Abstract
Infections with Bartonella spp. have been recognized as emerging zoonotic diseases in humans. Large knowledge gaps exist, however, relating to reservoirs, vectors, and transmission of these bacteria. We describe identification by culture, PCR, and housekeeping gene sequencing of Bartonella spp. in fed, wingless deer keds (Lipoptena cervi), deer ked pupae, and blood samples collected from moose, Alces alces, sampled within the deer ked distribution range in Norway. Direct sequencing from moose blood sampled in a deer ked-free area also indicated Bartonella infection but at a much lower prevalence. The sequencing data suggested the presence of mixed infections involving two species of Bartonella within the deer ked range, while moose outside the range appeared to be infected with a single species. Bartonella were not detected or cultured from unfed winged deer keds. The results may indicate that long-term bacteremia in the moose represents a reservoir of infection and that L. cervi acts as a vector for the spread of infection of Bartonella spp. Further research is needed to evaluate the role of L. cervi in the transmission of Bartonella to animals and humans and the possible pathogenicity of these bacteria for humans and animals.
INTRODUCTION
The deer ked (Lipoptena cervi) is a blood-sucking ectoparasitic fly prevalent in Europe and Asia which has been introduced to North America (1). In the Nordic countries, it was until recently restricted to Denmark and the southernmost part of Sweden, but during the last few decades L. cervi has shown a remarkable increase in abundance and is currently rapidly expanding its range northward in Finland, Sweden, and Norway (2). Its predominant hosts are cervids, but the insect may attack a wide range of animals (3). They are generally considered a nuisance due to their habit of swarming in large numbers and landing on humans, whom they not infrequently bite. While several authors have reported the presence of persistent pruritic papules on humans bitten by the insect (4–6), their role as vector of pathogens/disease has been poorly elucidated.
Over the years, several insect vectors and mammal hosts have been associated with Bartonella sp. infections (See Tsai and coauthors for a review [7]). Dehio et al. isolated Bartonella schoenbuchensis from the blood of roe deer (Capreolus capreolus) (8). Subsequently, the same workers also found that deer keds collected from individual roe deer or red deer (Cervus elaphus) were either negative or positive for this bacterium, indicating that the keds became infected when feeding on bacteremic individuals (9). The midgut of infected insects contains large numbers of B. schoenbuchensis bacteria (9). In a concurrent study (10), B. schoenbuchensis was detected in deer ked collected from roe deer in France, and the flies were again suggested as a vector. Since then, Bartonella DNA of closely related species has been reported from ticks (Ixodes ricinus) parasitizing roe deer in Poland (11), southern deer keds (Lipoptena mazamae) from white-tailed deer (Odocoileus virginianus) in Georgia and South Carolina (12), deer ked from white-tailed deer in Massachusetts (13), and forest flies (Hippobosca equina) and blood from rusa deer (Cervus timorensis russa) in New Caledonia (14). A single report has also described B. schoenbuchensis in blood from a French cow (15).
Several bartonellae are regarded as potential or established emerging zoonotic infections (see Chomel and Kasten for a recent review [16]). Although deer keds have not been directly associated with human bartonellosis, Dehio and coauthors (9) suggested that a risk of transmission of B. schoenbuchensis to humans exists through the bite of the insect and that Bartonella infection could be the cause of deer ked dermatitis.
The main objective of this study was to determine whether deer keds could be a candidate vector of Bartonella infection in moose (Alces alces) in Norway. Data from culture, PCR screening, and sequence analysis of Bartonella DNA from moose blood and deer keds at different developmental stages is presented.
MATERIALS AND METHODS
Collection of material.
A total of 41 moose (Alces alces) were sampled within the distribution range of deer ked in southeastern Norway. The studied material comprised 11 carcasses submitted for necropsy in conjunction with an outbreak of deer ked-associated alopecia (thoroughly described by Madslien and coauthors [17]) and free-ranging, presumably healthy moose chemically immobilized in association with radio-collaring. Wingless (fed) deer ked imagines and pupae were collected from the carcasses. Samples were taken from the liver, spleen, and (if available) blood at necropsy, while blood samples were taken from all live animals (Table 1). In addition, blood samples taken from 28 free-ranging, presumably healthy moose immobilized in association with radio-collaring in the Stor-Elvdal area (approximately 38 km north of the recognized deer ked distribution front) were included in the analyses. The collection also included winged deer keds (i.e., imagines that have not yet fed), caught in two localities: (i) Østfold, within a well-established deer ked area, and (ii) Akershus, at the deer ked expansion front (see Välimäki [18] for a more thorough description). The deer keds were captured as they settled on a study person walking slowly through a forested area. Blood samples from live animals were collected into EDTA plastic tubes (Becton, Dickinson, Franklin Lakes, NJ) and frozen at −80°C together with the other sampled materials on arrival in the laboratory.
Table 1.
Screening for Bartonella from moose and deer ked samples by culturing and PCR
| Sampling site | Sample type (no. of moose tested) | No. of samples cultivated (in pools of n) | No. of samples DNA extracted (in pools of n) | No. (%) of positive samples |
|
|---|---|---|---|---|---|
| Culture | PCRb | ||||
| Inside deer ked zone | Moose carcasses (11) | ||||
| Liver | 11 | 11 | 0 | 0 | |
| Spleen | 10 | 10 | 0 | 0 | |
| Blood | 7 | 7 | 0 | 0 | |
| Wingless fed deer ked imagines | 35 (3–4) | 50 (5) | 1 (10) | 10 (100) | |
| Deer ked pupaea | 20 (2–4) | 50 (5) | 0 | 5 (50) | |
| Blood from live moose (30) | 29 | 29 | 17 (57) | 21 (70) | |
| Winged unfed deer ked imagines | 44 (7–8) | 50 (5) | 0 | 0 | |
| Outside deer ked zone | Blood from live moose (28) | 28 | 28 | 0 | 10 (37) |
Sampled from only eight carcasses.
Both seminested and qPCR detection.
Bacterial culture.
Culture was attempted from samples obtained from necropsied moose carcasses, live moose, and winged unfed deer ked imagines as mentioned above by using Columbia agar medium with 5% horse blood (CA) and incubated for a total of 6 weeks with 5% CO2 at 37°C. Deer ked imagines and pupae were previously surface sterilized in 70% ethanol. While culture was attempted on all tissue samples and imagines from all carcasses, pupae from only eight carcasses were investigated in this way due to scarcity of material. Presumptive Bartonella isolates were identified by colony morphology, subsequently confirmed by DNA sequencing. Pure cultures were obtained by successive streaking on agar plates under the same culturing conditions as those described above.
DNA extraction.
Prior to DNA extraction, deer ked imagines and pupae were surface disinfected by immersion in 0.5% hypochlorite (5 min) and 70% ethanol (5 min), followed by three rinses in sterile water. These were transferred into a sterile 2-ml microcentrifuge tube containing one 3-mm tungsten carbide bead (Qiagen) and 0.5 g 0.1-mm glass beads (Biospec Products, Bartlesville, OK). To this was added 180 μl QIAamp DNA minikit tissue lysis buffer (Qiagen, Valencia, CA), and the flies were mechanically disrupted in a mini-bead beater (Biospec) for 2 min, with the speed set to “homogenize.” The homogenates were held at −20°C for 5 min to reduce foaming and centrifuged at 10,000 × g for 1 min to pellet debris. Proteinase K (1 mg/ml) was added to the supernatant and incubated overnight at 55°C. Thereafter, total genomic DNA was purified using the Qiagen QIAamp DNA minikit (Qiagen) according to the manufacturer's protocol. DNA templates from tissues and blood samples were either obtained directly using the same kit as mentioned above or from cultured isolates by boiling cell suspensions in a phosphate buffer for 10 min at 95°C.
PCR detection of Bartonella DNA.
DNA extracts from the individual samples were subjected to three different PCR strategies for the detection of Bartonella. Quantitative PCR (qPCR) and seminested PCR were initially used to screen the samples for Bartonella DNA, based on previously described protocols (19, 20). Both assays detect a region in the gltA gene. For further analysis of positive samples, conventional PCR was used to produce amplicons from five housekeeping genes (gltA, rpoB, ftsZ, ribC, and groEL) as described previously (21). Primers for ribC and groEL were, however, redesigned to amplify a wider range of Bartonella species. All primers were purchased from Invitrogen, and their respective sequences are listed in Table 2. Amplifications were performed in 25-μl reaction mixtures containing 1× Taq buffer, 0.4 μM forward and reverse primers, 0.2 mM deoxynucleoside triphosphate (dNTP) mix, 1.5 mM MgCl2, 1 U Taq DNA polymerase, and 2 to 3 μl DNA template. Each PCR was carried out in a PTC-100 programmable thermal controller (MJ Research Inc., Watertown, MA) with the following thermal cycling conditions: initial denaturation cycle at 95°C for 3 min; followed by 35 cycles of amplification at 95°C for 60s s, 55 or 60°C (depending on the gene) for 60 s, and 72°C for 60 s; and a final extension at 72°C for 10 min. With the exception of groEL (annealing at 60°C), all other genes were amplified at annealing temperature of 55°C. PCR products were identified by electrophoresis. In all analyses, positive and negative controls were included within each PCR assay.
Table 2.
Primers and probe used in this study
| Gene | Forward sequence (5′→3′) | Reverse sequence (5′-→3′) | Probe sequence (5′→3′)d | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| gltA | GGGGACCAGCTCATGGTGG | AATGCAAAAAGAACAGTAAACA | NA | ∼350 | 22 |
| gltAa | GGGGACCAGCTCATGGTGG | CGTGGATCATAATTTTTATA | CCAAAACCCATAAGGCGGAAAGGATCATTT | ∼143 | 19 |
| gltAb | GTTATCCTATTGACCAA | CCAAAACCCATAAGGCG | NA | ∼685 | 20 |
| AACTCTTGCCGCTATGG | TATTCTTCACAAGGAAC | ∼401/387c | |||
| rpoB | GCACGATTYGCATCATCTTTTCC | CGCATTATGGTCGTATTTGTCC | NA | ∼333 | 21 |
| groEL | ATGGACAAAGTTGGCAATGAA | TTCCACCACCAGCAACAATA | NA | ∼720 | This study |
| ribC | TAACCGATATTGGTTGTGTTGAAG | TAAAGCTAGAAAGTCTGGCAACATAACG | NA | ∼588 | This study |
| ftsZ | CATATGGTTTTCATTACTGCYGGTATGG | TTCTTCGCGAATACGATTAGCAGCTTC | NA | ∼515 | 21 |
qPCR.
Seminested PCR.
Seminested PCR product.
Dually labeled oligonucleotide probe with 5′-6-carboxyfluorescein and 3′-black hole quencher 1. NA, not applicable.
Multilocus sequence analysis (MLSA) and phylogeny of Bartonella DNA.
PCR products of the five examined housekeeping genes were purified with the Nucleospin purification kit (Macherey-Nagel, Düren, Germany), according to the manufacturer's protocol and sequenced in both directions on an automatic DNA sequencer (ABI 3130 Genetic Analyzer; Applied Biosystems, Foster City, CA). Sequencing reactions were performed in a PTC-100 programmable thermal cycler using the amplicon target PCR primers at a concentration of 2.5 μM. Cycling conditions for the sequencing reactions were as described by Platt and coauthors (23). Raw chromatograms (both directions) were assembled, inspected visually for errors, and edited using Sequencher 4.5 (Gene Codes, Ann Arbor, MI). Consensus individual gene sequences aligned in ClustalX 2.1 (24) were compared to reference sequences obtained from the GenBank database.
Phylogenies for individual genes generated using a neighbor-joining algorithm with the Kimura 2 parameter model (1,000 bootstrap replicates) in PAUP 4.0b10 (25) provided congruous tree topologies, and so the sequences were concatenated and reanalyzed using a maximum likelihood algorithm in PhyML (26) with a GTR substitution model (estimated using jModelTest [27]) implemented through the University of Oslo bioportal (www.uio.no/bioportal/).
Nucleotide sequence accession numbers.
Sequences obtained during the present study have been submitted to GenBank under the following accession numbers: gltA, JN990623 to JN990630; rpoB, JN990603 to JN990615; groEL, JN990631 to JN990639; ribC, JN990640 to JN990650; and ftsZ, JN990616 to JN990622.
RESULTS
Bacterial culture.
Bartonella spp. (as identified by subsequent MLSA) were successfully cultured from a single pooled sample of 10 pools of wingless/fed deer ked imagines and from 17 of 29 blood samples from live moose within the deer ked distribution range. No Bartonella spp. were cultured from deer ked pupae, winged/unfed imagines, moose tissues or blood from carcasses, or blood samples from live moose outside the deer ked distribution range (Table 1). Culture from necropsy samples was generally severely compromised by nonspecific bacterial contamination. With the exception of a single carcass, deer ked imagines and pupae were identified on all animals sampled within the deer ked distribution range but not on animals immobilized in the ked-free area (Stor-Elvdal).
Prevalence of Bartonella infection.
On qPCR screening, positive signals were obtained from 8 of 10 pools of wingless/fed deer keds and 1 of 10 pools of pupae. The cycle threshold (CT) values, representing the number of genome copies in the sample, were in the range of 20 to 30, indicative of a heavy Bartonella presence. In addition, weak positive signals (CT > 30) were identified from 2 of 10 pools of wingless/fed imagines and 4 of 10 pools of pupae examined, while the winged/unfed flies were negative for Bartonella DNA (Table 1). Although some inhibition was apparent, all tissues from the carcasses were also negative. An overall prevalence of 87% was detected directly from blood and blood-cultured samples from 30 individual moose within the deer ked range. Of these, 17 were positive by qPCR, and the remaining were detected on seminested PCR amplification and by culture. Generally, qPCR gave weak signals from blood (CT > 36.9), and some culture-positive samples were negative by qPCR. Positive but generally weak PCR signals (CT > 36.4) were also obtained from 10 of 28 blood samples from live moose in the presumed ked-free area, resulting in 36% prevalence (Table 1). The identity of these positive samples was confirmed by sequencing.
Multilocus sequence analysis.
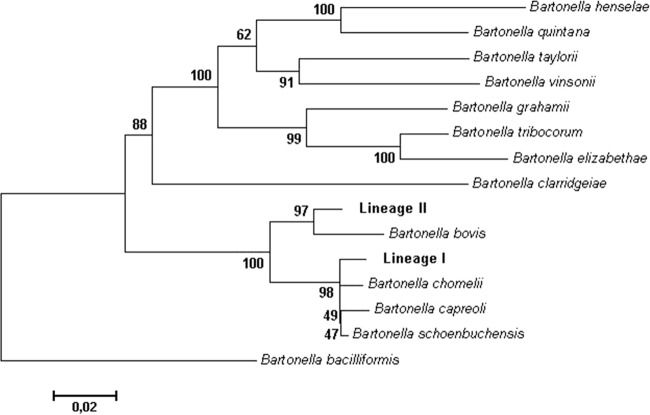
Good-quality sequences for all 5 genes were not obtained from all individual samples tested. However, a total of 16 (gltA), 17 (rpoB), 17(groEL), 18 (ribC), and 15 (ftsZ) sequences were retrieved from 7 wingless/fed imagine deer ked pools, 4 cultured bacterial isolates, and 10 moose blood extracts from within and outside the deer ked zone. Sequence chromatograms retrieved from some deer ked imagines and blood samples originating within the deer ked zone suggested the existence of a mixed infection of related Bartonella, as a small number of ambiguous bases were consistently identified in most genes, while all sequences from cultured isolates were identical with no ambiguous bases. Interestingly, most of the ambiguous bases observed in this study were at positions which are diagnostic for species identification in other ruminant-infecting Bartonella spp. (Table 3). Sequences obtained from moose blood sampled outside the deer ked zone displayed a very low level of ambiguity (Table 3). For phylogenetic placement, representative sequences obtained directly from blood and blood cultured isolates displaying no ambiguity in nucleotide sequence were used. The consensus maximum likelihood phylogeny based on concatenated sequences from all 5 studied genes resulted in two Bartonella lineages (Fig. 1; see also Fig. S1 in the supplemental material). Lineage I was confined within the deer ked zone and clustered closely with Bartonella chomelii, B. schoenbuchensis, and Bartonella capreoli, all infectious bacteria of ruminants within a single clade of Bartonella. Identity levels between examined samples and the type sequences of ruminant bartonellae deposited in GenBank were different for each of the 5 loci (see Table S1 in the supplemental material), and it proved difficult to subscribe the bacterial isolate concerned or other sequences generated to an individual Bartonella species. On the other hand, lineage II was identified both inside (in 10 of 26 infected moose) and outside (in 10 of 10 infected moose) the deer ked zone. Although the lineage II sequences were almost identical, sequences retrieved from samples originating outside the zone displayed very few ambiguous bases compared to those originating within the zone. As lineage II sequences displayed only limited identity to lineage I isolates/strains (approximately 95% at ftsZ, 97% at rpoB, 96% at gltA, 92% at ribC, and 97% at groEL) and to other sequences in GenBank (see Table S1), it should be considered a genetically distinct, previously undescribed clade of ruminant-infecting Bartonella.
Table 3.
Comparison of the sites with ambiguous basesa
| Gene | Position | Ambiguous base(s) (no. of samples) |
||||||
|---|---|---|---|---|---|---|---|---|
| Inside the ked zone | Outside the ked zone | B. chomelii | B. capreoli | B. schoenbuchensis | B. melophagi | B. bovis | ||
| ftsZ | 411 | A/G (2) | G | G | G | G | G | |
| 432 | A/G (2) | A | A | G | A | A | ||
| 453 | A | A/G (3) | A | A | A | A | ||
| 489 | A/G (5) | A | A | A | A | A | ||
| 492 | A | A/G (3) | A | A | A | A | ||
| 497 | G/T (1) | T | T | T | T | T | ||
| 624 | A/G (2) | A | G | G | G | A | ||
| gltA | 909 | T/C (4) | ND | C | T | C | C | T |
| 918 | T/C (5) | ND | C | C | C | C | T | |
| 936 | T/C (3) | ND | C | T | C | T | T | |
| 1077 | T/C (3) | ND | T | C | T | C | C | |
| groEL | 861 | A/G (4) | ND | A | A | G | ||
| ribC | 147 | T/C (3) | ND | C | C | C | C | G |
| 306 | T/C (2) | ND | T | T | T | T | T | |
| rpoB | 1833 | T/C (1) | T/C (2) | C | C | C | T | T |
| 1854 | G/T (3) | G/T (1) | G | G | G | A | G | |
| 1894 | A/G (4) | G | A | A | A | A | G | |
| 1902 | T/C (2) | T | T | T | T | T | T | |
| 1915 | A/G (9) | A/G (1) | G | G | G | G | G | |
| 1923 | A/G (3) | A | G | G | G | A | A | |
| 1974 | A or G | A/G (2) | G | G | G | G | A | |
| 2013 | A/G (7) | A | A | A | A | A | A | |
| 2019 | A/G (3) | A | A | A | A | A | A | |
Ambiguous bases within sequenced gene fragments of the examined samples and relevant nucleotides in the type sequences of ruminant-infecting bartonellae. The position in the gene is indicated by nucleotide number and with the number of samples in which they were found. Bold indicates the polymorphic sites of ruminant-infecting Bartonella spp., where the ambiguities in samples from moose occurred. ND, no data.
Fig 1.
Concatenated phylogenetic tree of Bartonella-type isolates and a representative isolate of this study, based on fragments of 3 genes, gltA, rpoB, and ribC (832 bp in total), generated using the maximum likelihood algorithm in PhyML with a GTR substitution model (1,000 replicates; bootstrap values indicated at the nodes).
DISCUSSION
The current report describes Bartonella infection in moose (Alces alces) and deer ked (Lipoptena cervi) feeding on this host. Our data indicated a higher prevalence of Bartonella DNA in moose within the deer ked zone than in animals outside the zone. Such variation may be due to levels of fly infestation, as there was no indication of deer ked, ticks, or other common cervid-parasitizing hippoboscids in the deer ked-free areas. The strong qPCR signals, equating to large numbers of Bartonella genomes in all but two pools of wingless/fed deer ked imagines, is consistent with the proliferation of Bartonella in the gut of this insect as described by Dehio and coworkers (9). The signal from moose blood was weak but positive, consistent with low numbers of circulating Bartonella-infected cells. Most interestingly, a strong signal was also obtained from a single pool of pupae and a weaker signal from a further four pools of pupae, suggesting the possibility of vertical transmission. Although this may have represented contamination of the pupae by bacteria from the genital tract of the female after pupal formation (hippoboscids produce fully developed pupae in the female reproductive tract), extensive hypochlorite and ethanol disinfection would have been expected to denature surviving bacterial DNA on the surface of the cuticle. The most likely explanation is, therefore, that bacteria are transferred from mother to larva in utero, as suggested by Zacharias (28).
Generally, species designation in Bartonella has been based on housekeeping gene sequence analysis. La Scola et al. (29) reported that gltA and rpoB were the most appropriate targets for species differentiation in the genus Bartonella. Cutoff values of ≤96.0% in gltA and ≤95.4% in rpoB gene sequences were proposed for the designation of novel Bartonella species. In the present study, we identified two Bartonella lineages, with lineage I showing high similarity to corresponding sequences from B. chomelii, B. schoenbuchensis, and B. capreoli, a clade of species primarily infecting ruminants (8, 30–32). The association between lineage I Bartonella infection in unwinged flies and moose exclusively within the deer ked range strongly suggests that deer keds may be potential vectors for the transmission of this Bartonella species in the sampled moose population. Lineage II sequences could be clearly differentiated as originating from a distinct Bartonella clade based on gltA and other housekeeping genes. Based on the concatenated sequences, sequences from lineage II form a distinct cluster separate from other known Bartonella species and therefore appear to constitute a novel, previously undescribed clade. As strains in lineage II were commonly found in the moose population both inside and outside the deer ked range, they may represent a long-standing endemic infection, for which the means of transmission is entirely unknown.
It is unknown whether these two bacterial species are pathogenic in moose. A single report has described B. bovis-associated endocarditis in cattle (33), but in another study, no effect of bacteremia was found on milk production or reproduction in cattle (34), suggesting that ruminant-infecting Bartonella species are of little clinical importance. In spite of a thorough examination, no pathological lesions associated with Bartonella were found in the necropsy cases in our study (17), and no signs of disease were reported in immobilized animals. The observed prevalence of Bartonella in moose blood, however, indicates that the detected strains are able to cause a persistent and systemic infection in this cervid host. As chronic, asymptomatic infection with long-term bacteremia is common for Bartonella spp. in their reservoir host (35–37), this may suggest that the moose is a primary host. This seems to be comparable with findings in roe deer, where ∼80% of the population was found to be positive for Bartonella DNA (8).
Many Bartonella spp. are important pathogens causing morbidity or mortality in humans (38–41). While Dehio and coauthors (9) suggested that B. schoenbuchensis transmitted with the bites of deer ked may establish a local infection in the skin and thereby contribute to the etiology of deer ked dermatitis, there is limited evidence to support a role for the ruminant-infecting Bartonella, such as B. schoenbuchensis, B. capreoli, B. melophagi, or B. chomelii, as zoonotic agents. Recently, Maggi and coauthors (42) reported isolation of the closely related “Candidatus Bartonella melophagi” from the blood of two diseased women, but no causal relationship between the disease and the infection was proven.
Interestingly, it was suggested that in Sweden, Bartonella-induced subacute myocarditis was the cause of sudden unexpected cardiac death (SUCD) in orienteers (43). PCR amplification of a short fragment of the gltA gene revealed sequences that could be consistent with B. quintana or B. henselae in samples from five orienteers who had succumbed to SUCD (43). Four of these five cases, as well as two other orienteers with cardiomyopathy and 31.3% of elite orienteers (compared with 6.8% of healthy blood donors), had antibodies against Bartonellaceae (43, 44). The specific identity of the Bartonella sp. concerned is, however, uncertain, as species determination was based on PCR and sequencing of a short fragment of the gltA gene alone, a gene that is prone to recombination (21). How the Swedish orienteers were exposed to Bartonella was not determined, although vector-borne transmission via blood-sucking arthropods was suspected (43). In light of our findings, it is pertinent to mention that the accumulation of SUCD among orienteers coincided with a major increase in the abundance and distribution range of the deer ked in Fennoscandia (2). Given the uncertainty over the identity of the Bartonella spp. identified from orienteers, it could be speculated that transmission of Bartonella spp. by deer keds, which frequently bite orienteers, may be one of the factors behind the observed high seroprevalence and disease among these sportsmen.
In conclusion, the presented findings show the presence of a potential vector-borne pathogen within a prevalent reservoir host (the moose) and a prevalent and geographically invasive vector which frequently attacks humans and other animals. Bartonella infection in the Norwegian moose population involves at least two different clades of Bartonella, one potentially transmitted by Lipoptena, the other almost certainly not. The high prevalence of infections both inside and outside the deer ked distribution range may suggest transmission of Bartonella by different vectors. This warrants further research on Lipoptena cervi and Bartonella spp. (45).
Supplementary Material
ACKNOWLEDGMENTS
We thank Anna Aspán at the National Veterinary Institute (NVI) in Uppsala for her help with extraction of DNA from tissues from the carcasses, Stina Vincentsson and Katarina Wallménius at the Section for Clinical Microbiology at the University Hospital in Uppsala for their help with the molecular biology work, Marthe Opland and Nina Brekke Tveit at NVI in Oslo for their help at necropsies and in the laboratory, and Ole Roer at Faun Naturforvaltning, Øystein Os and Silje Hvarnes at Veterinærconsult, Andreas Haga at the Norwegian School of Veterinary Science, and Jos Milner at Hedmark University College for their excellent help with collecting samples from immobilized moose.
Financial support was given by the National Health Surveillance Program for Cervids (HOP), Norwegian Research Council grant 185263, and grants from the University Hospital of Uppsala, Sweden.
Footnotes
Published ahead of print 26 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02632-12.
REFERENCES
- 1. Bequaert J. 1942. A monograph of the Melophaginae or ked flies of sheep, goats, deer and antelopes (Diptera, Hippoboscidae). Entomol. Am. 22:1–220 [Google Scholar]
- 2. Välimäki P, Madslien K, Malmsten J, Härkonen L, Härkonen S, Kaitala A, Kortet R, Laaksonen S, Mehl R, Redford L, Ylönen H, Ytrehus B. 2010. Fennoscandian distribution of an important parasite of cervids, the deer ked (Lipoptena cervi), revisited. Parasitol. Res. 107:117–125 [DOI] [PubMed] [Google Scholar]
- 3. Kadulski S. 1974. The dynamics of infestation of the Cervidae with bxLipoptena cervi L. (Diptera, Hippoboscidae) on the territory of Poland. Wiad. Parazytol. 20:703–707 [PubMed] [Google Scholar]
- 4. Chistyakov AF. 1968. Skin lesions in people due to bites of Lipoptena cervi. Vestn. Dermatol. Venerol. 42:59–62 [PubMed] [Google Scholar]
- 5. Ivanov VI. 1974. Injuriousness to deer of the louse fly Lipoptena cervi L. (Diptera, Hippoboscidae) in Belarus. Parazitologiia 8:252–253 (In Russian.) [PubMed] [Google Scholar]
- 6. Rantanen T, Reunala T, Vuojolahti P, Hackman W. 1982. Persistent pruritic papules from deer ked bites. Acta Derm. Venereol. 62:307–311 [PubMed] [Google Scholar]
- 7. Tsai YL, Chang CC, Chuang ST, Chomel BB. 2011. Bartonella species and their ectoparasites: selective host adaptation or strain selection between the vector and the mammalian host? Comp. Immunol. Microbiol. Infect. Dis. 34:299–314 [DOI] [PubMed] [Google Scholar]
- 8. Dehio C, Lanz C, Pohl R, Behrens P, Bermond D, Piemont Y, Pelz K, Sander A. 2001. Bartonella schoenbuchii sp. nov., isolated from the blood of wild roe deer. Int. J. Syst. Evol. Microbiol. 51:1557–1565 [DOI] [PubMed] [Google Scholar]
- 9. Dehio C, Sauder U, Hiestand R. 2004. Isolation of Bartonella schoenbuchensis from Lipoptena cervi, a blood-sucking arthropod causing deer ked dermatitis. J. Clin. Microbiol. 42:5320–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Halos L, Jamal T, Maillard R, Girard B, Guillot J, Chomel B, Vayssier-Taussat M, Boulouis HJ. 2004. Role of Hippoboscidae flies as potential vectors of Bartonella spp. infecting wild and domestic ruminants. Appl. Environ. Microbiol. 70:6302–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adamska M. 2008. Wild ruminants in the area of the North-Western Poland as potential reservoir hosts of Bartonella schoenbuchensis and B. bovis. Acta Parasitol. 53:407–410 [Google Scholar]
- 12. Reeves WK, Nelder MP, Cobb KD, Dasch GA. 2006. Bartonella spp. in deer keds, Lipoptena mazamae (Diptera: Hippoboscidae), from Georgia and South Carolina, U.S A. J. Wildl. Dis. 42:391–396 [DOI] [PubMed] [Google Scholar]
- 13. Matsumoto K, Berrada ZL, Klinger E, Goethert HK, Telford I., Sr 2008. Molecular detection of Bartonella schoenbuchensis from ectoparasites of deer in Massachusetts. Vector Borne Zoonotic Dis. 8:549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mediannikov O, Davoust B, Cabre O, Rolain JM, Raoult D. 2011. Bartonellae in animals and vectors in New Caledonia. Comp. Immunol. Microbiol. Infect. Dis. 34:497–501 [DOI] [PubMed] [Google Scholar]
- 15. Rolain JM, Rousset E, La SB, Duquesnel R, Raoult D. 2003. Bartonella schoenbuchensis isolated from the blood of a French cow. Ann. N. Y. Acad. Sci. 990:236–238 [DOI] [PubMed] [Google Scholar]
- 16. Chomel BB, Kasten RW. 2010. Bartonellosis, an increasingly recognized zoonosis. J. Appl. Microbiol. 109:743–750 [DOI] [PubMed] [Google Scholar]
- 17. Madslien K, Ytrehus B, Vikøren T, Malmsten J, Isaksen K, Hygen HO, Solberg EJ. 2011. Hair-loss epizootic in moose (Alces alces) associated with massive deer ked (Lipoptena cervi) infestation. J. Wildl. Dis. 47:893–906 [DOI] [PubMed] [Google Scholar]
- 18. Välimäki P, Kaitala A, Madslien K, Härkonen L, Varkonyi G, Heikkila J, Jaakola M, Ylönen H, Kortet R, Ytrehus B. 2011. Geographical variation in host use of a blood-feeding ectoparasitic fly: implications for population invasiveness. Oecologia 166:985–995 [DOI] [PubMed] [Google Scholar]
- 19. Ehrenborg C, Byström R, Hjelm E, Friman G, Holmberg M. 2008. High Bartonella spp. seroprevalence in a Swedish homeless population but no evidence of trench fever. Scand. J. Infect. Dis. 40:208–215 [DOI] [PubMed] [Google Scholar]
- 20. Holmberg M, McGill S, Ehrenborg C, Wesslen L, Hjelm E, Darelid J, Blad L, Engstrand L, Regnery R, Friman G. 1999. Evaluation of human seroreactivity to Bartonella species in Sweden. J. Clin. Microbiol. 37:1381–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paziewska A, Harris PD, Zwolinska L, Bajer A, Sinski E. 2011. Recombination within and between species of the alpha proteobacterium Bartonella infecting rodents. Microb. Ecol. 61:134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Norman AF, Reqnery R, Jameson P, Greene C, Krause DC. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33:1797–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Platt AR, Woodhall RW, George AL., Jr 2007. Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. Biotechniques 43:58–62 [DOI] [PubMed] [Google Scholar]
- 24. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 25. Swofford DL. 2002. PAUP* phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA [Google Scholar]
- 26. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 27. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 28. Zacharias Z. 1928. Studies on the intracellular symbiosis of the Pupiparous. Z. Morphol. Öekol. Tiere 10:676–737 [Google Scholar]
- 29. La Scola B, Zeaiter Z, Khamis A, Raoult D. 2003. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 11:318–321 [DOI] [PubMed] [Google Scholar]
- 30. Bai Y, Cross PC, Malania L, Kosoy M. 2011. Isolation of Bartonella capreoli from elk. Vet. Microbiol. 148:329–332 [DOI] [PubMed] [Google Scholar]
- 31. Bermond D, Boulouis HJ, Heller R, Van LG, Monteil H, Chomel BB, Sander A, Dehio C, Piemont Y. 2002. Bartonella bovis Bermond et al. sp. nov. and Bartonella capreoli sp. nov., isolated from European ruminants. Int. J. Syst. Evol. Microbiol. 52:383–390 [DOI] [PubMed] [Google Scholar]
- 32. Maillard R, Vayssier-Taussat M, Bouillin C, Gandoin C, Halos L, Chomel B, Piemont Y, Boulouis HJ. 2004. Identification of Bartonella strains isolated from wild and domestic ruminants by a single-step PCR analysis of the 16S-23S intergenic spacer region. Vet. Microbiol. 98:63–69 [DOI] [PubMed] [Google Scholar]
- 33. Maillard R, Petit E, Chomel B, Lacroux C, Schelcher F, Vayssier-Taussat M, Haddad N, Boulouis HJ. 2007. Endocarditis in cattle caused by Bartonella bovis. Emerg. Infect. Dis. 13:1383–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maillard R, Grimard B, Chastant-Maillard S, Chomel B, Delcroix T, Gandoin C, Bouillin C, Halos L, Vayssier-Taussat M, Boulouis HJ. 2006. Effects of cow age and pregnancy on Bartonella infection in a herd of dairy cattle. J. Clin. Microbiol. 44:42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chomel BB, Kasten RW, Sykes JE, Boulouis HJ, Breitschwerdt EB. 2003. Clinical impact of persistent Bartonella bacteremia in humans and animals. Ann. N. Y. Acad. Sci. 990:267–278 [DOI] [PubMed] [Google Scholar]
- 36. Dehio C. 2004. Molecular and cellular basis of Bartonella pathogenesis. Annu. Rev. Microbiol. 58:365–390 [DOI] [PubMed] [Google Scholar]
- 37. Jacomo V, Kelly PJ, Raoult D. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 9:8–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boulouis HJ, Chang CC, Henn JB, Kasten RW, Chomel BB. 2005. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet. Res. 36:383–410 [DOI] [PubMed] [Google Scholar]
- 39. Breitschwerdt EB, Kordick DL. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for humans. Infect. Clin. Microbiol. Rev. 13:428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, Birtles RJ, Koehler JE, Dehio C. 2009. Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet. Res. 40:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. 2006. Bartonella spp. in pets and effect on human health. Emerg. Infect. Dis. 12:389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maggi RG, Kosoy M, Mintzer M, Breitschwerdt EB. 2009. Isolation of Candidatus Bartonella melophagi from human blood. Emerg. Infect. Dis. 15:66–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wesslen L, Ehrenborg C, Holmberg M, McGill S, Hjelm E, Lindquist O, Henriksen E, Rolf C, Larsson E, Friman G. 2001. Subacute Bartonella infection in Swedish orienteers succumbing to sudden unexpected cardiac death or having malignant arrhythmias. Scand. J. Infect. Dis. 33:429–438 [DOI] [PubMed] [Google Scholar]
- 44. McGill S, Wesslen L, Hjelm E, Holmberg M, Rolf C, Friman G. 2001. Serological and epidemiological analysis of the prevalence of Bartonella spp. antibodies in Swedish elite orienteers 1992-93. Scand. J. Infect. Dis. 33:423–428 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.