Abstract
Lower vertebrates have been found to possess genes that have similar homology to both interleukin (IL)-17A and IL-17F, which have been termed IL-17A/F. In fish species, several of these genes can be present, but, to date, very little is known about their functional activity. This article describes the discovery and sequence analysis of a rainbow trout (Oncorhynchus mykiss) IL-17A/F2 molecule and an IL-17RA receptor. In addition, the bioactivity of the trout IL-17A/F2 is investigated for the first time in any species. The predicted IL-17A/F2 and IL-17RA proteins consist of 146 and 966 amino acids (aa), respectively, with both molecules containing conserved family motifs. Expression analysis revealed high constitutive expression of trout IL-17A/F2 in mucosal tissues from healthy fish, suggesting a potential role in mucosal immunity. When the modulation of IL-17A/F2 and IL-17RA in vitro was analyzed, it was observed that the two molecules were similarly affected. The expression of IL-17A/F2 was also induced in head kidney during bacterial, parasitic, and viral infections, revealing a possible function in defense against such pathogens. However, downregulation of IL-17RA was seen in some tissues and infections. The recombinant IL-17A/F2 protein was produced in Escherichia coli and was found to affect the expression of an antimicrobial peptide and the proinflammatory cytokines IL-6 and IL-8 in splenocytes. Consistent with mammalian IL-17 homologues, our expression and bioactivity results imply that trout IL-17A/F2 plays an important role in promoting inflammatory and host innate immune responses directed against different pathogen groups.
INTRODUCTION
Interleukin-17A (IL-17A) belongs to the IL-17 family, which includes five other members in humans, IL-17B to IL-17F (1). Within the IL-17 family, IL-17A shows the highest homology to IL-17F and the lowest to IL-17E, also known as IL-25 (2). The human IL-17A protein precursor is composed of 155 amino acids (aa), with a molecular mass of 17.7 kDa. It is secreted as a disulfide-linked homodimer exerting its activities as a 35-kDa protein (2, 3). IL-17A is mainly produced by CD4+ T cells belonging to a T helper (TH) cell lineage recently characterized as a consequence of its production of IL-17A and thus termed TH17 cells (4–6). TH17 cells differentiate under the control of a unique transcription factor, retinoid-related orphan receptor (ROR)-γt (4, 6–8). TH17-type cytokines are known to be involved in inflammatory responses and host defense against extracellular pathogens, such as extracellular bacterial and fungal infections in mucosal surfaces (4, 9, 10). Recent findings have revealed that IL-17A is also secreted by other cell types such as CD8+ T cells, natural killer (NK) cells, and γδT cells (11). TH17 cells are also known to produce IL-17F (4, 5, 12), which is encoded as a 163-aa precursor and is secreted as a polypeptide of 133 aa (13, 14). In addition to being secreted in a homodimeric form as seen with IL-17A and IL-17F, an IL-17A/IL-17F (IL-17A/F) heterodimeric cytokine has also been reported (15). The two cytokines share the same locus in chromosomes 6 and 1 in humans and mice, respectively (12–14). Initially, in lower vertebrates, three molecules were identified in zebrafish (Danio rerio) that had homology to IL-17A and IL-17F and were thus designated IL-17A/F-1 to -3 (16). This was followed by a similar report in fugu (Takifugu rubripes) (17) and, most recently, in medaka (Oryzias latipes) (18). Teleost IL-17s have also been reviewed recently (19).
IL-17A and IL-17F signal through a heterodimeric receptor complex composed of IL-17 receptor A (IL-17RA) and IL-17RC, belonging to the IL-17R family (20–23). Members of the IL-17R family include five molecules (IL-17RA to E) known to possess particular features not present in other receptor families (1, 24). Most recently, a new member of this family (IL-17RE-like, or IL-17REL) has also been characterized (25). IL-17 cytokines mediate intracellular signaling through activation of the adaptor protein ACT1, tumor necrosis factor (TNF) receptor-associated factor (TRAF)-6, and transcription nuclear factor (NF)-κB (7, 24). The presence of conserved motifs, such as two fibronectin type III (FNIII)-like domains in the extracellular region, has been described. In addition, a domain located in the cytoplasmic tail with homology to the Toll/interleukin-1 receptor (TIR) domain has been predicted and therefore termed “SEFIR,” for SEF/IL-17R (26, 27). IL-17RA was identified first in mice and later in humans, with the molecules sharing 69% aa identity (7, 28). Both mouse IL-17RA and human IL-17RA are broadly expressed in various tissues and cell lines, particularly in hematopoietic organs (7, 28). The 866-aa human IL-17RA precursor corresponds to a single transmembrane receptor, containing a 31-aa signal peptide, a 293-aa extracellular domain, a 21-aa transmembrane domain, and a rather large 525-aa intracellular domain (28).
In terms of biological activity, IL-17A and IL-17F are proinflammatory molecules that induce the expression of a range of cytokines (e.g., IL-6 and granulocyte colony-stimulating factor [G-CSF]), chemokines (e.g., CXCL-1 and CXCL-8), and metalloproteinases, which act to recruit neutrophils at the inflamed site and to increase granulopoiesis (2, 9, 12, 13, 29–32). They are also involved in promoting innate immunity by increasing antimicrobial peptide (AMP) expression, as seen with β-defensins (BDs) and S100As (10, 33–35). Several studies have demonstrated the unequivocal protective role that these cytokines play in host defense against bacterial infections (9, 31, 35). For instance, Ye et al. (9) reported on the increase of IL-17 production upon bacterial infection with Klebsiella pneumoniae. Additionally, when administered, IL-17 significantly contributed to bacterial clearance (9). Other studies concluded that IL-17A- and IL-17F-deficient mice were more susceptible to opportunistic infections with Staphylococcus aureus (35). Given these findings, the crucial involvement of IL-17 in immune responses might be explained by the fact that it produces strong inflammatory responses by activation of neutrophils and granulocyte-attracting chemokines (such as IL-8) and induction of other proinflammatory cytokines and antimicrobial peptides (11). Although a large body of work confirms the importance of IL-17 in host protection against bacterial diseases, there are also reports of its potential involvement against intracellular pathogens and parasitic infections. For example, Ouyang et al. reviewed the potential role of IL-17 and/or its receptor in host defense against fungal and parasitic diseases. It was reported, for instance, that IL-17RA-deficient mice had a compromised immune system, being more prone to infections with Candida albicans or Toxoplasmosis gondii (32).
With the growing mammalian literature on the importance of IL-17 in host defense, we speculate that this molecule could also have a role in fish immune responses. Thus, we describe in this work the sequencing of an IL-17A/F molecule in rainbow trout together with expression and bioactivity studies to gain an insight into its function in fish. These findings provide a more detailed insight into the piscine IL-17 family. In addition, a putative receptor for IL-17A/F signaling (IL-17RA) has been characterized in this economically important species.
MATERIALS AND METHODS
Fish.
Rainbow trout (Oncorhynchus mykiss) were purchased from the Mill of Elrich Trout Fishery (Aberdeenshire, United Kingdom) and maintained in 1-m-diameter aerated fiberglass tanks supplied with a continuous flow of recirculating freshwater at 15 ± 1°C within the aquarium facility in the School of Biological Sciences. Fish were fed twice daily on standard commercial pellets (EWOS) and were given a 2-week acclimatization period prior to treatment. When used for pathogen challenge, the fish were kept in the freshwater aquarium pathogen containment facility and fed as described above, except on the day of the challenge.
Cloning and sequencing of interleukin (IL)-17A/F2.
The human IL-17A protein sequence (accession number NM_002190) was used to search for homologous sequences in the rainbow trout expressed sequence tag (EST) database using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome). This led to the discovery of a number of trout EST sequences at NCBI (http://www.ncbi.nlm.nih.gov/) which included the following: BX082888, BX302846, BX302847, CX139078, CX148911, CX148912, CX149535, CX149536, and CU068487. Further BLAST analysis revealed that the protein sequence translated from BX082888 had the highest homology with known vertebrate IL-17As. On the basis of this finding, EST primers (IL-17-F1 and IL-17-R1) were designed to amplify by PCR the trout cDNA sequence using a cDNA template generated from primary head kidney leukocytes, previously stimulated with 25 μg/ml of phytohemagglutinin (PHA; Sigma-Aldrich) for 4 h. The PCR product was cloned into pGEM T Easy vector (Promega) and sequenced by MWG Biotech Ltd. (Germany), as described by Monte et al. (36). This clone contained the 5′ untranslated region (5′-UTR), the open reading frame (ORF) for IL-17A/F, and a partial 3′-UTR. Primers (F3 and F4) were then designed closer to the 3′ end in order to obtain the full 3′-UTR sequence. The 3′ rapid amplification of cDNA ends (3′-RACE) cDNA was prepared from primary head kidney leukocytes stimulated with 100 μg/ml of polyinosinic-polycytidylic acid (PolyI:C), a synthetic analogue of double-stranded RNA, for 20 h. The extracted RNA was transcribed into cDNA using an oligo(dT) adaptor primer. A product of 237 bp was amplified and sequenced and contained the 3′-UTR. The cDNA sequence was deposited in the EMBL/NCBI nucleotide database (see below).
Genomic DNA (gDNA) was obtained from head kidney using a DNeasy tissue kit (Qiagen) following the manufacturer's instructions. A PCR was performed using the gDNA with primers F1 and R1, which amplified a 710-bp product, and with primers F2 and R2, which amplified a product of 836 bp. Both PCR products were ligated into pGEM-T Easy vector and sequenced, revealing 2 introns. To confirm the full genomic and cDNA sequences of trout IL-17A/F, primers F1 and R2 were used to perform a single PCR. Performing the PCR with cDNA obtained from gut tissue of a healthy fish, the size of the product was 640 bp, whereas using gDNA the product was 1,553 bp. Sequences of all primers used are given in Table 1.
Table 1.
Oligonucleotide primers used for cloning and expression analysis
| Primer name | Primer sequence (5′ → 3′) | Application or reference |
|---|---|---|
| IL-17F1 | ACAGAGATCAGAGATGGAGC | Reverse transcription-PCR |
| IL-17R1 | CATATGTCAATGGGGATCTT | |
| F1 | ACAGAGATCAGAGATGGAGCTCAAAAGC | Primers used to obtain trout IL-17A/F2 |
| R1 | CCTCTGATTCCTCTGTGGGAATCTTG | |
| F2 | GAACATTCACACACGCTCTCTGTCAC | |
| R2 | CATATGTCAATGGGGATCTTTTTCTTGAA | |
| F3 | GCTACAGCGCCTCCTTCCTGTCT | 3′-RACE |
| F4 | GCCTGCAGTACACTACATCTATGGCCTT | |
| IL-17-RF2 | GGATCCaAAAGGAATGAAGGTGACAAAGG | Recombinant protein production |
| IL-17-RR3 | CTACATCAAGTAGTCCTTGCCCA | |
| EF-1α F | CAAGGATATCCGTCGTGGCA | Real-time PCR |
| EF-1α R | ACAGCGAAACGACCAAGAGG | |
| IL-17A/F2 F | CGTGTCGAAGTACCTGGTTGTGT | Real-time PCR |
| IL-17A/F2 R | GGTTCTCCACTGTAGTGCTTTTCCA | |
| IL-17RA F | CGTAATCCAGCATATGAAAAGTGGTCTT | Real-time PCR |
| IL-17RA R | CTCCAAACACACCTTGCTCCTCT | |
| BD-3 F | GCTTGTGGAATACAAGAGTCATCTGC | Real-time PCR (37) |
| BD-3 R | GCATACATTCGGCCATGTACATCC | |
| IL-6 F | CCTTGCGGAACCAACAGTTTG | Real-time PCR |
| IL-6 R | CCTCAGCAACCTTCATCTGGTC | |
| IL-8 F | AGAATGTCAGCCAGCCTTGT | Real-time PCR |
| IL-8 R | TCTCAGACTCATCCCCTCAGT | |
| TNF-α1 F | GTGGGGTCCTCTTAATAGCAGGTC | Real-time PCR (38) |
| TNF-α1 R | ATTTCATCCTGCATCGTTGACG |
A BamHI restriction site is underlined.
Cloning and sequencing of IL-17RA.
Full-length SMART cDNA libraries were prepared from liver, gills, head kidney, and spleen of fish after 24- and 48-h challenges with the fish bacterial pathogen Aeromonas salmonicida (strain MT423), as described previously (39). The libraries were pooled and subtracted with a mix of RNA from unchallenged fish. Random end sequencing of the resultant clones identified one with homology to IL-17RA. This clone was fully sequenced using internal primers which contained the 5′-UTR, an ORF for IL-17RA, and the 3′-UTR. The mRNA sequence obtained was submitted to the EMBL/NCBI database (see below).
Bioinformatics.
The generated sequences were analyzed for similarity to known molecules and used to identify homologous sequences in GenBank using BLAST (40), including a salmon IL-17RA sequence (accession number NP_001158836) that has not been described to date. Comparisons between three or more sequences were performed using the CLUSTAL W multiple sequence alignment package (http://www.genome.jp/tools/clustalw/) (41), and conserved residues were shaded using the BOXSHADE server (version 3.21) (http://www.ch.embnet.org/software/BOX_form.html). Signal peptide predictions were made using Signal P4.0 software (http://www.cbs.dtu.dk/services/SignalP) according to Petersen et al. (42), and N-glycosylation sites were predicted using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc) (R. Gupta, E. Jung, and S. Brunak, unpublished data). Amino acid homology comparisons were performed using the MatGAT package (version 2.02) (43). Phylogenetic trees were constructed from CLUSTAL W-generated alignments using the neighbor-joining (N-J) method within the MEGA (version 4.1) package (44). Domain structures were predicted using the SMART 6 program (http://smart.embl-heidelberg.de) (45) and DomPred (http://bioinf.cs.ucl.ac.uk/dompred) (46).
Tissue distribution.
Six rainbow trout were killed and tissues (liver, caudal kidney, spleen, heart, head kidney, skin, thymus, scales, brain, muscle, gonad, gills, tail fins, and intestine) collected for RNA extraction using TRI reagent (Applied Biosciences) following the manufacturer's instructions. The first-strand cDNA was synthesized using BioScript (Bioline, United Kingdom). The synthesized cDNA was diluted in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and stored at −20°C. Real-time PCR was performed using IMMOLASE (Bioline) and a SYBR green fluorescent tag (Invitrogen) in a LightCycler 480 system (Roche Applied Science, United Kingdom), as described previously (36). The relative expression levels of IL-17A/F2 and IL-17RA were calculated as arbitrary units and normalized to the expression level of rainbow trout elongation factor (EF)-1α, a housekeeping gene. Primer sequences are listed in Table 1.
Preparation of primary cultures.
Head kidneys were collected aseptically from 4 fish, and cells were pushed through a 100-μm-pore-size nylon mesh (John Stanier) with incomplete Leibovitz L-15 medium (Gibco), i.e., supplemented with penicillin and streptomycin (P/S) at 100 U/ml and 100 μg/ml, respectively (Gibco), 0.5% fetal bovine serum (FBS; Biosera), and 10 U/ml of heparin (Sigma-Aldrich). The suspensions were then centrifuged at 200 × g for 5 min and washed once with complete medium (same constituents as incomplete medium but with 10% FBS). Cells were counted, and 5 ml of 1 × 106 to 1.5 × 106 cells/ml was seeded into 25-cm2 flasks for use.
Spleen cells were used for the bioactivity studies and were collected and homogenized as described above but using L-15 supplemented with P/S and 2% FBS. The resultant cell suspension was enriched for leukocytes by layering onto 51% Percoll (Sigma-Aldrich) and centrifuging at 400 × g for 20 min at 4°C (without a brake). The layer of cells at the interface was collected and washed once with L-15 medium under the same conditions.
Modulation of IL-17A/F2 and IL-17RA expression in vitro.
Head kidney primary cultures, prepared as described above, were incubated with known stimulatory concentrations (47) of Escherichia coli lipopolysaccharide (LPS; 25 μg/ml), PolyI:C (50 μg/ml), phorbol 12-myristate 13-acetate (PMA; 100 ng/ml), calcium ionophore (CI; 0.5 μg/ml), PMA (100 ng/ml) plus CI (0.5 μg/ml), the mitogen phytohemagglutinin (PHA; 10 μg/ml), or the immunosuppressant dexamethasone (DM; 0.5 μg/ml) for 4, 8, and 24 h or were left untreated (control). All chemicals were purchased from Sigma-Aldrich. After the incubation periods, cells attached to the flasks and in suspension were dissolved in TRI reagent. The RNA extraction and cDNA synthesis were performed as described above. Gene expression data were normalized to EF-1α gene expression and analyzed against controls using the Pfaffl method (48).
Bacterial challenge by injection.
A pathogenic strain (MT3072) of the Gram-negative salmonid pathogen Yersinia ruckeri was injected intraperitoneally (i.p.) (2 × 106 CFU/ml in phosphate-buffered saline [PBS]; 0.5 ml/fish) into rainbow trout of ∼100 g, as described previously (49). A separate group was injected with PBS (0.5 ml/fish) as a control. Five fish from each of the groups (infected and control) were killed at different time points (6, 24, 48, and 72 h) postinfection and head kidneys collected for total RNA extraction. The expression level of IL-17A/F2 and IL-17RA was analyzed by real-time PCR, as described above. The fish injected with the bacteria which were kept until the last time point died or were euthanized when they showed at least two external symptoms of the disease, such as hemorrhages in the oral cavity and reddening at the base of the fins. To confirm that the fish were infected, swabs from kidney were placed onto tryptic soy agar (TSA) plates and incubated for 48 h at 22°C for bacterial growth as described by Harun et al. (49), and in each case colonies were apparent. The spleen burden was also analyzed by PCR using Y. ruckeri 16S RNA-specific primers in total DNA samples, where the presence of this bacterium was detected 24 h postinfection (49).
Bacterial challenge by immersion.
To further investigate the modulation of both IL-17A/F2 and IL-17RA expression, particularly at mucosal sites, a bacterial challenge was performed by immersion (bath) exposure. For this, Y. ruckeri strain MT3072 was adjusted to a concentration of 2 × 107 CFU/ml in aquarium tanks and 12 trout of ∼300 g were exposed by immersion for 1 h, when the tank water was turned on. This dose was used in previous studies, while doing immersion challenges, and was shown to cause infection (50). A separate group of 12 trout was subjected to the same procedure, but without the bacteria added, as a control. Twelve fish (control and infected) were killed 24 and 48 h postinfection and mucosal tissues (gill and gut) as well as head kidney tissues collected for total RNA extraction. The expression level of IL-17A/F2 and IL-17RA was analyzed by real-time PCR, as described above.
Parasitic infection.
The modulation of trout IL-17A/F2 and IL-17RA expression was also analyzed in trout infected with the myxozoan parasite Tetracapsuloides bryosalmonae, during a natural outbreak of proliferative kidney disease (PKD), as described previously (51). Briefly, two groups of fish, obtained from the same egg source, were used. The control group was maintained and sampled under parasite-free conditions, whereas the infection group was kept in parasite-infected water. Tissue corresponding to the posterior region of the kidney immediately below the dorsal fin, an area associated with the onset of clinical PKD, was collected from both control and infected fish. Routine checks for other parasite infestations and opportunistic bacterial pathogens were conducted as described by Wang et al. (51). The severity of the clinical disease of each fish was analyzed and a kidney swelling index value assigned to each sample using the Clifton-Hadley system, a reliable and accurate scoring system to describe the infection level of PKD (52). Briefly, the kidney samples were divided into 4 grades (0 to 3) according to the clinical signs observed, such as abdominal distension and exophthalmia. Unexposed fish (control) had no signs of pathology and were classified as 0, while parasite-infected fish exhibited various degrees of stress ranging between kidney swelling index values of 1 and 3, with the latter value corresponding to the most severe pathology. Five fish from the control group and each infection group were examined for the expression of IL-17A/F2 and IL-17RA, as described above. Kidney samples were placed into 1 ml of RNAlater (Ambion) and stored at −80°C until further use.
Viral infection.
To investigate the regulation of both genes upon viral exposure, the expression of IL-17A/F2 and IL-17RA was analyzed in fish infected with the pathogenic viral hemorrhagic septicemia virus (VHSV), strain DK-F1. The challenge was performed as described by Wang et al. (47) and Campbell et al. (53). Briefly, four fish were killed at 1, 2, 4, and 6 days postinfection with DK-F1 (1 × 108 50% tissue culture infective dose [TCID50]/fish) or injection with control media as a control. Before challenge, fish were screened for the presence of pathogens. It was confirmed they were free of VHSV, infectious pancreatic necrosis, PKD, enteric redmouth disease (ERM), bacterial kidney disease, salmonid alphavirus, and furuculosis as described by Wang et al. (47). The head kidneys were collected for RNA extraction and cDNA synthesis. The expression level of IL-17A/F2 and IL-17RA was analyzed by real-time PCR, as described above. To analyze the viral infection, an assay of the N gene which uses the intrinsic properties of the N protein, abundant in negative single-stranded RNA viruses, was performed as described by Campbell et al. (53). Viral gene transcription including the N gene was analyzed by real-time PCR detecting two distinct peaks 2 and 6 days postinfection, as described in detail by Campbell et al. (53). Additionally, cumulative mortality of 87.5% was observed, which confirmed the infectivity of the strain used (53).
Production of rIL-17A/F2.
The production and purification of the recombinant IL-17A/F2 protein (rIL-17A/F2) was performed using a protocol similar to that described by Monte et al. (36) for rainbow trout rIL-22. Briefly, the putative mature peptide was predicted from the full-length protein using Signal P4.0 software and amplified by PCR using primers IL-17-RF2 and IL-17-RR3 (Table 1). The PCR product was ligated into expression vector pQE30UA (Qiagen) and sequenced. The correct pQE30UA-IL-17 plasmid was selected and then digested with BamHI (Promega) to remove extra vector sequence and purified using a PCR purification kit (Invitrogen). The linearized plasmid DNA was then religated at 4°C overnight, and positive clones containing the IL-17 insertion were again sent to be sequenced. The constructed plasmid (termed pQE30-IL-17) yielded an IL-17 fusion protein with an N-terminal tag of MRGSHHHHHHGS when transformed into E. coli JM109 cells (Promega). The purified protein was then washed with polymyxin B resin (Sigma-Aldrich), following the manufacturer's instructions, to remove any contaminating endotoxin, as described by Zou et al. (54). By analyzing the bioactivity of the recombinant protein, it was confirmed that the purified rIL-17A was ineffective at inducing the expression of tumor necrosis factor alpha 1 (TNF-α1), a classic proinflammatory cytokine sensitive to the presence of bacterial endotoxins (55).
Bioactivity analysis of rIL-17A/F2: dose response.
Spleen cells, isolated from 6 trout as described above, were counted, and 5 ml, containing 2 × 106 cells/ml, was seeded into 25-cm2 cell culture flasks. The cells were cultured for 6 h with EB-2 buffer (containing 20 mM Tris-Cl [pH 8.0], 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 5 mM 2-mercaptoethanol, 20% glycerol, 0.1% NP-40, and 500 mM imidazole [Sigma-Aldrich]) (as a control) and rIL-17A/F2 protein in concentrations ranging from 0.1 to 100 ng/ml. The cells were then harvested and RNA extracted as described above. An initial screening using real-time PCR analysis was undertaken to detect the expression of a range of genes, including those corresponding to antimicrobial peptides (AMPs), cytokines, cytokine receptors, transcription factors, and other immune-related molecules. From this initial study, the inflammatory cytokines TNF-α1 (55), IL-6 (56), and IL-8 (57) were selected for further analysis. Additionally, AMP β-defensin 3 (BD-3) (37) was analyzed, as well as the putative IL-17RA. The sequences of the primers used are listed in Table 1.
Bioactivity analysis of rIL-17A/F2: time course.
Using a single dose of rIL-17A/F2, a time course experiment was undertaken using spleen primary cultures isolated from 6 freshly killed rainbow trout. Five milliliters of cells (1.5 × 106 cells/ml) was seeded into 25-cm2 cell culture flasks and incubated with EB-2 buffer or rIL-17A/F2 (10 ng/ml) for 2, 4, 6, and 8 h. After incubation, the cells were harvested and RNAs extracted as described above. Real-time PCR analysis was undertaken to determine the expression level of BD-3, one of the genes most highly modulated by rIL-17A/F2 in the dose-response experiment.
Statistical analysis.
Data were analyzed statistically using the Student t test and one-way analysis of variance (ANOVA), with the least significant difference (LSD) post hoc test used for comparison of means where appropriate. Differences were considered statistically significant when P < 0.05.
Nucleotide sequence accession numbers.
The IL-17A/F2 cDNA and gDNA sequences determined in this work were deposited in the EMBL/NCBI nucleotide database under accession numbers AJ580842 and HE663463. The IL-17RA mRNA sequence determined in this work was submitted to the EMBL/NCBI database under accession number AJ634727.
RESULTS
Cloning and sequencing of IL-17A/F2 and IL-17RA.
The trout IL-17A/F2 coding region (accession number AJ580842) consists of 441 nucleotides which translate into a 146-aa peptide, which was sequenced from a single PCR product of 640 bp using head kidney cDNA as the template. The 3′ untranslated region (UTR) contained two mRNA instability motifs (ATTTA) and a polyadenylation signal (AATAAA) located 15 bp upstream of the poly(A) tail, suggesting that this is the authentic site (see Fig. S1 in the supplemental material). Two putative N-glycosylation sites were identified in the protein sequence. Additionally, upon amplification of gDNA from head kidney by PCR with primers designed according to the available mRNA sequence (accession number HE663463), the IL-17A/F2 coding regions were found to be separated by two introns. The first and second introns were comprised of 534 and 379 nucleotides, respectively.
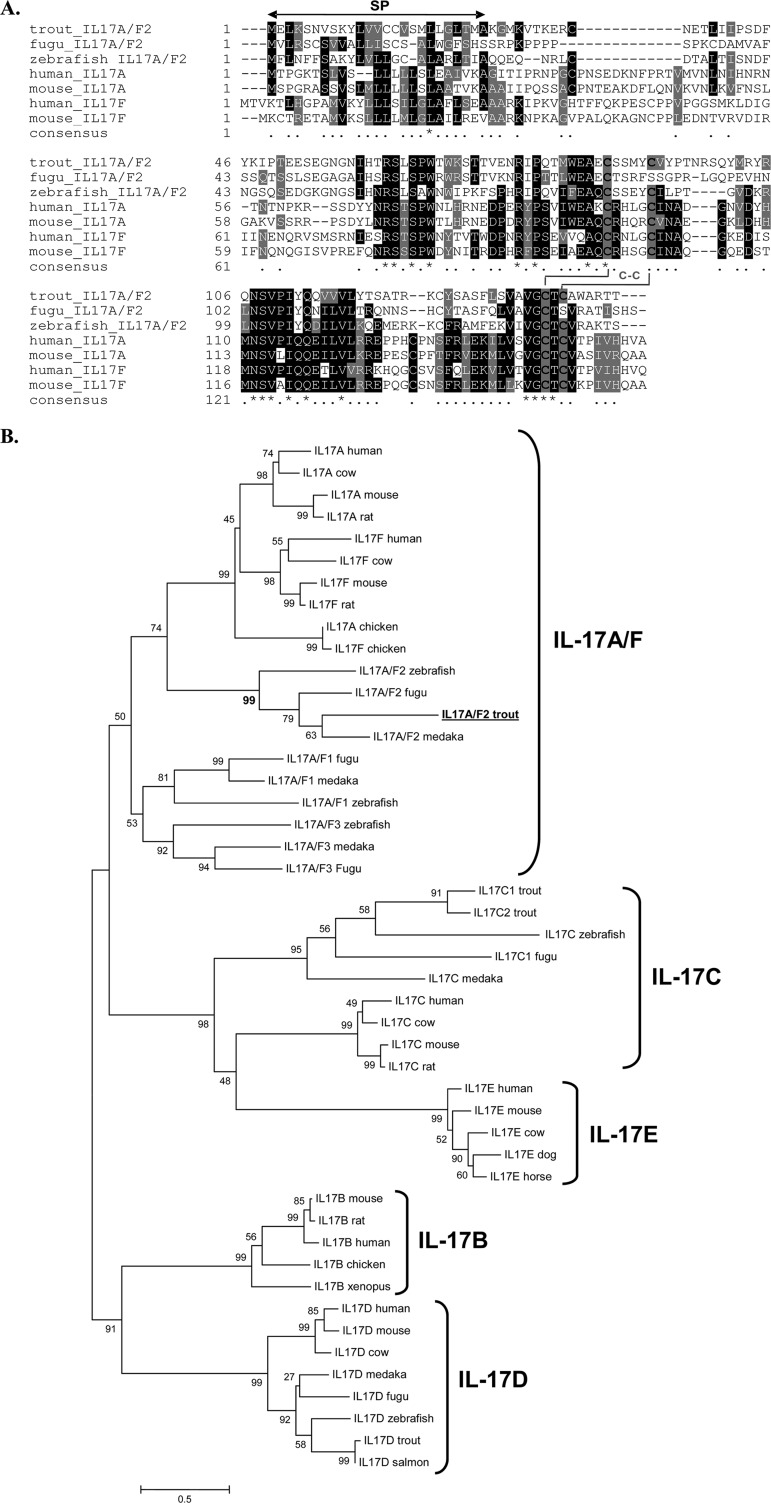
Multiple alignment with other fish IL-17A/F molecules and mammalian IL-17A and IL-17F molecules revealed conservation of amino acid sequences between the different vertebrate groups (Fig. 1A). Trout IL-17A/F2 showed conservation of four cysteine residues among all vertebrate species analyzed, with the exception of fugu IL-17A/F2, which had only two cysteine residues. The four conserved cysteines may be involved in the formation of two disulfide bridges. The relationship of trout IL-17A/F2 to other members of the IL-17 family was further investigated with the construction of a phylogenetic tree using the N-J method. Analysis of the tree revealed that the trout sequence grouped closely with those of other fish IL-17A/F2 molecules, supported by a high bootstrap value of 99%, branching with other known vertebrate IL-17A/F molecules within the IL-17A and IL-17F cluster (Fig. 1B). Analysis of homology between trout IL-17A/F2 and other known IL-17A/F proteins revealed a high degree of similarity to IL-17A/F2s from zebrafish (58.9%) and fugu (56.8%) (Table 2). Similarity to mammalian and bird IL-17A proteins ranged from 37.3% to 47.7%, while the similarity to IL-17F molecules in these species ranged from 42% to 44.2%.
Fig 1.
(A) Multiple alignment of selected fish IL-17A/F2 proteins with representative mammalian IL-17A and IL-17F molecules. Conserved residues are shaded using BOXSHADE (v3.21), with Cys residues indicated by black characters on a gray background. The predicted signal peptide (SP) is indicated by a black solid double-headed arrow above the alignment. The four cysteine residues that form two potential disulfide bonds are also indicated. Identical residues are identified by stars, whereas similar residues are identified by single dots. (B) Phylogenetic tree showing the relationship of the trout IL-17A/F2 amino acid sequence with those of other known IL-17 family members. The amino acid sequences were aligned using CLUSTAL W, and the tree was constructed by the N-J method supported with 10,000 bootstrap replications using MEGA 4.1 software. The trout IL-17A/F2 protein is in bold and underlined. Accession numbers of the IL-17 family members used are as follows: for IL-17A, human, NP_002181; cow, NP_001008412; mouse, NP_034682; rat, Q61453; and chicken, CAO79600; for IL-17F, human, NP_443104; cow, NP_001179011; mouse, NP_665855; rat, NP_001015011; and chicken, NP_989791; for IL-17A/F2, zebrafish, NP_00101863; fugu, BAI82579; and medaka, NP_001191713; for IL-17A/F1, fugu, BAI82578; medaka, NP_001191714; and zebrafish, NP_00101862; for IL-17A/F3, zebrafish, NP_00101862; medaka, NP_001191715; and fugu, BAI82580; for IL-17C, C1 trout, CAW30794; C2 trout, NP_001171959; zebrafish, NP_001018624; fugu, BAI82581; medaka, NP_001191723; human, NP_037410; cow, DAA20250; mouse, NP_665833; and rat, NP_0011780; for IL-17E, human, AAQ89484; mouse, NP_542767; cow, DAA25796; dog, XP_537375; and horse, XP_001918360; for IL-17B, mouse, NP_062381; rat, NP_446241; human, CAG33473; chicken, XP_425192; and xenopus, AAH75405; and for IL-17D, human, NP_612141; mouse, NP_665836; cow, DAA23895; medaka, NP_001191716; fugu, AAI58483; zebrafish, AAI62897; trout, CAE45584; and salmon, NP_001134365.
Table 2.
Protein homology between trout IL-17A/F2 and other known IL-17A and IL-17F molecules
| Molecule | Species/isoform | Identity (%) | Similarity (%) |
|---|---|---|---|
| Zebrafish IL-17A/F | IL-17A/F1 | 26.8 | 47.1 |
| IL-17A/F2 | 42.7 | 58.9 | |
| IL-17A/F3 | 24.7 | 41.8 | |
| Fugu IL-17A/F | IL-17A/F1 | 21.3 | 36.3 |
| IL-17A/F2 | 41.3 | 56.8 | |
| IL-17A/F3 | 13.5 | 24.7 | |
| Vertebrate IL-17A | Chicken | 23.7 | 46 |
| Mouse | 24.4 | 37.3 | |
| Human | 23.9 | 47.7 | |
| Vertebrate IL-17F | Chicken | 23.7 | 42 |
| Mouse | 24.4 | 42.2 | |
| Human | 25.6 | 44.2 |
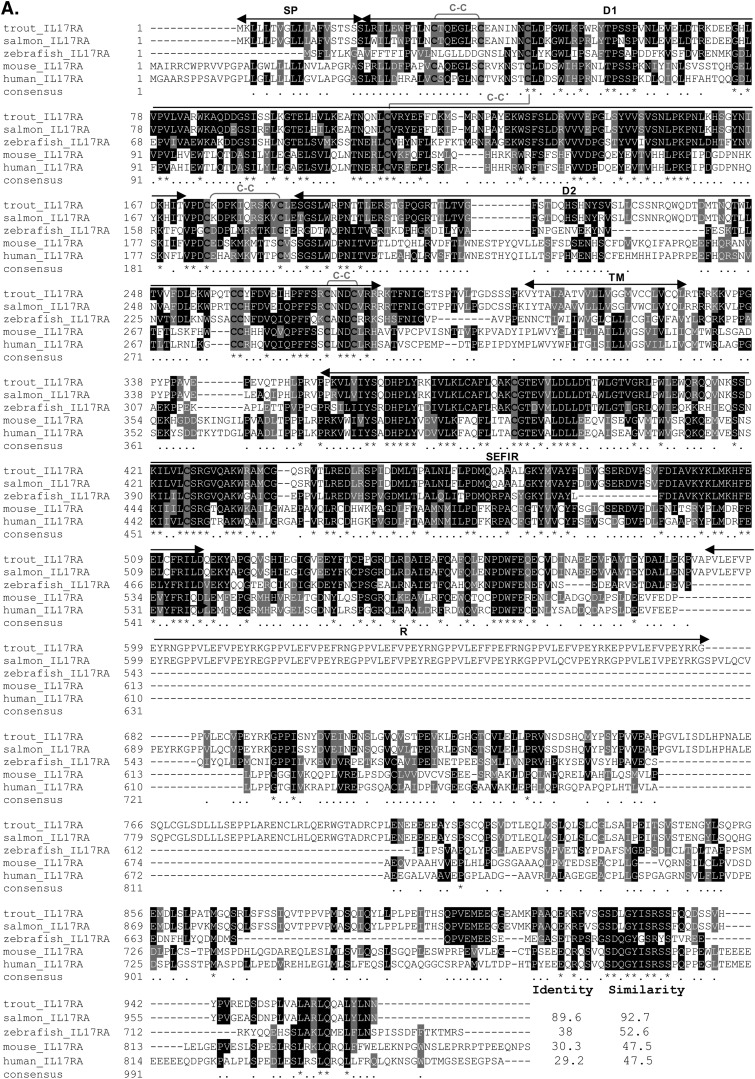
Complete sequencing of the IL-17RA cDNA clone (accession number AJ634727) revealed that it comprised 3,482 bp and contained a 5′-UTR of 267 bp, an ORF of 2,091 bp, and a 3′-UTR of 314 bp, with a polyadenylation signal (AATAAA) 16 bp upstream of the poly(A) tail (see Fig. S2 in the supplemental material). In the 5′-UTR, there are seven upstream ATGs before the main ORF (all with in-frame downstream stop codons within the 5′-UTR), suggesting posttranscriptional control of this gene (58). Translation of the major ORF gave a protein of 966 aa, with a predicted signal peptide of 19 aa, an extracellular region of 284 aa, a 24-aa transmembrane domain, and a large intracellular region of 639 aa.
A multiple alignment was performed with various IL-17RA molecules, revealing conservation of 12 cysteine residues among almost all the species analyzed, the exception being the zebrafish sequence, where 10 were present (Fig. 2A). Eight of the conserved cysteines were predicted to be involved in the formation of 4 disulfide bridges. Two putative FNIII domains (D1 and D2), as well as a SEFIR domain of 157 aa within the intracellular region, were also present in the trout IL-17RA protein. Most noticeable was the presence of 2 long insertions in the intracellular domain of the trout (and salmon) sequence, one of which contained 2 repeats of 57 aa. In terms of homology, the highest identity and similarity were with salmon (89.6 and 92.7%) and zebrafish (38% and 52.6%) IL-17RA. In order to analyze the relationship of trout IL-17RA with known IL-17R family members, a phylogenetic tree was constructed using the N-J method and bootstrapped 1,000 times (Fig. 2B). Trout IL-17RA was found to group with other fish IL-17RA proteins with high bootstrap support (83%) and away from the IL-17RB to -E clades.
Fig 2.
(A) Multiple alignment of mammalian and fish IL-17RA protein sequences. Conserved residues are shaded using BOXSHADE (v3.21), with Cys residues indicated by black characters on a gray background. The predicted signal peptide (SP), D1 and D2 domains, transmembrane region (TM), SEFIR domain, and repeats (R) are indicated by black solid double-headed arrows above the alignment. The eight cysteine residues that form four potential disulfide bonds are also indicated. Identical residues are identified by stars, whereas similar residues are identified by single dots. (B) Phylogenetic tree showing the relationship of trout receptors with other known IL-17R family members. The amino acid sequences were aligned using CLUSTAL W, and the tree was constructed by the N-J method supported with 1,000 bootstrap replications using MEGA 4.1 software. The trout IL-17RA protein is in bold and underlined. Accession numbers of the sequences used in the analysis are as follows: for IL-17RA, salmon, NP_001158836; zebrafish, XP_001921444; chicken, XP_416389; human, CAJ86450; and mouse, NP_032385; for IL-17RD, zebrafish, AAI63933; xenopus, XP_002940076; chicken, NP_989846; human, NP_060033; and mouse, NP_602319; for IL-17RB, cow, AAI33637; human, NP_061195; mouse, NP_062529, and rat, NP_001100760; for IL-17RC, rat, NP_001164036; human, NP_116121; mouse, NP_598920; and cow, NP_001068646; and for IL-17RE, rat, NP_001004091; mouse, AAH69861; human, NP_705613; and cow, XP_001251391.
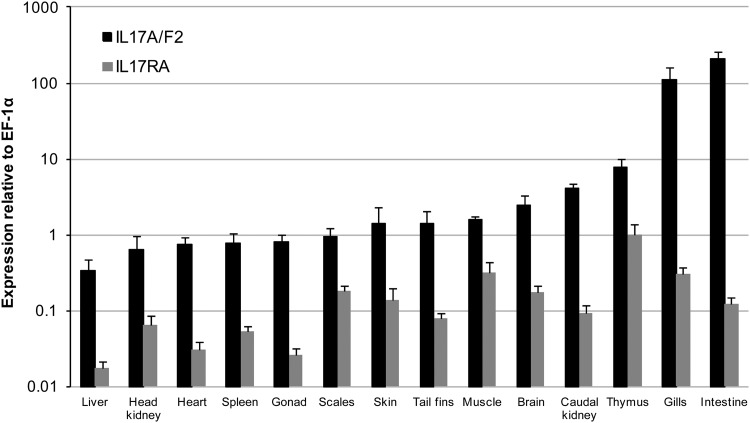
Constitutive expression of IL-17A/F2 and IL-17RA.
The expression of trout IL-17A/F2 and IL-17RA was examined in 14 tissues from 6 healthy fish (Fig. 3). Real-time PCR analysis indicated that IL-17A/F2 expression was detected in all tissues. However, high constitutive expression was observed in mucosal tissues such as intestine and gills, suggesting a potentially important role in mucosal immunity. The expression of IL-17RA was lower than that of IL-17A/F, with the highest expression seen in the thymus.
Fig 3.
Expression analysis of trout IL-17A/F2 and IL-17RA in tissues (liver, head kidney, heart, spleen, gonad, scales, skin, tail fins, muscle, brain, caudal kidney, thymus, gills, and intestine) obtained from six healthy fish as detected by real-time PCR analysis. Data represent averages + standard errors of the results of determinations of IL-17A/F2 and IL-17RA expression after normalization to that of elongation factor (EF)-1α.
Modulation of IL-17A/F2 and IL-17RA expression in vitro.
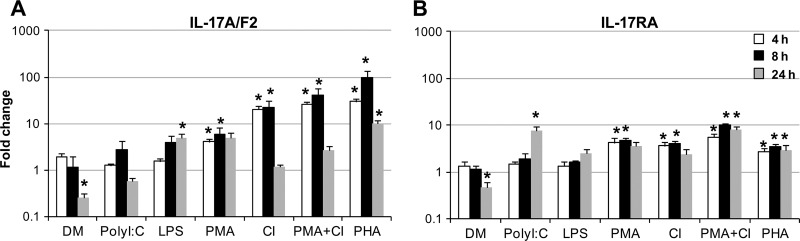
To investigate whether the expression of IL-17A/F2 and IL-17RA can be modulated in trout, primary leukocytes were freshly isolated from a major lymphoid tissue, the head kidney. They were cultured for 4, 8, and 24 h in the presence of E. coli lipopolysaccharide (LPS; bacterial endotoxin), polyinosinic-polycytidylic acid (PolyI:C; synthetic double-stranded RNA), dexamethasone (DM; immunosuppressant), and stimulators of the intracellular signaling pathways phytohemagglutinin (PHA), phorbol 12-myristate 13-acetate (PMA), and calcium ionophore (CI) (Fig. 4). In line with previous immune gene studies, cell stimulations with PHA, PMA, CI, or PMA plus CI strongly induced IL-17A/F2 and IL-17RA expression. In particular, PHA was a potent inducer of IL-17A/F2 and IL-17RA transcription, significantly (P < 0.05) upregulating their expression at all time points analyzed. The highest induction of both genes was observed after 8 h of incubation, with the upregulation of IL-17A/F2 expression increased almost 100-fold in the presence of PHA. PolyI:C and LPS were less potent, upregulating the transcript level of either IL-17A/F2 (LPS) or IL-17RA (PolyI:C) only after 24 h of incubation. In contrast, DM had a negative effect on both genes, with significant (P < 0.05) downregulation observed for both molecules after 24 h of incubation. Additionally, no variation between controls was seen (data not shown).
Fig 4.
Expression analysis of trout IL-17A/F2 (A) and IL-17RA (B) in head kidney primary leukocytes after incubation with the following stimulants: DM (0.5 μg/ml), PolyI:C (50 μg/ml), LPS (25 μg/ml), PMA (100 ng/ml), CI (0.5 μg/ml), PMA (100 ng/ml) and CI (0.5 μg/ml), or PHA (10 μg/ml). Expression of IL-17A/F2 (A) and IL-17RA (B) was detected by real-time PCR analysis and normalized to the expression of EF-1α and controls, using the Pfaffl method (48), and data are presented as fold changes. Data represent averages + standard errors of the results (n = 4). Asterisks indicate significant differences (P < 0.05) relative to the control samples. Data were analyzed using the Student T test.
Modulation of IL-17A/F2 and IL-17RA expression upon infection in vivo.
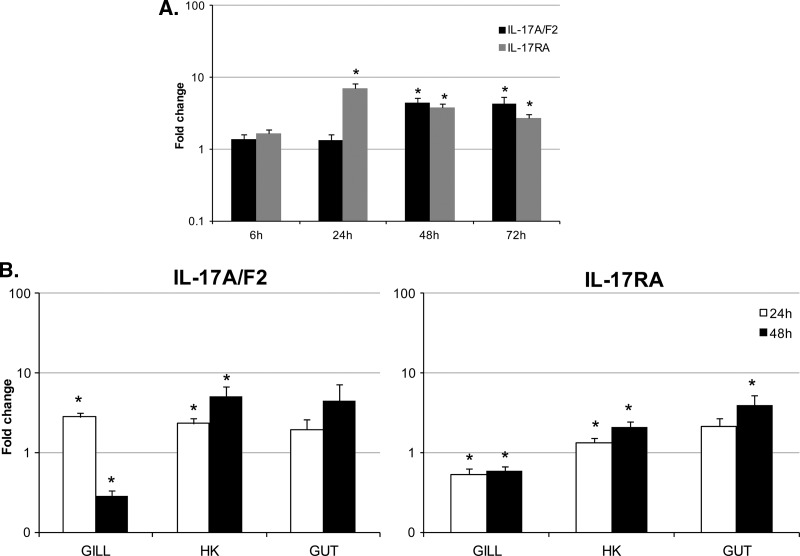
The expression of IL-17A/F2 and IL-17RA was analyzed in vivo after an i.p. bacterial challenge with Yersinia ruckeri (∼106 CFU/fish), in a major lymphoid organ, the head kidney (Fig. 5A). This bacterium belongs to the Enterobacteriaceae family and is the causative agent of yersiniosis, also known as enteric redmouth (ERM) disease. Transmission of Y. ruckeri into the host normally occurs by direct contact with infected fish or carriers. It has been suggested that the bacteria first adhere to the gill mucous, afterward invading the branchial vascular system which facilitates the colonization of internal organs. Histological examination in previous studies has confirmed that infected rainbow trout show a systemic inflammation of tissues, with major colonization of internal organs, including the kidney, spleen, and gills (50, 59). Since the kidney is one of the main tissues colonized by this pathogen, it was selected for analysis. The results revealed that the expression of IL-17RA was upregulated significantly (P < 0.05) at 24, 48, and 72 h postinfection, with 7-, 3.7-, and 2.7-fold changes, respectively. The expression of IL-17A/F2 was also induced at the latter two time points, with 4.5- and 4.3-fold changes. Additionally, no variation between controls was observed over time (data not shown).
Fig 5.
Expression analysis of trout IL-17A/F2 and IL-17RA during a bacterial infection. (A) Five rainbow trout were injected intraperitoneally with Yersinia ruckeri (0.5 ml; 1 × 106 CFU/fish) or with PBS (0.5 ml/fish). Head kidney tissue was collected at 6, 24, 48, and 72 h after challenge, and RNA was extracted for real-time PCR analysis. (B) Six rainbow trout were bath immersed with Y. ruckeri (2 × 107 CFU/fish) or with control media for 1 h. Gill, head kidney (HK), and gut tissues were collected 24 and 48 h after challenge, and RNA was extracted for real-time PCR analysis. Data were analyzed by the Pfaffl method (48) and are expressed as fold change. Data represent averages + standard errors of the results. Asterisks indicate significant differences (P < 0.05) relative to control. Data were analyzed using the Student T test.
To examine the regulation of IL-17A/F2 and IL-17RA in mucosal tissues, a further bacterial challenge was performed. Although the same pathogen was used, an immersion infection procedure was performed, as this was considered a more natural exposure that would target the mucosal sites. Head kidney tissue was again collected for comparison to the injection infection. The expression of IL-17A/F2 and IL-17RA in head kidney was again induced significantly (P < 0.05) by infection, at both time points sampled (Fig. 5B). In the mucosal tissues analyzed, the expression of IL-17A/F was upregulated significantly (P < 0.05) in the gills at 24 h postinfection but downregulated at 48 h. IL-17RA expression was induced in the gut (4-fold) at 48 h postinfection but downregulated in the gills at both time points, with fold changes of approximately 0.5.
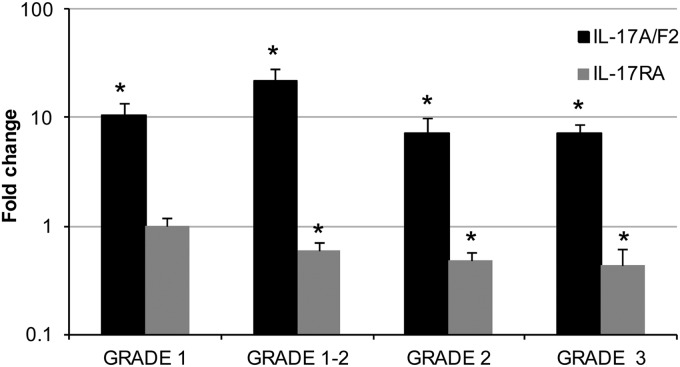
In addition to the analysis of the impact of a bacterial infection on the transcript level of IL-17A/F2 and IL-17RA, the modulation of both genes upon a natural parasitic infection with Tetracapsuloides bryosalmonae, the causative agent of salmonid proliferative kidney disease (PKD), was also investigated (Fig. 6). The disease occurs as a consequence of parasite spore development and release from infected bryozoans, a process closely linked to the increasing water temperatures. Following attachment to fish epithelial surfaces, single extrasporogonic amoeboid cells invade primarily via the gills and migrate through the vascular system to the kidney as the main target organ for further development. Extrasporogonic proliferation in the kidney interstitium provokes a chronic kidney pathology characterized by a lymphoid hyperplasia, formation of granulomatous lesions, renal atrophy, and hypersecretion of immunoglobulins (60). Since the kidney is the main organ targeted by this pathogen, it was selected to analyze regulation of immune-related genes. The results revealed that all fish with clinical pathology had a high induction of IL-17A/F2 expression, relative to the control (noninfected) fish, with fold changes > 7 being observed (with a maximum of a 22-fold increase in grade 1-2 infected fish). In contrast, the expression of IL-17RA was significantly downregulated (P < 0.05) in fish that had a pathology grade of 1-2 or more, with fold changes of 0.6 or below. Regarding the controls, there was no variation detected.
Fig 6.
Expression analysis of trout IL-17A/F2 and IL-17RA during a parasitic infection. Kidneys from rainbow trout infected with T. bryosalmonae or from unexposed fish (control) were collected during a natural infection. Kidney samples from control and parasite-infected fish exhibiting low kidney pathology (swelling indexes of 1 and 1-2) and advanced clinical pathology (swelling indexes of 2 and 3) were analyzed. RNA was extracted for real-time PCR analysis, and data were analyzed by the Pfaffl method (48) and expressed as fold change. Data represent averages + standard errors of the results (n = 5). Asterisks indicate significant differences (P < 0.05) relative to the control. Data were analyzed using the Student T test.
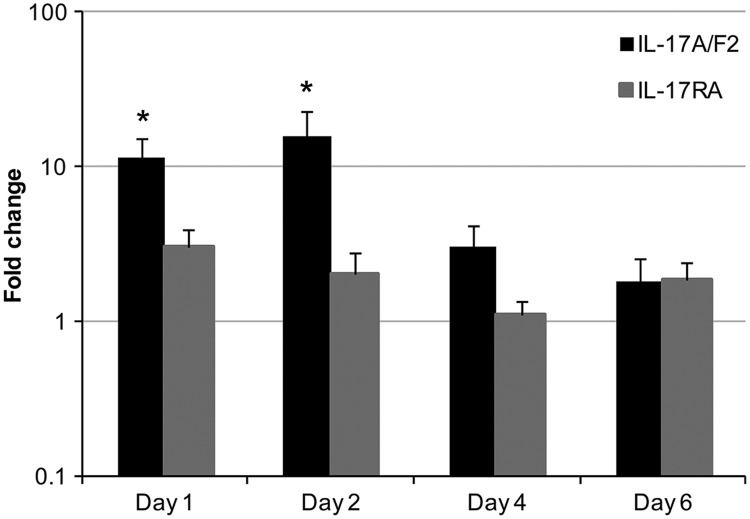
To assess the involvement of both molecules upon a viral infection, the expression of IL-17A/F2 and IL-17RA was analyzed in head kidney samples from 4 fish at several time points (days 1, 2, 4, and 6) after a viral hemorrhagic septicemia virus (VHSV) challenge (Fig. 7). The causative agent of VHS is a negative single-stranded RNA virus that belongs to the Rhabdoviridae family (53). The virus targets tissues such as the head kidney, heart, and spleen and is readily detectable at such sites (61). Therefore, once again, the kidney was chosen for gene expression analysis. The expression of trout IL-17A/F2 was upregulated significantly (P < 0.05) at days 1 and 2 postinfection, with fold changes of around 11 and 15, respectively. However, no effect on the IL-17RA transcript level, and also between controls, was observed.
Fig 7.
Expression analysis of trout IL-17A/F2 and IL-17RA during a viral infection. Four rainbow trout were injected intraperitoneally with VHSV (strain DK-F1; 1 × 108 TCID50/fish) or control media. Head kidney tissue was collected at days 1, 2, 4, and 6 after challenge and RNA extracted for real-time PCR analysis. Data were analyzed by the Pfaffl method (48) and expressed as fold change. Data represent averages + standard errors of the results. Asterisks indicate significant differences (P < 0.05) relative to the control. Data were analyzed using the Student T test.
Bioactivity studies.
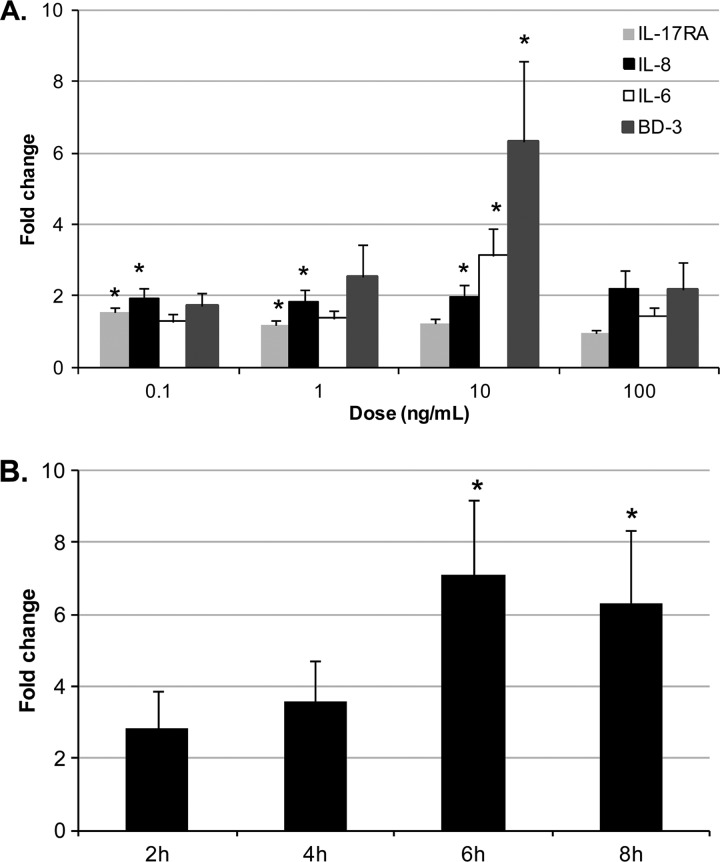
In order to determine the biological function of trout IL-17A/F2, a putative mature peptide was expressed as an N-terminal 6-histidine-tagged fusion protein in E. coli and purified with nickel-nitrilotriacetic acid (Ni-NTA) metal affinity chromatography. Splenocyte primary cultures were selected to investigate the bioactivity of trout rIL-17A/F2. A dose-response experiment was performed initially by incubating splenocyte primary cultures with a range of doses of rIL-17A/F2 from 0.1 to 100 ng/ml for 6 h (Fig. 8A). The results showed that trout rIL-17A/F2 significantly (P < 0.05) upregulated (6.3-fold) the expression of β-defensin 3 (BD-3) at one of the doses (10 ng/ml) tested. The presence of this cytokine also significantly (P < 0.05) upregulated the expression of IL-6 (3.1-fold) and IL-8 at this concentration. In addition, IL-8 was upregulated at 0.1 and 1 ng/ml rIL-17A/F2, suggesting that IL-8 gene expression is highly sensitive to this cytokine. Transcription of IL-17RA exhibited a moderate level of induced expression upon incubation with the 2 lowest doses of rIL-17A/F2. Incubation with a dose of 100 ng/ml had no significant effects. Furthermore, TNF-α1 expression was not modulated at any of the doses used (data not shown).
Fig 8.
Bioactivity of trout rIL-17A/F2. (A) Expression analysis of proinflammatory cytokines (IL-6 and IL-8), an antimicrobial peptide (BD-3), and IL-17RA in splenocytes after 6 h of incubation with a range of concentrations (0.1, 1, 10, and 100 ng/ml) of rIL-17A/F or EB-2 buffer (control). (B) Expression analysis of trout BD-3 in splenocytes incubated with rIL-17A/F2 (10 ng/ml) or EB-2 buffer (as a control) for 2, 4, 6, and 8 h. After the incubation periods, RNA was extracted and cDNA synthesized for real-time PCR analysis. Data were analyzed using the Pfaffl method (48) and expressed as fold change. Data represent averages + standard errors of the results (n = 6). Asterisks indicate significant differences (P < 0.05) relative to the control. Data were analyzed using the Student T test and one-way analysis of variance (ANOVA).
To further investigate the optimal incubation period for the activity of trout rIL-17A/F2, splenocytes were subjected to a time course experiment using a single (optimal) dose of rIL-17A/F2 (10 ng/ml) (Fig. 8B). At various times (2, 4, 6, and 8 h) postincubation, the cells were analyzed for an effect on expression of BD-3, which was the most highly modulated gene in the dose-response experiment. Cells were also incubated with elution buffer as a control. The results agreed with those of the dose-response experiment, confirming that a dose of 10 ng/ml rIL-17A/F significantly upregulated BD-3 expression after 6 and 8 h of stimulation but not at the earlier time points. Additionally, no variation between controls was detected at the different time points (data not shown).
DISCUSSION
For this study, the cloning and characterization of IL-17A/F2 and its putative receptor IL-17RA in rainbow trout are reported. The expression of these molecules during infection and stimulation in vitro has been studied and the bioactivity of the IL-17A/F protein investigated for the first time in a nonmammalian species. The putative IL-17A/F protein consists of 146 aa and contains a predicted 25-aa signal peptide, suggesting that it is a secreted protein, as reported in other vertebrate species (7, 16, 17). Two copies of the mRNA instability motif ATTTA were present in the 3′ untranslated region (UTR), as found in IL-17 molecules of other species, such as fugu (17), trout (62), and humans (7). These motifs are typical of proinflammatory molecules, which show transitory expression to reduce host damage (63), illustrating a potential role of the trout IL-17A/F in inflammatory responses. A multiple amino acid alignment revealed conservation of 4 cysteine residues for almost all the species considered, except for fugu IL-17A/F2 (17), which are signature residues in the IL-17 family (12, 16–18). The presence of 4 cysteine residues in trout and other IL-17A/F teleost molecules (16, 17) suggests that they might be involved in the formation of 2 disulfide bonds, which form the cysteine knot characteristic of the IL-17 family (12, 13). Relative to glycosylation, 2 putative sites were found in trout, with potential glycosylation sites also reported in other IL-17A and IL-17F molecules (7, 13, 63). In terms of homology, trout IL-17A/F2 showed the highest amino acid similarity to zebrafish IL-17A/F2 (58.9%), followed by fugu IL-17A/F2 (56.8%) and human IL-17A (47.7%). A phylogenetic tree constructed using several sequences belonging to members of the IL-17 family also confirmed that trout IL-17A/F shared a close relationship with teleost IL-17A/F2 molecules, supported by a high bootstrap value of 99%.
The trout IL-17RA consists of a transmembrane protein with a 284-aa extracellular region and a long intracellular domain composed of 639 aa. According to analysis of the phylogenetic relationship between the various IL-17R protein family members, trout IL-17RA clustered with the other piscine and homeotherm IL-17RA molecules. Multiple alignment of this protein sequence with those of other vertebrate IL-17RA molecules revealed a high degree of conservation within the extracellular and intracellular SEFIR domains, which are characteristic of the IL-17RA family (18, 24, 25). Several conserved cysteines, eight of which are potentially involved in disulfide bridge formation, as seen in mammalian molecules (64), were also present in trout IL-17RA. Two putative FNIII domains which are known to be crucial in the binding of the ligand to its receptor (25) were also identified within the extracellular domain of trout IL-17RA.
Tissue distribution analysis of trout IL-17A/F2 revealed a high transcript level in the intestine and gills. This finding was in agreement with previous work performed in zebrafish (16) and suggests a potential involvement of the trout molecule in mucosal immunity. The IL-17RA homologue was highly expressed in the thymus, which is in concordance with the high constitutive expression of the mammalian IL-17RA homologue in lymphoid tissues (35). In order to investigate whether the expression of trout IL-17A/F2 and IL-17RA could be modulated, primary cultures were prepared from the head kidney and stimulated in vitro. Of the stimulants used, phytohemagglutinin (PHA), phorbol 12-myristate 13-acetate (PMA), and calcium ionophore (CI) all increased the transcript level of both molecules. PHA, a known T cell stimulant, had the greatest impact on IL-17A/F2 expression. Since PHA can induce T cell proliferation, the expression profiles determined in this study imply that these molecules are modulated during T cell activation. Similarly, both PMA and PHA have been reported to induce detectable levels of IL-17A in human studies (3). In contrast, dexamethasone (DM) had a negative effect on the transcript levels of both IL-17A/F2 and IL-17RA. Glucocorticoids, such as DM, suppress cytokine expression in activated immune cells as a negative-feedback mechanism to regulate the extent of inflammatory reactions (65). The immunosuppressing activity of DM has been previously reported in the characterization of fish IL-2, IL-6 (47), and IL-20 (66). Moreover, the presence of this glucocorticoid inhibits the ability of IL-17 to induce IL-6 in human bronchial fibroblasts (65).
Yersinia ruckeri is the etiological agent of salmonid enteric redmouth disease (ERM), a disease that can potentially cause significant economic losses in the trout-farming industry (59, 67). This pathogen is known to colonize vascularized tissues, including the spleen and head kidney (59), and has been reported to induce a trout homologue of the T helper (TH) 17 cytokine, IL-22 (36). In this study, we found that both IL-17A/F2 and IL-17RA were induced in the head kidney of Y. ruckeri-infected fish. Since trout IL-17A/F2 was highly expressed in mucosal sites (gills and intestine) relative to other trout tissues, we investigated the expression of these molecules following Y. ruckeri immersion challenge. The results confirmed that both molecules were induced in the head kidney at the time points examined (24 and 48 h). The expression of IL-17A/F2 in the gill was upregulated 24 h after exposure to the pathogen but was downregulated at 48 h. The gills are the first site of entry of Y. ruckeri in a natural exposure, where its presence is detected within hours after infection. However, bacterial abundance in this tissue also seems to decrease rapidly, being almost absent 48 h postexposure (50). Thus, suppression of both IL-17A/F2 and IL-17RA expression indicates that the responses induced by this cytokine at this site may no longer be required and that a rapid negative-feedback loop is in place to control IL-17A/F2 secretion, perhaps due to the multifunctionality of this tissue where enhanced immune function may compromise oxygen or ion transport. In the gut, only the expression of IL-17RA was affected (at 48 h). Thus, the importance of trout IL-17A/F2 for mucosal immunity is not clear, despite the crucial role of IL-17 and IL-17RA in mucosae of bacterially infected mammalian hosts (9, 31, 68, 69). In addition, since the level of upregulation is not high, this leads us to speculate that the IL-17A/F2 already secreted in mucosal tissues is sufficient to control infection. However, further studies on other bacterial pathogens should help clarify the role of this pathway in fish.
Proliferative kidney disease is a chronic parasitic infection caused by the presence of the myxozoan parasite Tetracapsuloides bryosalmonae. Parasite development in infected salmonid kidney is characterized by an abnormal in situ proliferation of lymphomyeloid tissue within the kidney interstitium (60). Immune gene expression profiles in parasite-infected kidney tissue are consistent with a TH1-like immune response, although lacking the classical signs of a proinflammatory response (51). In this study, we observed upregulation of trout IL-17A/F2 at all stages of disease pathology, indicating that IL-17A/F2 is highly sensitive to the presence of this parasite. Similarly, Trypanoplasma carassii, the fish protozoan parasite, was found to induce carp IL-17A/F2 expression in head kidney tissue (70). Thus, modulation of the IL-17A/F2 ligand and the IL-17RA receptor in fish-parasite interactions reinforces the idea of the potential importance of this molecule in the immune response to eukaryotic fish parasites.
Viral hemorrhagic septicemia virus (VHSV) is an aquatic rhabdovirus, the etiological agent of viral hemorrhagic septicemia, which is responsible for severe economic losses of rainbow trout in aquaculture practices (71). The virus causes severe internal hemorrhaging, with the kidney being one of the tissues targeted in rainbow trout (71). In this study, the expression of IL-17A/F2 was upregulated at days 1 and 2 after viral infection. The role of IL-17A in viral infections is still not completely clear, with studies revealing a potential role of this molecule in protection against intracellular pathogens (32, 69). However, TH17 cytokines may have a more significant role in protection against extracellular pathogens, with only a minor involvement in the immune response to intracellular viral or bacterial pathogens. Indeed, it is known that IL-17RA-deficient mice are not susceptible to infections with intracellular pathogens, such as Mycobacterium tuberculosis (5). Since no VHSV-dependent modulation of IL-17RA was detected, it is possible that signaling via this receptor is not required for the primary control of VHSV in fish.
Even though several IL-17A/F molecules have been identified in different fish species (16–18), no functional data have been ascribed to IL-17A/F activity in lower vertebrates to date. Here, for the first time in a nonmammalian model, we have produced and tested the bioactivity of trout rIL-17A/F2. The effect of trout rIL-17A/F on the expression of antimicrobial peptides (AMPs) was studied with a particular focus on the β-defensins, since they are known to be modulated by TH17 cytokines in mammals (10, 33, 35, 72). β-defensins have been well characterized in previous rainbow trout reports (37, 73). In this study, β-defensin 3 (BD-3) was selected as a marker gene, since previous work (36) demonstrated a clear effect of fish recombinant IL-22 on BD-3 expression. Following incubation with rIL-17A/F2, trout splenocytes displayed a dose- and time-dependent induction of BD-3 expression. In mammalian studies, IL-17A induced the expression of human BD-2 and mouse BD-3 at a dose of 1 ng/ml (33). However, it is worth noting that mammalian BD-2 and BD-3 molecules are not equivalent homologues of trout BD-2 and BD-3 (37). Mammalian studies on the importance of IL-17A and IL-17F in inducing expression of inflammatory cytokines such as IL-6 and IL-8 have also been reported (2, 13, 29, 74). The results obtained with trout rIL-17A/F2 were in agreement with these studies, with 10 ng/ml of rIL-17A/F2 inducing a ∼3-fold increase in IL-6 expression and ∼2-fold increase in IL-8 expression. IL-6 is induced by the presence of inflammatory cytokines such as tumor necrosis factor alpha (TNF-α) (75) and IL-1β (76). Trout studies have revealed IL-6 to be upregulated in vitro upon incubation of leukocytes with bacterial LPS (56), PolyI:C, and IL-1β (77) or in vivo in bacterially infected trout (49, 78), implying an important role for IL-6 in fish inflammatory processes, a premise that is corroborated by IL-6 data in this study. The upregulation of IL-8 by rIL-17A/F2 is also in accordance with mammalian studies where IL-17 has been shown to induce the expression of IL-8 in a variety of cell types, including human fibroblasts (3) and bronchial epithelial and venous endothelial cells (2, 29, 30). IL-8 is widely known for its key role in recruiting and activating neutrophils in areas of acute inflammation, with functions similar to those ascribed to this cytokine in fish (79). When analyzing the impact of trout rIL-17A/F2 on IL-17RA expression, only the lowest doses were stimulatory, with no effect or a downward trend at higher doses. This might occur as a means of controlling IL-17A/F signaling via the internalization of IL-17A/F and reduction in IL-17RA surface expression (24). The functionality of rIL-17A/F2 was also confirmed in a time course experiment, where expression of trout BD-3 at a dose of 10 ng/ml was induced by 6 h poststimulation but not earlier.
In summary, this report describes the characterization of trout IL-17A/F2 and IL-17RA homologues and presents for the first time data on IL-17A/F2 bioactivity in a nonmammalian species. Analysis of the trout IL-17A/F2 and IL-17RA sequences revealed the presence of important features characteristic of their particular gene families. Expression of both ligand and putative receptor was modulated in trout lymphoid tissues upon bacterial, parasitic, and viral infections, which implicates IL-17A/F2 as a key molecule in fish immunity. Our investigations into the bioactivity of trout rIL-17A/F2 revealed the modulation of β-defensin and proinflammatory cytokine expression, further reinforcing its role in promoting the link between adaptive and innate immune responses, as seen with the mammalian molecules.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported financially by a Ph.D. studentship from the FCT (Foundation for Science and Technology, Portugal) and POPH/FSE (grant no. SFRH/BD/38236/2007) to Milena M. Monte and the European Commission (contract no. 007103; IMAQUANIM-Improved immunity of aquacultured animals).
We thank Marine Scotland staff for providing samples from VHSV-infected trout generated within Scottish government-funded research project FC1996. We also acknowledge Davide Pacitti for help in preparing the bacterially infected samples and Maria M. Costa for preparing head kidney stimulated leukocyte samples.
Footnotes
Published ahead of print 12 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00599-12.
REFERENCES
- 1. Aggarwal S, Gurney AL. 2002. IL-17: prototype member of an emerging cytokine family. J. Leukoc. Biol. 71:1–8 [PubMed] [Google Scholar]
- 2. Kolls JK, Lindén A. 2004. Interleukin-17 family members and inflammation. Immunity 21:467–476 [DOI] [PubMed] [Google Scholar]
- 3. Yao Z, Painter SL, Fanslow WC, Ulrich D, Macduff BM, Spriggs MK, Armitage RJ. 1995. Human IL-17: a novel cytokine derived from T cells. J. Immunol. 155:5483–5486 [PubMed] [Google Scholar]
- 4. Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6:1123–1132 [DOI] [PubMed] [Google Scholar]
- 5. Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. 2005. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201:233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6:1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yao Z, Fanslow WC, Seldin MF, Rousseau AM, Painter SL, Comeau MR, Cohen JI, Spriggs MK. 1995. Herpesvirus saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity 3:811–821 [DOI] [PubMed] [Google Scholar]
- 8. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. 2006. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17 T helper cells. Cell 126:1121–1133 [DOI] [PubMed] [Google Scholar]
- 9. Ye P, Garvey PB, Zhang P, Nelson S, Bagby G, Summer WR, Schwarzenberger P, Shellito JE, Kolls JK. 2001. Interleukin-17 and lung host defense against Klebsiella pneumoniae infection. Am. J. Respir. Cell Mol. Biol. 25:335–340 [DOI] [PubMed] [Google Scholar]
- 10. Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. 2006. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203:2271–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eyerich S, Eyerich K, Cavani A, Schmidt-Weber C. 2010. IL-17 and IL-22: siblings, not twins. Trends Immunol. 31:354–361 [DOI] [PubMed] [Google Scholar]
- 12. Weaver CT, Hatton RD, Mangan PR, Harrington LE. 2007. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25:821–852 [DOI] [PubMed] [Google Scholar]
- 13. Hymowitz SG, Filvaroff EH, Yin JP, Lee J, Cai L, Risser P, Maruoka M, Mao W, Foster J, Kelley RF, Pan G, Gurney AL, de Vos AM, Starovasnik MA. 2001. IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding. EMBO J. 20:5332–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Starnes T, Robertson MJ, Sledge G, Kelich S, Nakshatri H, Broxmeyer HE, Hromas R. 2001. Cutting edge: IL-17F, a novel cytokine selectively expressed in activated T cells and monocytes, regulates angiogenesis and endothelial cell cytokine production. J. Immunol. 167:4137–4140 [DOI] [PubMed] [Google Scholar]
- 15. Chang SH, Dong C. 2007. A novel heterodimeric cytokine consisting of IL-17 and IL-17F regulates inflammatory responses. Cell Res. 17:435–440 [DOI] [PubMed] [Google Scholar]
- 16. Gunimaladevi I, Savan R, Sakai M. 2006. Identification, cloning and characterization of interleukin-17 and its family from zebrafish. Fish Shellfish Immunol. 21:393–403 [DOI] [PubMed] [Google Scholar]
- 17. Korenaga H, Kono T, Sakai M. 2010. Isolation of seven IL-17 family genes from the Japanese pufferfish Takifugu rubripes. Fish Shellfish Immunol. 28:809–818 [DOI] [PubMed] [Google Scholar]
- 18. Kono T, Korenaga H, Sakai M. 2011. Genomics of fish IL-17 ligand and receptors: a review. Fish Shellfish Immunol. 31:635–643 [DOI] [PubMed] [Google Scholar]
- 19. Secombes CJ, Wang T, Bird S. 2011. The interleukins of fish. Dev. Comp. Immunol. 35:1336–1345 [DOI] [PubMed] [Google Scholar]
- 20. McAllister F, Henry A, Kreindler JL, Dubin PJ, Ulrich L, Steele C, Finder JD, Pilewski JM, Carreno BM, Goldman SJ, Pirhonen J, Kolls JK. 2005. Role of IL-17A, IL-17F, and the IL-17 receptor in regulating growth-related oncogene-alpha and granulocyte colony-stimulating factor in bronchial epithelium: implications for airway inflammation in cystic fibrosis. J. Immunol. 175:404–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kramer JM, Yi L, Shen F, Maitra A, Jiao X, Jin T, Gaffen SL. 2006. Cutting edge: evidence for ligand-independent multimerization of the IL-17 receptor. J. Immunol. 176:711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Toy D, Kugler D, Wolfson M, Vandel Bos T, Gurgel J, Derry J, Tocker J, Peschon J. 2006. Cutting edge: interleukin 17 signals through a heteromeric receptor complex. J. Immunol. 177:36–39 [DOI] [PubMed] [Google Scholar]
- 23. Kuestner RE, Taft DW, Haran A, Brandt CS, Brender T, Lum K, Harder B, Okada S, Ostrander CD, Kreindler JL, Aujla SJ, Reardon B, Moore M, Shea P, Schreckhise R, Bukowski TR, Presnell S, Guerra-Lewis P, Parrish-Novak J, Ellsworth JL, Jaspers S, Lewis KE, Appleby M, Kolls JK, Rixon M, West JW, Gao Z, Levin SD. 2007. Identification of the IL-17 receptor related molecule IL-17RC as the receptor for IL-17F. J. Immunol. 179:5462–5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaffen SL. 2009. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 9:556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu B, Jin M, Zhang Y, Wei T, Bai Z. 2011. Evolution of the IL17 receptor family in chordates: a new subfamily IL17REL. Immunogenetics 63:835–845 [DOI] [PubMed] [Google Scholar]
- 26. Novatchkova M, Leibbrandt A, Werzowa J, Neubüser A, Eisenhaber F. 2003. The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem. Sci. 28:226–229 [DOI] [PubMed] [Google Scholar]
- 27. Gaffen SL. 2008. An overview of IL-17 function and signaling. Cytokine 43:402–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yao Z, Spriggs MK, Derry JM, Strockbine L, Park LS, VandenBos T, Zappone JD, Painter SL, Armitage RJ. 1997. Molecular characterization of the human interleukin (IL)-17 receptor. Cytokine 9:794–800 [DOI] [PubMed] [Google Scholar]
- 29. Laan M, Cui ZH, Hoshino H, Lötvall J, Sjöstrand M, Gruenert DC, Skoogh BE, Lindén A. 1999. Neutrophil recruitment by human IL-17 via CXC chemokine release in the airways. J. Immunol. 162:2347–2352 [PubMed] [Google Scholar]
- 30. Kawaguchi M, Adachi M, Oda N, Kokubu F, Huang SK. 2004. IL-17 cytokine family. J. Allergy Clin. Immunol. 114:1265–1273 [DOI] [PubMed] [Google Scholar]
- 31. Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK. 2008. IL-22 mediates mucosal host defense against gram-negative bacterial pneumonia. Nat. Med. 14:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ouyang W, Kolls JK, Zheng Y. 2008. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 28:454–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kao CY, Chen Y, Thai P, Wachi S, Huang F, Kim C, Harper RW, Wu R. 2004. IL-17 markedly up-regulates beta-defensin-2 expression in human airway epithelium via JAK and NF-kappaB signaling pathways. J. Immunol. 173:3482–3491 [DOI] [PubMed] [Google Scholar]
- 34. Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, Basham B, Smith K, Chen T, Morel F, Lecron JC, Kastelein RA, Cua DJ, McClanahan TK, Bowman EP, de Waal Malefyt R. 2007. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat. Immunol. 8:950–957 [DOI] [PubMed] [Google Scholar]
- 35. Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, Iwakura Y. 2009. Differential roles of interleukin-17A and-17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30:108–119 [DOI] [PubMed] [Google Scholar]
- 36. Monte MM, Zou J, Wang T, Carrington A, Secombes CJ. 2011. Cloning, expression analysis and bioactivity studies of rainbow trout (Oncorhynchus mykiss) interleukin-22. Cytokine 55:62–73 [DOI] [PubMed] [Google Scholar]
- 37. Casadei E, Wang T, Zou J, González Vecino JL, Wadsworth S, Secombes CJ. 2009. Characterization of three novel beta-defensin antimicrobial peptides in rainbow trout (Oncorhynchus mykiss). Mol. Immunol. 46:3358–3366 [DOI] [PubMed] [Google Scholar]
- 38. Komatsu K, Tsutsui S, Hino K, Araki K, Yoshiura Y, Yamamoto A, Nakamura O, Watanabe T. 2009. Expression profiles of cytokines released in intestinal epithelial cells of the rainbow trout, Oncorhynchus mykiss, in response to bacterial infection. Dev. Comp. Immunol. 33:499–506 [DOI] [PubMed] [Google Scholar]
- 39. Wang T, Holland JW, Bols N, Secombes CJ. 2005. Cloning and expression of the first nonmammalian interleukin-11 gene in rainbow trout Oncorhynchus mykiss. FEBS J. 272:1136–1147 [DOI] [PubMed] [Google Scholar]
- 40. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 43. Campanella JJ, Bitincka L, Smalley J. 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4:29 doi:10.1186/1471-2105-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 45. Letunic I, Doerks T, Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37(Database issue):D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marsden RL, McGuffin LJ, Jones DT. 2002. Rapid protein domain assignment from amino acid sequence using predicted secondary structure. Protein Sci. 11:2814–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang T, Díaz-Rosales Costa MM, Campbell S, Snow M, Collet B, Martin SA, Secombes CJ. 2011. Functional characterization of a nonmammalian IL-21: rainbow trout Oncorhynchus mykiss IL-21 upregulates the expression of the Th cell signature cytokines IFN-gamma, IL-10, and IL-22. J. Immunol. 186:708–721 [DOI] [PubMed] [Google Scholar]
- 48. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 doi:10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Harun NO, Wang T, Secombes CJ. 2011. Gene expression profiling in naïve and vaccinated rainbow trout after Yersinia ruckeri infection: insights into the mechanisms of protection seen in vaccinated fish. Vaccine 29:4388–4399 [DOI] [PubMed] [Google Scholar]
- 50. Tobback E, Decostere A, Hermans K, Ryckaert J, Duchateau L, Haesebrouck F, Chiers K. 2009. Route of entry and tissue distribution of Yersinia ruckeri in experimentally infected rainbow trout Oncorhynchus mykiss. Dis. Aquat. Organ. 84:219–228 [DOI] [PubMed] [Google Scholar]
- 51. Wang T, Holland JW, Martin SAM, Secombes CJ. 2010. Sequence and expression analysis of two T helper master transcription factors, T-bet and GATA3, in rainbow trout Oncorhynchus mykiss and analysis of their expression during bacterial and parasitic infection. Fish Shellfish Immunol. 29:705–715 [DOI] [PubMed] [Google Scholar]
- 52. Clifton-Hadley RS, Bucke D, Richards RH. 1987. A study of the sequential clinical and pathological changes during proliferative kidney disease in rainbow trout, Salmo gairdneri Richardson. J. Fish Dis. 10:335–352 [Google Scholar]
- 53. Campbell S, McBeath A, Secombes C, Snow M, Collet B. 2011. Interferon response following infection with genetically similar isolates of viral haemorrhagic septicaemia virus (VHSV) exhibiting contrasting virulence in rainbow trout. Fish Shellfish Immunol. 30:287–294 [DOI] [PubMed] [Google Scholar]
- 54. Zou J, Carrington A, Collet B, Dijkstra JM, Yoshiura Y, Bols N, Secombes C. 2005. Identification and bioactivities of IFN-gamma in rainbow trout Oncorhynchus mykiss: the first Th1-type cytokine characterized functionally in fish. J. Immunol. 175:2484–2494 [DOI] [PubMed] [Google Scholar]
- 55. Laing KJ, Wang T, Zou J, Holland J, Hong S, Bols N, Hirono I, Aoki T, Secombes CJ. 2001. Cloning and expression analysis of rainbow trout Oncorhynchus mykiss tumour necrosis factor-alpha. Eur. J. Biochem. 268:1315–1322 [DOI] [PubMed] [Google Scholar]
- 56. Iliev DB, Castellana B, MacKenzie S, Planas JV, Goetz FW. 2007. Cloning and expression analysis of an IL-6 homolog in rainbow trout (Oncorhynchus mykiss). Mol. Immunol. 44:1803–1807 [DOI] [PubMed] [Google Scholar]
- 57. Laing KJ, Zou JJ, Wang T, Bols N, Hirono I, Aoki T, Secombes CJ. 2002. Identification and analysis of an interleukin 8-like molecule in rainbow trout Oncorhynchus mykiss. Dev. Comp. Immunol. 26:433–444 [DOI] [PubMed] [Google Scholar]
- 58. Morris DR, Geballe AP. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20:8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tobback E, Decostere A, Hermans K, Haesebrouck F, Chiers K. 2007. Yersinia ruckeri infections in Salmonid fish. J. Fish Dis. 30:257–268 [DOI] [PubMed] [Google Scholar]
- 60. Okamura B, Hartikainen H, Schmidt-Posthaus H, Wahli T. 2011. Life cycle complexity, environmental change and the emerging status of salmonid proliferative kidney disease. Freshwater Biol. 56:735–753 [Google Scholar]
- 61. Tafalla C, Coll J, Secombes C. 2005. Expression of genes related to the early immune response in rainbow trout (Oncorhynchus mykiss) after viral haemorrhagic septicemia virus (VHSV) infection. Dev. Comp. Immunol. 29:615–626 [DOI] [PubMed] [Google Scholar]
- 62. Wang T, Martin SAM, Secombes CJ. 2010. Two interleukin-17C-like genes exist in rainbow trout Oncorhynchus mykiss that are differentially expressed and modulated. Dev. Comp. Immunol. 34:491–500 [DOI] [PubMed] [Google Scholar]
- 63. Rouvier E, Luciani M, Mattei M, Denizot F, Golstein P. 1993. CTLA-8, cloned from an activated T cell, bearing AU-rich messenger RNA instability sequences, and homologous to a herpesvirus saimiri gene. J. Immunol. 150:5445–5456 [PubMed] [Google Scholar]
- 64. Ely LK, Fischer S, Garcia KC. 2009. Structural basis of receptor sharing by interleukin 17 cytokines. Nat. Immunol. 10:1245–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Molet S, Hamid Q, Davoine F, Nutku E, Taha R, PagÉ N, Olivenstein R, Elias J, Chakir J. 2001. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J. Allergy Clin. Immunol. 108:430–438 [DOI] [PubMed] [Google Scholar]
- 66. Wang T, Díaz-Rosales Martin SA, Secombes CJ. 2010. Cloning of a novel interleukin (IL)-20-like gene in rainbow trout Oncorhynchus mykiss gives an insight into the evolution of the IL-10 family. Dev. Comp. Immunol. 34:158–167 [DOI] [PubMed] [Google Scholar]
- 67. Fernández L, Méndez J, Guijarro JA. 2007. Molecular virulence mechanisms of the fish pathogen Yersinia ruckeri. Vet. Microbiol. 125:1–10 [DOI] [PubMed] [Google Scholar]
- 68. Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. 2001. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 194:519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Matsuzaki G, Umemura M. 2007. Interleukin-17 as an effector molecule of innate and acquired immunity against infections. Microbiol. Immunol. 51:1139–1147 [DOI] [PubMed] [Google Scholar]
- 70. Ribeiro CMS, Pontes MJSL, Bird S, Chadzinska M, Scheer M, Kemenade BMLV, Savelkoul HFJ, Wiegertjes GF. 2010. Trypanosomiasis-induced Th17-like immune responses in carp. PLoS One 5:e13012 doi:10.1371/journal.pone.0013012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Skall HF, Olesen NJ, Mellergaard S. 2005. Viral haemorrhagic septicaemia virus in marine fish and its implications for fish farming—a review. J. Fish Dis. 28:509–529 [DOI] [PubMed] [Google Scholar]
- 72. Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. 2004. IL-22 increases the innate immunity of tissues. Immunity 21:241–254 [DOI] [PubMed] [Google Scholar]
- 73. Falco A, Chico V, Marroqui L, Perez L, Coll J, Estepa A. 2008. Expression and antiviral activity of a beta-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol. Immunol. 45:757–765 [DOI] [PubMed] [Google Scholar]
- 74. Kawaguchi M, Kokubu F, Kuga H, Matsukura S, Hoshino H, Ieki K, Imai T, Adachi M, Huang SK. 2001. Modulation of bronchial epithelial cells by IL-17. J. Allergy Clin. Immunol. 108:804–809 [DOI] [PubMed] [Google Scholar]
- 75. Ammit AJ, Lazaar AL, Irani C, O'Neill GM, Gordon ND, Amrani Y, Penn RB, Panettieri RA., Jr 2002. Tumor necrosis factor-alpha-induced secretion of RANTES and interleukin-6 from human airway smooth muscle cells: modulation by glucocorticoids and beta-agonists. Am. J. Respir. Cell Mol. Biol. 26:465–474 [DOI] [PubMed] [Google Scholar]
- 76. Bergamaschi A, Corsi M, Garnier MJ. 2006. Synergistic effects of cAMP-dependent signalling pathways and IL-1 on IL-6 production by H19-7/IGF-IR neuronal cells. Cell Signal 18:1679–1684 [DOI] [PubMed] [Google Scholar]
- 77. Costa MM, Maehr T, Díaz-Rosales P, Secombes CJ, Wang T. 2011. Bioactivity studies of rainbow trout (Oncorhynchus mykiss) interleukin-6: effects on macrophage growth and antimicrobial peptide gene expression. Mol. Immunol. 48:1903–1916 [DOI] [PubMed] [Google Scholar]
- 78. Raida MK, Buchmann K. 2009. Innate immune response in rainbow trout (Oncorhynchus mykiss) against primary and secondary infections with Yersinia ruckeri O1. Dev. Comp. Immunol. 33:35–45 [DOI] [PubMed] [Google Scholar]
- 79. Harun NO, Zou J, Zhang YA, Nie P, Secombes CJ. 2008. The biological effects of rainbow trout (Oncorhynchus mykiss) recombinant interleukin-8. Dev. Comp. Immunol. 32:673–681 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.