Abstract
Upon growth on n-hexadecane (C16), n-tetracosane (C24), and n-hexatriacontane (C36), Dietzia sp. strain DQ12-45-1b could produce different glycolipids, phospholipids, and lipopeptides. Interestingly, cultivation with C36 increased cell surface hydrophobic activity, which attenuated the negative effect of the decline of the emulsification activity. These results suggest that the mechanisms of biosurfactant production and cell surface hydrophobicity are dependent upon the chain lengths of the n-alkanes used as carbon sources.
TEXT
Recently, the biodegradation of crude oil constituents, such as alkanes, through bioremediation of oil-polluted environments (1–3) and microbial enhanced oil recovery (MEOR) technology (4) has received worldwide attention. To date, a number of microorganisms have been reported to degrade alkanes of different chain lengths (5). A critical step in the biodegradation process requires microorganisms to access hydrophobic alkanes by at least two possible mechanisms. First, microorganisms produce surface-active materials, including glycolipids, phospholipids, and lipopeptides, to emulsify alkanes and achieve surfactant-mediated access (6–12). Second, they increase the hydrophobic activity of the cell surface to directly interact with alkanes (5, 13).
Although extensive research has been conducted on the production of different biosurfactants and the cell surface hydrophobic activities, these studies were mainly restricted to alkanes with chain lengths shorter than 18 carbon atoms (C18) (14–18). This raises the question of how bacteria, such as those belonging to the genus Dietzia, access hydrocarbons with chain lengths longer than C18. It is unclear whether accessing longer alkanes requires the production of surface-active materials that are similar to those produced when shorter alkanes (i.e., <C18) are degraded. In addition, whether cell surface hydrophobicity contributes to the accession of longer alkanes is unknown. The aim of this study was, therefore, to address these questions because degradation of alkanes longer than C18 is important for effective MEOR and bioremediation. We used a broad-spectrum alkane-degrading Dietzia sp. strain, DQ12-45-1b (19, 20), and the results of our study revealed that biosurfactant production and cell surface hydrophobic activity changed when different-chain-length n-alkanes were used as the sole carbon sources.
After Dietzia sp. strain DQ12-45-1b was incubated in mineral salt medium (MSM) (21) amended with 0.3% (vol/vol) n-hexadecane (C16) and 0.05% (wt/vol) n-tetracosane (C24) and n-hexatriacontane (C36) as the sole carbon sources, respectively, the cultures were sampled at different time points and analyzed for bacterial growth, alkane degradation, cell surface hydrophobic activities, and emulsifying capacity of the culture broth. The biosurfactants were also extracted from the culture broth, and the moieties of the glycolipid-like biosurfactant were additionally analyzed. The transcripts of the glycolipid synthesis-related genes were also analyzed by real-time reverse transcription-PCR (RT-PCR). All experiments were performed in triplicate with various controls. Detailed experimental procedures and methods are described in the supplemental material.
Strain DQ12-45-1b could degrade C16, C24, and C36 n-alkanes for growth (see Table S1 in the supplemental material), as was previously reported (19, 20). Along with the growth and degradation of alkanes, the surface tension of the culture broth decreased from approximately 62 mN m−1 to 27.78 ± 1.97 and 45.97 ± 2.19 mN m−1 at day 30 for C16 and C24 cultures, respectively, and to 48.66 ± 0.51 mN m−1 at day 45 for the C36 culture (see Fig. S1 in the supplemental material); this finding suggested that shorter alkanes resulted in a higher emulsifying activity, which allowed cells to access alkanes more easily. The C16, C24, and C36 culture broths yielded 95.7, 25.4, and 15.8 mg liter−1 of crude biosurfactant, respectively. Among them, 2 glycolipid compounds (16-A and 16-B) (Fig. 1A) were detected in the C16 culture, while levels of phospholipid and lipopeptide materials were negligible in the C16 culture (Fig. 1B and C). In the C24 culture, glycolipid (24-C) (Fig. 1A) and phospholipid (Fig. 1B) compounds were detected at relatively equal amounts, with no instance of lipopeptide being detected (Fig. 1C). One glycolipid (36-D) (Fig. 1A) and 2 lipopeptide (Fig. 1C) compounds were detected in the C36 culture; however, no phospholipids were detected (Fig. 1B). These results suggested that the production of biosurfactant by strain DQ12-45-1b was related to the length of the hydrocarbons. The biosurfactant activities of these materials were confirmed by oil displacement tests performed after they had been scraped out of the preparative thin-layer chromatography (TLC) plates (see the supplemental material).
Fig 1.
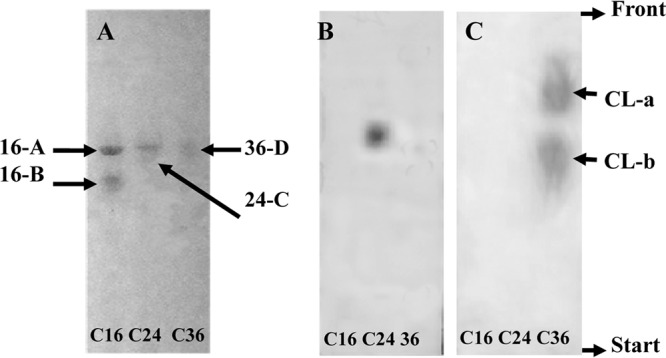
Thin-layer chromatography (TLC) analysis of crude biosurfactant extract from the cultures sampled at day 15. Panels A, B, and C show the presence and relative amounts of glycolipids, phospholipids, and lipopeptides from the n-hexadecane (C16), n-tetracosane (C24), and n-hexatriacontane (C36) cultures, respectively. In panel A, 2 glycolipids (16-A, Rf, 0.59; 16-B, Rf, 0.51) were detected from the C16 culture, whereas glycolipids 24-C (Rf, 0.60) and 36-D (Rf, 0.59) were detected from the C24 and C36 cultures, respectively. In panel B, phospholipids (Rf, 0.62) were detected only in the C24 culture. In panel C, lipopeptides were detectable only in the C36 culture, with 2 compounds detected: CL-a (Rf, 0.79) and CL-b (Rf, 0.56).
Since glycolipid-like materials were detected in all 3 cultures, the structures of the 4 glycolipids (16-A, 16-B, 24-C, and 36-D) were analyzed by gas chromatography-mass spectrometry (GC-MS). Three saccharide moieties were detected, including α-d-glucopyranoside-β-d-fructofuranose (disaccharide unit) for both 16-A and 16-B, 6-deoxy-mannose for 24-C, and 2-methoxime-gluconic acid for 36-D. The fatty acid moieties were also different among the glycolipids. In glycolipid 16-A, only hexadecanoic acid was detected, in contrast to the 5 acids detected in glycolipid 16-B, some of which had unsaturated bonds. Six fatty acids with chain lengths ranging from C12 to C19 and 11 fatty acids with chain lengths ranging from C12 to C24 were detected in 24-C and 36-D, respectively. These fatty acids also contained some unsaturated bonds (Table 1). Of note, the fatty acid moieties in 16-B contained fatty acids with chain lengths longer than C16, indicating that they have originated from fatty acid synthesis processes.
Table 1.
Moieties of the glycolipids produced from C16, C24, and C36 by strain DQ12-45-1b
| Fatty acid | Moiety ofa: |
|||
|---|---|---|---|---|
|
n-C16 |
n-C24 24-C | n-C36 36-D | ||
| 16-A | 16-B | |||
| C12H24O2 | ND | ND | + | + |
| C14H28O2 | ND | + | + | + |
| C15H30O2 | ND | ND | + | + |
| C16H30O2 | ND | ND | ND | + (7:1) |
| C16H32O2 | + | + | + | + |
| C17H34O2 | ND | ND | ND | + |
| C18H32O2 | ND | + (9:1, 12:1 and 9:1, 15:1) | ND | + (9:1, 11:1) |
| C18H34O2 | ND | + (9:1) | ND | + (8:1) |
| C18H36O2 | ND | + | + | + |
| C19H38O2 | ND | ND | + | + (10-methyl) |
| C24H48O2 | ND | ND | ND | + |
The saccharide moieties are as follows: n-C16 16-A and 16-B, α-d-glucopyranoside-β-d-fructofuranose; n-C24 24-C, 6-deoxymannose; and n-C36 36-D, 2-methoxime-gluconic acid. ND, not detected. The format “n:1” represents an unsaturated bond at the “nth” carbon atom.
The transcripts of phosphomannomutase (AlgC)-encoding gene homolog YMF1348 (accession no. JQ414011) and NADPH-dependent ketoacyl reductase (RhlG)-encoding gene homolog YMF0365 (accession no. JQ414010), which are key genes in glycolipid biosynthesis (see Table S2 in the supplemental material), were detected by real-time RT-PCR (see Fig. S2 in the supplemental material). Both YMF1348 and YMF0365 were significantly induced by n-alkanes compared to when cells were grown in a medium containing glucose. However, the 2 genes had different patterns of expression. In general, the transcriptional levels of both YMF1348 and YMF0365 were higher in cells grown on C16, with the lowest values being obtained for cells grown on C36, corresponding to the different amounts of glycolipids detected (see Fig. S2). The different transcription levels might be related to the different amounts of the precursors acetyl coenzyme A (acetyl-CoA) and malate in cells, which may suggest the different upregulation of genes in fatty acid biosynthesis, as detected in Alcanivorax borkumensis SK2 (22).
Although glycolipids, phospholipids, and lipopeptides have been reported as key microbial biosurfactants (6, 8, 10, 12), only 1 or 2 types (e.g., glycolipids, phospholipids, or lipopeptides) were simultaneously detected in the degradation of hydrocarbons with chain lengths of <C18 (15, 16, 23). However, strain DQ12-45-1b could produce all the 3 types in various amounts when C16, C24, and C36 were used as the sole carbon sources. Moreover, the simultaneous detection of different glycolipids, both with different saccharide and acid moieties, in a Dietzia strain has not been reported before.
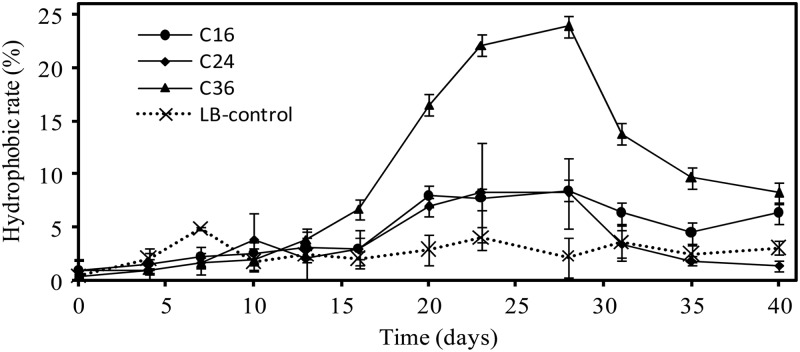
The hydrophobicity of the cell surface, which is measured as the surface hydrophobicity rate, plays an important role in the microbial attachment onto other hydrophobic surfaces, such as solid n-alkanes (24). In this study, the cell surface hydrophobicity rates slightly increased with the incubation time from approximately 0.5% to 8.5% ± 5.5% and 8.3% ± 4.6% for C16 and C24 cultures, respectively, compared to approximately 4% in the control without any hydrocarbon. In contrast, the hydrophobicity rate in C36 cultures could be as high as 23.9% ± 3.7% (Fig. 2), which caused more cells to attach to the solid surface of C36 than to C24 (see Fig. S3 in the supplemental material). These results could be attributed to the fact that glycolipids produced by C36 contain longer organic acid moieties, resulting in higher hydrophobic activities, or that lipopeptides (including the surfactin, iturin, and fengycin classes) changed the cell hydrophobicity and cell contact with hydrocarbons (6); therefore, when C36 was used as the sole carbon source, the decline in the emulsification activity (see Fig. S1 in the supplemental material) was compensated for by an increase in the cell surface hydrophobic activity (Fig. 2), which attenuated the negative impact of the long-chain length of C36 and maintained cell growth, as indicated in our previous studies (19, 20).
Fig 2.
Change in cell surface hydrophobicity rates of Dietzia sp. strain DQ12-45-1b grown on n-hexadecane (C16), n-tetracosane (C24), and n-hexatriacontane (C36), or grown on LB as the control.
On the basis of these results, we hypothesized that bacteria capable of degrading hydrocarbons of various chain lengths have similar functions for producing different biosurfactants and changing cell surface hydrophobic activity. These functions could be attributed to the unique regulation of different genes and pathways. Similar results were reported when investigating the influence of other environmental factors on biosurfactant production (22, 25–28). However, further investigation is required to verify this hypothesis.
Nucleotide sequence accession numbers.
The nucleotide sequences of YMF0365 and YMF1348 have been deposited in the GenBank database under accession no. JQ414010 and JQ414011, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31070107) and the National High Technology Research and Development Program (863 Program: 2012AA02A703).
Footnotes
Published ahead of print 26 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02497-12.
REFERENCES
- 1. Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D'haeseleer P, Holman HY, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. 2010. Deep-sea oil plume enriched indigenous oil-degrading bacteria. Science 330:204–208 [DOI] [PubMed] [Google Scholar]
- 2. Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A, Lee YJ, Mason OU, Dubinsky EA, Chavarria KL, Tom LM, Fortney JL, Lamendella R, Jansson JK, D'haeseleer P, Hazen TC, Zhou J. 2012. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J. 6:451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodríguez-Blanco A, Antoine V, Pelletier E, Delille D, Ghiglione JF. 2010. Effect of temperature and fertilization on total vs. active bacterial communities exposed to crude and diesel oil pollution in NW Mediterranean Sea. Environ. Pollut. 158:663–673 [DOI] [PubMed] [Google Scholar]
- 4. Lazar I, Petrisor IG, Yen TF. 2007. Microbial enhanced oil recovery (MEOR). Petrol. Sci. Technol. 25:1353–1366 [Google Scholar]
- 5. Wentzel A, Ellingsen T, Kotlar H-K, Zotchev S, Throne-Holst M. 2007. Bacterial metabolism of long-chain n-alkanes. Appl. Microbiol. Biotechnol. 76:1209–1211 [DOI] [PubMed] [Google Scholar]
- 6. Banat IM, Franzetti A, Gandolfi I, Bestetti G, Martinotti MG, Fracchia L, Smyth TJ, Marchant R. 2010. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 87:427–444 [DOI] [PubMed] [Google Scholar]
- 7. Desai JD, Banat IM. 1997. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61:47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mukherjee S, Das P, Sen R. 2006. Towards commercial production of microbial surfactants. Trends Biotechnol. 24:509–519 [DOI] [PubMed] [Google Scholar]
- 9. Neu TR. 1996. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60:151–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pacwa-Plociniczak M, Plaza GA, Piotrowska-Seget Z, Cameotra SS. 2011. Environmental applications of biosurfactants: recent advances. Int. J. Mol. Sci. 12:633–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosenberg E, Ron EZ. 1999. High- and low-molecular-mass microbial surfactants. Appl. Microbiol. Biotechnol. 52:154–162 [DOI] [PubMed] [Google Scholar]
- 12. Singh A, Hamme JD, Ward OP. 2007. Surfactants in microbiology and biotechnology. Part 2. Application aspects. Biotechnol. Adv. 25:99–121 [DOI] [PubMed] [Google Scholar]
- 13. Rosenberg RO, Mikkilineni R, Berne BJ. 1982. Hydrophobic effect on chain folding. The trans to gauche isomerization of n-butane in water. J. Am. Chem. Sci. 104:7647–7649 [Google Scholar]
- 14. Dastgheib SMM, Amoozegar MA, Khajeh K, Ventosa A. 2011. A halotolerant Alcanivorax sp. strain with potential application in saline soil remediation. Appl. Microbiol. Biotechnol. 90:305–312 [DOI] [PubMed] [Google Scholar]
- 15. Hua X, Wu Z, Zhang H, Lu D, Wang M, Liu Y, Liu Z. 2010. Degradation of hexadecane by Enterobacter cloacae strain TU that secretes an exopolysaccharide as a bioemulsifier. Chemosphere 8:951–956 [DOI] [PubMed] [Google Scholar]
- 16. Peng F, Liu Z, Wang L, Shao Z. 2007. An oil-degrading bacterium: Rhodococcus erythropolis strain 3C-9 and its biosurfactants. J. Appl. Microbiol. 6:1603–1611 [DOI] [PubMed] [Google Scholar]
- 17. Puntus IF, Sakharovsky VG, Filonov AE, Boronin AM. 2005. Surface activity and metabolism of hydrocarbon-degrading microorganisms growing on hexadecane and naphthalene. Proc. Biochem. 40:2643–2648 [Google Scholar]
- 18. Zheng C, Li S, Yu L, Huang L, Wu Q. 2009. Study of the biosurfactant-producing profile in a newly isolated Rhodococcus ruber strain. Ann. Microbiol. 4:771–776 [Google Scholar]
- 19. Nie Y, Liang JL, Fang H, Tang YQ, Wu XL. 2011. Two novel alkane hydroxylase-rubredoxin fusion genes isolated from a Dietzia bacterium and the functions of fused rubredoxin domains in long-chain n-alkane degradation. Appl. Environ. Microbiol. 77:7279–7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang XB, Chi CQ, Nie Y, Tang YQ, Tan Y, Wu G, Wu XL. 2011. Degradation of petroleum hydrocarbons (C6-C40) and crude oil by a novel Dietzia strain. Bioresour. Technol. 102:7755–7761 [DOI] [PubMed] [Google Scholar]
- 21. Rizzo ACL, da Cunha CD, Santos RLC, Santos RM, Magalhaes HM, Leite SGF, Soriano AU. 2008. Preliminary identification of the bioremediation limiting factors of a clay bearing soil contaminated with crude oil. J. Braz. Chem. Soc. 19:169–174 [Google Scholar]
- 22. Sabirova JS, Ferrer M, Regenhardt D, Timmis KN, Golyshin PN. 2006. Proteomic insights into metabolic adaptations in Alcanivorax borkumensis induced by alkane utilization. J. Bacteriol. 188:3763–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakano MM, Corbell N, Besson J, Zuber P. 1992. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol. Gen. Genet. 2:313–321 [DOI] [PubMed] [Google Scholar]
- 24. Liu Y, Yang S, Liu Q, Tay J. 2003. The role of cell hydrophobicity in the formation of aerobic granules. Curr. Microbiol. 46:270–274 [DOI] [PubMed] [Google Scholar]
- 25. Edmonds P, Cooney JJ. 1969. Lipids of Pseudomonas aeruginosa cells grown on hydrocarbons and on Trypticase soybean broth. J. Bacteriol. 98:16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Finnerty WR, Singer ME. 1985. A microbial biosurfactant—physiology, biochemistry and applications. Dev. Ind. Microbiol. 25:31–46 [Google Scholar]
- 27. Neidleman SL, Geigert J. 1984. Biotechnology and oleochemicals: changing patterns. J. Am. Oil Chem. Soc. 61:290–297 [Google Scholar]
- 28. Syldatk C, Lang S, Wagner F, Wray V, Witte L. 1985. Chemical and physical characterization of four interfacial-active rhamnolipids from Pseudomonas sp. DSM 2874 grown on n-alkanes. Z. Naturforsch. 40:51–60 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.