Abstract
Enterotoxigenic Escherichia coli (ETEC) strains are a leading cause of morbidity and mortality due to diarrheal illness in developing countries. There is currently no effective vaccine against these important pathogens. Because genes modulated by pathogen-host interactions potentially encode putative vaccine targets, we investigated changes in gene expression and surface morphology of ETEC upon interaction with intestinal epithelial cells in vitro. Pan-genome microarrays, quantitative reverse transcriptase PCR (qRT-PCR), and transcriptional reporter fusions of selected promoters were used to study changes in ETEC transcriptomes. Flow cytometry, immunofluorescence microscopy, and scanning electron microscopy were used to investigate alterations in surface antigen expression and morphology following pathogen-host interactions. Following host cell contact, genes for motility, adhesion, toxin production, immunodominant peptides, and key regulatory molecules, including cyclic AMP (cAMP) receptor protein (CRP) and c-di-GMP, were substantially modulated. These changes were accompanied by visible changes in both ETEC architecture and the expression of surface antigens, including a novel highly conserved adhesin molecule, EaeH. The studies reported here suggest that pathogen-host interactions are finely orchestrated by ETEC and are characterized by coordinated responses involving the sequential deployment of multiple virulence molecules. Elucidation of the molecular details of these interactions could highlight novel strategies for development of vaccines for these important pathogens.
INTRODUCTION
The enterotoxigenic Escherichia coli (ETEC) strains are among the leading causes of diarrheal illness in developing countries. Each year, these organisms account for hundreds of thousands of deaths, particularly in young nonimmune children (1–3). Presently there is no vaccine for ETEC that offers sustained broad-based protection (4).
While these organisms share the ability to produce and effectively deliver heat-stable (ST) and/or heat-labile (LT) enterotoxins, many aspects of the pathogenesis of these organisms remain unexplored. Much of the work on ETEC pathogenesis, and consequently ETEC vaccine development, has focused intensively on the known enterotoxins, and a heterogeneous collection of plasmid-encoded colonization factors (CFs) (5). More recent studies have suggested that the pathogenesis of ETEC is considerably more complex than previously appreciated, involving additional virulence molecules. These include the EtpA exoprotein adhesin (6–8), and EatA (9), a member of the serine protease autotransporter of the Enterobacteriaceae (SPATE) family, which has recently been shown to moderate EtpA-mediated adhesion and accelerate delivery of LT (10). Moreover, many proteins, including EtpA and EatA are recognized following infection, suggesting that there may be additional vaccine targets in addition to LT and CFs (11).
While modulation of virulence genes following host cell contact is well described for a number of pathogens (12, 13), and pilus-mediated adherence has been shown to induce gene expression in E. coli (14), little is known regarding transcriptional or translational modification of ETEC on interaction with the intestinal epithelium. Because the identification of genes modulated during pathogen-host interactions can potentially be used to identify additional previously unheralded targets for vaccine development (15–17), we investigated transcriptional changes in ETEC following attachment to host cells. The studies reported here demonstrate that in response to interactions with intestinal epithelial cells, ETEC strains modulate a large number of genes, including those encoding recently described novel virulence proteins, putative virulence factors, and important virulence regulators, including cyclic AMP (cAMP) receptor protein-cAMP complex (CRP-cAMP), and cyclic-di-GMP. Paralleling these changes, we observed significant alteration in surface molecules and the architecture of ETEC following host cell attachment.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A complete list of the strains and plasmids used in these studies is included in Table 1. ETEC strain E24377A was kindly provided by Stephen Savarino at the National Naval Medical Center and was obtained from good manufacturing practice (GMP) lots of bacteria maintained at Walter Reed Army Institute of Research (WRAIR). The ETEC H10407 isolate used in these studies was also originally obtained from Marcia Wolf at WRAIR from a GMP lot used in volunteer studies.
Table 1.
Plasmids and bacterial strains used in these studies
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strains | ||
| H10407 | ETEC (origin, Bangladesh) O78:H11 CFA/I LT/ST-1a/ST-1b | 61 |
| E24377A | ETEC (origin, Egypt) O139:H28 CFA/II (CS1/CS3) LT/ST | 62 |
| BL21-AI | F− ompT hsdSB(rB− mB−) gal dcm araB::T7 RNAP-tetA | |
| GPM1746a | H10407 hsdR1::lacZ::Tn10 | G. P. Munson, unpublished data |
| jf2624 | GPM1746a containing single-copy integrant of eaeH promoter-lacZYA transcriptional fusion at attBHK022 | This study |
| jf2575 | GPM1746a containing promoterless lacZYA reporter integrated at attBHK022 | This study |
| jf2450 | H10407(pGFPmut3.1) | This study |
| Plasmids | ||
| pAH69 | CRIM integrase expression helper plasmid | 23 |
| pHKLac1 | Reporter with promoterless lacZYA, R6Kγ, aadA, attPHK022 | 22 |
| pQL002 | eaeH promoter cloned into BamHI/EcoRI sites of pHKLac1 | This study |
| pGFPmut3.1 | pUC18-based gfpmut3* expression plasmid, Ampr | 63 |
| pDONOR221 | Entry cloning plasmid containing ccdB negative-selection gene and Cmr cassette between lambda attP sites for recombination with PCR products containing attB regions, Kmr | Invitrogen |
| pET-DEST42 | T7 lac promoter/IPTG-inducible expression plasmid, ccdB, Cmr genes flanked by attR sites for recombination with entry plasmid, C-terminal V5 and 6-His tags | Invitrogen |
| pSS001 | 4,254-bp eaeH amplicon cloned into pDONOR221 | This study |
| pSS002 | EaeH expression plasmid derived from LR recombination reaction of pSS001 and pET-DEST42 placing eaeH in frame with V5 and 6-His epitope tags | This study |
Ampr, ampicillin resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance; aadA, aminoglycoside-3-adenyltransferase gene encoding streptomycin and spectinomycin resistance; attB, bacterial chromosome phage integration site; attP, complementary phage-derived integration site; R6Kγ, pir-dependent origin of replication.
Caco-2 cell culture conditions.
Caco-2 intestinal epithelial cells (ATCC HTB-37) were propagated in Eagle's minimal essential medium supplemented with fetal bovine serum to a final concentration of 20% and maintained in a 5% CO2 atmosphere at 37°C. Caco-2 cell monolayers were grown to near confluence in 20- by 100-mm dishes.
Bacterial growth conditions and infection of intestinal epithelial cells.
Cultures of ETEC strains H10407 and E24377A were grown from frozen glycerol stocks overnight in 2 ml Luria broth (LB) at 37°C with shaking at 225 rpm. The following morning, cultures were diluted 1:100 in fresh LB and grown at 37°C at 225 rpm for an additional 90 min. Bacteria were then centrifuged at 10,000 × g for 1 min and resuspended in tissue culture medium at a concentration of ∼2 × 108 CFU/ml. Caco-2 monolayer dishes were each inoculated with ∼109 bacteria and allowed to adhere for 15, 30, 60, or 120 min. At the end of the incubation period, nonadherent (planktonic) bacteria were collected from the supernatant media by centrifugation at 10,000 × g and washed once in ice-cold Hanks' balanced salt solution (HBSS), followed by HBSS containing 1% saponin, and after centrifugation, bacterial pellets were saved at −80°C for further processing. Monolayers with adherent bacteria were washed twice with 5 ml of ice-cold Hanks' balanced salt solution (HBSS) and then lysed with 5 ml of 1% saponin in HBSS for 5 min on ice to release adherent bacteria. Bacteria released by lysis of Caco-2 cells were recovered as pellets following centrifugation at 4°C and then saved at −80°C for subsequent RNA extraction.
Isolation of ETEC RNA.
Total RNA was isolated from adherent and planktonic (nonadherent) bacterial fractions using an RNeasy minikit (Qiagen; catalog no. 74104) and was treated with DNase (Ambion; catalog no. AM1907). Conventional PCR for arcA (a housekeeping gene) was used to confirm the removal of DNA (data not shown). The RNA obtained from the adherent bacterial population is significantly contaminated with host RNA. Using 10 μg total RNA as the starting material, samples were processed with MICROBEnrich (Ambion; catalog no. AM1901) to remove host RNA, followed by depletion of the bacterial rRNA using MICROBExpress (Ambion; catalog no. AM1905). The remaining RNA, primarily bacterial mRNA, was ethanol precipitated and dissolved in 30 μl RNase-free water.
Microarray hybridization.
To compare the transcriptomes of adherent and planktonic ETEC E24377A following infection of Caco-2 intestinal epithelial cells, we used the FDA-E. coli-Shigella (FDA-ECSG) Affymetrix array (http://pfgrc.jcvi.org/index.php/microarray/affy_array_description/ecoli_shigella/version1.html), which represents genes from multiple E. coli pathogens, including those from ETEC strain E24377A (18). Array hybridizations were carried out as previously described (19). Briefly, cDNA was synthesized with random hexamers and partially digested with DNase to obtain 20- to 200-bp fragments, end labeled with biotin-11-ddATP, and hybridized for 16 h at 45°C. Microarrays for processing were allowed to warm to room temperature for 30 min. The OligoB2 mixture (Affymetrix; catalog no. P/N 900301) is heated to 65°C for 5 min then mixed with the labeled and verified gDNA described above, dimethyl sulfoxide (DMSO), and hybridization buffer included in the Affymetrix hybridization, wash, and stain kit (Affymetrix; catalog no. P/N 900720). This solution is further denatured at 94°C for 5 min. During this incubation, the microarrays are equilibrated with prehybridization buffer and placed in the Affymetrix hybridization oven at 45°C for 10 min with rotation at 60 rpm. The labeled DNA is then hybridized to the microarray for 16 h at 45°C with rotation at 60 rpm. Microarrays were then washed and stained according to manufacturer's specifications using two stain cycles and the Affymetrix prokaryotic washing protocol (http://www.affymetrix.com).
Microarray data analysis.
Array analysis was carried out a previously described (19). Briefly, array scans were acquired in Affymetrix GCOS software, and expression was normalized using the simpleaffy Bioconductor R package (20). Comparisons were completed by double filtering using fold change values of ≥2 and Student's t test values with P ≤ 0.05. Two biological replicates for the different conditions were obtained for each biological condition. Custom Perl and Python scripts were then used to identify transcriptionally altered genes. To construct a heat map representing genes of interest with significant changes in transcription, data were imported into and analyzed in R (version 2.15.1; http://www.R-project.org/) (21).
qRT-PCR.
For quantitative reverse transcriptase PCR (qRT-PCR) analysis, total RNA was converted to cDNA by use of reverse transcriptase and random hexamers using the Transcriptor first-strand cDNA synthesis kit from Roche (catalog no. 04379012001). Gene expression was quantified on an ABI-PRISM 7900HT sequence detection system with gene-specific primers in PCR buffer containing SYBR green. Gene-specific transcripts were normalized to the housekeeping gene arcA. Gene-specific primers were designed using Primer Express Software v3.0 (Applied Biosystems) and are shown in Table 2.
Table 2.
Primers used in quantitative reverse transcriptase PCR
| Gene | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| crp | CCGTCAGGAAATCGGTCAGA | TGCGTCCCACGGTTTCA |
| fimH | GATGCGGGCAACTCGATT | GCCCTGCGCTGGTGAA |
| fimA | TGCGGGTAGCGCAACAA | |
| fimB | GGAGATTCATCCGCACATGTT | TCCCATATTCGCCAAAGCA |
| fimC | CGTTGCCACCCGATCAG | AATTCGCGCTACGACGAAA |
| fimD | CAGTGCCAGCTACAGCATGTC | TACCGGCCAGATTGGTCATC |
| fimE | CGCGGGAGTCGGCTTT | ACCGGCATCACGAATAATGC |
| fimF | CCCTGTGGTAACGCCGTTT | GGCTATCTGCAACGCCAGTAA |
| fimG | AATCAGCGCACTTCCCGTTA | TGAGTGGCTCCGCCATTTAC |
| fimH | GATGCGGGCAACTCGATT | GCCCTGCGCTGGTGAA |
| fimI | CGGAAACTTGCCGGATTG | GCCCCATATTGACCGTCATT |
| cfaA | CAGGAGCGAGTAAATCAATACGTTT | CTATCTGGTTTTACCGCCTCAAA |
| cfaB | AAATGGCGTATCATCTTCTCAAGAG | GGCGGTACCGGCAGTTT |
| cfaC | GGTTGGAGTGGATGCTACGAA | CAGAGTACTTGTCCATCCTAATAAAGGA |
| eatA | AAAGCTCCAATGCCTGATTTTAGTA | GTGTATGGTCTCCTGGTGGTAATG |
| etpB | GAACAGGGATAATGCCACAAA | TCCCAGATTCACCTCGTTTT |
| etpA | ATGCCGGTGGCTATATTGTG | GTACGGATGGTGCCACTGTT |
| etpC | CCACCTGGTCTGAGCATATTG | CCGGGTCTCCCAGGTAGT |
| eaeH | GAAGGATGCGTACGGGAAC | CCGCTAAACACTGGTGCAT |
| tia | ACAGGCTTTTATGTGACCGGTAA | GACGGAAGCGCTGGTCAGT |
| arcA | ATCAATCTGCCGGGTAAGAACGGT | TCCAGATCACCGCAGAAGCGATAA |
| cexE | GCGTGTAATGAGACAGTTGCTAAAG | CCTCATAATTTACAGTCCGATGCA |
| aatD | CAAAACCACGGAGGAACTGTACT | GCGATGTTGCGAATACGCTAT |
| aatP | CAACTTTTTTGGATAGCCTTGGA | GCGCCGTTTCTAATGGGATA |
| aatB | CAGCGGTATGGAGGGTGTAAA | AGATCTGATGATAGGTTTCCATTGTTT |
| aatA | CTTGAGGAGGCGAATATTAATTTTGA | GGCGTCAGCCCCAAAGA |
| aatC | TGGCATATCTCCAGAAGCATCA | CTTGCAAGCGCAACTCGTT |
| gspC | GTCGCCACGCCGGTAA | GACGCGTTTCTGCCACAGA |
| gspD | CGGATACCAACGGCGATCT | CCGCTAAAGCCGGAAAGAA |
| gspE | TGCGCGCGATTTTGC | CGGATTTCACCGACCATCA |
| gspF | AAAGAGCTTATCCCCGTGCAT | AACATCCCCCCTGACGAAGT |
| gspG | GCAAGTCTGGTGGTGCCTAAC | TTTTGCCGATCCGCTTTCT |
| gspH | CAGATCCGTTTTTCGCCTTTT | CGAGTAGAAGCGCAGCGTAA |
| gspI | GCTCCAGGCGGTGGATATT | TGACTGAATGACCGACGAAAAG |
| gspJ | GCCGACAATGCAAAAGTTGA | CCGTCGTAGAACTGCAAACG |
| gspK | TGCTGGTAACCATCACGCTTT | GCGTTCGCCCAAGTTGTT |
| gspL | CTCCAGCAATGGCTGCAA | GATCCATTCGCCGGGAAT |
| gspM | CGCTGCGGGTGACACAA | ACATTCACCATCCCAGTCTTCTC |
| eltA | TTCATCAAGAACAATTACAGGTGATA | TGATATTTCCTGAGATATATTGTGCTC |
| yebT | TCGTAAACTCACCAGCAAAGG | AGAAACTCAACGCCATCCAG |
Construction of single-copy transcriptional fusions.
To construct transcriptional reporter fusions, regions upstream of the gene of interest were amplified by PCR and cloned into pHKLac1, a promoterless lacZYA reporter plasmid (22) containing an R6Kγ origin of replication and HK022 phage attP site for integration into the respective HK022 chromosomal attB site (23). Briefly, a 935-bp amplicon containing the eaeH promoter region (from −594 to +341 of eaeH) was PCR-amplified using primers jf032812.1 (5′-GATCGGATCCGATCCCATAGTTTATCCGG-3′) and jf032812.2 (5′-GATCGAATTCTTCCCGAGCCACTCCTG-3′) and directionally cloned into the corresponding BamHI and EcoRI sites (respectively underlined in sequences listed above) of pHKLac1, resulting in plasmid pQL002. pQL002 was then introduced by electroporation into strain GPM1746a containing the pAH69 CRIM integrase helper plasmid. After initial growth at 30°C on plates containing ampicillin (100 μg/ml), individual colonies were streak purified onto Luria agar plates containing streptomycin (Sm) (30 μg/ml) and spectinomycin (Sp) (30 μg/ml) and incubated overnight at 37°C. Ampicillin-sensitive, Smr/Spr colonies were then tested by PCR to verify the presence of stable single-copy integrants containing the reporter as previously described (23).
β-Galactosidase assays.
Assessment of β-galactosidase activity in intestinal epithelial cell monolayers infected with reporter strains was adopted from methodology previously reported by Klarsfeld, et al. (24). Briefly, Caco-2 cells were seeded in black-walled 96-well tissue culture plates and grown to semiconfluent monolayers. Cultures of reporter strains were grown overnight in Luria broth containing Sm (30 μg/ml) and Sp (30 μg/ml), diluted 1:100 into fresh LB the following morning, and then grown for an additional 2 h at 37°C at 225 rpm. Monolayers were then infected at a multiplicity of infection (MOI) of ∼100 and incubated for the indicated time period. Planktonic bacteria were resuspended in 200 μl of assay buffer containing Na2HPO4 · 7H2O (60 mM), NaH2PO4 (40 mM), KCl (10 mM), MgSO4 (1 mM), and β-mercaptoethanol (50 mM). Twenty microliters of resuspended bacteria was saved from each sample to determine the number of CFU present by plating dilutions. One hundred microliters of the remaining suspension was saved for analysis. Monolayers with attached bacteria were washed four times with phosphate-buffered saline (PBS), and 100 μl of assay buffer was added to each well. To quantitate the number of attached bacteria, parallel wells containing infected monolayers were treated in an identical fashion, with 100 μl of MUG (4-methylumbelliferyl β-d-galactopyranoside) assay buffer containing 0.1% Triton X-100. Dilutions of these cell lysates were plated onto Luria agar and incubated overnight at 37°C. To detect β-galactosidase activity in planktonic and attached bacteria, MUG was added at a final concentration of 0.2 mg/ml and the mixture was incubated for 1 h at 37°C. One hundred microliters of 1 M Na2CO3 stop buffer was added, the plate was read at absorbance and emission wavelengths of 365 and 450 nm, respectively, and endpoint data were acquired using Gen5 v 2.0 software. β-Galactosidase activity was then expressed as units of fluorescence/min/104 CFU.
Cloning, expression, and purification of recombinant EaeH.
In constructing an eaeH expression plasmid, primers jf051010.1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGGGAAGGAGATAGAACCATGTCACATTATAAAACAGGT-3′) and jf051010.2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTGGCATCTCCTCCTCGCCATT-3′) were first used to amplify a 4,254-bp eaeH gene fragment (lacking only the stop codon) from H10407 genomic DNA. The resulting amplicon, containing 5′- and 3′-terminal attB sites, was then cloned by bacteriophage λ-mediated recombination with corresponding attP sites on pDONR221, yielding pSS001. After confirmation of the construction of pSS001 by restriction digests and DNA sequencing, this plasmid was recombined with pET-DEST42, placing the eaeH gene in frame with C-terminal V5 and polyhistidine tags to create the pSS002 expression plasmid. The pSS002 expression plasmid encoding the EaeH–V5–6-His fusion protein was introduced into BL21A1 (Table 1), and recombinants were selected on ampicillin (100 μg/ml). Following induction of BL21A1(pSS002) with IPTG (isopropyl-β-d-1-thiogalactopyranoside), recombinant polyhistidine-tagged protein was recovered from bacterial lysates (bacterial protein extraction reagent [B-PER]; Pierce/Thermo Scientific) by nickel metal affinity chromatography. Western blotting using monoclonal antibody against the V5 epitope (Gly-Lys-Pro-Ile-Pro-Asn-Pro-Leu-Leu-Gly-Leu-Asp-Ser-Thr) was then used to confirm expression of the fusion protein.
Production and purification of polyclonal anti-EaeH antibodies.
Polyclonal antiserum was produced in rabbits as previously described (6). Briefly, two New Zealand White rabbits were immunized with recombinant V5 polyhistidine-tagged EaeH. The resulting polyclonal antisera were preabsorbed using an E. coli lysate column (Pierce) and lyophilized strain AAEC191-A (25). Protein A agarose (Protein A Plus; Thermo Scientific) was used to separate antibodies from serum components, and antibodies were affinity purified against rEaeH-V5.
Confocal immunofluorescence microscopy of ETEC-infected Caco-2 cells.
Caco-2 cells were seeded onto poly-d-lysine-coated glass coverslips, incubated overnight, and infected with cultures of H10407 expressing green fluorescent protein (GFP). At 5 h postinfection, samples were fixed with 2% paraformaldehyde for 30 min, permeabilized with 0.1% saponin for 10 min, and blocked with PBS supplemented with 1% bovine serum albumin (BSA) for 30 min. Samples were incubated with affinity-purified anti-EaeH primary antibody for 1 h at 37°C and then goat anti-rabbit secondary antibody conjugated to Alexa Fluor 594 (Invitrogen), DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen), and phalloidin 647 (Invitrogen) for 1 h at 37°C. Samples were mounted in ProLong reagent (Invitrogen). Images were captured with a Zeiss LSM 510 META confocal laser scanning microscope using Axiovision software.
Scanning electron microscopy.
To examine the surface of ETEC cells adherent to cultured epithelial cells, bacteria were added to nonconfluent Caco-2 cells seeded onto glass coverslips at a multiplicity of infection (MOI) of ∼100 organisms/cell and incubated for 15 to 60 min. At the end of the indicated time period, infected supernatants were removed, and the cells were washed with PBS and then processed for scanning electron microscopy (SEM) as described previously (7). All images were acquired on a Philips environmental scanning electron microscope (FEIC Philips XL30 ESEM) and saved as TIFF files. Images were then processed in Image J (v1.45s) augmented with the DeconvolutionLab plug-in (26).
Flow cytometry.
To examine the quantity of flagellin associated with the bacterial surface of H10407 (O78:H11), we performed flow cytometry as previously described (27). Briefly, suspensions of bacteria were fixed with 2% paraformaldehyde for 15 min, washed twice phosphate-buffered saline (PBS), pH 7.2, blocked with 1% BSA in PBS for 30 min, followed by flagellin detection with affinity-purified mouse polyclonal anti-H11 antisera (28), and labeled with Alexa Fluor 488 anti-mouse (IgG) secondary antibody (Invitrogen). Data were acquired on a BD FACSCalibur 4-color dual-laser flow cytometer and processed using FlowJo software (v9.4.1).
Supernatant protein precipitation and immunoblotting.
Proteins in culture supernatants were precipitated as previously described (6) with trichloroacetic acid (TCA). Briefly, 750 μl of ice-cold TCA was added to an equal volume of bacterial culture supernatant, incubated on ice for 30 min, centrifuged at 15,000 × g at 4°C, acetone washed, dried, and resuspended in 1 M Tris, prior to separation by SDS-PAGE and immunoblotting for EtpA and flagellin (6, 7).
Microarray data accession number.
All microarray data for this project have been deposited in the GEO (Gene Expression Omnibus) repository (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession no. GSE40427.
RESULTS
Global changes in the ETEC transcriptome with pathogen-cell contact.
Using the E. coli-Shigella pan-genome array, we interrogated the transcriptomes of planktonic and adherent ETEC strains. Comparative analysis of the genes that were transcriptionally altered >2-fold and had a P value of <0.05 are presented in Fig. 1. A total of 214 gene features on the array displayed a consistently altered transcriptional pattern at all three time points examined, suggesting an early and sustained response to the interaction of E. coli E24377A with epithelial cells (Fig. 1; see Tables S1 to S3 in the supplemental material). Fewer genes exhibited demonstrable increases in transcription with cell contact relative to the number that were negatively modulated. Figure 2 illustrates transcriptional regulation of known and potential virulence genes following attachment and encompasses a number of gene features exhibiting significant responses at early, middle, and late time points following cellular attachment. As depicted in Fig. 2, the majority of genes demonstrating significant increases in transcription following attachment are currently annotated as hypothetical genes. However, included among genes upregulated on attachment was marA, which encodes a regulatory protein involved in E. coli stress responses (29). In contrast, the majority of genes for known and/or putative virulence factors or those encoding immunodominant antigens were transcriptionally repressed following cell contact.
Fig 1.
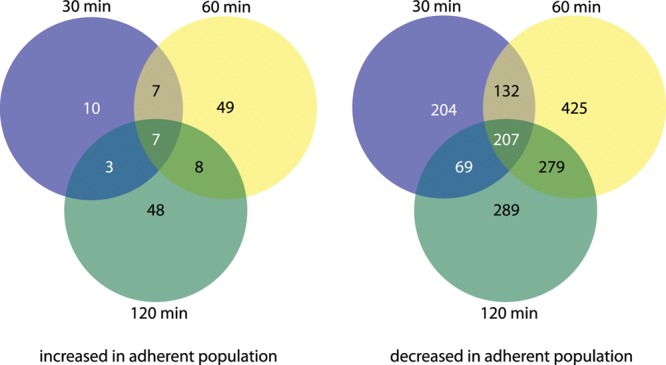
Global alteration in ETEC transcriptome over time, comparing planktonic and attached organisms. The total number of genes with significantly increased transcripts in adherent organisms at each time point is shown to the left. Gene transcripts that were increased in planktonic organisms (decreased in adherent organisms) are shown to the right. Complete lists of individual genes modulated at each time point are contained in Tables S1 to S3 in the supplemental material.
Fig 2.
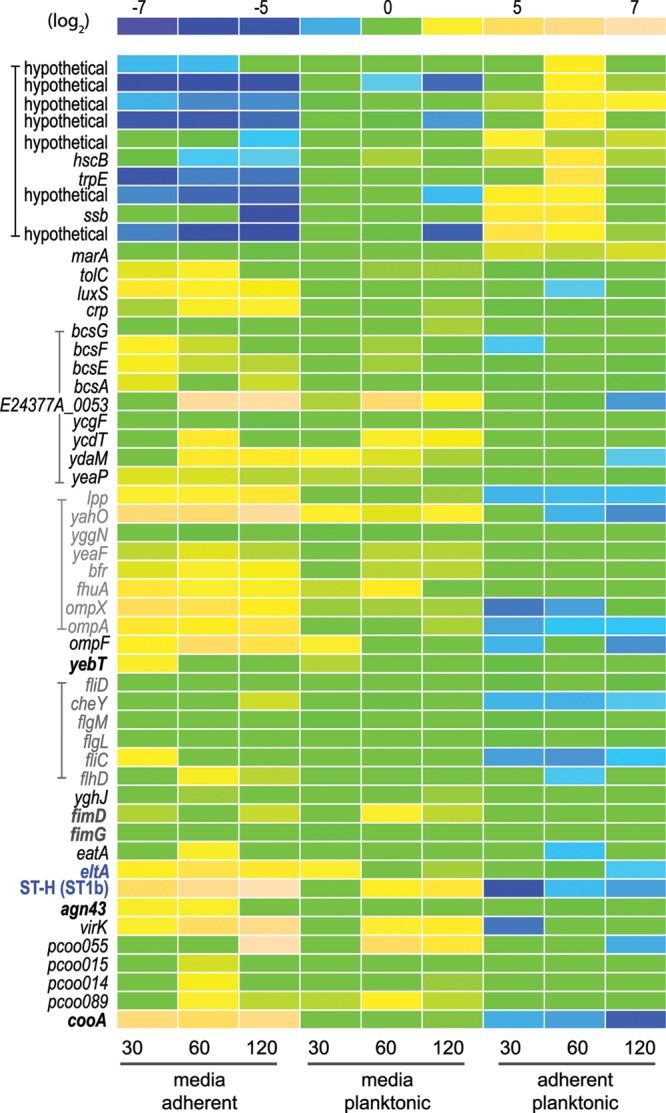
Modulation of ETEC gene expression upon interaction with intestinal epithelial cells. A relative expression intensity scale (log2 values) appears at the top of the figure. Gene features are shown to the left, with related features depicted with brackets/color. Included are known/candidate virulence genes, immunogens, or regulatory genes. The genes most responsive (n = 10) to bacterial adhesion are depicted at the top of the figure. The toxin gene eltA, encoding the LT-A subunit, and the gene coding for heat-stable toxin ST-H (ST-1b) are shown in blue print. Adhesins or putative adhesins are depicted in boldface letters. pCoo plasmid gene designations refer to plasmid-encoded sequences from the pCoo plasmid (64), for which the complete sequence is available at NCBI accession no. NC_007635.1 The conditions and time points (minutes) for each column of data are shown at the bottom of the figure. Data in the far left column depict the fold changes in transcription for bacteria grown in unconditioned tissue culture media relative to the adherent population at 30 min, while those in the far right column represent changes in adherent bacteria relative to planktonic organisms after 120 min. Elements for which there was no significant difference in the microarray data are depicted as 0-fold change (green).
crp and cAMP-CRP-dependent virulence genes are modulated following host contact.
Among the genes transcriptionally altered in ETEC strain E24377A upon contact with epithelial cells was crp, encoding the cAMP receptor protein (CRP), a global regulator involved in modulating the transcription of many E. coli genes (30), including a number of known ETEC virulence factors, such as the colonization factor antigens and the heat-labile and heat-stable enterotoxin genes (31). Likewise, genes previously shown to be crp dependent, including those governing flagellar assembly and chemotaxis and those coding for CFs, LT, and the heat-stable ST-1b toxin (ST-H), were significantly altered relative to planktonic organisms following attachment (Table 3), with both toxins transcriptionally repressed in the attached bacteria. Similarly, qRT-PCR studies of ETEC H10407 demonstrated that in comparison to planktonic organisms, crp expression was depressed in attached bacteria (Fig. 3). However, because an earlier analysis of H10407 demonstrated that the cAMP-CRP complex represses transcription of heat-labile toxin genes (31), we predicted that in contrast to E24377A, cell contact by H10407 should enhance transcription genes encoding LT. Indeed, consistent with the repression of CRP following cell contact in this strain, we found that transcription of the gene encoding the A subunit of the toxin (eltA) was increased relative to planktonic organisms (average, 7.2 ± 3.8-fold; P = 0.047). Together, these findings suggest that while the cAMP-CRP complex plays a substantial role in governing ETEC bacterium-host interactions, there are likely to be significant interstrain differences in transcription of essential virulence genes modulated by this key regulator.
Table 3.
Alteration of crp- and cAMP-CRP-dependent genes on cell contacta
| Gene name | Protein annotation | Time (min) | Fold change (log2)b | P value |
|---|---|---|---|---|
| crp | cAMP regulatory protein | 30 | −1.60 | 0.019 |
| 60 | −1.84 | 0.036 | ||
| 120 | −2.16 | 0.038 | ||
| Flagellar genes | ||||
| flgK | Flagellar hook-associated protein 1 | 30 | −1.22 | 0.042 |
| 60 | − | NSc | ||
| 120 | NS | |||
| fliD | Flagellar hook-associated protein 2 | 30 | −1.169 | 0.015 |
| 60 | NS | |||
| 120 | −1.106 | 0.0017 | ||
| flit | Flagellar biosynthesis protein | 30 | NS | |
| 60 | NS | |||
| 120 | −1.12 | 0.028 | ||
| flgL | Flagellar hook-associated protein 3 | 30 | −1.55 | 0.017 |
| 60 | NS | |||
| 120 | −1.44 | 0.016 | ||
| fliC | Flagellin | 30 | −3.19 | 0.02 |
| 60 | −3.33 | 0.0006 | ||
| 120 | −2.61 | 0.0019 | ||
| flgM | Anti-sigma 28 factor | 30 | −1.096 | 0.0082 |
| 60 | −1.488 | 0.0310 | ||
| 120 | NS | |||
| cheY | Chemotaxis regulatory protein | 30 | −2.894 | 0.0495 |
| 60 | −2.979 | 0.0004 | ||
| 120 | −2.379 | 0.0051 | ||
| cheW | Purine-binding chemotaxis protein | 30 | NS | |
| 60 | −1.176 | 0.0011 | ||
| 120 | NS | |||
| luxS | S-Ribosylhomocysteine lyase | 30 | NS | |
| 60 | −2.3 | 0.0003 | ||
| 120 | −1.93 | 0.016 | ||
| cooA | CS1 type fimbrial major subunit | 30 | −3.008 | 0.0241 |
| 60 | −3.212 | 0.0001 | ||
| 120 | −4.580 | 0.0043 | ||
| ompA | Outer membrane protein A | 30 | −3.137 | 0.0106 |
| 60 | −2.71 | 0.0040 | ||
| 120 | −2.64 | 0.016 | ||
| tolC | TolC outer membrane channel protein | 30 | −1.74 | 0.022 |
| 60 | −1.8267 | 0.013 | ||
| 120 | NS | |||
| eltA | Heat-labile toxin, A subunit | 30 | NS | |
| 60 | −1.19 | 0.017 | ||
| 120 | −2.46 | 0.003 | ||
| ST-1b | Heat-stable toxin (ST-H) | 30 | −5.14 | 0.036 |
| 60 | −2.79 | 0.015 | ||
| 120 | NS |
The data reported in the table depict all genes with previously established CRP-cAMP dependence for which there were statistically significant changes in gene expression comparing attached and planktonic organisms using the FDA-E. coli-Shigella (FDA-ECSG) Affymetrix array.
Adherent versus planktonic (log2 values).
NS, nonsignificant change.
Fig 3.
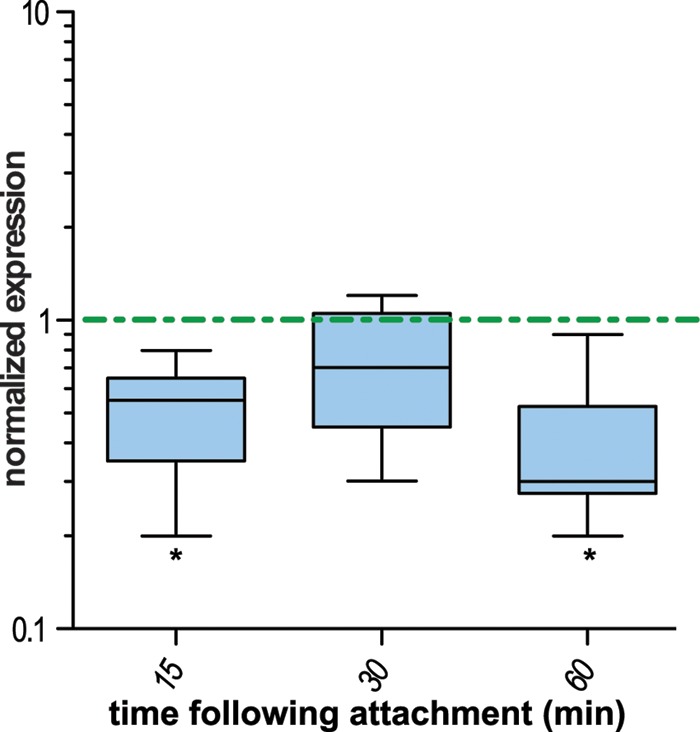
crp expression is depressed in ETEC H10407 following contact with epithelial cells. Data represent the summary of six independent experiments in which ETEC H10407 was used to inoculate Caco-2 intestinal epithelial cell cultures and then harvested from either the supernatant (planktonic organisms) or from the cell surface (attached), followed by comparison of crp transcripts determined by qRT-PCR for attached bacteria relative to planktonic organisms. All data are normalized to the housekeeping gene arcA and represent the ratio of attached to planktonic organisms. Whisker plots depict the range of data obtained over 6 experiments, with the horizontal line representing the mean value. A dashed horizontal line is drawn at a normalized expression value of 1 (no difference between attached and planktonic bacteria). *, P ≤ 0.05 by Wilcoxon's signed-rank test.
Influence of pathogen-host interaction on c-di-GMP signaling.
In addition to cAMP, gene expression in E. coli and other bacteria is also known to be governed by another second messenger, cyclic-di(3′→5′)-guanylic acid (c-di-GMP) (32). Microarray data (Fig. 2) indicated that multiple genes associated with c-di-GMP signaling, including several known or putative diguanylate cyclase proteins containing GGDEF domains, as well as known and a novel putative phosphodiesterase encoded on the pETEC_74 virulence plasmid from E24377A (E24377A_0053) were transcriptionally altered on ETEC-host interaction (Table 4). Likewise, we noted modulation of several genes associated with cellulose biosynthesis, which has been demonstrated to be influenced by alteration in c-di-GMP levels (33). Collectively, these data are congruent with previous studies in other pathogens that have demonstrated that c-di-GMP-coordinated changes are associated with the transition between the planktonic and adherent states (32).
Table 4.
Transcriptional modulation of genes related to c-di-GMP metabolism and sensinga
| Gene | Product descriptionb | Transcriptional modulation comparisonc | Time point (min) | Maximal difference (log2)d | P value |
|---|---|---|---|---|---|
| Cyclic-di-GMP metabolism | |||||
| yeaP | GAF/GGDEF-domain diguanylate cyclase | Medium vs adherent | 30 | 2.44 | 0.0001 |
| ydaM | Putative GGDEF-domain protein, cgsD regulator | Medium vs adherent | 60 | 3.781 | 0.028 |
| ycdT | Putative GGDEF-domain protein diguanylate cyclase | Medium vs adherent | 60 | 3.748 | 0.002 |
| ycgF | Cyclic-di-GMP phosphodiesterase | Medium vs adherent | 120 | −1.56 | 0.02 |
| Locus tag E0053 | Novel EAL-domain protein pETEC_74 virulence plasmid | Medium vs adherent | 60 | 6.64 | 5.32 × 10−6 |
| Cellulose biosynthesis | |||||
| bcsA | Cellulose synthase catalytic subunit | Medium vs adherent | 30 | 2.50 | 0.002 |
| bcsE | Cellulose biosynthesis protein | Medium vs adherent | 30 | 2.67 | 3.90 × 10−5 |
| bcsF | Cellulose biosynthesis protein | Medium vs adherent | 30 | 3.33 | 2.49 × 10−5 |
| bcsG | Cellulose biosynthesis protein (predicted endoglucanase) | Medium vs planktonic | 60 | 1.399 | 0.012 |
The data reported in the table depict all genes with previously established or putative association with c-di-GMP metabolism/sensing for which there were statistically significant changes in gene expression under the conditions reported using the FDA-E. coli-Shigella (FDA-ECSG) Affymetrix array.
The GAF domain refers to pfam01590, a domain present in cGMP-specific phosphodiesterases, the GGDEF domain refers to cd01949, a domain present diguanylate cyclases, and the EAL domain refers to cd01948, a second domain present in c-di-GMP phosphodiesterases.
“Medium” refers to unconditioned tissue culture medium used to grow Caco-2 epithelial cells.
A negative value reflects a decrease in transcription under the first condition relative to the comparison group.
Temporal activation of multiple bacterial adhesins.
Data from the DNA microarray studies suggested that the transcription of multiple virulence genes, including a number of adhesin genes, was modulated either upon exposure to media conditioned by host cells or by host cell contact. Therefore, we conducted additional experiments with ETEC H10407 to examine modulation of adhesion-related genes. These studies, summarized in Fig. 4, demonstrated that at early time points following attachment of ETEC to Caco-2 cells, multiple genes associated with the biogenesis of type 1 fimbriae were transcriptionally activated (Fig. 4a). In contrast, transcription of cfaABC genes involved in assembly of the plasmid-encoded colonization factor antigen 1 fimbriae (CFA/I), as well as those involved in synthesis and export of the EtpA two-partner exoprotein adhesin, was repressed following contact. We also examined the transcription of the gene encoding EatA, a secreted autotransporter recently shown to accelerate delivery of the heat-labile toxin by preventing accumulation of the EtpA adhesin (10). Interestingly, consistent with its function in modulating EtpA-mediated adhesion, eatA transcription was activated at early time points following cell contact.
Fig 4.
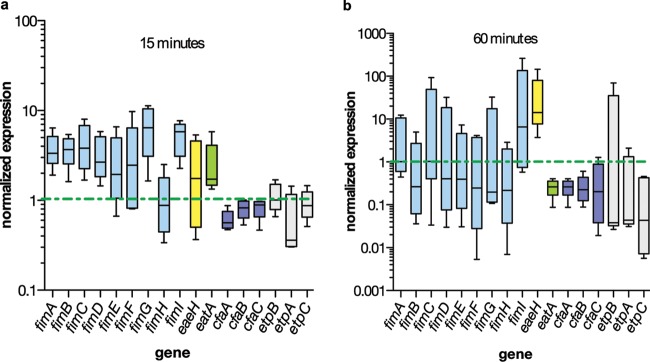
Modulation of ETEC H10407 adhesion-related genes on cell contact qRT-PCR for transcripts of adhesion-related genes in attached bacteria relative to planktonic organisms. Whisker plots depict the range of data obtained over five experiments, with the horizontal line representing the mean value. In each panel, a dashed horizontal line is drawn at normalized expression value of 1 (no difference between attached and planktonic bacteria). Panels a and b show the relative transcription at 15 min (a) and 60 min (b) following infection of Caco-2 cells with ETEC H10407.
Intriguingly, eaeH, a putative adhesin gene originally discovered by subtractive hybridization of ETEC H10407 and E. coli K-12 (34), which is highly conserved in pathogenic E. coli, was strongly upregulated at later time points following contact with intestinal epithelial cells (Fig. 4b). The predicted EaeH peptide shares a number of features in common with known adhesins, such as intimin from enteropathogenic E. coli (EPEC), as well as the Air protein from enteroaggregative E. coli (EAEC) (35), including multiple bacterial immunoglobulin-like (Big) domains (pfam02369). In support of the data obtained by quantitative PCR, we examined gene expression using single-copy eaeH promoter β-galactosidase fusion constructs. Again, these data demonstrated a significant increase in eaeH expression in attached bacteria relative to planktonic cells (Fig. 5a). We also identified EaeH on the surface of ETEC H10407 attached to intestinal cells in vitro, both at the point of cell contact (Fig. 5b to e) and diffusely distributed on the surface of H10407 (Fig. 5f to i).
Fig 5.
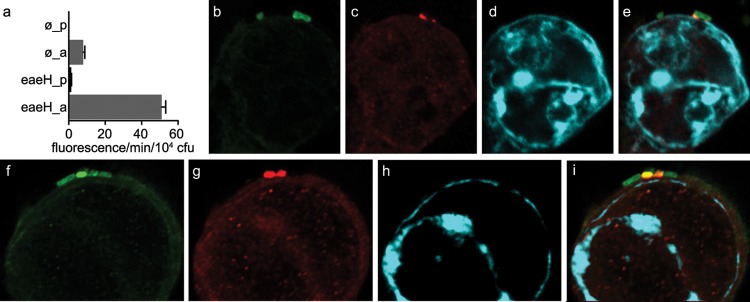
The putative adhesin gene eaeH is preferentially expressed in the context of epithelial cell attachment. (a) Activation of the eaeH promoter in attached bacteria relative to planktonic ETEC strains carrying an integrated single-copy eaeH promoter-lacZYA transcriptional fusion. ø_p and ø_a refer to control strain jf2575 with promoterless lacZYA in planktonic and adherent states, respectively, while eaeH_p and eaeH_a refer to data generated with strain jf2624 carrying the eaeH promoter-lacZYA fusion. MUG–β-galactosidase determinations are expressed in fluorescence units/min/104 CFU, with bars representing the mean of data from triplicate wells. EaeH expression following bacterial attachment was identified at the interface of ETEC H10407 and Caco-2 epithelial cells (b to e) and in a diffuse distribution on the surface of adherent bacteria (f to i). Confocal microscopy image panels from left to right show GFP-expressing ETEC (green), EaeH immunofluorescence (red), host cell actin (phalloidin; far-red, cyan), and the composite merged image.
Interestingly, we observed comparatively little transcriptional modulation following adhesion in yebT, a gene encoding another putative outer membrane protein adhesin highly conserved in E. coli and a number of other pathogens. Real-time quantitative PCR to examine yebT expression in ETEC H10407 suggested that this gene is similarly expressed in both planktonic and adherent ETEC strains (Fig. 4a and b). This is consistent with the proposed constitutive expression of yebT, potentially making its gene product, Mam7, available for participation in early phases of adhesion in a number of pathogenic bacteria, including other E. coli pathovars (36).
Collectively, these data suggest that ETEC strains engage host cells by sequentially altering multiple adhesin molecules as the interaction matures. Furthermore, they suggest that pathovar-specific virulence factors, such as the CFs and EtpA, function within a contextual framework of highly conserved adhesins, including type 1 fimbriae, EaeH, and Mam7.
ETEC undergoes morphological changes following cell contact.
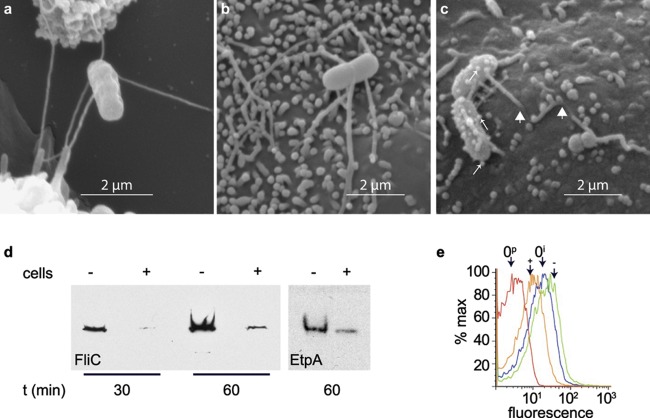
In the course of recent studies to investigate the involvement of flagella in attachment of ETEC to epithelial cells (7), we noted that at very early time points (5 to 15 min), ETEC cells appear to engage host cells at a distance via their long peritrichous flagella (Fig. 6a), while at later time points (Fig. 6b and c), flagella appear shortened and/or engulfed by the host cell, accompanied by the emergence of bleb- or vesicle-like structures on the bacterial surface. Consistent with these apparent changes in bacterial surface structures, we demonstrated that the production of two secreted proteins, flagellin (the major subunit of flagella), and EtpA, a flagellin-binding adhesin, were both reduced following exposure to host cell-conditioned media (Fig. 6d), as demonstrated by immunoblotting. In support of these data, flow cytometry studies also indicated that flagellin on the surface of H10407 decreased with exposure to conditioned media (Fig. 6e).
Fig 6.
ETEC strains undergo changes in surface architecture after engaging intestinal epithelial cells. Scanning electron micrographs obtained at 15 (a), 30 (b), and 60 (c) min following addition of ETEC H10407 to intestinal epithelial cells in vitro. Small arrows in panel c indicate the appearance of blebs on the surface of H10407, while large arrowheads show the course of flagellum that has been partially engulfed by the cell surface. Panels b and c were enhanced using deconvolution. (d) Treatment of ETEC with Caco-2 cell-conditioned media leads to decreased amounts of flagellin (FliC) and EtpA in bacterial culture supernatants. FliC and EtpA immunoblots are shown to the left and right, respectively. −, tissue culture medium alone; +, medium that has been conditioned by growth of Caco-2 epithelial cells. (e) Flow cytometry used to detect FliC H11 on the surface of ETEC H10407 (serotype O78:H11) following growth in ordinary Luria broth prior to incubation with tissue culture media. 0i, detection of FliC expression using anti-FliC antibody at time zero prior to incubation in media; 0p, preimmune sera control at time zero; +, FliC expression after 60 min in conditioned media; −, FliC expression after 60 min in tissue culture media not preconditioned with Caco-2 cells.
The appearance of bleb-like structures on the surface of ETEC is intriguing in that previous studies have demonstrated that much of the LT produced by ETEC is associated with these outer membrane vesicles (37, 38) and that these structures can effectively deliver toxin (39) to target epithelial cells. Consistent with vesicle structure formation and the membrane remodeling observed following attachment, we also observed concomitant transcriptional modulation of hns encoding the global DNA binding transcriptional repressor H-NS, previously associated with vesicle formation in E. coli. Interestingly, mutation of hns has been associated with an increase in vesicle formation (37), and following attachment hns transcription was significantly repressed (over 12-fold; P = 0.002) relative to that of planktonic organisms (see Table S2 in the supplemental material), suggesting that attachment of ETEC to the host cells may promote the formation of vesicles.
ETEC suppresses production of immunogenic proteins on cell contact.
Recent immunoproteomic studies of ETEC using human sera obtained following natural infections or from experimentally infected mice demonstrated that multiple proteins are recognized during ETEC infections (11). Interestingly, we found that genes encoding many of these immunoreactive proteins were transcriptionally repressed upon interaction with host cells or conditioned media (Table 5), suggesting that ETEC may transcriptionally modulate these genes as a strategy to avoid elimination by the host.
Table 5.
Transcriptional modulation of previously identified ETEC immunoreactive proteinsa
| Gene | Product description | Transcriptional modulation comparison | Time point (min) | Maximal difference (log2)b | P value |
|---|---|---|---|---|---|
| fliC | Flagellin monomer | Adherent vs planktonic | 60 | −3.33 | 0.0006 |
| eatA | Secreted ETEC autotransporter virulence protein | Adherent vs planktonic | 60 | −2.71 | 0.002 |
| agn43.2 | Surface autotransporter | Adherent vs medium | 60 | −2.96 | 0.0007 |
| cooAc | CS1 major fimbrial subunit | Medium vs adherent | 120 | 5.98 | 1.8 × 10−5 |
| fimD | Fimbrial usher, type 1 fimbriae | Medium vs planktonic | 60 | 3.13 | 0.003 |
| eltA | Heat-labile toxin subunit | Medium vs adherent | 60 | 4.66 | 0.0002 |
| ompA | OmpA porin | Medium vs adherent | 120 | 4.16 | 0.0007 |
| fecA | Iron transport membrane receptor | Medium vs adherent | 60 | 1.06 | 0.03 |
| ompX | OmpX porin | Medium vs adherent | 30 | 4.84 | 0.001 |
| fhuA | Ferrichrome outer membrane transporter | Medium vs adherent | 30 | 4.06 | 0.009 |
| bfr | Bacterioferritin | Medium vs adherent | 60 | 2.88 | 0.017 |
| yggN | Hypothetical | Medium vs planktonic | 60 | 1.92 | 0.0009 |
| yahO | Hypothetical | Medium vs adherent | 120 | 6.60 | 7.42 × 10−5 |
| lpp | Hypothetical | Medium vs adherent | 30 | 3.22 | 0.001 |
| yghJ | Secreted type II secretion effector | Medium vs adherent | 60 | 1.14 | 0.006 |
| yeaF | Hypothetical; MipA domain protein/envelope biogenesis | Medium vs adherent | 60 | 2.483 | 0.03 |
The data reported in the table depict all genes identified in prior immunoproteomic analysis (11) for which there were statistically significant changes in gene expression under the conditions reported using the FDA-E. coli-Shigella (FDA-ECSG) Affymetrix array.
Negative values reflect decreases in transcription under the first condition relative to the comparison group.
Earlier immunoproteomic analysis performed with H10407 identified the CfaB protein as the major structural subunit of CFA/I fimbriae.
DISCUSSION
The enterotoxigenic E. coli strains are an important focus of current enteric vaccine development efforts. ETEC vaccine attempts to date have been largely directed at heat-labile toxin and/or colonization factor antigens (2). Unfortunately, these approaches have been hindered by poor conservation of colonization factor antigens and the lack of complete, sustained protection afforded by LT immunization (5). The more recent discovery of additional novel virulence genes, even among prototypical ETEC strains, implies that our understanding of the pathogenesis of these organisms is incomplete (40) and also raises the possibility that recent technologic advances may be applied to the discovery of previously unheralded vaccine targets. One approach that has been applied to other pathogens is the use of transcriptional profiling to identify genes modulated during host-pathogen interactions (16).
Here we demonstrate that ETEC-host interactions promote pronounced changes in the ETEC transcriptome that are accompanied by significant modification of the architecture of these organisms at the pathogen-host interface. Collectively, the data suggest that ETEC interactions with host cells are finely orchestrated, with the sequential deployment of multiple adhesins as the bacteria engage their epithelial targets culminating in effective toxin delivery.
The precise stimuli for these changes remain to be elucidated. However, the studies do appear to affirm a central role of cAMP and the cAMP receptor protein (CRP) in modulation (41) of known (22) and putative (42) virulence genes, including genes involved in flagellar biosynthesis (7), whose expression is known to be under the control of crp (43).
Interestingly, the crp gene is itself negatively autoregulated by the cAMP-CRP complex (44). One possible explanation for the modulation of genes under the control of cAMP-CRP and crp in response to interaction with host cells is that ETEC strains are able to sense cAMP generated by host epithelial cells in response to stimulation by heat-labile toxin. Bacteria have long been known to respond to extracellular cAMP, and more recently, host cyclic nucleotide efflux systems that are activated in response to increasing intracellular concentrations of cAMP have been described in some detail (45, 46). Therefore, it is plausible to suggest that ETEC strains may not only provoke increases in cAMP as they successfully deliver heat-labile toxin, but respond to pathogen-induced extracellular increases in this critical second messenger by modulating the expression of essential virulence genes.
Among the genes whose expression was significantly affected by cell contact were those associated with bacterial cell signaling, including the autoinducer 2 (AI-2) synthase gene luxS, which has previously been shown to be dependent on cAMP-CRP (47). Interestingly, we also noted significant alteration of other bacterial cell signaling genes, including a number of genes coding for known or putative diguanylate cyclases (e.g., yeaP) needed for synthesis of cyclic di-GMP (48, 49), a highly conserved bacterial second messenger involved in regulating a host of virulence traits, including biofilm formation, motility, and expression of virulence genes (50). In addition, there was appreciable modulation of genes encoding di-GMP phosphodiesterases that typically contain EAL domains (51). These included ycgF, which encodes a known E. coli c-di-GMP phosphodiesterase as well as a novel, yet uncharacterized gene located on an E24377A virulence plasmid encoding a putative EAL domain protein. Likewise, we noted the modulation of putative and known c-di-GMP binding proteins, including YcgR, which has been shown to be essential for E. coli motility (50). These findings are in keeping with the known central role of c-di-GMP sensing in orchestrating adaptation of bacteria from a planktonic to an attached or sessile lifestyle (32).
Interestingly, we found significant overlap in the assemblage of genes that were negatively modulated by either direct cell contact or exposure to media conditioned by host cells and genes encoding proteins previously identified in recent immunoproteomic studies of ETEC using convalescent-phase sera (11). This may suggest that ETEC strains are programmed to specifically extinguish production of immunodominant proteins once they have reached their epithelial targets, a phenomenon that could have significant implications for the design of effective mucosal vaccine strategies for ETEC.
The modulation of multiple genes involved in formation of outer membrane vesicles (OMV) following successful attachment is also intriguing for several reasons. Enhancement of the production of vesicles in the context of intimate interaction with the host may provide an important means of countering innate defenses as most Gram-negative OMV serve as potent stimuli of innate immune responses (52, 53). More importantly, the apparent formation of outer membrane vesicles following attachment would potentially provide an important link between earlier studies demonstrating that ETEC vesicles are laden with LT and that purified vesicles can serve as effective means of toxin delivery (38). Programmed vesicle formation upon pathogen-host interaction could theoretically enhance delivery of OMV-associated virulence proteins such as LT. While vesicle biogenesis in the context of epithelial cells requires additional study, the formation of vesicle-like structures by other E. coli pathotypes (54) upon cell contact might suggest that ETEC strains have coopted a more general response to host-pathogen interactions for toxin delivery.
One of the gene transcripts that significantly increased following adhesion was marA, which encodes an AraC-type transcriptional regulator involved in the modulation of a variety of genes which impact virulence in uropathogenic E. coli, including the ability to form biofilms, and effectively colonize the urinary tract (55). MarA acts to indirectly reduce levels of OmpF (56); correspondingly, ompF transcription was noted to be decreased in bacteria adherent to host cells. Interestingly, MarA also governs resistance to cationic antimicrobial peptides (CAMPs), thought to be part of the innate immune response to enteric pathogens, such as ETEC. Human β-defensin 1 (HBD-1), which is constitutively expressed by Caco-2 intestinal epithelial cells (57) and was used here as the target for bacterial adhesion, has been shown to induce marA transcription (58).
While the modulation of genes following cell contact has been described for a number of other pathogens (12–14, 59), these are the first studies to systematically examine alteration in ETEC gene transcription upon interaction with host epithelial cells. Most prior studies have focused narrowly on transcriptional changes in genes encoding colonization factors or the toxins. We should note that one recent genome-wide transcriptome study of ETEC that examined the effect of exposure to bile salts (19) revealed significant interstrain variation in transcriptional responses of key virulence genes. Indeed, the demonstration here that transcription of genes encoding heat-labile toxin is differentially modulated in response to cell contact in E24377A and H10407 appears to support this concept and add another layer of complexity to the study of this genetically diverse population of pathogens.
Collectively, however, the data presented here would suggest that ETEC interactions with host cells are finely orchestrated events during which the bacteria engage their epithelial targets by concerted action of a number of different bacterial adhesins. Early predominance of pathoype-specific adhesins, including EtpA, may set the stage for more intimate adhesion events mediated by more highly conserved structures, including type 1 fimbriae, and outer membrane proteins, including EaeH.
Ultimately, ETEC strains must be able to effectively deliver their toxin payload to epithelial cells. Remarkably, the essential elements required for toxin delivery by these complex pathogens to host cells are still very poorly understood. Although we have demonstrated that the EtpA adhesin (60) and the EatA protease that modulates EtpA-mediated adhesion (10) are both essential for optimal delivery of LT to the target intestinal epithelial cells, the studies included here intimate a complex series of events that would appear to include significant alteration of bacterial surface structures. Unraveling the molecular details underlying these events could provide novel approaches to the rational design of ETEC vaccines.
Supplementary Material
ACKNOWLEDGMENTS
The project described here was supported by grant no. 1R01AI089894-01 from the National Institute of Allergy and Infectious Diseases and by Merit Review funding from the Department of Veterans Affairs.
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the Department of Veterans Affairs.
Footnotes
Published ahead of print 31 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00919-12.
REFERENCES
- 1. Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, Rudan I, Campbell H, Cibulskis R, Li M, Mathers C, Black RE. 2012. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 379:2151–2161 [DOI] [PubMed] [Google Scholar]
- 2. Walker RI, Steele D, Aguado T. 2007. Analysis of strategies to successfully vaccinate infants in developing countries against enterotoxigenic E. coli (ETEC) disease. Vaccine 25:2545–2566 [DOI] [PubMed] [Google Scholar]
- 3. Wenneras C, Erling V. 2004. Prevalence of enterotoxigenic Escherichia coli-associated diarrhoea and carrier state in the developing world. J. Health Popul. Nutr. 22:370–382 [PubMed] [Google Scholar]
- 4. Svennerholm AM, Tobias J. 2008. Vaccines against enterotoxigenic Escherichia coli. Expert Rev. Vaccines 7:795–804 [DOI] [PubMed] [Google Scholar]
- 5. Boedeker EC. 2005. Vaccines for enterotoxigenic Escherichia coli: current status. Curr. Opin. Gastroenterol. 21:15–19 [PubMed] [Google Scholar]
- 6. Fleckenstein JM, Roy K, Fischer JF, Burkitt M. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74:2245–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roy K, Hilliard GM, Hamilton DJ, Luo J, Ostmann MM, Fleckenstein JM. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roy K, Hamilton D, Allen KP, Randolph MP, Fleckenstein JM. 2008. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect. Immun. 76:2106–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patel SK, Dotson J, Allen KP, Fleckenstein JM. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roy K, Kansal R, Bartels SR, Hamilton DJ, Shaaban S, Fleckenstein JM. 2011. Adhesin degradation accelerates delivery of heat-labile toxin by enterotoxigenic Escherichia coli. J. Biol. Chem. 286:29771–29779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roy K, Bartels S, Qadri F, Fleckenstein JM. 2010. Enterotoxigenic Escherichia coli elicits immune responses to multiple surface proteins. Infect. Immun. 78:3027–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson KE, Wolf-Watz H. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231–1233 [DOI] [PubMed] [Google Scholar]
- 13. Cotter PA, Miller JF. 1996. Triggering bacterial virulence. Science 273:1183–1184 [DOI] [PubMed] [Google Scholar]
- 14. Zhang JP, Normark S. 1996. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science 273:1234–1236 [DOI] [PubMed] [Google Scholar]
- 15. Rinaudo CD, Telford JL, Rappuoli R, Seib KL. 2009. Vaccinology in the genome era. J. Clin. Invest. 119:2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grifantini R, Bartolini E, Muzzi A, Draghi M, Frigimelica E, Berger J, Ratti G, Petracca R, Galli G, Agnusdei M, Giuliani MM, Santini L, Brunelli B, Tettelin H, Rappuoli R, Randazzo F, Grandi G. 2002. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat. Biotechnol. 20:914–921 [DOI] [PubMed] [Google Scholar]
- 17. Echenique-Rivera H, Muzzi A, Del Tordello E, Seib KL, Francois P, Rappuoli R, Pizza M, Serruto D. 2011. Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathog. 7:e1002027 doi:10.1371/journal.ppat.1002027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang H, Xu J, Ding D, Jackson SA, Patel IR, Frye JG, Zou W, Nayak R, Foley S, Chen J, Su Z, Ye Y, Turner S, Harris S, Zhou G, Cerniglia C, Tong W. 2010. An FDA bioinformatics tool for microbial genomics research on molecular characterization of bacterial foodborne pathogens using microarrays. BMC Bioinformatics 11(Suppl 6):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sahl JW, Rasko DA. 2012. Analysis of global transcriptional profiles of enterotoxigenic Escherichia coli isolate E24377A. Infect. Immun. 80:1232–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilson CL, Miller CJ. 2005. Simpleaffy: a BioConductor package for Affymetrix quality control and data analysis. Bioinformatics 21:3683–3685 [DOI] [PubMed] [Google Scholar]
- 21. R Core Team 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 22. Espert SM, Elsinghorst EA, Munson GP. 2011. The tib adherence locus of enterotoxigenic Escherichia coli is regulated by cyclic AMP receptor protein. J. Bacteriol. 193:1369–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haldimann A, Wanner BL. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klarsfeld AD, Goossens PL, Cossart P. 1994. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol. Microbiol. 13:585–597 [DOI] [PubMed] [Google Scholar]
- 25. Blomfield IC, McClain MS, Eisenstein BI. 1991. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol. Microbiol. 5:1439–1445 [DOI] [PubMed] [Google Scholar]
- 26. Vonesch C, Unser M. 2008. A fast thresholded landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE Trans. Image Process. 17:539–549 [DOI] [PubMed] [Google Scholar]
- 27. Harris JA, Roy K, Woo-Rasberry V, Hamilton DJ, Kansal R, Qadri F, Fleckenstein JM. 2011. Directed evaluation of enterotoxigenic Escherichia coli autotransporter proteins as putative vaccine candidates. PLoS Neglected Trop. Dis. 5:e1428 doi:10.1371/journal.pntd.0001428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roy K, Hamilton D, Ostmann MM, Fleckenstein JM. 2009. Vaccination with EtpA glycoprotein or flagellin protects against colonization with enterotoxigenic Escherichia coli in a murine model. Vaccine 27:4601–4608 [DOI] [PubMed] [Google Scholar]
- 29. Yang J, Tauschek M, Robins-Browne RM. 2011. Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends Microbiol. 19:128–135 [DOI] [PubMed] [Google Scholar]
- 30. Zheng D, Constantinidou C, Hobman JL, Minchin SD. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32:5874–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bodero MD, Munson GP. 2009. Cyclic AMP receptor protein-dependent repression of heat-labile enterotoxin. Infect. Immun. 77:791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 33. Camilli A, Bassler BL. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Q, Savarino SJ, Venkatesan MM. 2006. Subtractive hybridization and optical mapping of the enterotoxigenic Escherichia coli H10407 chromosome: isolation of unique sequences and demonstration of significant similarity to the chromosome of E. coli K-12. Microbiology 152:1041–1054 [DOI] [PubMed] [Google Scholar]
- 35. Sheikh J, Dudley EG, Sui B, Tamboura B, Suleman A, Nataro JP. 2006. EilA, a HilA-like regulator in enteroaggregative Escherichia coli. Mol. Microbiol. 61:338–350 [DOI] [PubMed] [Google Scholar]
- 36. Krachler AM, Ham H, Orth K. 2011. Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. Proc. Natl. Acad. Sci. U. S. A. 108:11614–11619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Horstman AL, Kuehn MJ. 2002. Bacterial surface association of heat-labile enterotoxin through lipopolysaccharide after secretion via the general secretory pathway. J. Biol. Chem. 277:32538–32545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horstman AL, Kuehn MJ. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. 2004. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23:4538–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fleckenstein JM, Hardwidge PR, Munson GP, Rasko DA, Sommerfelt H, Steinsland H. 2010. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 12:89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McDonough KA, Rodriguez A. 2012. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat. Rev. Microbiol. 10:27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin HH, Hsu CC, Yang CD, Ju YW, Chen YP, Tseng CP. 2011. Negative effect of glucose on ompA mRNA stability: a potential role of cyclic AMP in the repression of hfq in Escherichia coli. J. Bacteriol. 193:5833–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Okamoto K, Freundlich M. 1986. Mechanism for the autogenous control of the crp operon: transcriptional inhibition by a divergent RNA transcript. Proc. Natl. Acad. Sci. U. S. A. 83:5000–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wielinga PR, van der Heijden I, Reid G, Beijnen JH, Wijnholds J, Borst P. 2003. Characterization of the MRP4- and MRP5-mediated transport of cyclic nucleotides from intact cells. J. Biol. Chem. 278:17664–17671 [DOI] [PubMed] [Google Scholar]
- 46. Li C, Krishnamurthy PC, Penmatsa H, Marrs KL, Wang XQ, Zaccolo M, Jalink K, Li M, Nelson DJ, Schuetz JD, Naren AP. 2007. Spatiotemporal coupling of cAMP transporter to CFTR chloride channel function in the gut epithelia. Cell 131:940–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang L, Hashimoto Y, Tsao CY, Valdes JJ, Bentley WE. 2005. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187:2066–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. D'Argenio DA, Miller SI. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497–2502 [DOI] [PubMed] [Google Scholar]
- 49. Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7:724–735 [DOI] [PubMed] [Google Scholar]
- 52. Ellis TN, Leiman SA, Kuehn MJ. 2010. Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect. Immun. 78:3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bielig H, Rompikuntal PK, Dongre M, Zurek B, Lindmark B, Ramstedt M, Wai SN, Kufer TA. 2011. NOD-like receptor activation by outer membrane vesicles from Vibrio cholerae non-O1 non-O139 strains is modulated by the quorum-sensing regulator HapR. Infect. Immun. 79:1418–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Giron JA, Torres AG, Freer E, Kaper JB. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361–379 [DOI] [PubMed] [Google Scholar]
- 55. Casaz P, Garrity-Ryan LK, McKenney D, Jackson C, Levy SB, Tanaka SK, Alekshun MN. 2006. MarA, SoxS and Rob function as virulence factors in an Escherichia coli murine model of ascending pyelonephritis. Microbiology 152:3643–3650 [DOI] [PubMed] [Google Scholar]
- 56. Cohen SP, McMurry LM, Levy SB. 1988. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J. Bacteriol. 170:5416–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O'Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, Ganz T, Kagnoff MF. 1999. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163:6718–6724 [PubMed] [Google Scholar]
- 58. Warner DM, Levy SB. 2010. Different effects of transcriptional regulators MarA, SoxS and Rob on susceptibility of Escherichia coli to cationic antimicrobial peptides (CAMPs): Rob-dependent CAMP induction of the marRAB operon. Microbiology 156:570–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aldon D, Brito B, Boucher C, Genin S. 2000. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 19:2304–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roy K, Hamilton DJ, Fleckenstein JM. 2012. Cooperative role of antibodies against heat-labile toxin and the EtpA adhesin in preventing toxin delivery and intestinal colonization by enterotoxigenic Escherichia coli. Clin. Vaccine Immunol. 19:1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Evans DG, Silver RP, Evans DJ, Jr, Chase DG, Gorbach SL. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Levine MM, Ristaino P, Marley G, Smyth C, Knutton S, Boedeker E, Black R, Young C, Clements ML, Cheney C, Patnaik R. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44:409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Froehlich B, Parkhill J, Sanders M, Quail MA, Scott JR. 2005. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J. Bacteriol. 187:6509–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.