Abstract
Aristolochic acids (AAs) are a structurally-related family of nephrotoxic and carcinogenic nitrophenanthrene compounds found in Aristolochia herbaceous plants, many of which have been used worldwide for medicinal purposes. AAs have been implicated in the etiology of so-called Chinese herbs nephropathy and of Balkan endemic nephropathy. Both of these disease syndromes are associated with carcinomas of the upper urinary tract (UUC). 8-Methoxy-6-nitrophenanthro-[3,4-d]-1,3-dioxolo-5-carboxylic acid (AA-I) is a principal component of Aristolochia herbs. Following metabolic activation, AA-I reacts with DNA to form aristolactam (AL-I)-DNA adducts. We have developed a sensitive analytical method, using ultra-performance liquid chromatography-electrospray ionization/multistage mass spectrometry (UPLC-ESI/MSn) with a linear quadrupole ion-trap mass spectrometer, to measure 7-(deoxyadenosin-N6-yl) aristolactam I (dA-AL-I) and 7-(deoxyguanosin-N2-yl) aristolactam I (dG-AL-I) adducts. Using 10 μg of DNA for measurements, the lower limits of quantitation of dA-AL-I and dG-AL-I are, respectively, 0.3 and 1.0 adducts per 108 DNA bases. We have used UPLC-ESI/MSn to quantify AL-DNA adducts in tissues of rodents exposed to AA, and in the renal cortex of patients with UUC who reside in Taiwan, where the incidence of this uncommon cancer is the highest reported for any country in the world. In human tissues, dA-AL-I was detected at levels ranging from 9 to 338 adducts per 108 DNA bases, whereas dG-AL-I was not found. We conclude that UPLC-ESI/MSn is a highly sensitive, specific and robust analytical method, positioned to supplant 32P-postlabeling techniques currently used for biomonitoring of DNA adducts in human tissues. Importantly, UPLC-ESI/MSn could be used to document exposure to AA, the toxicant responsible for AA nephropathy and its associated UUC.
Introduction
The Aristolochiaceae family of herbaceous plants, specifically members of the genus Aristolochia, have been used for medicinal purposes for more than 2500 years.1,2 Recently, aristolochic acid (AA), a principal component of all Aristolochia sp, was shown to be responsible for the clinical syndromes known as Chinese herbs nephropathy3 and Balkan endemic nephropathy (BEN),4,5 a devastating environmental disease. Both disorders are associated with a high incidence of urothelial carcinomas of the upper urinary tract (UUC),6,7 constituting a disease entity now designated aristolochic acid nephropathy (AAN).8
Aristolochia herbal remedies have been used widely in the practice of traditional Chinese medicine; moreover, the incidence of UUC in Taiwan, where usage has been carefully documented, is the highest of any country in the world.9 Aristolactam (AL) DNA adducts are present in the renal cortex of many Taiwanese patients with UUC and the documented TP53 mutational signature in these carcinomas is almost identical to that observed in BEN.10,11 These molecular epidemiologic studies implicate AA as the causative factor in AAN, now recognized as both an environmental and long-overlooked iatrogenic disease.2,8
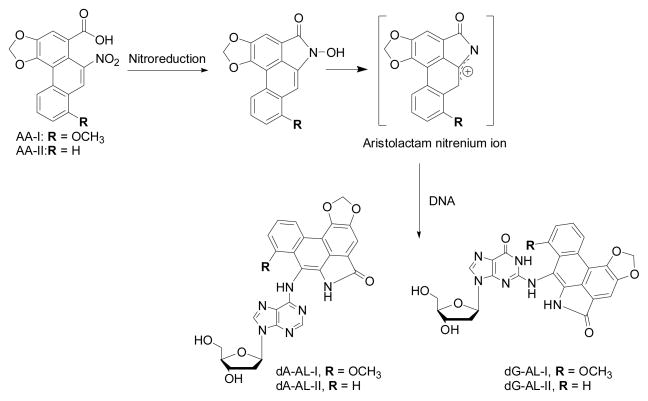
8-Methoxy-6-nitrophenanthro-[3,4-d]-1,3-dioxolo-5-carboxylic acid (AA-I) and 6-nitrophenathrene-[3,4-d]-1,3-dioxolo-5-carboxylic acid (AA-II) undergo bioactivation via enzymatic reduction of the nitro moieties of the phenanthrene rings to form N-hydroxyaristolactams and the postulated nitrenium intermediates, which bind covalently to dA and dG residues in DNA (Figure 1).12,13 The major AL-DNA adducts identified in rodent tissues are 7-(deoxyadenosin-N6-yl)aristolactam I (dA-AL-I), 7-(deoxyguanosin-N2-yl) aristolactam I (dG-AL-I), 7-(deoxyadenosin-N6-yl)aristolactam II (dA-AL-II), and 7-(deoxyguanosin-N2-yl)aristolactam II (dG-AL-II) (Figure 1).13,14 In humans, the highest levels of dA-AL-I are found in the renal cortex.15,16 This mutagenic lesion is responsible, via translesion synthesis, for generating A → T transversions in AA-I-exposed fibroblasts prepared from Hupki (human TP53 knock-in) mice17 and in the TP53 tumor suppressor gene in urothelial carcinomas of patients with AA-induced UUC.10,11,18
Figure 1.
Metabolic activation of AA and its reaction with DNA. Adduct formation occurs after reduction of the nitro moiety of the phenanthrene rings of AA-I and AA-II to form the N-hydroxyaristolactams and proposed nitrenium ions, which react with dG and dA to form dA-AL-I, dG-AL-I, dA-AL-II, and dG-AL-II.
To date, biomonitoring of AL-DNA adducts in rodents and humans has been limited to 32P-postlabeling techniques.19,20 However, this sensitive method of detection does not provide spectral data to establish the chemical identity of the lesion. Because humans are exposed to a plethora of genotoxicants,9,21 the potential for mis-characterizing DNA adducts when 32P-postlabeling methods are used is quite high.22 Moreover, 32P-postlabeling methods utilize high specific-activity radioactive phosphorus, and require labor-intensive chromatographic manipulations to separate DNA adducts from excess γ-[32P]-ATP. As a consequence, implementation of 32P-postlabeling techniques for population studies is problematic. Recent advances in liquid chromatography/mass spectrometric (LC/MS) methods,23,24 including several methods developed in our laboratory,25,26 have shown that the identification and quantitation of certain classes of DNA adducts by LC/MS techniques can be achieved, using small amounts of DNA, at a level of sensitivity that approaches the limit of adduct detection by 32P-postlabeling methods.19,20
Both triple-stage quadrupole mass spectrometry (TSQ MS) and quadrupole time-of-flight mass spectrometry have been used to measure AL-DNA adducts in rodent tissues.27,28 Our previous studies demonstrated that capillary liquid chromatography-electrospray ionization/multistage mass spectrometry (LC-ESI/MSn), using a linear quadrupole ion trap mass spectrometer, had the requisite sensitivity to identify AL-DNA adducts in the renal cortex of a woman who developed end-stage renal failure after using an herbal remedy containing AAs.4 Similar results were obtained in studies of patients with AAN/UUC from Balkan countries5 and in Taiwan.10 The objective of the present study was to establish and validate a robust, high-throughput LC-ESI/MSn procedure employing the stable, isotope dilution method and to measure AL-DNA adducts in human tissues.
Experimental Section
Caution: Aristolochic acids are human carcinogens and should be handled with caution in a well-ventilated fume hood with the appropriate protective clothing. Human tissues specimens were processed in a biohazard hood and all unused tissue material was treated with bleach prior to discarding the material in biohazard waste receptacles.
Materials
8-Methoxy-6-nitrophenanthro-(3,4-d)-1,3-dioxolo-5-carboxylic acid (AA-I), aristolactam-I (AL-I), 7-(deoxyadenosin-N6-yl) aristolactam-II (dA-AL-II), [N1,N3,N6–15N3]-dA-AL-II), 7-(deoxyguanosin-N2-yl) aristolactam II (dG-AL-II), and [N1,N2,N3-15N3]-dG-AL-II were kind gifts from Dr. Horacio Priestap, Department of Pharmacological Science, Stony Brook University. Uniformly 15N-labeled 2′-deoxyguanosine, [15N5]-dG and 2′-deoxyadenosine, [15N5]-dA (isotopic purities ≥ 98.5%) were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). Calf thymus DNA (CT DNA), DNase I (Type IV, from bovine pancreas), alkaline phosphatase (from Escherichia coli), and nuclease P1 (from Penicillium citrinum) were purchased from Sigma (St. Louis, MO). Phosphodiesterase I (from Crotalus adamanteus venom) was purchased from GE Healthcare (Piscataway, NJ). γ-32P-ATP (6000 Ci/mmol) was purchased from PerkinElmer (Boston, USA). Enzymes used for digestion of DNA were obtained from Worthington (Newark, NJ, USA). 3′-Phosphatase-free OptiKinase was purchased from Affymetrix Inc. (Santa Clara, CA). Dimethyl sulfoxide (DMSO) (>99.9%), N,N-dimethylformamide (DMF) (>99%) and ACS reagent-grade formic acid (98%) were purchased from Sigma-Aldrich (St. Louis, MO). All solvents were high-purity B & J Brand from Honeywell Burdick and Jackson (Muskegon, MI) or Optima LC/MS brand from Fisher Scientific (Fair Lawn, NJ).
Synthesis and Purification of dA-AL-I and dG-AL-I Adducts
The syntheses of 7-(deoxyadenosin-N6-yl)aristolactam-I (dA-AL-I), ([15N5]-dA-AL-I), 7-(deoxyguanosin-N2-yl) aristolactam I (dG-AL-I), and ([15N5]-dG-AL-I) were performed by the reaction of AA-I with dA or dG (or [15N5]-dA or [15N5]-dG), using a modification of the protocol described by Schmeiser.29 AA-I (1 mg) in DMF (100 μL) was mixed with 20 mg of Zn dust pre-activated by treatment with 1% HCl. Then, dA or dG (or [15N5]-labeled deoxynucleosides) (2 mg) in 50 mM potassium phosphate buffer (1 mL, pH 5.8) was added to the AA-I/Zn dust mixture. The reaction was conducted at 37 oC for 16 h in the dark. Thereafter, the solution was placed on ice for 30 min. The mixture was centrifuged at 15,000g for 5 min, and the supernatant was carefully removed from the Zn pellet and evaporated to dryness by vacuum centrifugation. The dried supernatants were dissolved in CH3OH (1 mL), followed by dilution with H2O (4 mL) and applied to Isolute C18 cartridges (100 mg) (Biotage, LLC, Charlotte, NC), which were preconditioned with CH3OH (2 mL), followed by H2O (2 mL). Then, the cartridges were washed with H2O (2 mL), followed by sequential washing with 40% and 50% CH3OH in H2O (1 mL). The desired adducts were then eluted from the cartridges with CH3OH (2 × 1 mL) and evaporated to dryness by vacuum centrifugation. The extracts were dissolved in DMSO (100 μL) and diluted to 1 mL with H2O. The Zn pellet was washed with DMSO (100 μL) and diluted to 1 mL with H2O, to further recover some of the precipitated adduct. Both extracts were then directly purified by HPLC (vide infra).
dA-AL-I, dG-AL-I and their [15N5]-labelled internal standards were purified by HPLC with an Agilent 1100 Chemstation system (Palo Alto, CA) equipped with a semi-preparative XBridge Prep Phenyl Column (250 × 10 mm, 5 μm) (Waters Corp., Milford, MA). A linear gradient commenced at 90% solvent A (0.1% HCO2H in H2O) and 10% solvent B (95% CH3CN, 4.9% H2O, and 0.1% HCO2H) and arrived at 90% solvent B at 30 min. The flow rate was 3 mL/min. The adducts were monitored at 410 nm. The fractions of dG-AL-I (tR= 11.5 min) and dA-AL-I (tR = 19.5 min) were collected in 0.5 M ammonium acetate (pH 6.8), to prevent hydrolysis of the N-glycosidic linkages by the acidic mobile phase solvents. The effluents were evaporated to dryness by vacuum centrifugation and purified by a second HPLC cycle to obtain adducts of the desired purity (>99%, based on HPLC with UV detection). The isotopic purity of 15N-labelled dA/dG-AL-I/II (≥98.5%) was determined by LC-ESI/MS (vide infra). The approximate overall yields of adducts were ~2% of the AA-I starting material.
Subsequent to the completion of the biomimetic syntheses of dG-AL-I and dA-AL-I and their internal standards, an unambiguous multi-step synthesis of dA-AL-I was achieved. The final coupling step that produced the adduct and 1H NMR spectrum are reported in Supporting Information, Figures S1 and S2, and the details of the complete synthesis will be published elsewhere.
UV/Vis Spectra and Molar Extinction Coefficients of dA-AL-I and dG-AL-I
The dA-AL-I adduct was insoluble in water and most organic solvents. Therefore, the UV/Vis spectrum of dA-AL-I was acquired in DMSO (Supporting Information, Figure S3). The molar extinction coefficient of the adduct was determined at the maxima at 305 nm (16,140 M−1 cm−1) and 415 nm (5,400 M−1 cm−1). The yield of dG-AL-I obtained by reaction of AA-I with Zn was insufficient to weigh accurately for the direct determination of its molar extinction coefficient by UV spectroscopy. Therefore, the molar extinction coefficient for dG-AL-I was estimated by comparison of its UV/vis spectrum to the spectrum of the synthetic dA-AL-I adduct, which contains the same phenanthrene ring structure. The UV/Vis spectra of dG-AL-I and dA-AL-I are very similar, and we assumed that the molar extinction coefficients of the adducts at their maxima are similar (Supporting Information, Figure S3). Throughout this study, these molar extinction coefficients were used to determine concentrations of unlabeled and labeled dG-AL-I and dA-AL-I adducts.
Rodent Experiments
The guidelines established by the National Institutes of Health Office of Laboratory Animal Welfare, were adhered to for the use of animals. Male C57BL/6J mice (Jackson Laboratories, Bar Harbor, Maine), 8–10 weeks old, were acclimatized to temperature (22 ± 2 °C) and humidity (55 ± 5%) in controlled rooms with a 12 h light/dark cycle for at least 1 week before the experiment. Laboratory chow and tap water were given ad libitum. The mice were dosed (i.p.) with AA-I (0, 0.1, 1 or 2 mg/kg, in phosphate buffered saline). After 20 h, mice were euthanized by asphyxiation with CO2, followed by cervical dislocation. This mouse strain is particularly resistant to the nephrotoxic effects of AA, which enabled us to give large doses of AA required for the DNA adduct validation study
Mouse tissues were homogenized and digested overnight in 10 mM Tris-HCl (pH 8.3) containing 10 mM EDTA, 0.1% (w/v) sodium dodecyl sulphate (1 mL), proteinase K (0.5 mg/mL), and RNase A (0.1 mg/ml), at 55 °C. DNA was purified by sequential phenol:CHCl3 (2×) and CHCl3 extractions. DNA was recovered from the aqueous phase by precipitation in 50% isopropanol. The DNA pellet was rinsed with 70% ethanol and resuspended in Tris-HCl, pH 7.5, 0.1 mM EDTA. The concentration of DNA was determined by UV, assuming that 50 μg/ml of DNA gives an absorbance at 260 nm of 1.0.
Human Tissues
The research protocol was reviewed and approved by the Institutional Review Boards at Stony Brook University, the Wadsworth Center, and the National Taiwan University Hospital, Taipei, Taiwan. Matched tissue samples from both renal cortex and UUC tissue of the same subjects from Taiwan were obtained following nephroureterectomy. The samples were snap-frozen in liquid nitrogen and stored at −80 °C. The human tissues were thawed on ice, diluted with 3 vol 10 mM Tris-HCl, pH 8.3, 10 mM EDTA, and homogenized with a Potter Elvehjem homogenizer. The homogenates were centrifuged at 3000g for 10 min. The nuclear pellets containing DNA were processed as described above for mouse tissue.
Enzymatic Digestion of AL-DNA and 32P-Postlabeling/PAGE Analysis
Human DNA (10–20 μg) or mouse DNA (5–10 μg) was digested with micrococcal nuclease (30 units) and spleen phosphodiesterase (0.15 unit) by incubating for 16 h at 37°C h in 17 mM sodium succinate buffer (100 μL, pH 6.0) containing 8 mM CaCl2, followed by enrichment of the adducts with n-butanol.14 DNA digests (5 or 20 μg) were then incubated with 20 μCi of [γ-32P]-ATP (6000 Ci/mmol) and 10 units of 3′-phosphatase-free OptiKinase at 37°C for 40 min, followed by treatment with apyrase (50 mU) for 30 min, as described previously.14,19,20 32P-labeled adducts were subjected to electrophoresis for 5 h on a nondenaturing 30% polyacrylamide gel at 1500 to 1800 V/20 to 40 mA. The gels were exposed for 12 h, and AL-DNA adducts were measured using a s-phosphorimager with Image QuaNT v5.2 software (Molecular Dynamics Inc. Piscataway, NJ). The sensitivity of the 32P-postlabeling/PAGE assay was assessed using kidney DNA obtained from a mouse treated with AA-I (2 mg/kg). The levels of dG- and dA-AL-I adducts in the mouse sample, predetermined in three independent digestion experiments, were estimated at 1 and 4 adducts/106 DNA bases, respectively. The DNA was diluted with unmodified DNA (20 μg total) to establish levels of modification of 0, 0.3, 0.7, 2.2 and 10.8 dA-AL-I adducts per 108 bases. Oligonucleotides containing dG-AL-II and dA-AL-II were synthesized as described, and employed as standards.18 The recovery and labeling efficiency of dA-AA-II- and dG-AA-II- adducts from the oligonucleotides exceeded 70%.14
Enzymatic Digestion of AL-DNA for Ultra-Performance Liquid Chromatography-Electrospray Ionization/Multistage Mass Spectrometry (UPLC-ESI/MSn) Measurements
The enzymatic digestion conditions used for the hydrolysis of DNA (20 μg) to 2′-deoxynucleosides have been described previously26 and employed DNase I for 1.5 h, followed by incubation with nuclease P1 for 3 h, and lastly by incubation with alkaline phosphatase and spleen phosphodiesterase for 18 h at 37 °C. We have shown that these conditions are highly efficient in the enzymatic digestion of DNA modified with bulky carcinogens of diverse structures.25,26 Isotopically labeled internal standards were added to the DNA prior to enzymatic digestion at a level of 5 adducts per 108 DNA bases for genomic mouse DNA, human renal cortex DNA, and DNA from human urothelial tumor tissue. After enzyme digestion, 2 vol of chilled C2H5OH (100%) was added, and the solution was kept on ice bath for 1 h to precipitate protein and salts. The solution was centrifuged at 15,000g for 5 min at room temperature. The supernatant containing the DNA adducts and internal standards was transferred into cap LC vials containing silylated inserts (350 μL) from Microliter Analytical Supplies, Inc. (Suwanee, GA). The extracts were concentrated to dryness by vacuum centrifugation and dissolved in a solvent of 1:1 H2O:DMSO (20 μL). The efficiency of DNA digestion was assessed by HPLC analysis of the unmodified DNA bases as previously reported.22
Analyses of AL-DNA Adducts by Online Solid Phase Extraction Coupled UPLC-ESI/MSn
Analyses were performed with a NanoAcquity™ UPLC system (Waters Corporation, Milford, MA) interfaced with an Advance CaptiveSpray™ source from Michrom Bioresource Inc. (Auburn, CA) and linear quadrupole ion-trap mass spectrometer (LTQ Velos, Thermo Fisher, San Jose, CA). A Waters Symmetry trap column (180 μm × 20 mm, 5 μm particle size) was employed for online solid phase enrichment of the DNA adducts. The analytical column was a C18 AQ (0.3 × 150 mm, 3 μm particle size) from Michrom Bioresources Inc. The DNA digests were loaded onto the trap column and washed with 0.01% HCO2H in 20% CH3CN at a flow rate of 12 μL/min for 4 min. Thereafter, the DNA adducts were back-flushed onto the C18 AQ column and a linear gradient was employed to resolve the DNA adducts, starting at 0.01% HCO2H containing 20% solvent B (95% CH3CN containing 4.99% H2O and 0.01% HCO2H) and arriving at 53% solvent B at 12 min, and then reaching 99% B at 13 min, employing a flow rate of 5 μL/min. The solvent mixture was held at 99% B for 2 min, then returned to the starting solvent conditions (20% B) over 2.5 min, and held for a 4 min period to allow the column to re-equilibrate at the starting solvent conditions.
Xcalibur version 2.1.0 software was used for data manipulations. The adducts were measured at the MS2 (dG-AL-I and dG-AL-II) or MS3 (dA-AL-I and dA-AL-II) scan stages in the positive ionization mode. The temperature of the capillary tube was set at 305 °C, the spray voltage was 2 kV, and the skimmer offset voltage was set at 10 V for dA-AL-I/II, and 40 V for dG-AL-I/II adducts. The skimmer offset voltage settings for the dG-AL adducts was elevated to achieve complete in-source fragmentation of dG-AL-I/II to their respective aglycone adducts [BH2]+. Helium was used as the collision and damping gas in the ion trap and was set at a pressure of 1 mTorr. There was no sheath or auxiliary gas. The automatic gain control settings were full MS target 30000 and MSn target 10000, and the maximum injection time was 10 ms. One μscan was used for data acquisition. An isolation width of 1 m/z and a normalized collision energy of 40 eV were employed for dG-AL-I/II at the MS2 scan stage. The MS2 transitions employed for quantitative measurements of the aglycone adducts of dG-AL were: dG-AL-I ([BH2]+) at m/z 443.2 → 292.3 and 293.2; [15N5]-dG-AL-I ([BH2]+) at m/z 448.2 → 292.3 and 293.2; and dG-AL-II ([BH2]+) at m/z 413.2 → 305.2 and 368.2; and [15N3]-dG-AL-II ([BH2]+) at m/z 416.2 → 306.2 and 370.2. Isolation widths of 4 m/z and 1 m/z and normalized collision energies 28 and 36 eV were used for dA-AL-I/II adducts, respectively at the MS2 and MS3 scan stages. The MS3 transitions employed for quantitative measurements dA-AL adducts were: dA-AL-I at m/z 543.3 → 427.2 → 292.3, 293.2, and 412.2; [15N5]-dA-AL-I at m/z 548.3 → 432.2 → 292.3, 293.2, and 417.2; dA-AL-II at m/z 513.3 → 397.2 → 262.1, 263.1, and 354.2; [15N3]-dA-AL-II at m/z 516.2 → 400.2 → 262.1, 263.1, and 357.2. The activation Q was 0.35, and the activation time was 10 ms for both MS2 and MS3 scan stages.
Validation of AL-Modified Mouse Kidney DNA Analysis by UPLC-ESI-MSn
Kidney DNA samples from a mouse treated with AA-I (2 mg/kg) were used for method validation. The levels of dA-AL-I and dG-AL-I adducts, estimated by 32P-postlabeling, were serially diluted with CT DNA so as, to arrive at different levels of AL-adduct modification (0.3, 0.5, 1.0, 5.0, or 10.0 adducts per 108 DNA bases), in 20 μg DNA. 15N-labeled internal standards were added at a level of 5.0 adducts per 108 bases, and DNA samples (20 μg) underwent enzyme hydrolysis, followed by online solid phase enrichment of the adducts and UPLC-ESI-MSn analysis.
Calibration Curves for AL-DNA Adducts
CT DNA (20 μg) samples were spiked with increasing amounts of a mixture of dG-AL-I/II and dA-AL-I/II adducts (0, 0.3, 0.6, 1.0, 3.0, 6.0, 10.0, and 30.0 adducts per 108 DNA bases) and a fixed amount of the four 15N-labeled internal standards set at a level of 5.0 adducts per 108 DNA bases. Calibration curves were generated at the MS2 scan stage for dG-AL-I/II and at the MS3 scan stage for dA-AL-I/II adducts. The calibration curves were generated from the combined integrated areas of the selected transitions employed for each adduct (vide supra). The integrated peak area ratio of unlabeled AL-DNA adducts over the respective 15N-labeled AL-DNA adducts was plotted as the ordinate, and the varying amounts of unlabeled AL-DNA adducts per 108 DNA bases were plotted as the x-coordinate (Supporting Information, Figure S4). The calibration curves were conducted in quadruplicate at each calibrant level, and the data were fitted to a straight line, using ordinary least-squares with equal weightings for linear regression. The coefficient of determination (r2) values of the slopes equaled or exceeded 0.999.
Results
Characterization of the Product Ion Spectra of AL-DNA Adducts by ESI-MS3
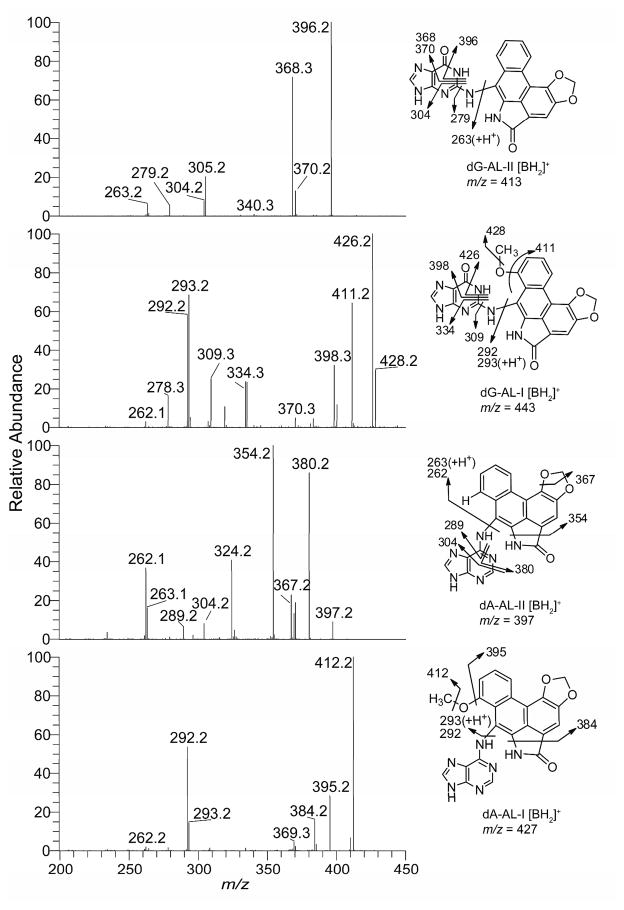
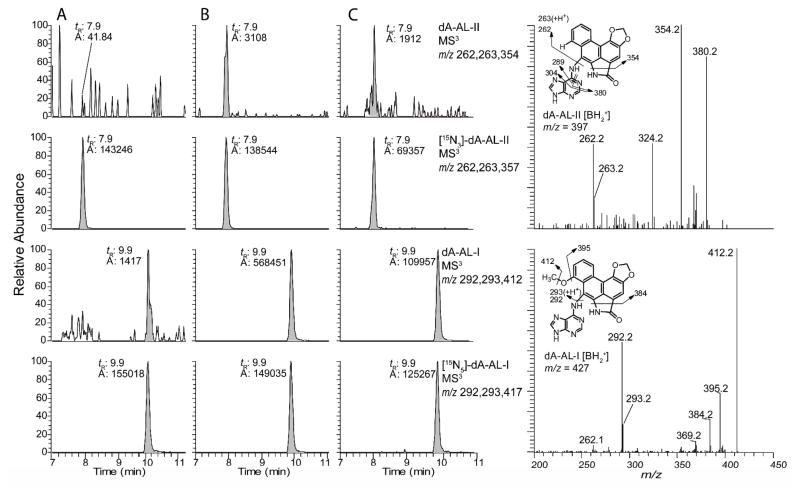
The product ion spectra of the dG-AL-I, dG-AL-II, dA-AL-I, and dA-AL-II at the MS3 scan stage and several proposed fragmentation patterns are depicted in Figure 2. The product ion spectra at the MS3 scan stage provide rich structural information about the aglycone adducts and were used to corroborate identities of dA-AL adducts present in mouse and human DNA samples. The product ion spectra of the 15N-labeled internal standards served as an aid in the interpretation of several the proposed fragment ions. The spectra of the 15N-labeled AL-DNA adducts are shown in Supporting Information, Figure S5.
Figure 2.
Product ion spectra of AL-DNA adducts acquired at the MS3 scan stage, representing the fragmentation of the aglycone adducts [BH2]+ of dG-AL-II (m/z 413), dG-AL-I (m/z 443), dA-AL-II (m/z 397), and dA-AL-I (m/z 427). Based upon the comparison of the product ion spectra of the dA-AL-II and [15N3]-dA-AL-II adducts (Supporting Information, Figure S5), the fragment ion at m/z 324.2 (m/z 327.2 for [15N3]-dA-AL-II) may form by the loss of CH2O and HNCO from the substituted phenanthrene ring.
UPLC-ESI/MSn Analysis of AL-Adduct Formation in Mouse Liver and Kidney DNA
A dual switching valve system and a Waters Symmetry trap column (180 μm × 20 mm, 5 μm particle size) were employed to enrich the dG-AL-I/II and dA-AL-I/II adducts in the presence of unmodified DNA bases. The signals of the synthetic dG-AL-I/II and dA-AL-I/II standards spiked into the CT DNA at levels ranging from 0.3 to 30 adducts per 108 DNA bases were comparable to the responses obtained for the pure standards (data not shown). Thus, the online enrichment procedure was highly effective in the removal of unmodified DNA bases, which were present at 106 – 108-fold higher concentrations than the AL-DNA adducts, as well as other constituents in the DNA digests that could interfere with the MS analysis and/or provoke ion suppression effects. The efficacy of the enzymatic hydrolysis of DNA was quantitative, as judged by the HPLC analysis of the unmodified DNA bases (BH Yun, unpublished observations).22
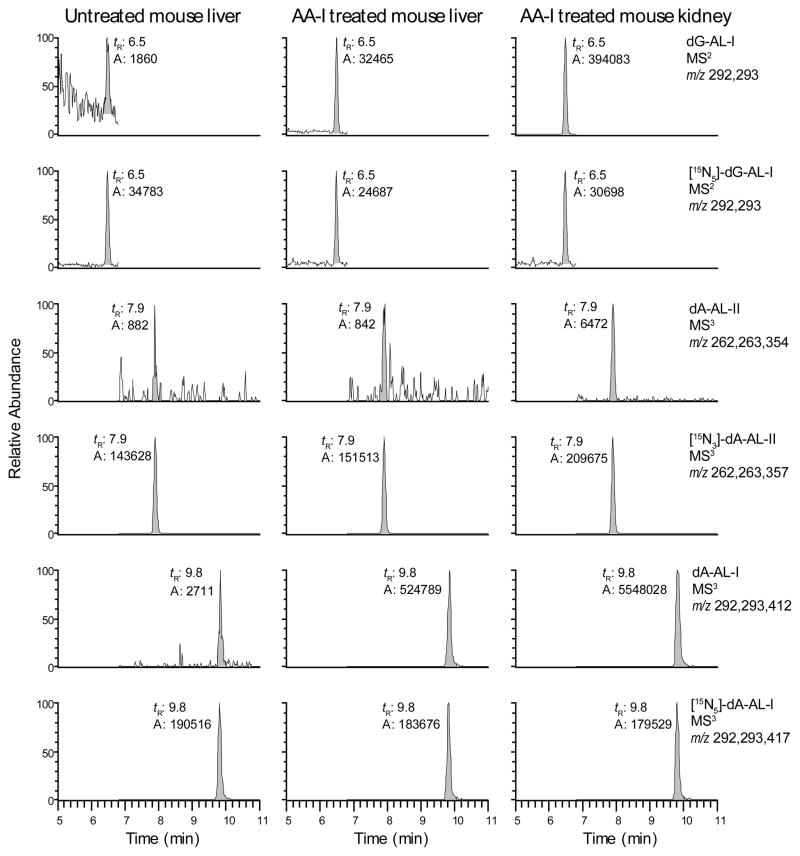
The mass chromatograms of the dG-AL-I and dA-AL-I/II adducts from liver DNA of an unexposed mouse, and liver and kidney DNA of a mouse given AA-I (1 mg/kg) are shown in Figure 3, and the product ion spectra of these adducts are presented in Supplementary Information, Figure S6. dA-AL-I is the most abundant adduct, followed by dG-AL-I. Surprisingly, dA-AL-II also was detected in the kidney of the treated mouse, occurring at a level of ~2 adducts per 109 bases. dA-AL-II does not contain the 8-methoxy substituent of the phenanthrene ring and, therefore, cannot be formed from a reactive metabolite of AA-I. The identification of dA-AL-II in the DNA sample suggests that the AA-I administered to the mouse was contaminated with trace levels of AA-II. Indeed, 0.4% AA-II was found to be present in the sample of AA-I used to inject mice. (T. Rosenquist, unpublished observations).
Figure 3.
The mass chromatograms of the AL-modified genomic DNA from a mouse dosed with AA-I (1 mg/kg). Mouse liver and kidney DNA were diluted with unmodified CT DNA by a factor of 10 and 5, respectively. The level of 15N-labeled internal standards was 5 adducts per 108 bases. The levels of AL-DNA adducts expressed per 108 bases in undiluted mouse liver and kidney were dG-AL-I: 58.1 (liver) and 300 (kidney); dA-AL-II: 0.7 (liver) and 0.8 (kidney); dA-AL-I: 176 (liver) and 1017 (kidney). The combined ions presented in the mass chromatograms of the adducts and internal standards were employed for quantitative measurements. The trace amounts of AL-DNA adducts detected in untreated DNA are attributed to the residual unlabeled dG and dA present in the isotopically labelled internal standards.
Performance of the Analytical Method
The calibration curves constructed for dG-AL-I/II and dA-AL-I/II adducts, spiked prior to enzymatic digestion with CT DNA at levels ranging from 0, 0.3 to 30.0 adducts per 108 DNA bases, were highly linear (coefficient of determination was r2 ≥ 0.999 for all adducts) (Supporting Information, Figure S4). AL-I-DNA from the kidney of a mouse treated with AA-I (2 mg/kg), which was also analyzed by 32P-postlabeling, was diluted with CT DNA and used for validation of the analytical UPLC-ESI/MSn method. Data on the performance of this method are summarized in Table 1. The within-a-day accuracy and precision of the method (coefficient of variation, CV) were obtained from quadruplicate DNA samples assayed on the same day. The between-day accuracy and precision of the method was determined from repetitive analyses of the DNA samples conducted over a period of 6 weeks. Estimates of AL-I adduct levels measured at the MS3 scan stage were on average within 10% of the target values obtained by 32P-postlabeling. However, the estimated level of the dG-AL-I adduct was ~40% lower than the values determined by 32P-postlabeling with the set of DNA samples employed for method validation.
Table 1.
Performance of the Analytical UPLC-ESI/MSn Method to Measure AL-DNA Adducts
| Adducts per 109 bases
|
||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Target level dA-AL-l | Day 1 | Day 2 | Day 3 | Day 4 | Overall mean | CV(%) within-day | CV(%) between-day | |
| 3 adducts per 109 bases | Mean | 3.3 | 2.6 | 3.3 | ND | 3.1 | 7.8 | 14.3 |
| SD | 0.2 | 0.3 | 0.2 | |||||
| RSD(%) | 5.5 | 10.9 | 7.4 | |||||
| 5 adducts per 109 bases | Mean | 5.6 | 5.0 | 4.7 | ND | 5.1 | 13.8 | 15.2 |
| SD | 0.6 | 0.8 | 0.6 | |||||
| RSD(%) | 11.1 | 16.4 | 12.9 | |||||
| 10 adducts per 109 bases | Mean | 11.0 | 8.1 | 8.8 | ND | 9.3 | 14.3 | 20.5 |
| SD | 2.1 | 0.7 | 0.6 | |||||
| RSD(%) | 19.1 | 8.9 | 6.4 | |||||
| 50 adducts per 109 bases | Mean | 50.8 | 43.5 | 50.0 | 48.0 | 48.0 | 6.2 | 8.6 |
| SD | 1.3 | 4.4 | 1.6 | 3.4 | ||||
| RSD(%) | 2.5 | 10.2 | 3.3 | 7.0 | ||||
| 100 adducts per 109 bases | Mean | 94.5 | 81.8 | 79.3 | ND | 85.2 | 6.2 | 11.2 |
| SD | 2.9 | 7.9 | 3.0 | |||||
| RSD(%) | 3.1 | 9.7 | 3.8 | |||||
| Adducts per 109 bases
|
||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Target level dG-AL-l | Day 1 | Day 2 | Day 3 | Day 4 | Overall mean | CV(%) within-day | CV(%) between-day | |
| 20 adducts per 109 bases | Mean | 12.5 | 12.3 | 11.0 | 11.5 | 11.8 | 9.4 | 10.0 |
| SD | 0.6 | 0.2 | 1.5 | 0.6 | ||||
| RSD(%) | 4.6 | 12.2 | 13.4 | 5.0 | ||||
| 40 adducts per 109 bases | Mean | 26.0 | 19.0 | 19.3 | ND | 19.8 | 9.8 | 21.6 |
| SD | 2.3 | 2.2 | 1.3 | |||||
| RSD(%) | 8.9 | 11.4 | 6.5 | |||||
N = 4 analyses per dose
ND, not determined
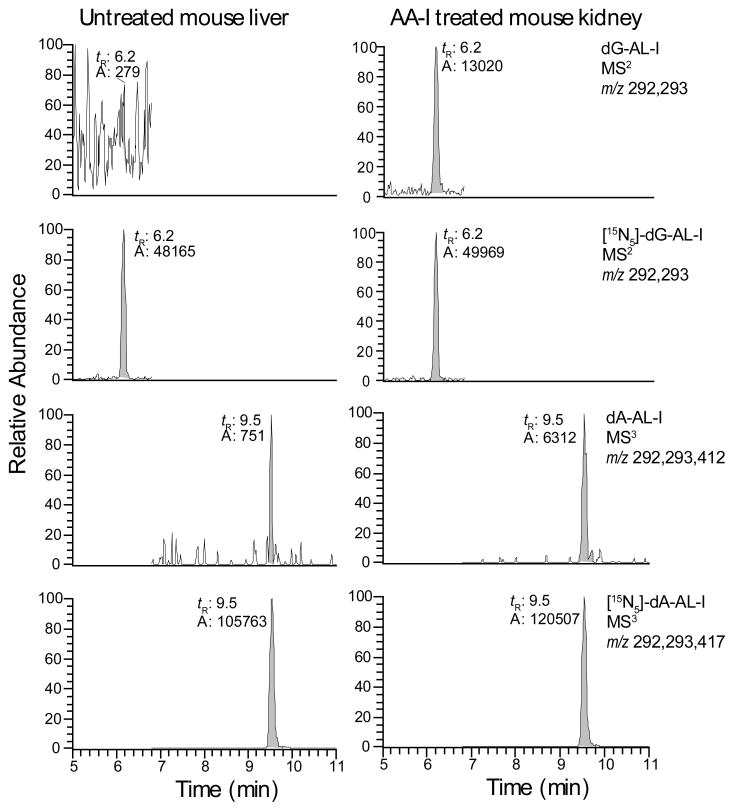
Values for the limit of detection (LOD) and limit of adduct quantitation (LOQ) of AL-DNA adducts, defined, respectively, as the average background signals + 3*SD and + 10*SD,30 were determined with CT DNA and untreated liver DNA of mice. The LOD and LOQ values of dG-AL-I were, respectively, ~0.3 and 1.0 adducts per 108 bases, and the LOD and LOQ values of dA-AL-I were ~0.1 and 0.3 adducts per 108 bases. We attribute the discrepancy in sensitivity between dG-AL-I and dA-AL-I adducts to the different MSn scan stages used to measure the adducts. The dG-AL adduct was somewhat unstable and underwent hydrolysis to form the aglycone [BH2]+ in the MS source. Hence, the dG-AL-I was measured as the aglycone at the MS2 scan stage, whereas the dA-AL-I adduct was measured as the aglycone at the MS3 scan stage. A mass chromatogram of AL-I-DNA adducts formed in kidney DNA of a mouse dosed with AA-I (2 mg/kg), followed by dilution of the DNA with unmodified CT DNA to achieve a level of AL-DNA modification approaching the LOQ values of AL-DNA adducts, is shown in Figure 4. The product-ion spectrum of the aglycone adduct of dG-AL-I acquired at the MS2 scan stage, displays the principal fragment ions of the modified base; however, a number of fragment ions, attributed to the solvent or constituents of the DNA digest matrix, also are observed (Supporting Information, Figure S7). A high quality product ion spectrum of the aglycone of dA-AL-I was obtained at the MS3 scan stage with relatively minor interfering fragment ions (Supporting Information, Figure S7). Thus, consecutive reaction monitoring at the MS3 scan stage effectively filters out many of the isobaric interferences observed in MS2 scan stage and results in an improved signal-to-noise ratio, and superior product ion spectra of the aglycone adducts.26
Figure 4.
The mass chromatograms of dG-AL-I and dA-AL-I adducts in mouse kidney DNA diluted with unmodified CT DNA to levels approaching the LOQ. Mass chromatograms of untreated and undiluted mouse liver DNA (left panel); AA-I treated mouse kidney (right panel) with dG-AL-I estimated at 1.3 adduct per 108 bases and dA-AL-I estimated at 0.3 adducts per 108 bases. The internal standards were added at a level of 5.0 adducts per 108 bases.
Comparison of AL-DNA Adduct Analysis by UPLC-ESI/MSn and 32P-Postlabeling/PAGE
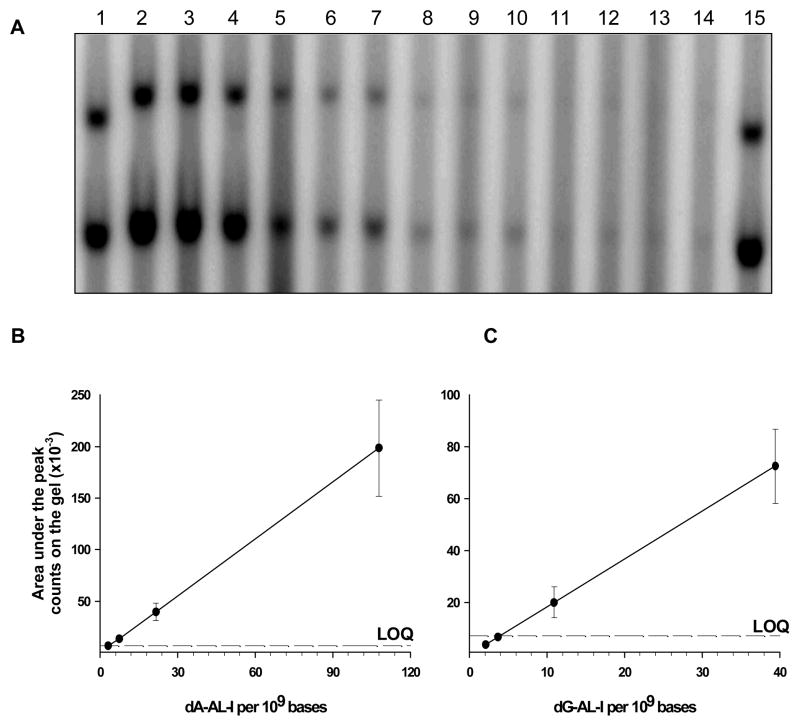
The mouse DNA samples employed for validating AL-DNA adduct measurements by UPLC-ESI/MS3 (Figure 4), were assayed by 32P-postlabeling/PAGE. AL-DNA adducts were diluted with unmodified DNA (20 μg total) to arrive at adduct levels ranging from 0.3 to 10.8 dA-AL-I adducts per 108 bases. The levels of dG-AL-I adducts in this experiment are 3 times less than dA-AL-I. The entire postlabeled DNA digest was applied to the gel. Adducts were visualized by the s-phosphorimager (Figure 5A), and the levels of adduct modification were estimated using an external calibration curve.14 At 0.3 AL-I adducts per 108 DNA bases, the counts on the gel were significantly higher than the background. The dose-response signals of the dA- and dG-AL-I adducts were linear (Figure 5B and C). The LOQ for 32P-postlabeling/PAGE was ~0.3 adducts per 108 DNA bases.
Figure 5.
32P-postlabeling/PAGE analysis of AL-DNA adducts. DNA was obtained from the renal cortex of a mouse treated with AA-I (2 mg/kg) and diluted with unmodified DNA (20 μg total) to arrive at levels of modification of 3, 7, 21 and 108 dA-AL-I adducts per 109 bases. For this experiment, the levels of dG-AL-I adducts are 3 times less than dA-AL-I. The experiment was performed in quadruplicate for the lowest adduct level and in triplicate for higher levels. (A) Lanes 1 and 15 (dA-AL-II and dG-AL-II standards, 80 adducts/109 bases); lanes 2–4 (100 dA-AL-I adducts per 109 bases); lanes 5–7 (21 dA-AL-I adducts per 109 bases); lanes 8–10 (7 dA-AL-I adducts per 109 bases); lanes 11–14 (3 dA-AL-I adducts per 109 bases). Upper and lower bands correspond to dG-AL and dA-AL adducts, respectively. (B) Dose response for dA-AL-I and (C) dose response for dG-AL-I levels measured with Image QuaNT v5.2 software were plotted as a function of adduct modification.
Interlaboratory estimates of AL-DNA adducts, by UPLC-ESI/MSn and 32P-postlabeling measurements were conducted with liver and kidney samples of mice exposed to AA-I (0.1 or 1.0 mg/kg). The levels of adducts in these tissues are reported in Table 2. The measured values of dA-AL-I adduct levels were within 0.6 to 1.4-fold, and the values of dG-AL-I were between 0.4 to 1.2-fold, when assayed by these two analytical methods.
Table 2.
dA-AL-I and dG-AL-I Adduct Estimates in Mouse Liver and Kidney: UPLC-ESI/MSn versus 32P-Postlabeling Measurements
| AA-I Dose (mg/kg) | Tissue | dA-AL-I per 108 bases
|
dG-AL-I per 108 bases
|
||||
|---|---|---|---|---|---|---|---|
| UPLC-ESI/MS/MSn | 32P-Postlabeling | UPLC-ESI/MS/MSn | 32P-Postlabeling | ||||
| 0.1 | Liver | Avg | 52.7 | 55.1 | Avg | 19.0 | 14.6 |
| SD | 7.4 | SD | 2.8 | ||||
| CV(%) | 14.1 | CV(%) | 14.6 | ||||
| 1.0 | Liver | Avg | 176 | 113.0 | Avg | 58.1 | 35.7 |
| SD | 3.5 | SD | 4.6 | ||||
| CV(%) | 2.0 | CV(%) | 7.9 | ||||
|
| |||||||
| 0.1 | Kidney | Avg | 43.9 | 61.6 | Avg | 22.8 | 27.1 |
| SD | 3.4 | SD | 1.8 | ||||
| CV(%) | 7.8 | CV(%) | 7.7 | ||||
| 1.0 | Kidney | Avg | 1020 | 717 | Avg | 300 | 115 |
| SD | 97.2 | SD | 10.5 | ||||
| CV(%) | 9.6 | CV(%) | 3.5 | ||||
N = 4 independent measurements by UPLC-ESI/MS/MSn
Identification and Quantitation of dA-AL-I and dA-AL-II DNA adducts in the Renal Cortex and UUC Tissue of Patients with UUC
The mass chromatograms of dA-AL-I/II adducts present in the renal cortex from two Taiwanese patients with UUC are shown in Figure 6. On the basis of elevated levels of AL-DNA adduct estimated by 32P-postlabeling, the human DNA samples were diluted by 6 to 10-fold with CT DNA so that the AL DNA adduct levels were within the concentration range of adducts used for construction of the calibration curves (Supporting Information, Figure S4). The level of dA-AL-I was determined, by UPLC-ESI/MS3, at 142 adducts per 108 bases for subject 1 and 44.8 adducts per 108 bases for subject 2. The dA-AL-II adduct was detected at a level of < 1 adduct per 108 bases in the undiluted DNA samples; however, there was no evidence for dG-AL-I/II adducts. Product ion spectra acquired at the MS3 scan stage confirmed the identities of both dA-AL adducts.
Figure 6.
The mass chromatograms of dG-AL-I and dA-AL-I in renal cortex DNA from two patients with UUC. (A) unmodified CT DNA, (B) Subject C95, (C) Subject C112. The human DNA was diluted with unmodified CT DNA by a factor of 6 for subject 1 and and 10-fold for subject 2. The estimated levels of dA-AL-I were 142 adducts per 108 bases for subject 1 and 44.8 adducts per 108 bases for subject 2 in non-diluted DNA.
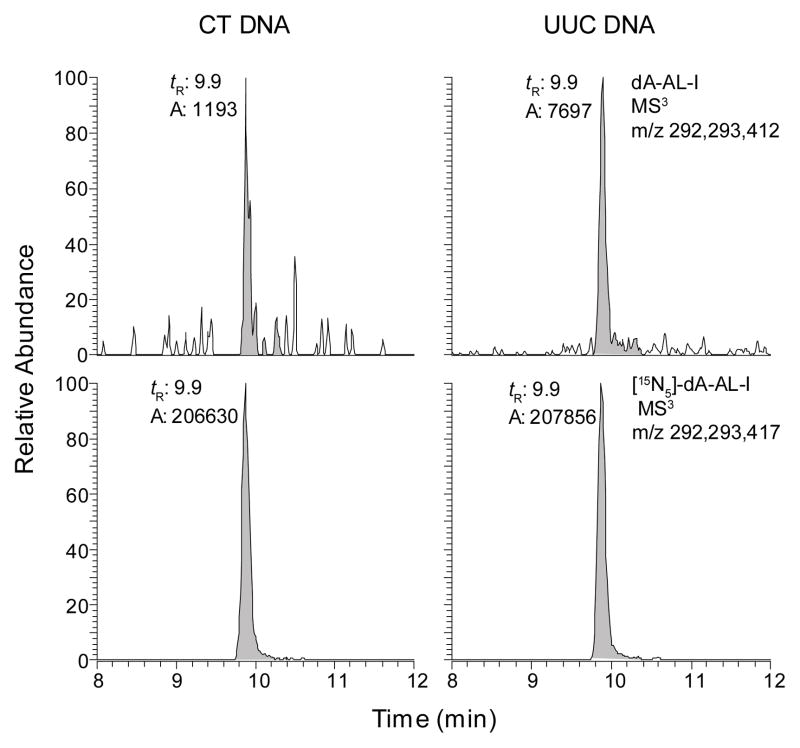
DNA obtained from two UUC tissues also contained the dA-AL-I adduct at levels more than 100-fold lower than that in the renal cortex of same subject (Figure 7). In contrast to these data, obtained by UPLC-ESI/MS3, the dA-AL-I adduct could not be detected in UUC tissue of the same patients by 32P-postlabeling/PAGE (data not shown).
Figure 7.
The mass chromatograms of dA-AL-I adduct in unmodified CT DNA and DNA from human UUC tissue. Human DNA (5 μg) was diluted with CT DNA (15 μg). The dA-AL-I adduct was estimated at 0.9 adducts per 108 DNA bases in the undiluted sample.
In Table 3, we summarize our estimates of AL-DNA adduct levels in human renal cortical and UUC tissue, as measured by UPLC-ESI/MS3 and 32P-postlabeling/PAGE. dG-AL adducts were not detected by either method.
Table 3.
dA-AL-I Adducts in Human Kidney: UPLC-ESI/MS3 versus 32P-Postlabeling Measurements
| Country | Age | Gender | Tissue | UPLC-ESI-MS/MS3 dA-AL-I adducts/108 bases | UPLC-ESI-MS/MS3 dA-AL-Il adducts/108 bases | 32P-Postlabeling dA-AL adducts/108 bases |
|---|---|---|---|---|---|---|
| Taiwan | 74 | Female | Cortex | 96.8 | 0.4 | 234 |
| Tumor | 1.2 | N.D. | N.D. | |||
| Taiwan | 60 | Male | Cortex | 388 | 1.0 | 192 |
| Tumor | 0.9 | N.D. | N.D. | |||
| Taiwan | 51 | Male | Cortex | 142 | 0.7 | 161 |
| Tumor | N.D. | N.D. | N.D. | |||
| Taiwan | 79 | Female | Cortex | 8.7 | N.D | 3.0 |
| Taiwan | 44 | Male | Cortex | 44.8 | N.D. | 3.9 |
| Taiwan | 63 | Male | Cortex | 170 | N.D | 73.8 |
| Tumor | N.D. | N.D. | N.A. | |||
| Taiwan | 52 | Female | Cortex | 28.0 | N.D. | 3.0 |
| Taiwan | 58 | Male | Cortex | 6.8 | N.D. | 7.8 |
| Tumor | N.D. | N.D. | N.A. | |||
| Taiwan | 67 | Male | Cortex | 9.6 | N.D. | 10.3 |
| Tumor | N.D. | N.D. | N.A. | |||
| Taiwan | 61 | Male | Cortex | 3.0 | N.D. | 7.5 |
| Tumor | N.D. | N.D. | N.A. |
Not detected (N.D.) = < 3 adducts per 109 bases by UPLC-ESI/MS3 or by 32P-Postlabeling/PAGE
Not analyzed (N.A)
Discussion
The goal of our study was to establish a rapid and quantitative method to measure AL-DNA adducts, employing UPLC-ESI/MSn, for implementation in human biomonitoring studies. DNA adducts represent internal dosimeters to measure exposure to environmental, dietary, and endogenous genotoxicants.31 Detection of DNA adducts has been used in molecular epidemiology studies to identify individuals at elevated risk to cancer due to high exposures to genotoxicants and may provide clues to the origin of human cancers for which an environmental cause is suspected.32,33 For the past 25 years, various modifications of the original 32P-postlabeling method have served as a mainstay for measuring DNA adducts, including AL-DNA adducts,4,14–16,34 in experimental animals and humans.19,21,35 Post-labeling methods are highly sensitive but suffer from several drawbacks. For example, the labeling efficiencies of DNA adducts are highly variable,36 which leads to tenuous estimates of adduct levels; a complex profile of poorly defined 32P-labeled lesions are often observed in human DNA samples;21 the identities of the carcinogen adducts are equivocal; and the technique is not a high through-put method. There is a critical need to establish rapid, yet specific methods of adduct detection which, at the same time can provide spectral data to corroborate the identities of the DNA lesions in humans. Currently, mass spectrometry is the sole method that can be employed in molecular epidemiology studies to quantify DNA adducts, while providing spectral data to corroborate the identity of the lesion.24
Only a few studies have directly compared estimates of DNA adducts obtained by 32P-postlabeling and quantitative LC/MS methods. We have shown previously that 32P-postlabeling underestimates the levels of dG-C8 adducts of bulky heterocyclic aromatic amines, when compared to LC/MS measurements. The discrepancy has been ascribed to incomplete 32P-labeling of the adducts.26,37 The 32P-postlabeling method also was reported to underestimate benzo[a]pyrene- and 4-aminobiphenyl-derived DNA adducts in comparison to quantitative LC/MS methods,24,38 although a third study reported good concordance in the estimates of a specific benzo[a]pyrene-DNA adduct by 32P-postlabeling and LC/MS.39
Our inter-laboratory comparison of AL-DNA measurements by UPLC-ESI/MSn and by 32P-postlabeling/PAGE techniques, using rodent or human DNA specimens, show satisfactory concordance (Tables 2 and 3). The estimates of dA-AL-I adducts, by these two analytical methods, are within about 2-fold of each other for eight out of the ten human biospecimens (Table 3). However, the levels of dA-AL-I in two human DNA samples, as determined by 32P-postlabeling/PAGE, are as much as 10-fold lower than the amounts measured by UPLC-ESI/MS3, suggesting that components of the DNA digest matrix of some samples decreased the efficiency of the polynucleotide kinase used to postlabel the AL-DNA adducts.
Both UPLC-ESI/MS3 and the 32P-postlabeling/PAGE methods are highly sensitive methods for the detection of dA-AL-I (Figures 4 and 5). The LOQ value for the dA-AL-I adduct is ~0.3 adducts per 108 DNA bases for UPLC-ESI/MS3, using 10 μg DNA, and ~0.3 adducts per 108 for 32P-postlabeling/PAGE, using 20 μg DNA. In this study, matching pairs of UUC and renal cortex samples were obtained from several subjects. The dA-AL-I adduct was detected by UPLC-ESI/MS3 in two UUC samples, albeit at levels more than 100-fold lower than adduct levels in the renal cortex. The low levels of AL-DNA adducts in UUC may be attributed, in part, to cell proliferation, which is expected to dilute adduct levels in growing tumors. However, the 32P-postlabeling/PAGE technique failed to detect any dA-AL-I in UUC, signifying that UPLC-ESI/MS3 is a superior method for the detection of AL-DNA adducts, particularly at trace levels. Importantly, our data show clearly that biomonitoring of AL-DNA adducts in human populations can be conducted by UPLC-ESI/MSn.
Historically, triple-stage quadrupole mass spectrometric (TSQ MS) instruments have been more commonly used than linear or circular ion trap-mass spectrometers (LIT MS) for the quantitation of DNA adducts.24,40 TSQ MS instruments are highly robust and provide excellent sensitivity and precision, when monitoring specific transitions of molecules under collision-induced dissociation conditions with selected reaction monitoring (SRM). However, the optimal collision-induced dissociation conditions employed with the TSQ MS in the SRM mode to measure DNA adducts usually produces a single transition: the loss of deoxyribose from the DNA adduct ([M+H]+ → [M+H-116]+).24,40 Thus, the criteria used for DNA adduct identification with TSQ MS instruments, are generally limited to the monitoring of this single transition combined with the characteristic tR of the adduct. A major advantage of the LIT MS over TSQ MS instruments is the capacity of the former to acquire product-ion spectra of the aglycone adducts [BH2]+ at trace levels due to its rapid scanning capability and the high ion transmission efficiency for many DNA adducts at the MS3 scan stage.26 Full product-ion scan spectra of DNA adducts can be acquired with TSQ MS instruments; however, this scan mode is accompanied by a significant loss in instrument sensitivity and cannot be used routinely for the analysis of DNA adducts at trace levels.24,40 The product-ion spectra obtained by LIT MS at the MS3 scan stage provides extensive mass fragmentation of the aglycone adduct, and the spectral data can be used to corroborate the identities of the adducts.26 We have employed ESI/MS3 to identify and quantify various carcinogen DNA adducts found in animal or human tissues and fluids.25,26,41,42 The powerful scanning capabilities of the LIT MS in acquiring high-quality, full product-ion spectra of DNA adducts formed at trace levels, as shown in this current study on AL-DNA adducts, represents a significant advance in using DNA adducts for human biomonitoring studies.
The performance of the analytical UPLC-ESI/MSn method in measuring AL-DNA adducts is influenced by the variation in MS instrument parameters, and also by any variation in the efficiency of enzyme hydrolysis of AL-modified DNA or the efficacy of the online solid-phase enrichment procedure. For improved precision, we performed quadruplicate measurements for these samples, rather than conducting a larger number of replicates, as our goal was to measure AL-DNA adducts in human tissues, for which the limited quantities of DNA available (generally 10 – 30 μg) preclude more than two replicate measurements. The performance of the analytical method reported here is representative of “real world” adduct analysis. The average within-a-day (CV ≤10%) and between-days (CV ≤15%) precision values, approach those obtained with TSQ MS instruments for DNA adduct measurements (Table 1).26,43–45
The level of dA-AL-I adducts formed in the renal cortex of certain patients with UUC were exceptionally high, being present at levels > 1 adduct per 107 bases. This level of adduct modification is far greater than that formed during environmental exposure to hazards such as tobacco and dietary carcinogens, or by endogenous electrophiles.21,24,46–48 The quantity of AAs ingested, over time, by individuals in our study are not known. Moreover, the amount of AA-I in Chinese medicinal herbs depends on the plant species. In some herbs, the estimates for AA-I have been reported to range from 17 – 8,800 μg/g, whereas the levels of AA-II are usually two to 5-fold lower.49,50 The failure of global genomic repair to excise dA-AL adducts51 explains their persistence in the human renal cortex for many years after the ingestion of Aristolochia herbs has ceased.4,15,16 As a result, AL-DNA adducts serve as a robust and reliable biomarker of exposure to AAs.4,5,6,15
Despite the occurrence of AA-II at appreciable levels in Aristolochia herbs,49 the dA-AL-I adduct is found in the renal cortex at levels about 100-fold higher than that of dA-AL-II. We observed a similar ratio of dA-AL-I to dA-AL-II adducts in the renal cortex in our study conducted in Taiwan,10 in a patient in the United States who developed end-stage renal failure after using an herbal remedy containing AA,4 and in patients with AAN/UUC in Balkan countries.5 Similar findings were obtained using the 32P-postlabeling method with renal cortex tissue samples from Belgian women who developed AAN after following a slimming regimen that included Chinese herbs.15,16 These differences may be attributed to differences in metabolism of AA-I and AA-II. The presence of the dA-AL-I adduct, in conjunction with its signature TP53 mutations, underscores the importance of dA-AL-I adducts in the carcinogenicity of AAs.4,5 The high levels of dA-AL-I in human renal cortex of subjects with UUC from different parts of the world,4,5,15 reinforces the notion that AA-I is a causative agent of AAN and UUC.
In summary, online solid-phase enrichment coupled with UPLC-ESI/MS3 is a powerful method that can be employed to measure AL-DNA adducts in human tissues, providing a sensitive and specific method for establishing exposure to aristolochic acids.
Supplementary Material
Acknowledgments
Funding
This research was supported by grants R01ES019564 (to RJT) and NIEHS ES04068 (to APG) from the National Institute of Environmental Health Sciences; by a grant PTH9807 (CHC) from Taoyuan General Hospital, and a grant DOH100-TD-C-111-001 (YSP) from the Department of Health, Taiwan. TR was the recipient of a Zickler Translational Research Scholar Award from the Zickler Family Foundation.
Abbreviations
- AL
aristolactam
- AA
aristolochic acids
- AAN
aristolochic acid nephropathy
- CV
coefficient of variation
- CT DNA
calf thymus DNA
- BEN
Balkan endemic nephropathy
- LOD
limit of detection
- LOQ
limit of quantitation
- LC-ESI/MSn
liquid chromatography-electrospray ionization/multistage mass spectrometry
- LIT MS
linear ion-trap mass spectrometry
- LC/MS
liquid chromatography/mass spectrometry
- TSQ
triple-stage quadrupole mass spectrometry
- UPLC
ultra-performance liquid chromatography
- UUC
upper tract urothelial carcinoma
- AA-I
8-methoxy-6-nitrophenanthro-[3,4-d]-1,3-dioxolo-5-carboxylic acid
- AA-II
6-nitrophenanthro-[3,4-d]-1,3-dioxolo-5-carboxylic acid
- dA-AL-I
7-(deoxyadenosin-N6-yl) aristolactam I
- dG-AL-I
7-(deoxyguanosin-N2-yl) aristolactam I
- dA-AL-II
7-(deoxyadenosin-N6-yl) aristolactam II
- dG-AL-II
7-(deoxyguanosin-N2-yl) aristolactam II
Footnotes
Supporting Information: Additional information is available as noted in text (Figures S1 – S7). This material is available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.Dawson DR. Birthwort: a study of the progress of medical botany through twenty-two centuries. Pharmaceutial J Pharmacist. 1927;396–397:427–430. [Google Scholar]
- 2.Grollman AP, Scarborough J, Jelaković B. Aristolochic acid nephropathy: an environmental and iatrogenic disease. In: Fishbein J, editor. Advances in Molecular Toxicology. Vol. 3. Elsevier; Amsterdam: 2009. pp. 211–222. [Google Scholar]
- 3.Vanherweghem JL, Debelle FD, Muniz Martinez MC, Nortier JL. Aristolochic acid nephropathy after Chinese herb remedies. In: De Broe ME, Porter GA, Bennett WM, Verpooten GA, editors. Clinical Nephrotoxins. 2. Kluwer; Dordrecht: 2003. pp. 579–603. [Google Scholar]
- 4.Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, Jakovina K, Brdar B, Slade N, Turesky RJ, Goodenough AK, Rieger R, Vukelic M, Jelakovic B. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci U S A. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jelaković B, Karanović S, Vuković-Lela I, Miller F, Edwards KL, Nikolić J, Tomić K, Slade N, Brdar B, Turesky RJ, Stipancić Z, Dittrich D, Grollman AP, Dickman KG. Aristolactam-DNA adducts are a biomarker of environmental exposure to aristolochic acid. Kidney Int. 2011;81:559–567. doi: 10.1038/ki.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nortier JL, Martinez MC, Schmeiser HH, Arlt VM, Bieler CA, Petein M, Depierreux MF, De PL, Abramowicz D, Vereerstraeten P, Vanherweghem JL. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi) N Engl J Med. 2000;342:1686–1692. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- 7.Petronić V. Tumors of the upper urothelium and endemic nephropathy. In: Radovanović Z, Sindić M, Polenaković M, Djukanović L, Petronić V, Petronić V, editors. Endemic Nephropathy Institute for Textbook Publishing. Institute for Text Book Publishing; Belgrade: 2000. pp. 350–439. [Google Scholar]
- 8.Debelle FD, Vanherweghem JL, Nortier JL. Aristolochic acid nephropathy: a worldwide problem. Kidney Int. 2008;74:158–169. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- 9.Lai MN, Wang SM, Chen PC, Chen YY, Wang JD. Population-based case-control study of Chinese herbal products containing aristolochic acid and urinary tract cancer risk. J Natl Cancer Inst. 2010;102:179–186. doi: 10.1093/jnci/djp467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C-H, Dickman KG, Moriya M, Zavadil J, Sidorenko V, Edwards KG, Gnatenko D, Wu L, Turesky RJ, Pu Y-S, Grollman AP. Aristolochic acid-associated urothelial carcinoma in Taiwan is a harbinger of a global disease. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1119920109. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moriya M, Slade N, Brdar B, Medverec Z, Tomic K, Jelakovic B, Wu L, Truong S, Fernandes A, Grollman AP. TP53 Mutational signature for aristolochic acid: an environmental carcinogen. Int J Cancer. 2011;129:1532–1536. doi: 10.1002/ijc.26077. [DOI] [PubMed] [Google Scholar]
- 12.Pfau W, Schmeiser HH, Wiessler M. Aristolochic acid binds covalently to the exocyclic amino group of purine nucleotides in DNA. Carcinogenesis. 1990;11:313–319. doi: 10.1093/carcin/11.2.313. [DOI] [PubMed] [Google Scholar]
- 13.Arlt VM, Stiborova M, Schmeiser HH. Aristolochic acid as a probable human cancer hazard in herbal remedies: a review. Mutagenesis. 2002;17:265–277. doi: 10.1093/mutage/17.4.265. [DOI] [PubMed] [Google Scholar]
- 14.Dong H, Suzuki N, Torres MC, Bonala RR, Johnson F, Grollman AP, Shibutani S. Quantitative determination of aristolochic acid-derived DNA adducts in rats using 32P-postlabeling/polyacrylamide gel electrophoresis analysis. Drug Metab Dispos. 2006;34:1122–1127. doi: 10.1124/dmd.105.008706. [DOI] [PubMed] [Google Scholar]
- 15.Bieler CA, Stiborova M, Wiessler M, Cosyns JP, van Ypersele De SC, Schmeiser HH. 32P-post-labelling analysis of DNA adducts formed by aristolochic acid in tissues from patients with Chinese herbs nephropathy. Carcinogenesis. 1997;18:1063–1067. doi: 10.1093/carcin/18.5.1063. [DOI] [PubMed] [Google Scholar]
- 16.Schmeiser HH, Bieler CA, Wiessler M, van Ypersele De SC, Cosyns JP. Detection of DNA adducts formed by aristolochic acid in renal tissue from patients with Chinese herbs nephropathy. Cancer Res. 1996;56:2025–2028. [PubMed] [Google Scholar]
- 17.Nedelko T, Arlt VM, Phillips DH, Hollstein M. TP53 mutation signature supports involvement of aristolochic acid in the aetiology of endemic nephropathy-associated tumours. Int J Cancer. 2009;124:987–990. doi: 10.1002/ijc.24006. [DOI] [PubMed] [Google Scholar]
- 18.Attaluri S, Bonala RR, Yang IY, Lukin MA, Wen Y, Grollman AP, Moriya M, Iden CR, Johnson F. DNA adducts of aristolochic acid II: total synthesis and site-specific mutagenesis studies in mammalian cells. Nucleic Acids Res. 2010;38:339–352. doi: 10.1093/nar/gkp815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RC. 32P-postlabelling analysis of bulky aromatic adducts. IARC Sci Publ. 1993;124:11–23. [PubMed] [Google Scholar]
- 20.Shibutani S, Kim SY, Suzuki N. 32P-Postlabeling DNA damage assay: PAGE, TLC and HPLC. In: Henderson DS, editor. DNA Repair Protocols: Eukaryotic systems, second edition. Humana Press Inc; Totawa, NJ: 2005. pp. 307–321. [Google Scholar]
- 21.Phillips DH. Smoking-related DNA and protein adducts in human tissues. Carcinogenesis. 2002;23:1979–2004. doi: 10.1093/carcin/23.12.1979. [DOI] [PubMed] [Google Scholar]
- 22.Gu D, Turesky RJ, Tao Y, Langouet SA, Nauwelaers GC, Yuan JM, Yee D, Yu MC. DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 4-aminobiphenyl are infrequently detected in human mammary tissue by liquid chromatography/tandem mass spectrometry. Carcinogenesis. 2012;33:124–130. doi: 10.1093/carcin/bgr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koc H, Swenberg JA. Applications of mass spectrometry for quantitation of DNA adducts. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;778:323–343. doi: 10.1016/s1570-0232(02)00135-6. [DOI] [PubMed] [Google Scholar]
- 24.Singh R, Farmer PB. Liquid chromatography-electrospray ionization-mass spectrometry: the future of DNA adduct detection. Carcinogenesis. 2006;27:178–196. doi: 10.1093/carcin/bgi260. [DOI] [PubMed] [Google Scholar]
- 25.Bessette EE, Spivack SD, Goodenough AK, Wang T, Pinto S, Kadlubar FF, Turesky RJ. Identification of carcinogen DNA adducts in human saliva by linear quadrupole ion trap/multistage tandem mass spectrometry. Chem Res Toxicol. 2010;23:1234–1244. doi: 10.1021/tx100098f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodenough AK, Schut HA, Turesky RJ. Novel LC-ESI/MS/MSn method for the characterization and quantification of 2′-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem Res Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan W, Yue H, Poon WT, Chan YW, Schmitz OJ, Kwong DW, Wong RN, Cai Z. Quantification of aristolochic acid-derived DNA adducts in rat kidney and liver by using liquid chromatography-electrospray ionization mass spectrometry. Mutat Res. 2008;646:17–24. doi: 10.1016/j.mrfmmm.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 28.Chan W, Zheng Y, Cai Z. Liquid chromatography-tandem mass spectrometry analysis of the DNA adducts of aristolochic acids. J Am Soc Mass Spectrom. 2007;18:642–650. doi: 10.1016/j.jasms.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Schmeiser HH, Frei E, Wiessler M, Stiborova M. Comparison of DNA adduct formation by aristolochic acids in various in vitro activation systems by 32P-post-labelling: evidence for reductive activation by peroxidases. Carcinogenesis. 1997;18:1055–1062. doi: 10.1093/carcin/18.5.1055. [DOI] [PubMed] [Google Scholar]
- 30.MacDougall D, Amore FJ, Cox GV, Crosby DG, Estes FL, Freeman DH, Gibbs WE, Gordon GE, Keith LH, Lal J, Langner RR, McClelland NI, Phillips WF, Pojasek RB, Sievers RE. Guidelines for data acquistion and data quality evaluation in environmental chemistry. Anal Chem. 1980;52:2242–2249. [Google Scholar]
- 31.Harris CC, Weston A, Willey JC, Trivers GE, Mann DL. Biochemical and molecular epidemiology of human cancer: indicators of carcinogen exposure, DNA damage, and genetic predisposition. Environ Health Perspect. 1987;75:109–119. doi: 10.1289/ehp.8775109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarabek AM, Pottenger LH, Andrews LS, Casciano D, Embry MR, Kim JH, Preston RJ, Reddy MV, Schoeny R, Shuker D, Skare J, Swenberg J, Williams GM, Zeiger E. Creating context for the use of DNA adduct data in cancer risk assessment: I. Data organization. Crit Rev Toxicol. 2009;39:659–678. doi: 10.1080/10408440903164155. [DOI] [PubMed] [Google Scholar]
- 33.Phillips DH. DNA adducts as markers of exposure and risk. Mutat Res. 2005;577:284–292. doi: 10.1016/j.mrfmmm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Pfau W, Schmeiser HH, Wiessler M. 32P-postlabelling analysis of the DNA adducts formed by aristolochic acid I and II. Carcinogenesis. 1990;11:1627–1633. doi: 10.1093/carcin/11.9.1627. [DOI] [PubMed] [Google Scholar]
- 35.Randerath K, Randerath E, Agrawal HP, Gupta RC, Schurda ME, Reddy MV. Postlabeling methods for carcinogen-DNA adduct analysis. Environ Health Perspect. 1985;62:57–65. doi: 10.1289/ehp.856257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips DH, Farmer PB, Beland FA, Nath RG, Poirier MC, Reddy MV, Turteltaub KW. Methods of DNA adduct determination and their application to testing compounds for genotoxicity. Environ Mol Mutagen. 2000;35:222–233. doi: 10.1002/(sici)1098-2280(2000)35:3<222::aid-em9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 37.Soglia JR, Turesky RJ, Paehler A, Vouros P. Quantification of the heterocyclic aromatic amine DNA adduct N-(deoxyguanosin-8-yl)-2-amino-3-methylimidazo[4,5-f]quinoline in livers of rats using capillary liquid chromatography/microelectrospray mass spectrometry: a dose-response study. Anal Chem. 2001;73:2819–2827. doi: 10.1021/ac010218j. [DOI] [PubMed] [Google Scholar]
- 38.Beland FA, Doerge DR, Churchwell MI, Poirier MC, Schoket B, Marques MM. Synthesis, characterization, and quantitation of a 4-aminobiphenyl-DNA adduct standard. Chem Res Toxicol. 1999;12:68–77. doi: 10.1021/tx980172y. [DOI] [PubMed] [Google Scholar]
- 39.Beland FA, Churchwell MI, Von Tungeln LS, Chen S, Fu PP, Culp SJ, Schoket B, Gyorffy E, Minarovits J, Poirier MC, Bowman ED, Weston A, Doerge DR. High-performance liquid chromatography electrospray ionization tandem mass spectrometry for the detection and quantitation of benzo[a]pyrene-DNA adducts. Chem Res Toxicol. 2005;18:1306–1315. doi: 10.1021/tx050068y. [DOI] [PubMed] [Google Scholar]
- 40.Turesky RJ, Vouros P. Formation and analysis of heterocyclic aromatic amine-DNA adducts in vitro and in vivo. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:155–166. doi: 10.1016/j.jchromb.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 41.Bessette EE, Goodenough AK, Langouet S, Yasa I, Kozekov ID, Spivack SD, Turesky RJ. Screening for DNA adducts by data-dependent constant neutral loss-triple stage mass spectrometry with a linear quadrupole ion trap mass spectrometer. Anal Chem. 2009;81:809–819. doi: 10.1021/ac802096p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turesky RJ, Bendaly J, Yasa I, Doll MA, Hein DW. The impact of NAT2 acetylator genotype on mutagenesis and DNA adducts from 2-amino-9H-pyrido[2,3-b]indole. Chem Res Toxicol. 2009;22:726–733. doi: 10.1021/tx800473w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S, Villalta PW, Wang M, Hecht SS. Analysis of crotonaldehyde- and acetaldehyde-derived 1,n(2)-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography electrospray ionization tandem mass spectrometry. Chem Res Toxicol. 2006;19:1386–1392. doi: 10.1021/tx060154d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang M, Yu N, Chen L, Villalta PW, Hochalter JB, Hecht SS. Identification of an acetaldehyde adduct in human liver DNA and quantitation as N2-ethyldeoxyguanosine. Chem Res Toxicol. 2006;19:319–324. doi: 10.1021/tx0502948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S, Villalta PW, Wang M, Hecht SS. Detection and quantitation of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem Res Toxicol. 2007;20:565–571. doi: 10.1021/tx700023z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medeiros MH. Exocyclic DNA adducts as biomarkers of lipid oxidation and predictors of disease. Challenges in developing sensitive and specific methods for clinical studies. Chem Res Toxicol. 2009;22:419–425. doi: 10.1021/tx800367d. [DOI] [PubMed] [Google Scholar]
- 47.Ricicki EM, Soglia JR, Teitel C, Kane R, Kadlubar F, Vouros P. Detection and quantification of N-(deoxyguanosin-8-yl)-4-aminobiphenyl adducts in human pancreas tissue using capillary liquid chromatography-microelectrospray mass spectrometry. Chem Res Toxicol. 2005;18:692–699. doi: 10.1021/tx049692l. [DOI] [PubMed] [Google Scholar]
- 48.Zayas B, Stillwell SW, Wishnok JS, Trudel LJ, Skipper P, Yu MC, Tannenbaum SR, Wogan GN. Detection and quantification of 4-ABP adducts in DNA from bladder cancer patients. Carcinogenesis. 2007;28:342–349. doi: 10.1093/carcin/bgl142. [DOI] [PubMed] [Google Scholar]
- 49.Chan W, Hui KM, Poon WT, Lee KC, Cai Z. Differentiation of herbs linked to “Chinese herb nephropathy” from the liquid chromatographic determination of aristolochic acids. Anal Chim Acta. 2006;576:112–116. doi: 10.1016/j.aca.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Aristolochic Acids, In National Toxicology Program Report on Carcinogens. 12. US Dept Health and Human Services, Public Health Service, Research Triangle Park; North Carolina: 2008. [Google Scholar]
- 51.Sidorenko VS, Yeo JE, Bonala RR, Johnson F, Scharer OD, Grollman AP. Nucleic Acids Res. 2011. Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactam-DNA adducts. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.