotaxime, ceftazidime, ticarcillin/clavulanate, and trimethoprim-sulfamethoxazole, to imipenem (BK31572), and to ertapenem (BK31572 and BK31567) and exhibited reduced susceptibility to piperacillin-tazobactam. In addition, BK31551 and BK31567 displayed intermediate resistance to imipenem (MIC 2 μg/ml), while BK31572 displayed intermediate resistance to doripenem (MIC 2 μg/ml) (25). The E. coli DH10B transformants displayed antimicrobial susceptibility profiles similar to those of their parental strains but were not as resistant to imipenem, ertapenem, and doripenem (Table 1). Notably, although some of the MIC values in Table 1 fall within the susceptibility range for carbapenems (≤1 μg/ml for imipenem, meropenem, and doripenem and ≤0.5 μg/ml for ertapenem), they are nevertheless higher than those reported previously for wild-type K. pneumoniae strains (without carbapenemase or porin loss) (37).
Table 1.
Characteristics of KPC-4- and KPC-5-producing K. pneumoniae strains and their E. coli DH10B transformants
| Isolatea | β-Lactamase(s) | MIC (μg/ml)b |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MER | ERT | DOR | CAZ | CTX | CEF | ATM | TIC-CLAV | PIP-TAZ | AMI | TOB | DOX | CIP | LEV | SXT | TGC | COL | PLB | ||
| BK31551 | TEM-1, SHV-11, KPC-4 | 2 | 0.5 | 1 | 1 | >16 | >32 | 4 | >16 | 128/2 | 64/4 | ≤4 | ≤1 | >16 | 1 | 2 | >4/76 | 1 | 0.5 | 1 |
| T-BK31551 | TEM-1, KPC-4 | 1 | 0.5 | 0.5 | 0.5 | >16 | 16 | 8 | >16 | >128/2 | >64/4 | ≤4 | 4 | ≤2 | ≤0.25 | ≤1 | >4/76 | ≤0.25 | ≤0.25 | 0.5 |
| BK31572 | TEM-1, SHV-11, KPC-4 | 4 | 1 | 4 | 2 | >16 | >32 | 4 | >16 | >128/2 | >64/4 | ≤4 | ≤1 | ≤2 | ≤0.25 | ≤1 | >4/76 | ≤0.25 | 0.5 | 1 |
| T-BK31572 | TEM-1, KPC-4 | 2 | 1 | 1 | 0.5 | >16 | >32 | 8 | >16 | >128/2 | 64/4 | ≤4 | 2 | 2 | ≤0.25 | ≤1 | >4/76 | ≤0.25 | ≤0.25 | 0.5 |
| BK31567 | SHV-1, KPC-5 | 2 | 1 | 4 | 0.5 | >16 | 32 | ≤2 | >16 | >128/2 | >64/4 | ≤4 | ≤1 | 4 | >2 | ≤1 | >4/76 | 1 | 0.5 | 1 |
| T-BK31567 | KPC-5 | 1 | 0.5 | 0.5 | 0.5 | >16 | 4 | ≤2 | >16 | >128/2 | 16/4 | ≤4 | ≤1 | ≤2 | ≤0.25 | ≤1 | >4/76 | ≤0.25 | ≤0.25 | 0.5 |
T-, E. coli DH10B transformant.
MICs were determined using broth microdilution; resistance is indicated in boldface. IMP, imipenem; MER, meropenem; ERT, ertapenem; DOR, doripenem; CAZ, ceftazidime; CTX, cefotaxime; CEF, cefepime; ATM, aztreonam; TIC-CLAV, ticarcillin-clavulanate; PIP-TAZ, piperacillin-tazobactam; AMI, amikacin; TOB, tobramycin; DOX, doxycycline; CIP, ciprofloxacin; LEV, levofloxacin; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; COL, colistin; PLB, polymyxin B. The 2012 CLSI breakpoints were used to interpret the MIC results for carbapenem-susceptible (S), -intermediate (I), and -resistant (R) isolates, as follows (S/I/R): imipenem, ≤1/2/≥4 μg/ml; meropenem, ≤1/2/≥4 μg/ml; ertapenem, ≤0.5/1/≥2 μg/ml; and doripenem ≤1/2/≥4 μg/ml (25).
Structure of blaKPC-4-harboring plasmid pBK31551.
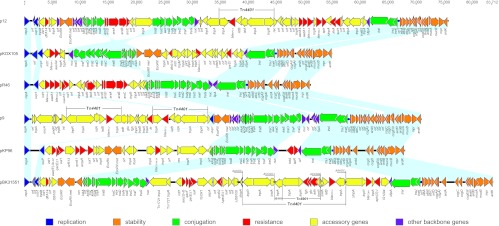
Plasmid pBK31551 is a circular molecule of 83,712 bp belonging to incompatibility group N, with an average GC content of 53.4%, and harboring 80 predicted ORFs (Fig. 1). The plasmid comprises a 37,343-bp core region and two discontinuous acquired regions (42,924 and 3,445 bp). The core region defines plasmid “housekeeping genes,” including a replication module, two transfer (tra) systems, a stability operon, and an antirestriction system (Fig. 1). The two discontinuous acquired regions are composed of numerous resistance genes, insertion sequences, and transposons (Fig. 1).
Fig 1.
Comparative analysis of IncN plasmids pBK31551 (JX193301), pKP96 (EU195449), p9 (FJ223607), pR46 (AY046276), pKOX105 (HM126012), and p12 (FJ223605). Light blue shading denotes shared regions of homology. It is noted that the region between kikA and nuc is inverted in pKOX105 relative to the other plasmids and is split into two regions in p12 due to the insertion of the ars operon. Open reading frames (ORFs) are portrayed by arrows and colored based on predicted gene function. Resistance genes are indicated in red arrows and include aminoglycoside resistance genes [aph (3′)-I, aac(3)-Ib, aac(6′)-Ib-cr, aph, aadA1, and aacA4], β-lactamase genes (blaKPC-2, blaKPC-3, blaKPC-4, blaKPC-5, blaVIM-1, blaCTX-M-24, blaOXA-2, blaOXA-9, blaTEM-1, blaSHV-12, and ampR), quinolone resistance genes (qnrA1, qnrB2, and qnrS1), arsenic resistance genes (arsA, arsB, arsC, arsD, and arsR), tetracycline resistance genes (tetA and tetR), and quaternary compound, trimethoprim, and sulfonamide resistance genes (sul1, qacF, qacEΔ1, and dfrA14), as well as glyoxalase/bleomycin resistance gene glo, alcohol dehydrogenase gene adh, and S-formylglutathione hydrolase gene fgh.
The core genes in pBK31551 and other IncN plasmids are clustered into different functional modules (Fig. 1). The replication region includes a 720-bp replication initiation protein gene (repA), flanked by direct repeat sequences and adjacent to a resolvase gene (uvp1 or resP) required for resolving cointegrates at the end of replication (38). pBK31551 also possesses two modules that are predicted to be involved in the conjugative transfer of plasmid DNA. The first module consists of 11 genes (traL, traM, traA, traB, traC, traD, traN, traE, traO, traF, and traG) flanked by a group of plasmid core genes involved in regulating transfer and replication (korA and korB) (39), excluding DNA entry (eex) (40), DNA degradation (nuc) (41), and killing of Klebsiella (kikA) (42, 43) (Fig. 1). The second module includes three genes (traI, traJ, and traK) that also function in replication and DNA transfer, while the fipA gene, located upstream of traI, functions in fertility inhibition of IncP plasmids (44).
Additional core genes common to IncN plasmids and found on pBK31551 contribute to plasmid stability. The restriction/antirestriction system encoded by the EcoRII and EcoRII met genes probably functions as a toxin/antitoxin system to lyse cells which have lost the plasmid (45). Genes located downstream of the traIJK modules for conjugative transfer were also predicted to play a role in plasmid stability. Specifically, the proteins encoded by the stbABC operon are believed to stabilize single-stranded DNA during conjugation (46), while ardAB and ardR encode proteins involved in regulating antirestriction functions (47, 48). pBK31551 also possesses ccg genes (cup-controlled genes), which encode products involved in the protection of plasmid DNA from type I restriction enzymes (48).
The pBK31551 plasmid also has two distinct regions harboring antibiotic resistance genes. A 3,445-bp region downstream of the uvp1 gene includes a class 1 integron with a 3′ conserved sequence (3′ CS) truncated by insertion of an IS6100 element. This integron contains the dfrA14 gene cassette, which confers trimethoprim resistance.
The primary cluster of resistance genes in pBK31551 is located in a 42,924-bp region between genes fipA and nuc and includes β-lactamase genes blaKPC-4 and blaTEM-1, aminoglycoside resistance genes aph(3′)-I and aac (3)-1b, quinolone resistance gene qnrB2, quaternary compound resistance genes qacF and qacEΔ1, sulfonamide resistance gene sul1, glyoxalase/bleomycin resistance gene glo, alcohol dehydrogenase gene adh, and S-formylglutathione hydrolase gene fgh.
Within this 42,924-bp region, an 18,282-bp fragment contains the Tn4401 element harboring the blaKPC-4 gene, which encodes the carbapenemase. The insertion of the element, which disrupted the Tn3 transposase gene, generated a 5-bp (AAAGC) target sequence duplication (TSD) at each end of Tn4401 (Fig. 1). Surprisingly, sequencing of the Tn4401 element identified a unique 8,271-bp insertion which disrupted the istA gene and generated a 5-bp TSD (ACCGG). This insertion contains two discrete elements: a 1,395-bp insertion sequence that belongs to the IS110 family and is 91% similar to an insertion sequence in Yersinia pestis plasmid pMT (www.ncbi.nlm.nih.gov/GenBank) and a 6,876-bp region that is 99.7% similar to the Tn3 family transposon Tn6901 (49). Tn6901 carries 6 genes, including a transposase gene, a resolvase gene, an alcohol dehydrogenase gene (adh), a glyoxalase/bleomycin resistance gene (glo), an S-formylglutathione hydrolase gene (fgh), and a regulator protein gene (frmR). The blaKPC-4 gene is located downstream of Tn6901 and displays 100% identity to previously reported blaKPC-4 sequences from Enterobacter cancerogenus strain E624 (FJ473382) and K. pneumoniae strain KpAM1(EU447304).
There are two additional genetic elements located upstream of Tn4401 (Fig. 1). Transposon Tn1721 is flanked by a class I integron, consisting of aac(3)-Ib, qacF, qacEΔ1, and sul1. The next element consists of a truncated class I integron containing sapA, qnrB2, ΔqacEΔ1, sul1, and two unknown ORFs, as well as an ISCR1 element inserted within the 5′ conserved sequence (5′ CS). Acquired genes located downstream of Tn4401 include IS1294, aph(3′)-I, blaTEM-1, and the transposase (tnpA) and resolvase (tnp) genes of Tn3.
Comparison of pBK31551 with IncN plasmids.
pBK31551 was compared to IncN prototype plasmid pR46 (AY046276) as well as several other bla gene-carrying IncN plasmids, including pKP96 (EU195449), p9 (FJ223607), pKOX105 (HM126012), and p12 (FJ223605) (Fig. 1). All plasmids shared similarities in core genes responsible for plasmid maintenance, including replication, transfer, and stability systems (Fig. 1). Plasmids p12 and pKOX105 lack the antirestriction genes ccgC, ccgD, and ardA, while p12 has a nonfunctional transfer region due to insertion of an arsenic resistance operon (ars) between traB and traA. In pKOX105, moreover, the region containing the traAB operon (from nuc to kikA) is inverted relative to that in other IncN plasmids.
The major differences among these plasmids are related to the number of acquired genes (Fig. 1). Interestingly, these plasmids possess similar integration sites. For example, with the exception of plasmid p9, the IncN plasmids all share integration of a class I integron downstream of the uvp1 gene (Fig. 1). Similarly, the 42-kb antibiotic resistance region inserted between the fipA and nuc genes in pBK31551 closely resembles the acquired resistance genes in p12 (also located between fipA and nuc), pKOX105 (between EcoRII and fipA), and pKP96 (between nuc and traI). Thus, although pBK31551 and other Inc N plasmids carry different antibiotic resistance genes, they share similar integration sites for the acquisition of variable resistance determinants, which may represent entry sites or “hot spots” for integration of transposable elements within the IncN plasmid scaffold (17).
Characterization of pBK31572.
Given the unusual Tn6901 and IS110 insertions within Tn4401 in pBK31551, we investigated additional insertions within other KPC-harboring isolates, including another KPC-4-producing strain (BK31572) isolated in 2006. PCR assays targeting both insertion junctions as well as istA showed negative results for all blaKPC-2- or blaKPC-3-harboring strains (data not shown). Interestingly, pBK31572 possesses the same Tn6901 and IS110 insertions as pBK31551, as demonstrated by PCR (data not shown). Comparative restriction enzyme digests of pBK31551 and pBK31572 using EcoRV displayed similar profiles, suggesting genetic relatedness (data not shown). Plasmid MLST (32) results also showed that both pBK31551 and pBK31572 belong to IncN ST6 (2-4-2), the same genotype described previously for KPC-3-producing strain p12, isolated in 2005 (10). Additional PCR assays used to detect blaTEM-1 (29), aac(3)-Ib (50), qnrB2 (51), and aph(3′)-I (52) in pBK31551 showed that pBK31572 was negative for qnrB2 but positive for other three targets. Nevertheless, in addition to Tn4401, pBK31572 and pBK31551 appear to share similar backbone genes, with the observed size differences likely due to variable content within acquired genetic elements.
Structure of blaKPC-5-harboring plasmid pBK31567.
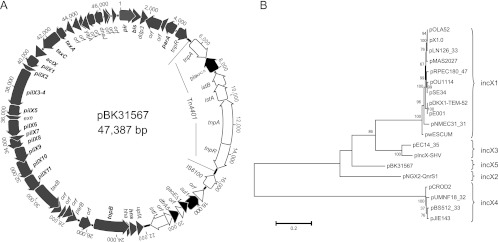
Plasmid pBK31567 is a circular molecule of 47,387 bp, with an average GC content of 49.0%, and harboring 54 predicted ORFs (Fig. 2A). Sequence analysis of pBK31567 classified it within plasmid incompatibility group X, and we have further typed it as a novel IncX subgroup (IncX5) (described below). The IncX group of plasmids are narrow-host-range plasmids common in Enterobacteriaceae, and they exist even in isolates from the preantibiotic era (53). They are known to encode type IV fimbriae enabling their own conjugative transfer, and they provide accessory functions to their host bacteria such as biofilm formation and resistance to antimicrobial agents (54). IncX plasmids possess highly syntenic plasmid backbones but are quite divergent with respect to nucleotide and amino acid similarity (54). Based on phylogenetic comparisons of sequenced IncX plasmids, the IncX plasmid group has been divided into at least four subgroups, IncX1 to IncX4 (54).
Fig 2.
(A) Structure of plasmid pBK31567. Open reading frames (ORFs) are portrayed by arrows, with backbone region ORFs in dark gray and acquired region ORFs in white. Core backbone genes in pBK31567 are shown in boldface. Resistance genes are depicted by black arrows, including β-lactamase genes (blaKPC-5) and quaternary compound, trimethoprim, and sulfonamide resistance genes (qacF, qacEΔ1, dfrA5, and sul1). (B) Phylogenetic maximum-likelihood tree of single nucleotide polymorphisms (SNPs) from backbone sequences of 19 IncX plasmids, including (GenBank accession numbers are shown in parentheses) pBK31567 (JX193302), p2ESCUM (CU928149), pBS512_33 (CP001059), pCROD2 (FN543504), pDKX1-TEM-52 (JQ269336), pE001 (JF776874), pEC14_35 (JN935899), pIncX-SHV (JN247852), pJIE143 (JN194214), pLN126_33 (HE578058), pMAS2027 (FJ666132), pNGX2_QnrS1(JQ269335), pNMEC31_31 (JN935897), pOLA52 (EU370913), pOU1114 (DQ115387), pRPEC180_47 (JN935898), pSE34 (EU219533), pUMNF18_32 (AGTD1000006), and pX1.0 (HM114226). All plasmids from a previous study (54), as well as pBK31567, have been included for phylogenetic analysis.
Previous studies indicated that the characteristic structure of the IncX plasmid group includes a set of core genes positioned in the following order: pir-bis-par-hns-topB-pilX-actX-taxCA (54, 55). These same core genes are found in the backbone region of pBK31567 that is involved in plasmid replication, conjugation and DNA transfer, and partitioning (Fig. 2A). A BLASTn comparison against the NCBI database revealed that the putative backbone region of pBK31567 is closely related to InX3 plasmids pEC14_35 (JN935899) (54) and pIncX-SHV (JN247852) (18), displaying approximately 80% nucleotide similarity.
The core region of pBK31567, which is common to other IncX plasmids, includes a 1.35-kb replicon fragment encoding the essential replication initiator (Pir) and the accessory protein (Bis), as well as the partition-determining proteins ParG and ParF, which are involved with plasmid segregation and stability. The other conserved modules involved in stabilizing horizontal transfer of plasmid DNA include the gene expression modulation (gem) region, which contains putative transcriptional regulator (hha) and DNA-binding protein (hns) genes, as well as the type IA topoisomerase gene (topB) (55). The putative tra region of pBK31567 is divided into two regions: mating pair formation (Mpf) genes and DNA transfer and replication (Dtr) genes. The Mpf region includes 10 pilx gene members (pilx1 to pilx11), while the Dtr complex consists of three relaxase genes (taxA, -B, and -C), which nick the plasmid DNA at the oriT site and initiate the formation of the transfer DNA strand (Fig. 2A).
Based on phylogenetic analysis of SNPs from conserved regions, Johnson et al. (54) divided 18 completely sequenced IncX plasmids into 4 subgroups (IncX1 to IncX4). taxC, one of the aforementioned Dtr genes, is conserved among different IncX plasmids, and comparative sequence analysis has been used to identify and group IncX plasmids into subgroups (IncX1 to IncX4) (28, 54). The taxC genes of plasmids from different incompatibility subgroups share between 37% and 92% nucleotide similarity, whereas those within a given incompatibility subgroup (e.g., IncX1 to IncX4) share between 92% and 99% nucleotide similarity (54). A comparison of the taxC gene in pBK31567 to those of the IncX plasmids depicted in Fig. 2B showed 82%, 78%, 73%, and 46% nucleotide similarity with the IncX3 (pIncX-SHV and pEC14-35), IncX1 (e.g., pEC14-35), IncX2 (pNGX2-QnrS1), and IncX4 (e.g., pJIE143) subgroups, respectively, effectively relegating pBK31567 to a novel IncX subgroup. Phylogenetic analysis of single nucleotide polymorphisms (SNPs) from the conserved regions of 19 plasmids was further used to explore the relationship between pBK31567 and other IncX plasmids (Fig. 2B). The results divided the IncX plasmids into five subgroups, with pBK31567 clustering separately from the aforementioned subgroups IncX1 to -4 (54), in agreement with the results of the taxC gene comparisons; pBK31567 is therefore best classified as a plasmid subgroup of IncX5.
Also common to IncX plasmids is a conserved integration region for acquired elements, located downstream of the serine resolvase gene, tnpR (nucleotides [nt] 4265 to 4912 in Fig. 2A). Several resistance genes have been observed directly downstream of the resolvase gene in variable IncX plasmids, including β-lactamase genes (blaTEM-52, blaTEM-1, blaSHV-11, and blaCTX-M-15), as well as genes encoding quinolone resistance (qnrS1 and oqxAB), aminoglycoside resistance (aphA1), and bleomycin resistance (blmS) (54). Similarly, a 16,727-bp acquired region, which includes a blaKPC-5-harboring Tn4401 element and a class I integron, has been found inserted downstream of the serine resolvase gene in pBK31567. The Tn4401 region of pBK31567 displayed 99.9% similarity with the sequences of previously described Tn4401 elements from p9 and p12 (10), with the only observed sequence variation associated with their respective blaKPC variants (blaKPC-5 versus blaKPC-2/-3). An intact class I integron, located downstream of the Tn4401 and IS6100 elements, harbors the trimethoprim resistance gene dfrA5, the quaternary compound resistance gene qacEΔ1, and the sulfonamide resistance gene sul1. To the best of our knowledge, this is the first description of a carbapenemase gene (blaKPC) and a class I integron carried on the same IncX plasmid.
DISCUSSION
Currently, KPC-2 and KPC-3 carbapenemases have been described in multiple Gram-negative species, including various Enterobacteriaceae, and several blaKPC-2- and blaKPC-3-harboring plasmids have been fully sequenced (2, 4, 6, 10, 16). In contrast, strains harboring blaKPC-4- and blaKPC-5-carrying plasmids are rarely reported, and these plasmids have not been completely analyzed. As of this writing, blaKPC-4 has been reported in three isolates: E. cancerogenus strain E624 from Scotland, isolated in 2003 (56), and K. pneumoniae strain KpAM1 (57) and Acinetobacter sp. strain M2AC9-31 (58), isolated in Puerto Rico in 2006 and 2009, respectively. Similarly, blaKPC-5 has been found only in Pseudomonas aeruginosa strain PS28 (PR280) and in isolates in Puerto Rico between October 2006 and March 2007 (14, 59).
In this study, we characterized complete sequences of blaKPC-4- and blaKPC-5-harboring plasmids pBK31551 and pBK31567, isolated from a single hospital in New Jersey, revealing a number of unusual findings. First, a large element carrying both IS110 and Tn6901 (8.3 kb) was found inserted within Tn4401 in pBK31551 (Fig. 1), and the same element was also identified in plasmid pBK31572. Both plasmids were also shown to have the same plasmid MLST profile, supporting their genetic relatedness, although pBK31572 is smaller and lacks certain antibiotic resistance genes present in pBK31551. Insertions and genetic alterations have been previously described in Tn4401 adjacent to the blaKPC genes (9, 13, 14), including a previous study in which we described a 5.3-kb deletion within Tn4401 encompassing all of ISKpn7 (istA and istB), 80% of tnpA, and 60% of blaKPC (33). The genetic diversity associated with Tn4401 underlines the notion that the region surrounding blaKPC undergoes significant recombination.
The insertion of Tn6901 and IS110 upstream of blaKPC raises the possibility that they may alter gene expression and the level of carbapenem resistance. In fact, both KPC-4-producing K. pneumoniae strains displayed low-level resistance to carbapenems, with MIC values of 1 to 4 μg/ml, 2 to 4 μg/ml, and 1 to 2 μg/ml for ertapenem, imipenem, and doripenem, respectively, and total susceptibility to meropenem (Table 1). In addition, the E. coli DH10B blaKPC-4 transformants were even more susceptible than the parental strains, providing further evidence that the host influences plasmid gene expression levels. In contrast, the KPC-4 resistance reported in E. cancerogenus strain E624 exhibited higher MIC levels against imipenem (>32 μg/ml), meropenem (>32 μg/ml), and ertapenem (>16 μg/ml) (14). Significantly, sequence analysis of the 363-bp region upstream of blaKPC-4 in pBK31551, including the putative promoter region (60), showed that these upstream sequences are identical to that found in the E. cancerogenus strain E624 plasmid, raising speculation that the difference in MICs could be related to the IS110 and Tn6901 insertions in Tn4401 or else to other factors such as a combination of porin loss/alteration and β-lactamases (extended-spectrum β-lactamases or AmpC enzymes) in E. cancerogenus strain E624.
The second unusual finding was the identification of a KPC-5-producing K. pneumoniae strain in which the blaKPC-5-harboring Tn4401 element was located on a plasmid belonging to a novel IncX subgroup (IncX5). IncX plasmids have been implicated in the acquisition and dissemination of transferrable drug resistance in different Enterobacteriaceae spp. (54) and have been shown to be transferrable from E. coli to P. aeruginosa (61). Although various resistance determinants have been described in IncX plasmids, carbapenemases have not been identified previously. It is therefore likely that pBK31567 originated from an IncX5 plasmid which acquired Tn4401 and the class I integron by horizontal transfer. Interestingly, the blaKPC-5 gene was initially described in P. aeruginosa strain PS28 and notably was not located on Tn4401 (14). Instead, the carbapenemase gene was located within a novel genetic environment, with Tn5563- and IS6100-related elements inserted upstream of blaKPC-5. The differences in the flanking regions of blaKPC-5 observed between PS28 and BK31567 indicate that blaKPC-5 is not associated with the unique genetic environment described in strain P. aeruginosa strain PS28. It is interesting that strain BK31567 was isolated in the same year (2006) as strain PS28 (14, 59); however, data on patient demographics are unknown for this isolate (PS28). Likewise, neither the complete plasmid sequence nor the Inc group of the PS28 plasmid is available, precluding direct sequence comparisons between these two plasmids. Consequently, the relationship between these two blaKPC-5-harboring plasmids remains unclear.
Previous sequence comparison between different blaKPC variants showed that blaKPC-3, blaKPC-5, and blaKPC-6 each differ from blaKPC-2 by a single nucleotide change, while blaKPC-5 and blaKPC-6 also differ from blaKPC-4 by a single nucleotide (14, 23). Accordingly, it was hypothesized that blaKPC-4 may have emerged through a sequential process, wherein blaKPC-5 and/or blaKPC-6 represents an intermediate step (14, 23). Interestingly, KPC-4 and -5 were both identified in the same hospital; nevertheless, evidence that the blaKPC-4 gene in pBK31551 or pBK31572 evolved directly from blaKPC-5 in pBK31567 is lacking, as these genes were found in distinct plasmids from diverse genetic backgrounds and different time periods.
In this study, we have described in detail the complete sequences of IncN and IncX plasmids harboring blaKPC-4 and blaKPC-5 carbapenemase genes. Comparative genomic analyses provide new insights into the evolution and dissemination of KPC-producing carbapenem resistance plasmids belonging to different incompatibility groups. Our studies and previous investigations highlight the essential roles of Tn4401 in KPC dissemination and evolution, including large-scale deletions, insertions, and replacements, as well as carriage of different blaKPC variants and horizontal transfer between different plasmids and host strains.
ACKNOWLEDGMENTS
This study was supported by a grant (to B.N.K.) from the National Institutes of Health (1R01AI090155). This work was supported in part by the Veterans Affairs Merit Review Program (R.A.B.), the National Institutes of Health (grants AI072219-05 and AI063517-07 to R.A.B.), and the Geriatric Research Education and Clinical Center VISN 10 (R.A.B.).
We thank the team from Genotyping of Pathogens and Public Health (Institut Pasteur, Paris, France) for curating the MLST profiles and isolates databases at www.pasteur.fr/mlst.
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53:60–67 [DOI] [PubMed] [Google Scholar]
- 2. Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 4. Andrade LN, Curiao T, Ferreira JC, Longo JM, Climaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, Ribeiro VB, Barth AL, Paula MC, Baquero F, Canton R, Darini AL, Coque TM. 2011. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob. Agents Chemother. 55:3579–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, Bojarska K, Zabicka D, Kania-Pudlo M, Mlynarczyk G, Zak-Pulawska Z, Hryniewicz W, Gniadkowski M. 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008-2009. Antimicrob. Agents Chemother. 55:5493–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Gales AC, Venezia SN, Quinn JP, Nordmann P. 2010. Worldwide diversity of Klebsiella pneumoniae that produce β-lactamase blaKPC-2 gene. Emerg. Infect. Dis. 16:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cuzon G, Naas T, Villegas MV, Correa A, Quinn JP, Nordmann P. 2011. Wide dissemination of Pseudomonas aeruginosa producing β-lactamase blaKPC-2 gene in Colombia. Antimicrob. Agents Chemother. 55:5350–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gomez SA, Pasteran FG, Faccone D, Tijet N, Rapoport M, Lucero C, Lastovetska O, Albornoz E, Galas M, Melano RG, Corso A, Petroni A. 2011. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin. Microbiol. Infect. 17:1520–1524 [DOI] [PubMed] [Google Scholar]
- 10. Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53:1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54:4201–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Naas T, Cuzon G, Villegas M-V, Lartigue M-F, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53:4333–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolter DJ, Kurpiel PM, Woodford N, Palepou MF, Goering RV, Hanson ND. 2009. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob. Agents Chemother. 53:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob. Agents Chemother. 55:5370–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mataseje LF, Boyd DA, Willey BM, Prayitno N, Kreiswirth N, Gelosia A, Poutanen SM, Low DE, Jenkins SG, Katz K, Mulvey MR. 2011. Plasmid comparison and molecular analysis of Klebsiella pneumoniae harbouring blaKPC from New York City and Toronto. J. Antimicrob. Chemother. 66:1273–1277 [DOI] [PubMed] [Google Scholar]
- 17. Carattoli A, Aschbacher R, March A, Larcher C, Livermore DM, Woodford N. 2010. Complete nucleotide sequence of the IncN plasmid pKOX105 encoding VIM-1, QnrS1 and SHV-12 proteins in Enterobacteriaceae from Bolzano, Italy compared with IncN plasmids encoding KPC enzymes in the USA. J. Antimicrob. Chemother. 65:2070–2075 [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56:2143–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang Y, Yu D, Wei Z, Shen P, Zhou Z, Yu Y. 2010. Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob. Agents Chemother. 54:3967–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob. Agents Chemother. 54:4493–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu P, Li P, Jiang X, Bi D, Xie Y, Tai C, Deng Z, Rajakumar K, Ou HY. 2012. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 194:1841–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L, Chavda KD, Mediavilla JR, Zhao Y, Fraimow HS, Jenkins SG, Levi MH, Hong T, Rojtman AD, Ginocchio CC, Bonomo RA, Kreiswirth BN. 2012. Multiplex real-time PCR for detection of an epidemic KPC-producing Klebsiella pneumoniae ST258 clone. Antimicrob. Agents Chemother. 56:3444–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J. Clin. Microbiol. 49:579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schechner V, Straus-Robinson K, Schwartz D, Pfeffer I, Tarabeia J, Moskovich R, Chmelnitsky I, Schwaber MJ, Carmeli Y, Navon-Venezia S. 2009. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 47:3261–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. CLSI 2012. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement, M100-S22. CLSI, Wayne, PA [Google Scholar]
- 26. Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 27. Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 29. Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 30. Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 31. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garcia-Fernandez A, Villa L, Moodley A, Hasman H, Miriagou V, Guardabassi L, Carattoli A. 2011. Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 66:1987–1991 [DOI] [PubMed] [Google Scholar]
- 33. Chen L, Chavda KD, Mediavilla JR, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. 2012. Partial excision of blaKPC from Tn4401 in carbapenem-resistant Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75 doi:10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147 doi:10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Endimiani A, Perez F, Bajaksouzian S, Windau AR, Good CE, Choudhary Y, Hujer AM, Bethel CR, Bonomo RA, Jacobs MR. 2010. Evaluation of updated interpretative criteria for categorizing Klebsiella pneumoniae with reduced carbapenem susceptibility. J. Clin. Microbiol. 48:4417–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Papp PP, Iyer VN. 1995. Determination of the binding sites of RepA, a replication initiator protein of the basic replicon of the IncN group plasmid pCU1. J. Mol. Biol. 246:595–608 [DOI] [PubMed] [Google Scholar]
- 39. More MI, Pohlman RF, Winans SC. 1996. Genes encoding the pKM101 conjugal mating pore are negatively regulated by the plasmid-encoded KorA and KorB proteins. J. Bacteriol. 178:4392–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pohlman RF, Genetti HD, Winans SC. 1994. Entry exclusion of the IncN plasmid pKM101 is mediated by a single hydrophilic protein containing a lipid attachment motif. Plasmid 31:158–165 [DOI] [PubMed] [Google Scholar]
- 41. Pohlman RF, Liu F, Wang L, More MI, Winans SC. 1993. Genetic and biochemical analysis of an endonuclease encoded by the IncN plasmid pKM101. Nucleic Acids Res. 21:4867–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hengen PN, Denicourt D, Iyer VN. 1992. Isolation and characterization of kikA, a region on IncN group plasmids that determines killing of Klebsiella oxytoca. J. Bacteriol. 174:3070–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodriguez M, Holcik M, Iyer VN. 1995. Lethality and survival of Klebsiella oxytoca evoked by conjugative IncN group plasmids. J. Bacteriol. 177:6352–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Santini JM, Stanisich VA. 1998. Both the fipA gene of pKM101 and the pifC gene of F inhibit conjugal transfer of RP1 by an effect on traG. J. Bacteriol. 180:4093–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kulakauskas S, Lubys A, Ehrlich SD. 1995. DNA restriction-modification systems mediate plasmid maintenance. J. Bacteriol. 177:3451–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paterson ES, More MI, Pillay G, Cellini C, Woodgate R, Walker GC, Iyer VN, Winans SC. 1999. Genetic analysis of the mobilization and leading regions of the IncN plasmids pKM101 and pCU1. J. Bacteriol. 181:2572–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Belogurov AA, Delver EP, Rodzevich OV. 1993. Plasmid pKM101 encodes two nonhomologous antirestriction proteins (ArdA and ArdB) whose expression is controlled by homologous regulatory sequences. J. Bacteriol. 175:4843–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Delver EP, Belogurov AA. 1997. Organization of the leading region of IncN plasmid pKM101 (R46): a regulation controlled by CUP sequence elements. J. Mol. Biol. 271:13–30 [DOI] [PubMed] [Google Scholar]
- 49. Murata T, Ohnishi M, Ara T, Kaneko J, Han CG, Li YF, Takashima K, Nojima H, Nakayama K, Kaji A, Kamio Y, Miki T, Mori H, Ohtsubo E, Terawaki Y, Hayashi T. 2002. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 184:3194–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwocho LR, Schaffner CP, Miller GH, Hare RS, Shaw KJ. 1995. Cloning and characterization of a 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ib, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1790–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60:394–397 [DOI] [PubMed] [Google Scholar]
- 52. Noppe-Leclercq I, Wallet F, Haentjens S, Courcol R, Simonet M. 1999. PCR detection of aminoglycoside resistance genes: a rapid molecular typing method for Acinetobacter baumannii. Res. Microbiol. 150:317–322 [DOI] [PubMed] [Google Scholar]
- 53. Datta N, Hughes VM. 1983. Plasmids of the same Inc groups in Enterobacteria before and after the medical use of antibiotics. Nature 306:616–617 [DOI] [PubMed] [Google Scholar]
- 54. Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50 [DOI] [PubMed] [Google Scholar]
- 55. Norman A, Hansen LH, She Q, Sorensen SJ. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74 [DOI] [PubMed] [Google Scholar]
- 56. Palepou MF, Woodford N, Hope R, Colman M, Glover J, Kaufmann ME, Lafong C, Reynolds R, Livermore DM. 2005. Novel class A carbapenemase, KPC-4, in an Enterobacter isolate from Scotland, abstr 1134_1101_1120. Abstr. 15th Eur. Cong. Clin. Microbiol. Infect. Dis., Copenhagen, Denmark [Google Scholar]
- 57. Robledo IE, Moland ES, Aquino EA, Vazquez GJ, Sante MI, Bertran J, Hanson ND. 2007. First report of a KPC-4 and CTX-M producing K. pneumoniae isolated from Puerto Rico (PR), abstr C2-1933. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 58. Robledo IE, Aquino EE, Sante MI, Santana JL, Otero DM, Leon CF, Vazquez GJ. 2010. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 54:1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolter DJ, Khalaf N, Robledo IE, Vazquez GJ, Sante MI, Aquino EE, Goering RV, Hanson ND. 2009. Surveillance of carbapenem-resistant Pseudomonas aeruginosa isolates from Puerto Rican medical center hospitals: dissemination of KPC and IMP-18 beta-lactamases. Antimicrob. Agents Chemother. 53:1660–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roth AL, Kurpiel PM, Lister PD, Hanson ND. 2011. blaKPC RNA expression correlates with two transcriptional start sites but not always with gene copy number in four genera of Gram-negative pathogens. Antimicrob. Agents Chemother. 55:3936–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tardif G, Grant RB. 1983. Transfer of plasmids from Escherichia coli to Pseudomonas aeruginosa: characterization of a Pseudomonas aeruginosa mutant with enhanced recipient ability for enterobacterial plasmids. Antimicrob. Agents Chemother. 24:201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]