Abstract
Thiosulfate dehydrogenase is known to play a significant role in thiosulfate oxidation in the acidophilic, obligately chemolithoautotroph, Acidithiobacillus ferrooxidans. Enzyme activity measured using ferricyanide as the electron acceptor was detected in cell extracts of A. ferrooxidans ATCC 23270 grown on tetrathionate or sulfur, but no activity was detected in ferrous iron-grown cells. The enzyme was enriched 63-fold from cell extracts of tetrathionate-grown cells. Maximum enzyme activity (13.8 U mg−1) was observed at pH 2.5 and 70°C. The end product of the enzyme reaction was tetrathionate. The enzyme reduced neither ubiquinone nor horse heart cytochrome c, which serves as an electron acceptor. A major protein with a molecular mass of ∼25 kDa was detected in the partially purified preparation. Heme was not detected in the preparation, according to the results of spectroscopic analysis and heme staining. The open reading frame of AFE_0042 was identified by BLAST by using the N-terminal amino acid sequence of the protein. The gene was found within a region that was previously noted for sulfur metabolism-related gene clustering. The recombinant protein produced in Escherichia coli had a molecular mass of ∼25 kDa and showed thiosulfate dehydrogenase activity, with maximum enzyme activity (6.5 U mg−1) observed at pH 2.5 and 50°C.
INTRODUCTION
Acidithiobacillus ferrooxidans is an acidophilic, chemolithoautotrophic bacterium that derives energy from the oxidation of ferrous iron and reduced inorganic sulfur compounds (RISCs). This bacterium is one of the key organisms used in industrial bioleaching applications (1, 2). The oxidation of ferrous iron by this bacterium has been previously examined in great detail (3–6). Some enzymes or proteins previously found and thought to be involved in the aerobic oxidation of RISCs by A. ferrooxidans include thiosulfate dehydrogenase (7, 8), thiosulfate:quinone reductase (9), sulfur dioxygenase (10), sulfur:ferric ion oxidoreductase (11), sulfite:ferric ion oxidoreductase (12), a rhodanese-like protein (2, 13–15), sulfide:quinone oxidoreductase (SQR) (16, 17), and tetrathionate hydrolase (TTH) (18). RISCs are chemically reactive, and thus, some reactions can occur nonenzymatically. The mechanism of the biological sulfur oxidation in this bacterium remains elusive.
We proposed that sulfide and tetrathionate are intermediates of sulfur oxidation in A. ferrooxidans and are further metabolized by TTH and SQR (16–18). A bd-type ubiquinol oxidase is also involved in sulfur oxidation (19). TTH catalyzes the hydrolysis of tetrathionate to thiosulfate, sulfur, and sulfate (18). Thiosulfate has also been postulated as a key intermediate compound in the oxidation of the sulfur moiety of pyrite (thiosulfate pathway) (20). Thus, thiosulfate is also thought to be a key intermediate of sulfur oxidation in A. ferrooxidans, and the elucidation of its mechanism is an important and relevant research problem.
Numerous chemolithotrophic sulfur-oxidizing microorganisms can utilize thiosulfate as an electron donor in their respiration (21, 22). At least four biochemical pathways associated with thiosulfate oxidation have been found in microorganisms (22). The first pathway was reported in Thiobacillus versutus (now known as Paracoccus versutus), which oxidizes thiosulfate by using the multi-enzyme complex system (Sox system). The Sox complex is widely distributed in bacteria (23). Genes encoding sox homologs have been found in acidophilic sulfur oxidizers Acidithiobacillus caldus (24, 25) and Acidithiobacillus thiooxidans (26), the neutrophilic sulfur oxidizer Thiobacillus denitrificans (27), and the iron- and/or sulfur-oxidizer Acidithiobacillus ferrivorans (28), although the sox genes are not necessarily complete. However, homologous genes have not been found in the A. ferrooxidans ATCC 23270 genome.
The second pathway has been reported in Starkeya novella, which possess a Sox complex, albeit an incomplete one. In addition to the Sox complex, it uses a membrane-bound multienzyme complex, which includes rhodanese, sulfur-oxidizing enzyme, sulfite:cytochrome c oxidoreductase, and cytochrome c oxidase (29).
The third pathway mainly occurs in Acidithiobacillus, Thermithiobacillus, Halothiobacillus, Acidiphilium, and Tetrathiobacter. These bacterial species oxidize thiosulfate through the formation of a tetrathionate intermediate (known as the S4 intermediate pathway) (9, 30–33). In this pathway, thiosulfate is oxidized in the periplasmic space by thiosulfate dehydrogenase, which requires a c-type cytochrome as an electron acceptor and/or a c-type heme molecule in the protein. Several thiosulfate-oxidizing and tetrathionate-forming thiosulfate dehydrogenases have been identified and characterized from the periplasmic or soluble fractions of both chemolithotrophic and phototrophic sulfur bacteria (7, 8, 32, 34–38), and genes encoding these proteins have not been identified until recently. The first identified gene (tsdA) responsible for the tetrathionate-forming thiosulfate dehydrogenase was reported in the purple sulfur bacterium Allochromatium vinosum (39). However, a homologous protein has not been found in any of the Acidithiobacillus genomes.
The fourth pathway was reported in the thermoacidophilic archaeon Acidianus ambivalens, in which thiosulfate is metabolized by a membrane-bound, tetrathionate-forming thiosulfate:quinone oxidoreductase (TQO) (40). Although a suite of genes potentially encoding TQO has been identified in A. ferrooxidans (5), A. caldus (24, 25), A. thiooxidans (26), and A. ferrivorans (28), the biochemical function of TQOs in these bacterial species remains elusive.
Two thiosulfate-oxidizing enzymes from A. ferrooxidans have been purified (7, 8). Silver and Lundgren reported that the thiosulfate dehydrogenase enzyme did not possess a heme molecule, and its optimal activity was observed at pH 5.0 (8); however, neither its subunit composition nor its natural state as membrane-bound or soluble have been established. Janiczek et al. (7), on the other hand, reported that the enzyme's optimal activity was observed at pH 3.0 and that it was composed of four identical subunits with molecular masses of ∼45 kDa, but its corresponding gene has not yet been identified. In this report, for the first time, we describe the genetic information on tetrathionate-forming thiosulfate dehydrogenase in A. ferrooxidans ATCC 23270.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
A. ferrooxidans ATCC 23270 was grown aerobically in 9K medium (pH 2.5) supplemented with 3% (wt/vol) FeSO4 · 7H2O, 1% (wt/vol) elemental sulfur or 5 mM tetrathionate (K2S4O6) at 30°C (18, 19). When growth was observed on tetrathionate, up to 3 mM additional tetrathionate was added to the culture after 8 days of cultivation. Escherichia coli strains DH5α and BL21(DE3) (Applied Biosystems, Inc., Carlsbad, CA), commonly used as host cells for cloning and recombinant gene expression, were aerobically grown in Luria-Bertani (LB) medium. Ampicillin (50 μg ml−1) was added to the medium as needed.
Enzyme assay.
Thiosulfate dehydrogenase (TSD) activity was indirectly measured by monitoring the reduction of ferricyanide. The reaction mixture contained 50 mM β-alanine buffer (pH 2.5), 1 mM K-ferricyanide, 10 mM Na-thiosulfate, 200 mM Na-sulfate, and the enzyme preparation. The reaction was initiated by adding thiosulfate at 40°C. The reduction of ferricyanide was monitored by measuring the absorbance of the reaction mixture at 420 nm. One unit of activity (U) is defined as 1 μmol of ferricyanide reduced per min. TQO activity was measured as the decrease in absorbance at 275 nm. The reaction mixture contained 50 mM β-alanine buffer (pH 2.5), 30 μM ubiquinone-2 (Eizai Co., Tokyo, Japan), 10 mM Na-thiosulfate, and the enzyme preparation. Thiosulfate:cytochrome c oxidoreductase activity was measured as the increase in absorbance at 550 nm. The reaction mixture contained 50 mM β-alanine buffer (pH 2.5), 0.1 mg ml−1 of an oxidized horse heart cytochrome c, 10 mM Na-thiosulfate, and the enzyme preparation. Sulfite:ferricyanide oxidoreductase was measured in the reaction mixture containing 50 mM β-alanine buffer (pH 2.5), 1 mM K-ferricyanide, 10 mM K-sulfite, and the enzyme preparation. All reaction rates were corrected for the nonenzymatic reaction by heat-inactivated enzyme (10 min at 100°C). All measurements were separately performed in triplicates. Each data point is given as the arithmetic mean value.
Purification of TSD from A. ferrooxidans.
After 15 days of cultivation in 9K medium supplemented with tetrathionate, the cells were harvested by centrifugation at 6,000 × g for 10 min. The cells were washed three times with 0.1 M K-phosphate buffer (pH 6.3) and disrupted by sonication on ice (Ultrasonic homogenizer VP-300 [Taitec, Koshigaya, Japan]; 23% intensity cycles of 30 s on and 60 s off for a total time of 30 min). Unbroken cells and cellular debris were removed by centrifugation at 10,000 × g for 10 min. The resulting supernatant (cell extract) was further centrifuged at 110,000 × g for 60 min to prepare total membrane (insoluble) and cytosolic/periplasmic (soluble) fractions. Ammonium sulfate was added to the soluble fraction at a final concentration of 3 M. The precipitate was recovered by centrifugation (20,000 × g, 10 min), and the resulting pellet was suspended in 20 mM citrate buffer (pH 4.0). After centrifugation of the suspension at 20,000 × g for 10 min, the supernatant was dialyzed against 20 mM citrate buffer (pH 4.0) at 4°C overnight, followed by centrifugation at 20,000 × g for 10 min. This fraction was called “soluble fraction at pH 4.” The fraction was applied to a cation-exchange column chromatography by using a CM-650M (Tosoh, Tokyo, Japan) equilibrated with 20 mM citrate buffer (pH 4.0), and the proteins were eluted with a linear gradient of 0 to 0.5 M NaCl in 20 mM citrate buffer (pH 4.0). Fractions showing TSD activity were pooled and dialyzed against 20 mM citrate buffer (pH 4.0) to remove NaCl. Ammonium sulfate was then added to the solution at a final concentration of 1.3 M. After centrifugation at 20,000 × g for 10 min, the supernatant was applied to a hydrophobic column chromatography by using a Butyl-650M (Tosoh) equilibrated with 20 mM citrate buffer (pH 4.0) containing 1.3 M ammonium sulfate, and the proteins were eluted with a linear gradient of 1.3 to 0 M ammonium sulfate in 20 mM citrate buffer (pH 4.0). Fractions showing TSD activity were pooled, concentrated by Centricut U-10 (Cosmo Bio, Tokyo, Japan), and applied on a prepacked TSKgel G3000 column (Tosoh) equilibrated with 20 mM citrate buffer (pH 4.0). The protein molecular mass was calculated by gel permeation chromatography using aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), RNase A (13.7 kDa), and myoglobin (17,000 Da) as size standards. All chromatographies were carried out by using ÄKTAprime plus (GE Healthcare, Buckinghamshire, United Kingdom).
Cloning and expression of gene encoding tsd from A. ferrooxidans in Escherichia coli.
The forward primer (5′-AATGCCTCCCATATGGCCGCCGGCATGAGC-3′) with a restriction enzyme site for NdeI (indicated by underlining) was constructed on the basis of the N-terminal amino acid sequence determined from the purified TSD of A. ferrooxidans ATCC 23270. The reverse primer (5′-CTCATTTCTCGAGAGTTATTTGGCGTACTG-3′) with a restriction enzyme site for XhoI (indicated by underlining) was designed from the complementary sequence corresponding to the C-terminal region of the open reading frame obtained from the whole genome database for A. ferrooxidans ATCC 23270. PCR was performed using the two primers (0.3 μM) and genomic DNA (0.01 μg μl−1) of A. ferrooxidans ATCC 23270 as a template. The amplified DNA fragment was purified on agarose gel by using an Illustra GFX PCR DNA and a gel band purification kit (GE Healthcare). The purified DNA fragment digested with NdeI and XhoI was inserted into the corresponding cloning site in the expression vector pET21a (Novagen, Madison, WI). The constructed vector, pET-tsd, was introduced into E. coli DH5α cells. After insertion of the nucleotide sequence into the plasmid was confirmed, the E. coli BL21(DE3) cells were transformed with pET-tsd. The recombinant clones were selected on LB solid medium containing 50 μg of ampicillin ml−1.
Purification of recombinant TSD.
E. coli BL21(DE3) cells with pET-tsd were cultured in LB medium containing 50 μg of ampicillin ml−1 at 30°C until the culture reached an optical density at 600 nm of 0.6. Gene expression was induced by adding 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubating the cells at 30°C for 24 h. The cells were harvested by centrifugation at 6,000 × g for 10 min. The cells were washed three times with 0.1 M K-phosphate buffer (pH 6.3) and disrupted by sonication on ice as previously described. The solution was centrifuged at 10,000 × g for 10 min to obtain a cell extract. Total membrane (insoluble) and cytosolic/periplasmic (soluble) fractions were prepared from the cell extract by centrifugation at 110,000 × g for 60 min. The recombinant protein from the soluble fraction was purified using the same protocol that was used for purifying TSD from A. ferrooxidans ATCC 23270, with the exception of the cation-exchange column chromatography using CM-650M.
Protein analysis.
Protein concentration and N-terminal amino acid sequences were determined and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) were carried out as described elsewhere (17). The molecular masses and isoelectric points of proteins were calculated using GENETYX (Genetic information analysis software, Genetyx Co., Tokyo, Japan). The InterProScan Sequence Search tool (http://www.ebi.ac.uk/Tools/pfa/iprscan/) was also used for the sequence analysis.
Analysis of sulfur compounds.
Thiosulfate and tetrathionate were determined colorimetrically as described by Kelly et al. (41).
Database analyses.
The sequence, annotation, and accession numbers (AFE numbers) of the complete A. ferrooxidans strain ATCC 23270 genome were derived from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) and the Comprehensive Microbial Resource (CMR; J. Craig Venter Institute [http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=gtf]).
RESULTS AND DISCUSSION
Detection of TSD activity in tetrathionate-grown A. ferrooxidans cells.
When TSD activity was measured in a reaction mixture containing a cell extract, ferricyanide, and thiosulfate, no activity was detected within the pH range of 1.5 to 6.0 at 40°C. TSD activity was detected by adding K2SO4 or Na2SO4 to the reaction mixture. The highest TSD activity was obtained at 200 mM Na-sulfate (see Fig. S1A in the supplemental material). The addition of KCl or NaCl did not show this effect, indicating a specific sulfate requirement for the enzyme activity (see Fig. S1B in the supplemental material). Our previous research on TTH from A. ferrooxidans ATCC 23270 also demonstrated the requirement of sulfate ion for its activity (18). Thiosulfate is unstable at a pH of <4.0 (38) and reacts chemically not only with ferricyanide but also with ferric iron at a pH of <3.5. TSD activity (0.085 U mg−1) was detected in the soluble fraction by subtracting the nonenzymatic values (obtained by using the heat-inactivated enzyme) from enzymatic values. Time-dependent ferricyanide reduction with thiosulfate and the enzyme (soluble fraction or soluble fraction at pH 4) is shown in Fig. S2 in the supplemental material.
Two optimal pH values (2.5 and 4.0) were shown to favor enzyme activity in cell extracts, suggesting the involvement of at least two different enzymes in thiosulfate oxidation in tetrathionate-grown cells (see Fig. S1C in the supplemental material). Enzyme activity (0.04 U mg−1) with optimum pH of 2.5 was detected in the soluble fraction, and enzyme activity (0.03 U mg−1) with an optimum pH of 4.0 was detected in the membrane fraction. However, these enzyme activities were not detected in the reaction mixture without the sulfate ions.
We have already reported that SQR and TTH are synthesized in cells grown in sulfur or tetrathionate medium but not in cells grown in ferrous iron medium (17, 18). Similarly, although TSD activity at pH 2.5 was detected in both soluble fractions prepared from sulfur-grown (0.063 U mg−1) and tetrathionate-grown (0.085 U mg−1) cells, it was not detected in the soluble fraction from iron-grown cells. The results suggested that the tsd gene was specifically expressed in the presence of RISCs.
Purification of TSD from soluble fraction prepared from tetrathionate-grown A. ferrooxidans.
Because A. ferrooxidans is an acidophilic bacterium, and the pH of its periplasm is within the 2.5 to 3.0 range (42), the soluble fraction was initially dialyzed against a 20 mM citrate buffer (pH 4.0). TSD activity was detected in the supernatant after centrifugation at 20,000 × g for 10 min. The purification procedure of the TSD resulted in a 63-fold enrichment, with a recovery rate of 3% (Table 1). When an active sample obtained from a hydrophobic column chromatography (Butyl-650M) was applied on a gel permeation chromatography (TSKgel G3000), the specific activity was reduced to ca. 35% for unknown reasons. The molecular mass of the active peak was calculated to be ∼25 kDa. As shown in Fig. 1, lanes 2 to 5, the content of the protein with a molecular mass of ∼25 kDa was increased in the sample from Butyl-650M chromatography (Fig. 1, lane 5) compared to the sample derived from CM-650M chromatography (Fig. 1, lane 4). The specific activity in the Butyl-650M fraction also increased compared to that in the CM-650M fraction (Table 1). Although the specific activity was reduced in the TSK gel fraction, only one protein band with a molecular mass of ∼25 kDa was detected by SDS-PAGE analysis of the TSK gel fraction (data not shown). Therefore, it was concluded that the 25-kDa protein was responsible for the TSD activity. Because the activity was significantly reduced in the TSK gel fraction, the sample from the Butyl-650M column chromatography was used to characterize the properties of the enzyme.
Table 1.
Purification of thiosulfate dehydrogenase at the optimal pH of 2.5 from the soluble fraction of tetrathionate-grown A. ferrooxidans ATCC 23270a
| Purification step | Total protein (mg) | Sp act (μmol min−1 mg−1) | Total activity (μmol min−1) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 200.8 | 0.05 | 10.0 | 100 | 1 |
| Soluble fraction | 55.1 | 0.09 | 4.9 | 49 | 1.8 |
| Soluble fraction at pH 4 | 11.1 | 0.20 | 2.2 | 22 | 4.0 |
| CM-650M fraction | 1.3 | 2.28 | 3.0 | 30 | 45.6 |
| Butyl-650M fraction | 0.1 | 3.16 | 0.3 | 3 | 63.2 |
Enzyme activity was measured at pH 2.5 and 40°C in a mixture containing 1 mM ferricyanide and 10 mM thiosulfate.
Fig 1.
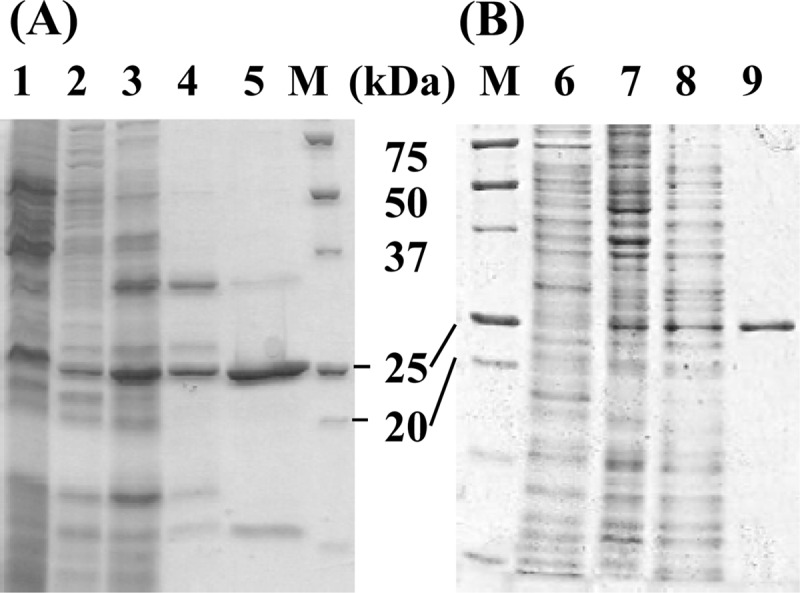
SDS-PAGE of partially purified thiosulfate dehydrogenase. (A) Lanes 1 to 5, samples from A. ferrooxidans ATCC 23270. (B) Lanes 7 to 9, samples from E. coli BL21(DE3) harboring pET-tsd. Lane 1, cell extract (20 μg); lane 2, soluble fraction (6.8 μg); lane 3, soluble fraction at pH 4 (11.35 μg); lane 4, CM-650M fraction (1.58 μg); lane 5, Butyl-650M fraction (1.04 μg); lane M, molecular markers; lane 6, cell extract from E. coli with pET21a (10 μg); lane 7, cell extract (9.9 μg); lane 8, soluble protein at pH 4.0 (9.9 μg); lane 9, Butyl-650M fraction (1.53 μg).
Properties of partially purified TSD.
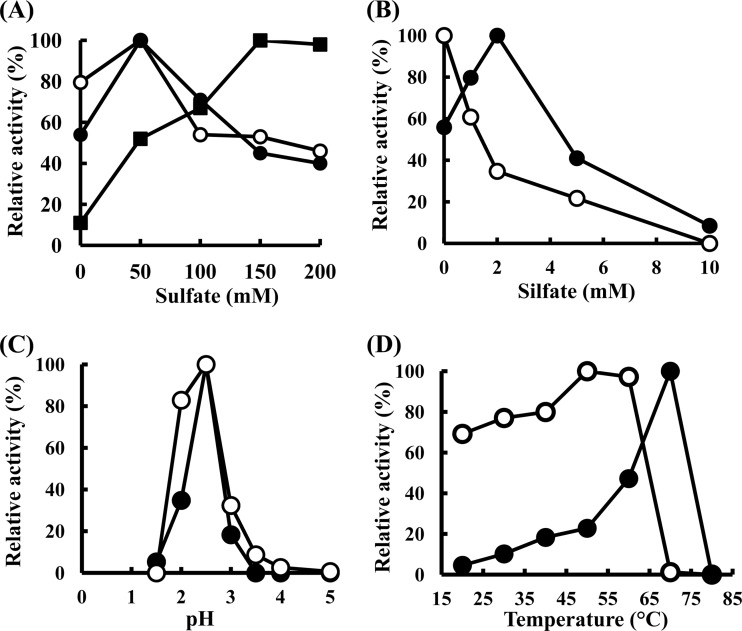
The partially purified TSD (Butyl-650M fraction) catalyzed the reduction of ferricyanide with thiosulfate but not with sulfite (Fig. 2). The nonenzymatic reduction of ferricyanide was observed with thiosulfate but not with sulfite (Fig. 2). TSD in the CM-650M fraction required 150 mM Na-sulfate to generate maximum activity (Fig. 3A, black squares). TSD activity in the Butyl-650M fraction was detected without the sulfate ion (Fig. 3A, black circles). In contrast, a 60% reduction in TSD activity was observed in the reaction mixture containing 200 mM Na-sulfate (Fig. 3A, black circles). It is possible that cofactors required for the sulfate-dependent enzyme activation dissociated from the enzyme complex during purification. The addition of sulfite (2 mM) resulted in the activation of the enzyme at a magnitude of 1.6-fold (Fig. 3B, black circles). However, higher concentration of sulfite ions resulted in inhibition of the enzyme, and almost all of the activity was inhibited in the presence of 10 mM sulfite. In contrast, previous reports have shown that TSD activity in Thiobacillus sp. strain W5 was inhibited by the addition of a low concentration of sulfite, with a Ki of 0.3 mM (38). The mechanisms for the sulfite-mediated activation of TSD in A. ferrooxidans ATCC 23270 are currently being examined. Ubiquinone and horse heart cytochrome c were not reduced by TSD. The optimal pH and temperature were pH 2.5 and 70°C, respectively (Fig. 3C and D, black circles). This pH profile indicated that the purified enzyme corresponds to the pH 2.5 peak of the total cell extract. The Km value obtained in the absence of Na-sulfate in the reaction mixture containing 1 mM ferricyanide as the electron acceptor was estimated as 15 mM (Fig. 4, black circles). When the concentration of thiosulfate was lower than 4 mM, no enzyme activity was detected. Because thiosulfate is unstable at a pH of <4.0, the Km value estimated for thiosulfate might have been higher. A Km value of 0.9 mM for a TSD isolated from A. ferrooxidans has been reported elsewhere (8). The relationship between the substrate concentration and reaction rate is represented by a sigmoidal curve (Fig. 4, black circles) similar to the characteristics of an allosteric enzyme, suggesting that allosteric activators are required for an enzyme reaction.
Fig 2.
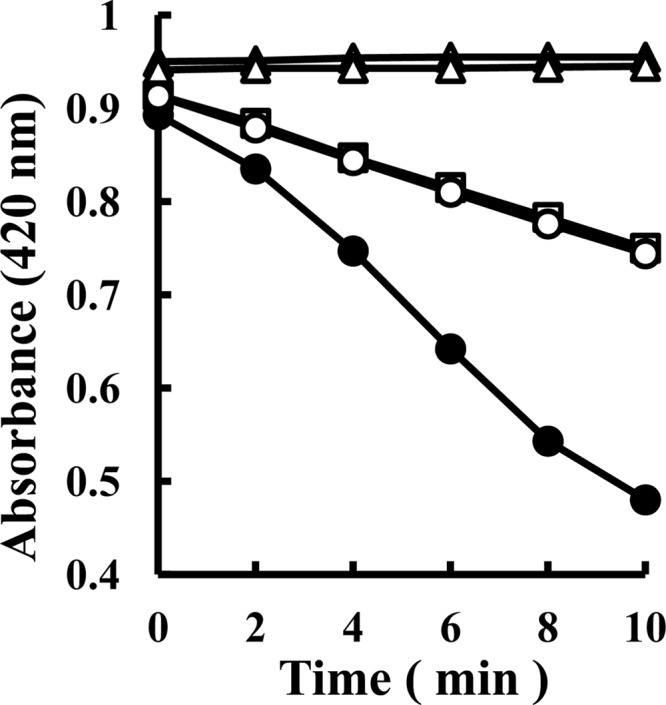
Time course of absorbance changes at 420 nm after mixing thiosulfate or sulfite and ferricyanide in the presence or absence of enzyme. Symbols: ●, thiosulfate and enzyme; ○, thiosulfate and heat-denatured enzyme; □, thiosulfate and water; ▲, sulfite and enzyme; △, only enzyme without substrates. Experiments were carried out at pH 2.5 and 40°C in triplicates in a mixture containing 1 mM ferricyanide, 10 mM Na-thiosulfate or 10 mM Na-sulfite, 50 mM Na-sulfate, and 7 μg of protein ml−1 (Butyl-650M fraction) from A. ferrooxidans ATCC 23270.
Fig 3.
Effect of sulfate (A), sulfite (B), pH (C), or temperature (D) on TSD activity in Butyl-650M fraction from A. ferrooxidans ATCC 23270 (●) or E. coli BL21(DE3) harboring pET-tsd (○). The effect of sulfate on TSD activity in CM-650M fraction is represented by a black square (■) in panel A. The specific activities (U mg−1) for the 100% value are indicated in the four panels as follows: 3.47 (●), 4.57 (○), and 2.34 (■) (A); 5.47 (●) and 4.57 (○) (B); 3.47 (●) and 4.57 (○) (C); and 13.79 (●) and 6.53 (○) (D).
Fig 4.
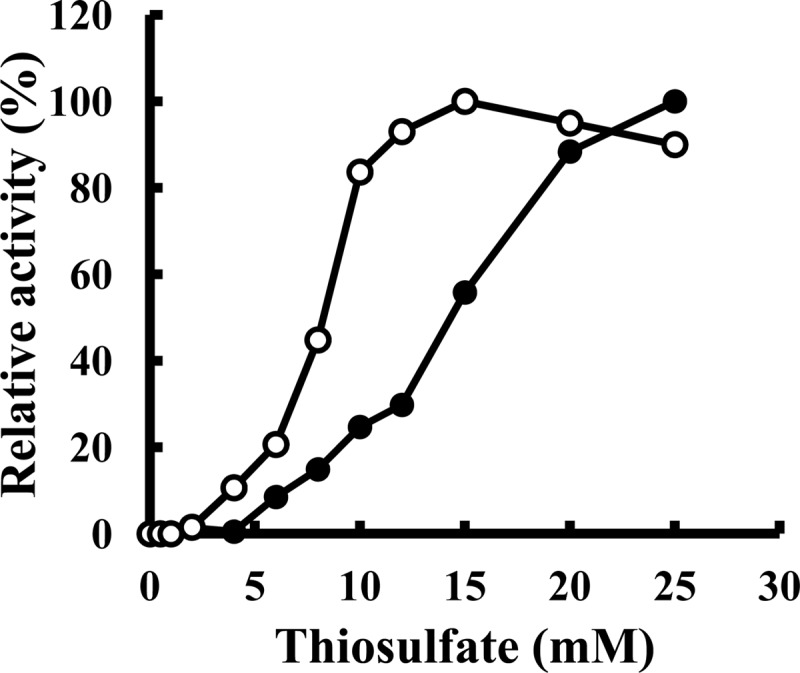
Effect of thiosulfate concentration on TSD activity in the Butyl-650M fraction of A. ferrooxidans ATCC 23270 (●) or E. coli BL21(DE3) harboring pET-tsd (○). Experiments were carried out at pH 2.5 and 40°C in a mixture containing 1 mM ferricyanide and 50 mM Na-sulfate.
Some thiosulfate dehydrogenases catalyzing the oxidation of thiosulfate to tetrathionate have been reported to contain a c-type heme molecule in their proteins (35, 37, 38). Because the visible absorption spectrum of the thiosulfate- or dithionite-reduced enzyme did not give any distinctive maxima (data not shown), the presence of heme groups in the protein was not suggested. Heme staining of the Butyl-650M fraction was also negative.
Stoichiometry of thiosulfate oxidation.
Because the rate of chemical reaction increases with the temperature, the stoichiometry of thiosulfate oxidation by TSD was measured at 20°C in reaction mixtures containing 5 mM thiosulfate, 1 mM ferricyanide, and 7 μg of enzyme ml−1 (Butyl-650M fraction) (Fig. 5). The end product of thiosulfate oxidation was tetrathionate. Cyanolysis revealed the stoichiometric conversion of 0.33 mM thiosulfate into 0.18 mM tetrathionate at 30 min and of 0.78 mM thiosulfate into 0.33 mM tetrathionate at 60 min. The results indicate that TSD catalyzed the formation of 1 mol of tetrathionate from 2 mol of thiosulfate.
Fig 5.
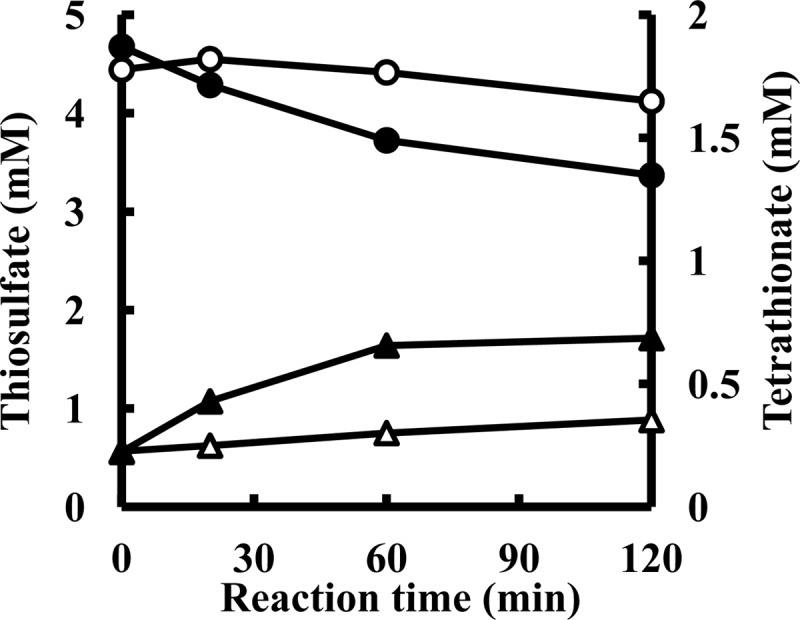
Stoichiometry of thiosulfate consumption and tetrathionate formation by TSD from A. ferrooxidans. Experiments were carried out at pH 2.5 and 20°C in a mixture containing 5 mM Na-thiosulfate, 1 mM ferricyanide, and 6.88 μg · ml−1 of TSD (Butyl-650M fraction) from A. ferrooxidans. Symbols: circles, thiosulfate concentrations in the presence (●) or absence (○) of the enzyme; triangles, tetrathionate concentrations in the presence (▲) or absence (△) of the enzyme.
Identification of the gene encoding TSD of A. ferrooxidans.
The N-terminal amino acid sequence of the 25-kDa protein was determined as AGMSGNPANLLPTGA. A BLASTP search using the National Center for Biotechnology Information nonredundant database revealed a hypothetical protein, Lferr_0043 (A. ferrooxidans ATCC 53993), including this 15-amino-acid sequence. The corresponding gene for A. ferrooxidans ATCC 23270, AFE_0042, was found in the GenBank database. This 810-bp gene encoded a 270-amino acid protein (see Fig. S4 in the supplemental material). According to the observed N-terminal amino acid sequence of the 25-kDa protein, the first 37 residues constituted the signal peptide. Therefore, the molecular mass and the isoelectric point of the mature 233-amino-acid protein without the signal peptide were calculated to be 25,796 Da and 6.2, respectively. Analysis using the InterProScan Sequence Search website showed conserved sequences for a transmembrane helix and a TAT signal at the N terminus. Although the TAT signal sequence was conserved in the other homologs of TSD (see Fig. S5 in the supplemental material), the transmembrane helix was not conserved. This type of TAT signal found in the TSD of A. ferrooxidans ATCC 23270 might be recognized by the Sec system.
As previously described, the results of spectroscopic analysis and heme staining suggested the absence of a heme molecule in the protein. No conserved domain for heme binding or metallic cofactor binding, such as the iron-sulfur cluster and molybdenum, was detected within the amino acid sequence of AFE_0042. AFE_0042 had already been registered as a sulfur-regulated gene (2, 13, 43), and microarray and bioinformatic analysis coupled with gene transcript profiling revealed that the AFE_0042 gene is upregulated by 19.0-fold in sulfur-grown cells and by 4.9-fold in thiosulfate-grown cells compared to iron-grown cells (13). Our semiquantitative reverse transcription-PCR analysis for the transcription of the AFE_0042 gene also demonstrated this upregulation (see Fig. S4 in the supplemental material). Although the involvement of AFE_0042 in sulfur metabolism has been suggested, its actual function has not been fully determined. The periplasmic localization of AFE_0042 (HypA1) has been reported elsewhere (44).
The genomic region surrounding the AFE_0042 gene in A. ferrooxidans ATCC 23270 has been previously reported for its clustering of sulfur metabolism-related genes (see Fig. S3 in the supplemental material) (5, 13–15). Genes encoding two TSD homologs, two solute-binding protein homologs, two DoxDA homologs, two rhodanese domain-containing proteins, and a thioredoxin homolog were found in this region. The rhodanese-like protein gene, p21 (AFE_0045), which encodes a putative thiosulfate sulfur transferase protein, was upregulated by 132.2-fold in sulfur-grown cells compared to iron-grown cells (13). Although the function of p21 remains elusive, its involvement in sulfur metabolism has been strongly suggested (13, 14). A similar gene organization (only for tsd1-modA1-doxDA1 and not for the remaining parts of the cluster) has been detected in Gluconobacter oxydans (6).
Homologs of AFE_0042 were identified in Acidithiobacillus ferrivorans, Acidiphilium multivorum, Thiomonas intermedis, Hydrogenobaculum sp., Burkholderia cenocepacia, G. oxydans, and Leptospirillum ferrodiazotrophum (see Fig. S5 in the supplemental material). With respect to utilization of RISCs, members of the genera Acidithiobacillus, Acidiphilium, Thiomonas, Hydrogenobaculum, and Burkholderia are able to oxidize thiosulfate (10, 28, 45–47). Although L. ferrodiazotrophum is an acidophilic iron oxidizer, sulfur oxidation for energy generation has been suggested by the results of genomic analysis (48). No information is available on the utilization of thiosulfate in G. oxydans (49). It is not known whether the bacteria possessing an AFE_0042 homolog show TSD activity.
Detection of a putative solute-binding protein in the partially purified TSD fraction.
BLASTP analysis revealed that bacteria carrying an AFE_0042 homolog also contained an AFE_0043 homolog (see Fig. S3, S5, and S6 in the supplemental material). The AFE_0043 encodes a putative periplasmic molybdate-binding protein (ModA) functioning as part of the ABC-type transporter (13). Although the TSD activity in CM-650M fraction required sulfate ion for its maximum activity, the requirement was not apparent in the Butyl-650M fraction (Fig. 3A). The results implied that cofactors involved in the sulfate-dependent enzyme activation were contained in the CM-650M fraction and disappeared after hydrophobic column chromatography. Although a major protein band with a molecular mass of ∼33 kDa was detected in the CM-650M fraction, the relative amount of the protein was reduced in the Butyl-650M fraction (Fig. 1A, lanes 4 and 5). The N-terminal amino acid sequence of the 33-kDa protein in the CM-650M fraction (Fig. 1A, lane 4) was determined as ADMGWNGKAEAPRYQ. The BLASTP search revealed a putative solute-binding protein, Lferr_0044 (A. ferrooxidans ATCC 53993), that included this 15-amino-acid sequence. The corresponding gene for A. ferrooxidans ATCC 23270 was found to be AFE_0043 encoding a putative ModA (see Fig. S3 and S6 in the supplemental material). According to the observed N-terminal amino acid sequence of this 33-kDa protein, the first 27 residues constituted the signal peptide. Therefore, the molecular mass and the isoelectric point of the mature protein without the signal peptide were calculated to be 33,983 and 9.2, respectively.
Considering that the AFE_0043 gene locates downstream of the tsd gene, and was cotranscribed with the tsd gene (see Fig. S4 in the supplemental material), the AFE_0043 protein may be composed of a hetero-oligomer that includes the TSD protein and may confer the sulfate-dependent enzyme activity to the complex in vivo. Mechanisms for the sulfate-mediated activation of TSD in A. ferrooxidans ATCC 23270 are currently being examined.
Purification and characterization of the recombinant TSD in E. coli harboring the tsd gene.
To determine whether AFE_0042 encodes TSD, we attempted to express the gene in E. coli. Because AFE_0042 carried a signal peptide in its N-terminal region, we constructed a truncated gene corresponding to a protein without the signal peptide. When E. coli BL21(DE3) cells harboring pET-tsd were grown in an LB medium containing 50 μg of ampicillin ml−1 at 30°C and induced with 0.2 mM IPTG for 24 h, a gene product with an apparent molecular mass of 25 kDa was detected by SDS-PAGE analysis of the cell extract (Fig. 1B, lane 7). Although the gene was not highly expressed in E. coli, the recombinant protein with TSD activity was purified to homogeneity following almost the same procedures as those used for the native TSD from A. ferrooxidans ATCC 23270 (Fig. 1B, lane 9). Purification of the recombinant TSD resulted in a 61-fold enrichment, with a recovery rate of 26% (see Table S1 in the supplemental material). TSD activity was detected without sulfate ions in the reaction mixture (Fig. 3A, white circles). The specific activity of the recombinant TSD (4.16 U mg−1) measured at pH 2.5 and 40°C was similar to that of the native TSD (3.16 U mg−1). The optimal pH for TSD activity of the recombinant protein was 2.5 (Fig. 3C, white circles), and the highest activity was obtained at 50°C (Fig. 3D, white circles). Although the activation of TSD activity by sulfite was observed with native TSD, the activation was not observed with the recombinant enzyme (Fig. 3B, white circles). The Km value (8 mM) for thiosulfate was lower than that for the native TSD (15 mM) (Fig. 4, white circles). Thus, some properties of the recombinant TSD were different from those of the native TSD. These differences may be due to the various pH levels at sites where TSD matured (periplasm and cytoplasm). On the basis of these results, we concluded that the 25-kDa protein encoded by the AFE_0042 gene catalyzed the TSD activity.
Concluding remarks.
To the best of our knowledge, this is the first report on the genetic information on the tetrathionate-forming TSD from A. ferrooxidans ATCC 23270. Because genes thought to be involved in thiosulfate metabolism surround the AFE_0042 gene (see Fig. S3 in the supplemental material), AFE_0042 is strongly suggested to play an essential role in thiosulfate oxidation in A. ferrooxidans ATCC 23270. However, a Km value of 15 mM, with no activity at 4 mM, indicated a very low affinity of the enzyme for thiosulfate and suggested the requirement for some cofactors or subunits for its activation. The same Km value (15 mM) for thiosulfate was obtained with the CM-650M fraction (containing both TSD and AFE_0043 protein), suggesting that the AFE_0043 protein was involved in the sulfate-dependent enzyme activation but not in the stimulation of TSD activity at a low concentration of thiosulfate. These observations raise the possibility that tetrathionate formation from thiosulfate may be a side reaction of TSD. The involvement of TQO in thiosulfate oxidation has been proposed in A. ferrooxidans (4, 6, 40). Although the membrane-bound TSD activity detected in the present study may be attributed to the TQO, it is currently premature to link the observed activity to the TQO. Therefore, more experiments are required to characterize these reactions. However, we believe that this research provides the possibility of increasing the understanding of thiosulfate oxidation in A. ferrooxidans ATCC 23270 and also reveals the presence of a new and unique thiosulfate-oxidizing system in some sulfur-oxidizing bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to H. Yamada (Graduate School of Natural Science and Technology, Okayama University) and Tsugumi Shiokawa (Advanced Science Research Center, Okayama University) for their assistance in the determination of the N-terminal amino acid sequences of TSD and SBP.
This study was supported by a Grant-in-Aid for Scientific Research (no. 2358045803) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print 12 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02251-12.
REFERENCES
- 1. Rawlings DE. 2005. Characterization and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microbiol. Cell Fact. 4:13 doi:10.1186/1475-2859-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Valenzuela L, Chi A, Beard S, Orell A, Guiliani N, Shabanowitz J, Hunt DF, Jerez CA. 2006. Genomics, metagenomics and proteomics in biomining microorganisms. Biotechnol. Adv. 24:197–211 [DOI] [PubMed] [Google Scholar]
- 3. Bonnefoy V, Holmes DS. 2011. Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environ. Microbiol 14:1597–1611 doi:10.111/j.1462-2920.2011.02626.x [DOI] [PubMed] [Google Scholar]
- 4. Brasseur G, Levican G, Bonnefoy V, Holmes D, Jedlicki E, Lemesle-Meunier D. 2004. Apparent redundancy of electron transfer pathways via bc1 complexes and terminal oxidases in the extremophilic chemolithoautotrophic Acidithiobacillus ferrooxidans. Biochim. Biophys. Acta 1656:114–126 [DOI] [PubMed] [Google Scholar]
- 5. Quatrini R, Appia-Ayme C, Denis Y, Jedlicki E, Holmes DS, Bonnefoy V. 2009. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394 doi:10.1186/1471-2164-10-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valdés J, Pedroso I, Quatrini R, Dodson RJ, Tettelin H, Blake RII, Eisen JA, Holmes DS. 2008. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597 doi:10.1186/1471-2164-9-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Janiczek O, Zemanova J, Mandl M. 2007. Purification and some properties of thiosulfate dehydrogenase from Acidithiobacillus ferrooxidans. Prep. Biochem. Biotechnol. 37:101–111 [DOI] [PubMed] [Google Scholar]
- 8. Silver M, Lundgren DG. 1968. Sulfur-oxidizing enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 46:457–461 [DOI] [PubMed] [Google Scholar]
- 9. Kelly DP, Shergill JK, Lu WP, Wood AP. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71:95–107 [DOI] [PubMed] [Google Scholar]
- 10. Rohwerder T, Sand W. 2003. The sulfane sulfur of persulfides is the actual substrates of the sulfur oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149:1699–1709 [DOI] [PubMed] [Google Scholar]
- 11. Sugio T, Mizunashi M, Inagaki K, Tano T. 1987. Purification and some properties of sulfur:ferric ion oxidoreductase from Thiobacillus ferrooxidans. J. Bacteriol. 169:4916–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sugio T, Hirose T, Zhen YL, Tano T. 1992. Purification and some properties of sulfite:ferric ion oxidoreductase from Thiobacillus ferrooxidans. J. Bacteriol. 174:4189–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Acosta M, Beard S, Ponce J, Vera M, Mobarec JC, Jerez CA. 2005. Identification of putative sulfotransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome: structural and functional characterization of the proteins. OMICS 9:13–29 [DOI] [PubMed] [Google Scholar]
- 14. Ramírez P, Toledo H, Guiliani N, Jerez CA. 2002. An exported rhodanese-like protein is induced during growth of Acidithiobacillus ferrooxidans in metal sulfides and different sulfur compounds. Appl. Environ. Microbiol. 68:1837–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramírez P, Guiliani N, Valenzuela L, Beard S, Jerez CA. 2004. Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds or metal sulfides. Appl. Environ. Microbiol. 70:4491–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wakai S, Kikumoto M, Kanao T, Kamimura K. 2004. Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans NASF-1. Biosci. Biotechnol. Biochem. 68:2519–2528 [DOI] [PubMed] [Google Scholar]
- 17. Wakai S, Tsujita M, Kikumoto M, Manchur MA, Kanao T, Kamimura K. 2007. Purification and characterization of sulfide:quinone oxidoreductase from an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans. Biosci. Biotechnol. Biochem. 71:2735–2742 [DOI] [PubMed] [Google Scholar]
- 18. Kanao T, Kamimura K, Sugio T. 2007. Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans. J. Biotechnol. 132:16–22 [DOI] [PubMed] [Google Scholar]
- 19. Kamimura K, Fujii S, Sugio T. 2001. Purification and some properties of ubiquinol oxidase from obligately chemolithotrophic iron-oxidizing bacterium, Thiobacillus ferrooxidans NASF-1. Biosci. Biotechnol. Biochem. 65:63–71 [DOI] [PubMed] [Google Scholar]
- 20. Rohwerder T, Gehrke T, Kinzler K, Sand W. 2003. Bioleaching review. A. Progress in bioleaching: fundamentals and mechanism of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 63:239–248 [DOI] [PubMed] [Google Scholar]
- 21. Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fisher J. 2005. Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8:253–259 [DOI] [PubMed] [Google Scholar]
- 22. Ghosh W, Dam B. 2009. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33:999–1043 [DOI] [PubMed] [Google Scholar]
- 23. Meyer B, Imhoff JF, Kuever J. 2007. Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria: evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 9:2957–2977 [DOI] [PubMed] [Google Scholar]
- 24. Mongold S, Valdes J, Holmes DS, Dopson M. 2011. Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2:17 doi:10.3389/fmicb.2011.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valdes J, Quatrini R, Hallberg K, Dopson M, Valenzuela PD, Holmes DS. 2009. Draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus. J. Bacteriol. 199:5877–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valdes J, Ossandon F, Quatrini R, Dopson M, Holmes DS. 2011. Draft genome sequence of the extremely acidophilic biomining bacterium Acidithiobacillus thiooxidans ATCC 19377 provides insight into the evolution of the Acidithiobacillus genus. J. Bacteriol. 193:7003–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beller HR, Chain PS, Letain TE, Chakicherla A, Larimer FW, Richardson PM, Coleman MA, Wood AP, Kelly DP. 2006. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J. Bacteriol. 188:1473–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liljeqvist M, Valdes J, Holmes DS, Dopson M. 2011. Draft genome of the psychrotolerant acidophile Acidithiobacillus ferrivorans SS3. J. Bacteriol. 193:4304–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kappler U, Friedrich CG, Trüper HG, Dahl C. 2001. Evidence for two pathways of thiosulfate oxidation in Starkeya novella (formerly Thiobacillus novellus). Arch. Microbiol. 175:102–111 [DOI] [PubMed] [Google Scholar]
- 30. Dam B, Mandal S, Ghosh W, Das Gupta SK, Roy P. 2007. The S4-intermediate pathway for the oxidation of thiosulfate by the chemolithoautotroph Tetrathiobacter kashmirensis and inhibition of tetrathionate oxidation by sulfite. Res. Microbiol. 158:330–338 [DOI] [PubMed] [Google Scholar]
- 31. Hallberg KB, Dopson M, Lindström EB. 1996. Reduced sulfur compound oxidation by Thiobacillus caldus. J. Bacteriol. 178:6–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meulenberg R, Scheer EJ, Pronk JT, Hazeu W, Bos P, Kuenen JG. 1993. Metabolism of tetrathionate in Thiobacillus acidophilus. FEMS Microbiol. Lett. 112:167–172 [Google Scholar]
- 33. Pronk JT, Meulenberg R, Hazeu W, Bos P, Kuenen JG. 1990. Oxidation of reduced inorganic sulfur compounds by acidophilic thiobacilli. FEMS Microbiol. Rev. 75:293–306 [Google Scholar]
- 34. Gregersen LH, Bryant DA, Frigaard NU. 2011. Mechanism and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front. Microbiol. 2:116 doi:10.3389/fmicb.2011.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hensen D, Sperling D, Trüper HG, Brune DC, Dahl C. 2006. Thiosulfate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 62:794–810 [DOI] [PubMed] [Google Scholar]
- 36. Lu W, Kelly DP. 1988. Kinetic and energetic aspects of inorganic sulphur compound oxidation by Thiobacillus tepidarius. J. Gen. Microbiol. 134:865–876 [Google Scholar]
- 37. Nakamura K, Nakamura M, Yoshikawa H, Amano Y. 2001. Purification and properties of thiosulfate dehydrogenase from Acidithiobacillus thiooxidans JCM7814. Biosci. Biotechnol. Biochem. 65:102–108 [DOI] [PubMed] [Google Scholar]
- 38. Visser JM, de Jong GAH, Robertson LA, Kuenen JG. 1997. Purification and characterization of a periplasmic thiosulfate dehydrogenase from the obligately autotrophic Thiobacillus sp. W5. Arch. Microbiol. 166:372–378 [DOI] [PubMed] [Google Scholar]
- 39. Denkmann K, Grein F, Zigann R, Siemen A, Bergmann J, van Helmont S, Nicolai A, Pereira IAC, Dahl C. 2012. Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ. Microbiol 14:2673–2688 doi:10.1111/j.1462-2920.2012.02820.x [DOI] [PubMed] [Google Scholar]
- 40. Müller FH, Bandeiras TM, Urich T, Teixeira M, Gomes CM, Kletzin A. 2004. Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53:1147–1160 [DOI] [PubMed] [Google Scholar]
- 41. Kelly DP, Chambers LA, Trudinger PA. 1969. Cyanolysis and spectrophotometric estimation of trithionate in mixture with thiosulfate and tetrathionate. Anal. Chem. 41:898–902 [Google Scholar]
- 42. Ingledew WJ. 1982. Thiobacillus ferrooxidans. The bioenergetics of an acidophilic chemolithotroph. Biochim. Biophys. Acta 683:89–117 [DOI] [PubMed] [Google Scholar]
- 43. Bouchal P, Zdráhal Z, Helánová S, Janiczek O, Hallberg KB, Mandl M. 2006. Proteomic and bioinformatic analysis of iron- and sulfur-oxidizing Acidithiobacillus ferrooxidans using immobilized pH gradients and mass spectrometry. Proteomics 6:4278–4285 [DOI] [PubMed] [Google Scholar]
- 44. Chi A, Valenzuela L, Beard S, Mackey AJ, Shabanowitz J, Hunt DF, Jerez CA. 2007. Periplasmic proteins of the extremophile Acidithiobacillus ferrooxidans: a high throughput proteomics analysis. Mol. Cell. Proteomics 6:2239–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bhowal S, Chakraborty R. 2011. Five novel acid-tolerant oligotrophic thiosulfate-metabolizing chemolithotrophic acid mine drainage strain affiliated with the genus Burkholderia of Betaproteobacteria and identification of two novel soxB gene homologues. Res. Microbiol. 162:436–445 [DOI] [PubMed] [Google Scholar]
- 46. Panda SK, Jyoti V, Bharda B, Nayak KC, Shivaji S, Rainey FA, Das SK. 2009. Thiomonas bdubaneswarensis sp. nov., an obligately mixotrophic, moderately thermophilic, thiosulfate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 59:2171–2175 [DOI] [PubMed] [Google Scholar]
- 47. Stöhr R, Waberski A, Völker H, Tindall BJ, Thomm M. 2001. Hydrogenother musmarinus gen. nov., sp. nov., a novel thermophilic hydrogen-oxidizing bacterium, recognition of Calderobacterium hydrogenophilum as a member of the genus Hydrogenobacter and proposal of the reclassification of Hydrogenobacter acidophilus as Hydrogenobaculum acidophilus gen. nov., comb. nov., in the phylum “Hydrogenobacter/Aquifex.” Int. J. Syst. Evol. Microbiol. 51:1853–1862 [DOI] [PubMed] [Google Scholar]
- 48. Goltsman DSA, Denef VJ, Singer SW, VerBerkmoes NC, Lefsrud M, Mueller RS, Dick GJ, Sun CL, Wheeler KE, Zemla A, Baker BJ, Hauser L, Land M, Shah MB, Thelen MP, Hettich RL, Banfield JF. 2009. Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing “Leptospirillum rubarum” (group II) and “Leptospirillum ferrodiazotrophum” (group III) bacteria in acid mine drainage biofilms. Appl. Environ. Microbiol. 75:4599–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deppenmeier U, Ehrenreich A. 2009. Physiology of acetic acid bacteria in light of the genome sequence of Gluconobacter oxydans. J. Mol. Microbiol. Biotechnol. 16:69–80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.