Abstract
Animal replication-dependent histone pre-mRNAs are processed at the 3′ end by endonucleolytic cleavage that is not followed by polyadenylation. The cleavage reaction is catalyzed by CPSF73 and depends on the U7 snRNP and its integral component, Lsm11. A critical role is also played by the 220-kDa protein FLASH, which interacts with Lsm11. Here we demonstrate that the N-terminal regions of these two proteins form a platform that tightly interacts with a unique combination of polyadenylation factors: symplekin, CstF64, and all CPSF subunits, including the endonuclease CPSF73. The interaction is inhibited by alterations in each component of the FLASH/Lsm11 complex, including point mutations in FLASH that are detrimental for processing. The same polyadenylation factors are associated with the endogenous U7 snRNP and are recruited in a U7-dependent manner to histone pre-mRNA. Collectively, our studies identify the molecular mechanism that recruits the CPSF73 endonuclease to histone pre-mRNAs, reveal an unexpected complexity of the U7 snRNP, and suggest that in animal cells polyadenylation factors assemble into two alternative complexes—one specifically crafted to generate polyadenylated mRNAs and the other to generate nonpolyadenylated histone mRNAs that end with the stem-loop.
INTRODUCTION
The vast majority of eukaryotic pre-mRNAs are processed at the 3′ end by cleavage coupled to polyadenylation (1–4). In this reaction, pre-mRNAs are cleaved 15 to 30 nucleotides after the highly conserved AAUAAA sequences and the upstream cleavage product is extended by addition of a poly(A) tail. Cleavage coupled to polyadenylation is carried out by a macromolecular machinery consisting of multiple proteins that assemble into at least four separate subcomplexes or factors. The AAUAAA sequence is recognized by cleavage and polyadenylation specificity factor (CPSF), which contains CPSF160, CPSF100, CPSF73, CPSF30, Fip1 (5), and the recently identified WDR33 (6). CPSF160 directly contacts the AAUAAA hexanucleotide, whereas CPSF73 is the endonuclease that catalyzes the cleavage reaction (7). Cleavage stimulation factor (CstF), consisting of CstF77, CstF64, and CstF50, recognizes the GU-rich sequence located downstream of the cleavage site. CstF64 makes direct contacts with this sequence and also interacts with CstF77, which in turn interacts with CstF50 (8). 3′-end processing by cleavage and polyadenylation additionally requires cleavage factor (CF) Im, consisting of 25-kDa and 68-kDa subunits (9), and cleavage factor IIm containing at least two subunits, Pcf11 and Clp1 (2, 10). Individual components of the cleavage and polyadenylation machinery are connected with each other through a dense network of protein-protein interactions that stabilizes the entire complex and juxtaposes CPSF73 with the cleavage site. An important role in forming this network is played by symplekin, a protein that interacts with a number of polyadenylation factors and likely functions as a scaffold in 3′-end processing (8, 11, 12) and other processes, including cytoplasmic polyadenylation (13).
Animal replication-dependent histone pre-mRNAs are processed at the 3′ end by a distinct processing reaction (14, 15). In this reaction, cleavage occurs after a conserved stem-loop structure and the upstream cleavage product is not polyadenylated. The cleavage reaction critically depends on U7 snRNP consisting of an approximately 60-nucleotide U7 snRNA (16–18) and an unusual ring of Sm proteins in which the two spliceosomal proteins SmD1 and SmD2 are replaced by the related Lsm10 and Lsm11 (19). The 5′ end of U7 snRNA recognizes histone pre-mRNAs by base pairing with the histone downstream element (HDE) located 3′ of the cleavage site (17, 20). The stem-loop structure interacts with the stem-loop binding protein (SLBP), also known as hairpin binding protein (HBP) (21, 22). This protein stabilizes the association of the U7 snRNP with histone pre-mRNA but is not essential for cleavage in mammalian nuclear extracts (23, 24).
Lsm11, one of the two U7-specific Sm subunits, contains an extended N terminus that is absolutely essential for processing (25). This region interacts with the N-terminal region of FLASH (26), a 220-kDa protein that localizes to histone locus bodies and is required for histone gene expression (27, 28) and transcriptional regulation of several essential genes, including oncogenes (29). Intriguingly, FLASH was initially discovered as a factor involved in Fas-mediated activation of caspase 8 (30). Cleavage of histone pre-mRNAs is catalyzed by CPSF73 (31), the same endonuclease that cleaves canonical pre-mRNAs (2), and requires at least two other factors shared with the cleavage/polyadenylation machinery: symplekin and CPSF100 (32–34).
How CPSF73 is recruited to the unique 3′-end processing machinery assembled on histone pre-mRNAs has not been determined. Here we show that FLASH and Lsm11 form a highly efficient platform that binds a unique combination of polyadenylation factors: symplekin, CstF64, and all subunits of CPSF, including the endonuclease CPSF73. These factors are likely preassembled into a complex specifically engaged in 3′-end processing of histone pre-mRNAs that we refer to as the histone pre-mRNA cleavage complex (HCC). The HCC conspicuously lacks the two remaining CstF subunits of 50 and 77 kDa and components of cleavage factors Im and IIm. The same combination of polyadenylation factors is associated with U7 snRNP purified from mammalian nuclear extracts. Hence, in mammalian cells at least a fraction of U7 snRNP and a number of polyadenylation factors exist as a preassembled complex that is directly delivered to histone pre-mRNA through the base-pairing interaction involving U7 snRNA and the HDE.
MATERIALS AND METHODS
3′-end processing.
The 86-nucleotide mouse H2a pre-mRNA substrate was generated by T7 RNA polymerase using an appropriate DNA template and was subsequently treated with calf intestinal phosphatase (NEB) to remove the 5′ triphosphate. The dephosphorylated H2a pre-mRNA was labeled at the 5′ end with 32P using T4 polynucleotide kinase (NEB). The 3′-end processing reaction was carried out in a nuclear extract prepared from mouse myeloma cells (Biovest International, Inc., MN), as previously described (35). The H2a/5m pre-mRNA, anti-U7, and anti-Mock oligonucleotides were synthesized by Dharmacon (Lafayette, CO) and had the following sequences: H2a/5m (61-mer), 5′-(biotin)(18S)(18S)CUCCCAAAAAGGCUCUUUUCAGAGCCACmCmCmAmCmUGAAUCAGAUAAAGAGCUGUGACACGGUA3-′; anti-U7 (17-mer), 5′-mAmAmAmGmAmGmCmUmGmUmAmAmCmAmCmUmU(18S)(18S)(biotin)-3′; and anti-Mock (15-mer), 5′-mCmGmAmGmCmUmCmGmAmUmUmCmGmCmC(18S)(18S)(biotin)-3′. Note that in these sequences, “(18S)” represents an 18-atom spacer and “m” represents the 2′-O-methyl group.
Mutagenesis and protein expression.
Mutations in the N-terminal fragments of FLASH (amino acids 29 to 139) and Lsm11 (amino acids 1 to 168) were generated using PCR and appropriately altered oligonucleotide primers. FLASH mutants were expressed in bacteria from the pET-42a vector, as described previously (26), and contained an N-terminal glutathione S-transferase (GST) tag. The wild-type N-terminal fragment of Lsm11 and its mutant versions were expressed in bacteria from the pDEST566 vector as fusions with an N-terminal maltose binding protein (MBP). Both FLASH and Lsm11 proteins additionally contained a 6×His tag that was used for purification on nickel beads (Qiagen), as described by the manufacturer.
GST pulldown assay.
The wild type and various deletion mutants of the N-terminal fragment of human Lsm11 (amino acids 1 to 168) were synthesized in the presence of [35S]methionine using the TnT kit (Promega), as recommended by the manufacturer. Their ability to interact with FLASH was tested using a GST-mediated pulldown assay, as described in detail previously (26). Briefly, the 35S-labeled Lsm11 proteins were mixed with the bacterially expressed GST-tagged N-terminal FLASH (amino acids 29 to 139) or with GST alone. Proteins bound to FLASH were adsorbed on glutathione (GSH) beads, separated on SDS-polyacrylamide gels, and detected by autoradiography.
Binding of nuclear proteins to the FLASH/Lsm11 complex.
The FLASH/Lsm11 complex was formed by mixing 100 pmol of each recombinant protein in 100 μl of buffer D (100 mM KCl, 20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA [pH 8], 0.5 mM dithiothreitol [DTT]). The complex was subsequently incubated for 20 min at room temperature with 100 to 300 μl of a nuclear extract from HeLa or mouse myeloma cells. Bound proteins were purified on glutathione-Sepharose beads via the GST-tagged FLASH, electrophoretically resolved on 8 to 12% SDS–polyacrylamide gels, and detected by Western blotting and/or staining with either Coomassie blue or silver. To identify protein bands that correspond to recombinant FLASH and Lsm11, the FLASH/Lsm11 complex was bound to glutathione beads in the absence of the nuclear extract and electrophoretically separated next to samples containing nuclear proteins.
Formation of processing complexes and purification of the U7 snRNP.
Processing complexes were assembled in a final volume of 1 ml containing 750 μl of a highly active mouse myeloma nuclear extract, 1.25 μg of the 61-nucleotide H2a/5m pre-mRNA, and 20 mM EDTA. The samples were incubated for 5 min at 22°C followed by a 1-h rotation at 4°C. The RNA substrate and associated proteins were bound to streptavidin beads, washed several times with buffer D containing 20 mM EDTA, and separated on an SDS-polyacrylamide gel. The same method was used for the single-step purification of the endogenous U7 snRNP, with the exception that the anti-U7 2′-O-methyl oligonucleotide (1 μg) containing biotin at the 3′ end was used instead of the H2a/5m pre-mRNA.
Antibodies.
Antibodies against Lsm11 and FLASH were described previously (26, 36). Antibodies against all polyadenylation factors were purchased from Bethyl Laboratories. Anti-viral protein R binding protein (anti-VprBP) and anti-damaged DNA binding protein 1 (anti-DDB1) were kindly provided by the laboratory of Y. Xiong (University of North Carolina at Chapel Hill).
Other methods.
For details on mass spectrometry and size exclusion chromatography, see the supplemental material.
RESULTS
Bacterially expressed FLASH and Lsm11 form a tight complex.
Although human FLASH contains almost 2,000 amino acids, a short N-terminal region encompassing amino acids 52 to 139 is sufficient to stimulate processing of histone pre-mRNAs in vitro (37) (Fig. 1D, lanes 3 and 4). Amino acids 100 to 135 interact with Lsm11, whereas the highly conserved LDLY sequence located between amino acids 55 and 58 plays an essential but undetermined role in processing (Fig. 1B). Mutant proteins lacking the LDLY motif inhibit rather than stimulate processing in vitro by sequestering Lsm11 (and hence U7 snRNP) into an inactive processing complex (37) (Fig. 1D, lane 5). The Lsm11 binding site and the LDLY motif are functionally conserved in Drosophila FLASH and are essential in vivo (38).
Fig 1.
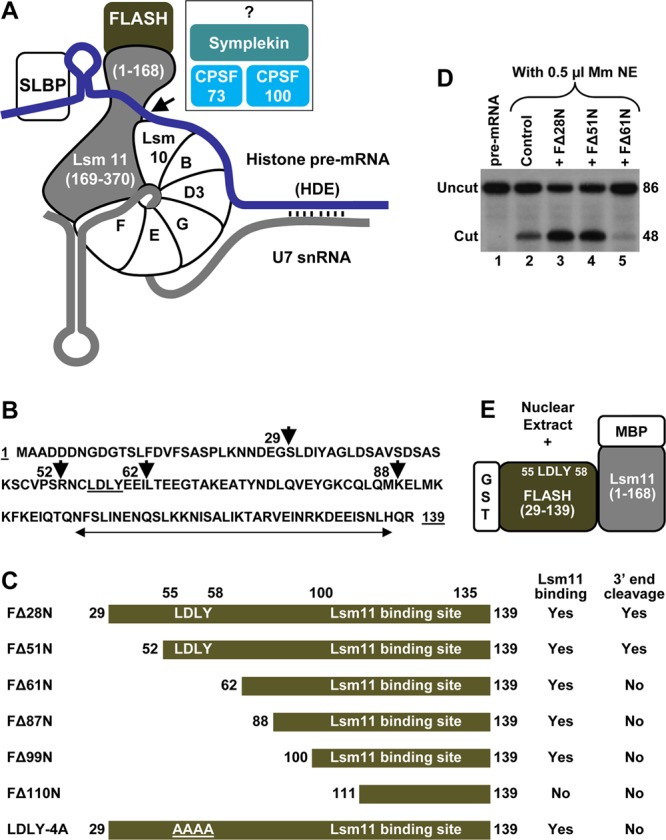
Biochemical assay to identify proteins interacting with a complex of FLASH and Lsm11. (A) The known critical factors in 3′-end processing of histone pre-mRNA (blue line) are shown. Vertical lines indicate base paring between the histone downstream element (HDE) and the 5′ end of U7 snRNA (gray line). Histone pre-mRNAs are cleaved by CPSF73 typically five nucleotides after the stem-loop (arrow). The reaction also requires CPSF100, symplekin, and potentially other factors (question mark). (B) Amino acid sequence of the N-terminal region of human FLASH (amino acids 1 to 139). The first amino acids of various N-terminal truncations are indicated with arrowheads. The Lsm11-binding site is indicated with a double-headed arrow, and the LDLY motif is underlined. (C) Diagram of FLASH mutants used in this study. The GST tag is fused N terminally to each protein and is not indicated. The ability of each FLASH protein to support processing or bind Lsm11 is indicated. (D) In vitro processing of histone pre-mRNA with a limiting amount of mouse nuclear extract (NE) in the absence of any recombinant protein (lane 2) or in the presence of 100 ng of various FLASH proteins, as indicated (lanes 3 to 5). Lane 1 contains the input substrate. (E) Schematic of the assay for isolation of factors that bind to the FLASH/Lsm11 complex. The N-terminal FLASH (amino acids 29 to 139) fused to GST and the N-terminal Lsm11 (amino acids 1 to 168) fused to MBP were incubated with a nuclear extract, and interacting proteins were purified on glutathione beads.
We first tested whether bacterially expressed N-terminal regions of FLASH and Lsm11 interact with the same efficiency and specificity as previously shown for radioactively labeled proteins generated using in vitro transcription and translation system (TnT) (37, 38). We bacterially expressed a series of GST-tagged FLASH deletions beginning at amino acid 52, 62, 88, 100, or 111 and ending at amino acid 139: FΔ51N, FΔ61N, FΔ87N, FΔ99N, and FΔ110N (Fig. 1B and C). Of these FLASH variants, only FΔ28N and FΔ51N contain the essential LDLY motif (amino acids 55 to 58) and are active in processing (Fig. 1D, lanes 3 and 4) (37). We also bacterially expressed Lsm11 (amino acids 1 to 168) tagged at the N terminus with MBP. With the exception of FΔ110N, which lacks a substantial portion of the Lsm11 binding site (Fig. 1C), all of the remaining FLASH deletion mutants strongly interact with Lsm11 and the two proteins can be recovered either on glutathione beads via the GST tag attached to FLASH (see below; see Fig. S1 in the supplemental material) or on amylose beads via the MPB tag attached to Lsm11 (not shown). Thus, bacterially expressed N-terminal fragments of FLASH and Lsm11 together form a tight complex and the interaction between these two proteins involves amino acids 100 to 135 of FLASH, as previously concluded (37).
We used size exclusion chromatography to analyze the oligomeric status of bacterially expressed N-terminal Lsm11 (amino acids 1 to 168), FΔ51N and their complex (see the supplemental material). These experiments demonstrated that Lsm11 exists in solution as a monomer, while FΔ51N alone under the same conditions forms a homo-oligomer, most likely a tetramer (see Fig. S2B in the supplemental material). The molecular mass of the FΔ51N/Lsm11 complex is consistent with four molecules of FΔ51N interacting with one molecule of Lsm11, although further detailed studies are required to confirm this stoichiometry in the complex.
The FLASH/Lsm11 complex interacts with multiple polyadenylation factors.
Our hypothesis was that a complex formed by FLASH and Lsm11 interacts with another processing factor and that the LDLY motif is essential for this interaction (37). To test this hypothesis, we mixed bacterially expressed N-terminal portions of human FLASH and Lsm11 and analyzed the ability of the FLASH/Lsm11 complex to interact with CPSF73 and/or other polyadenylation factors present in nuclear extracts (Fig. 1E). Initially, we used FΔ51N (the shortest FLASH active in processing in vitro) either alone or together with Lsm11. The recombinant proteins were incubated either alone or with a HeLa nuclear extract for 20 min and subsequently collected on glutathione beads via the GST tag attached to FΔ51N. The two components of the FΔ51N/Lsm11 complex were recovered on glutathione beads, indicating that the presence of the nuclear extract does not interfere with formation of the complex (Fig. 2A, compare lanes 2 and 3). The same nuclear extract was also incubated with FΔ51N alone (Fig. 2A, lane 4). As visualized by silver staining of SDS gels, only a small number of HeLa nuclear proteins nonspecifically bind to glutathione beads in the absence of FΔ51N or Lsm11. One of these HeLa proteins migrates slightly slower than FΔ51N (Fig. 2A, lane 1) and comigrates with the longer FLASH variant FΔ28N (see Fig. S3A in the supplemental material).
Fig 2.
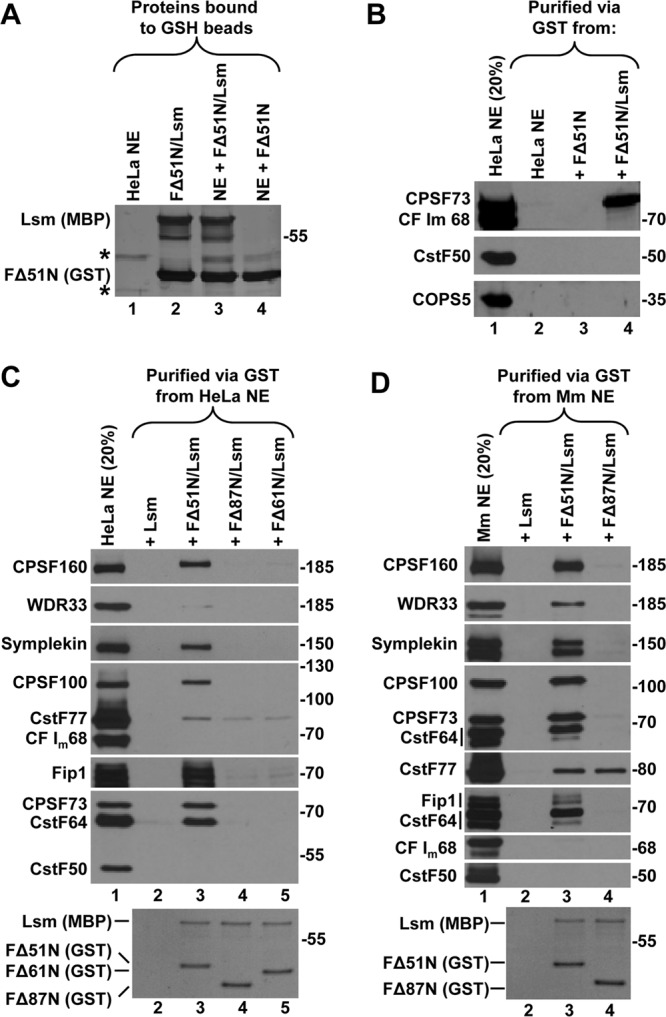
The FLASH/Lsm11 complex binds a subset of factors involved in cleavage and polyadenylation. (A) GST-mediated binding of the FΔ51N/Lsm11 complex to glutathione (GSH) beads in the absence (lane 2) or in the presence (lane 3) of a HeLa nuclear extract (NE). Binding of FΔ51N alone in the presence of the same nuclear extract is shown in lane 4. Proteins bound to GSH beads were resolved in an 8% SDS–polyacrylamide gel and stained with silver. Proteins in the nuclear extract bound to GSH beads in the absence of recombinant proteins are shown in lane 1 (indicated with asterisks). (B) FΔ51N (lane 3), alone or together with Lsm11 (lane 4), was incubated with a HeLa nuclear extract, and the material bound to GSH beads was tested for the presence of CPSF73, CF Im68, CstF50, and COPS5 using specific antibodies. Lane 2 contains proteins bound to GSH beads in the absence of recombinant proteins, and HeLa NE input (20%) is shown in lane 1. (C and D) Proteins of a HeLa nuclear extract (C) or mouse myeloma (Mm) nuclear extract (D) bound to GSH beads in the presence of indicated recombinant proteins were analyzed by Western blotting using specific antibodies. Coomassie blue staining of the two recombinant proteins bound to GSH beads is shown in the bottom panels. Lane 2 in each panel shows proteins collected on GSH beads in the presence of Lsm11, which is tagged with MBP and does not bind to GSH beads, hence serving as a negative control to measure the nonspecific background (see the bottom panels).
HeLa nuclear proteins bound to the FΔ51N/Lsm11 complex were screened by specific antibodies for the presence of CPSF73, CF Im68, and CstF50, each representing a separate class of cleavage and polyadenylation factors. Importantly, the FΔ51N/Lsm11 complex interacted with readily detectable amounts of CPSF73 but not with CF Im68 or CstF50 (Fig. 2B, lane 4). The precipitate also lacked COPS5, a 35-kDa component of the octameric COP9 signalosome (39), which served as a negative control for protein complexes unrelated to 3′-end processing. No CPSF73 bound to FΔ51N alone, emphasizing the requirement for the Lsm11 partner (Fig. 2B, lane 3).
We extended this experiment by using FΔ51N and two larger FLASH deletion mutants, FΔ61N and FΔ87N (Fig. 1C). We also used more antibodies to look for the presence of other polyadenylation factors. No detectable amount of polyadenylation factors accumulated on glutathione beads in the presence of Lsm11 alone, which is tagged with MBP and does not bind to GSH beads in the absence of GST-tagged FLASH, hence serving as a negative control to measure the nonspecific background of nuclear proteins (Fig. 2C, bottom panel, lane 2). Among HeLa proteins associated with the complex composed of Lsm11 and the processing-proficient FΔ51N FLASH, we identified CPSF73 and all of the remaining CPSF subunits larger than 50 kDa, including CPSF160, CPSF100, Fip1, and WDR33, although the latter subunit was only weakly detected by Western blotting (Fig. 2C, lane 3). CPSF30, which is the smallest subunit of CPSF, was detected in independent experiments after resolving proteins in a higher-percentage polyacrylamide gel (not shown) (Fig. 3A; see Fig. S3B in the supplemental material). The bound proteins also included symplekin and CstF64 (Fig. 2C, lane 3). Importantly, the binding of all these proteins was abolished when the complex contained the processing-deficient FLASH mutant FΔ61N or FΔ87N (Fig. 2C, lanes 4 and 5). This result clearly demonstrates that the strong binding of CPSF73 and other polyadenylation factors to the complex is not mediated solely by Lsm11, as the same amounts of Lsm11 were precipitated by all three FLASH deletion mutants (Fig. 2C, bottom panel, lanes 3 to 5).
Fig 3.
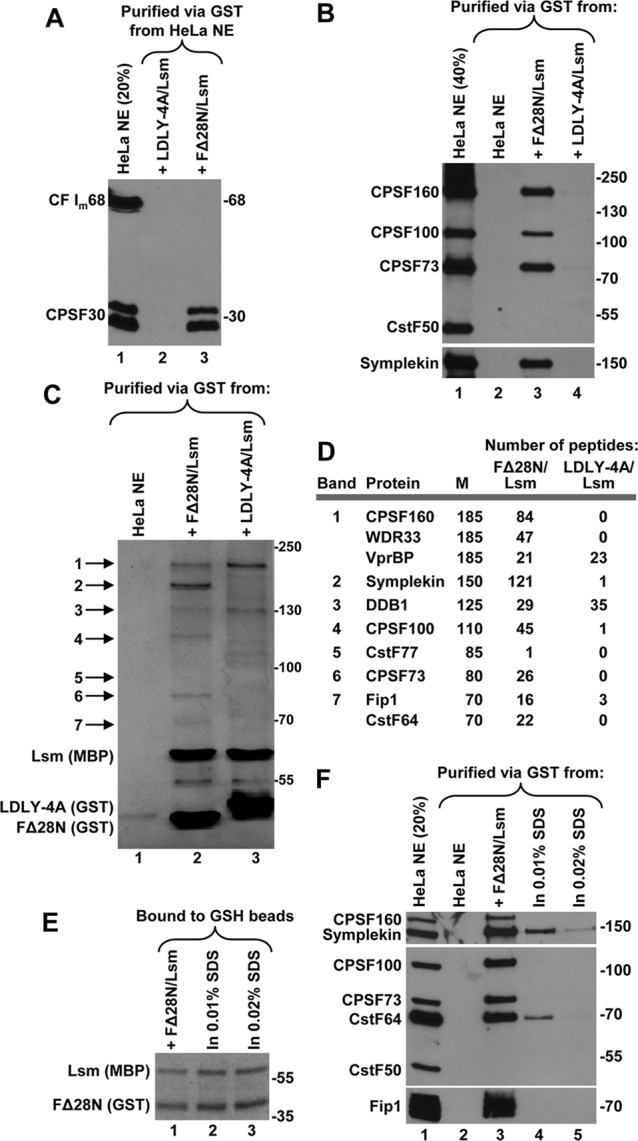
The LDLY motif in FLASH is required for the interaction of the complex with polyadenylation factors. (A and B) HeLa nuclear proteins bound to GSH beads in the presence of the indicated FLASH/Lsm11 complexes were analyzed using specific antibodies. Lane 2 in panel B contains HeLa nuclear proteins bound to GSH beads in the absence of recombinant proteins. (C and D) The identity of HeLa nuclear proteins bound to GSH beads in the presence of the indicated complexes (lanes 2 and 3) was determined by mass spectrometry. HeLa proteins were electrophoretically separated in a 10% SDS–polyacrylamide gel and stained with silver (C). Note that the LDLY-4A mutant migrates slower than the wild-type FΔ28N. Lanes 2 and 3 were divided into 11 equal sections (not shown) and treated with trypsin, and the complete proteome of each gel section was determined by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The most abundant proteins identified by mass spectrometry were assigned to individual silver-stained bands (indicated with arrows) and are listed in panel D. Lane 1 in panel C contains HeLa nuclear proteins bound to GSH beads in the absence of recombinant proteins, and it was not analyzed by mass spectrometry. Column M in panel D shows molecular masses in kDa. (E) Binding of recombinant FΔ28N and Lsm11 to GSH beads under control conditions (no SDS) (lane 1) or in the presence of the indicated concentrations of SDS (lanes 2 and 3). Proteins were detected by staining with Coomassie blue. (F) Binding of polyadenylation factors to the FΔ28N/Lsm11 complex under control conditions (no SDS) (lane 3) or in the presence of the indicated concentrations of SDS (lanes 4 and 5). Lane 1 contains 20% input of the HeLa NE, and lane 2 contains HeLa nuclear proteins bound to GSH beads in the absence of recombinant proteins. Polyadenylation factors were detected by Western blotting using specific antibodies.
The material bound to the FΔ51N/Lsm11 complex lacked CF Im68 and CstF50 and contained only weakly detectable amounts of CstF77 that were also present when the first 87 amino acids were deleted from FLASH (Fig. 2C, lanes 3 to 5). Subsequent studies demonstrated that CstF77 nonspecifically interacts with Lsm11 (not shown) and also has a weak affinity for streptavidin beads (described below and see Fig. 5A). The complex bound to the FΔ51N/Lsm11 complex also did not contain any detectable amounts of CstF64 Tau (not shown) (see Fig. S3B in the supplemental material). This paralogue of CstF64 associates with canonical pre-mRNAs (6) and can partially substitute for CstF64 in cleavage and polyadenylation and in 3′-end processing of histone pre-mRNAs (40).
Fig 5.
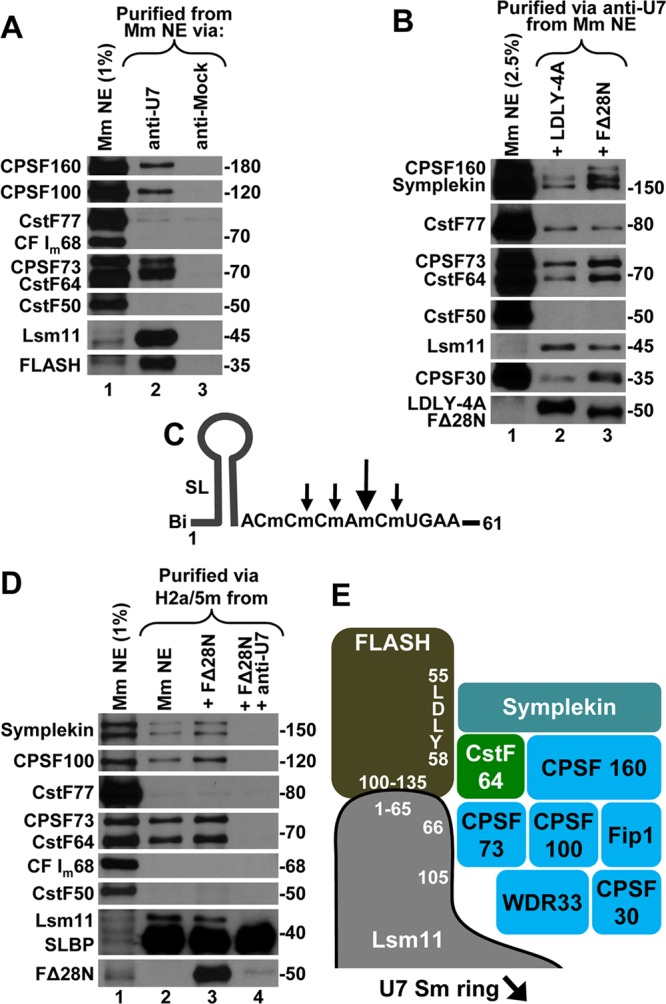
Polyadenylation factors bind to the endogenous U7 snRNP and histone pre-mRNA. (A) A mouse myeloma nuclear extract was incubated with the anti-U7 oligonucleotide (lane 2) or with an unrelated oligonucleotide (lane 3, anti-Mock), both containing a biotin tag at the 3′ end. Bound proteins were collected on streptavidin beads and analyzed by Western blotting using specific antibodies. (B) The anti-U7 oligonucleotide (lane 2) was used to purify the U7 snRNP from a mouse myeloma nuclear extract supplemented with either LDLY-4A (lane 2) or wild-type FΔ28N (lane 3). Purified proteins were analyzed using specific antibodies. Note that the recombinant FLASH is not present in the input nuclear extract. (C) A depiction of the 61-nucleotide H2a/5m histone pre-mRNA in which 5 nucleotides adjacent to the major cleavage site (large arrow) and minor cleavage sites (small arrows) are modified with a 2′-O-methyl group. The H2a/5m pre-mRNA contains the upstream stem-loop, the HDE fully complementary to the U7 snRNA (not shown), and biotin at the 5′ end. (D) The H2a/5m pre-mRNA was incubated with a mouse myeloma nuclear extract under control conditions (no recombinant FLASH added) (lane 2) or in the presence of recombinant FΔ28N without (lane 3) or with the anti-U7 oligonucleotide (lane 4). Proteins bound to the H2a/5m substrate were collected on streptavidin beads and analyzed by Western blotting using specific antibodies. FΔ28N is not present in the input nuclear extract, and the band detected in the same region in lane 1 is a result of cross-reactivity with an antibody used concomitantly with the anti-FLASH antibody. (E) Composition of the polyadenylation complex recruited to the U7 snRNP and histone pre-mRNA by the FLASH/Lsm11 complex. Amino acid regions in FLASH and Lsm11 involved in formation of the complex and binding the polyadenylation factors are indicated. The arrangement of polyadenylation subunits is arbitrary. The interaction between CstF64 and symplekin is mutually exclusive with the formation of the CstF complex, (CstF64, CstF77, and CstF50), which is involved in 3′-end processing of canonical pre-mRNAs by cleavage and polyadenylation.
The FΔ51N/Lsm11 complex bound the same set of proteins in a nuclear extract prepared from mouse myeloma cells (Fig. 2D, lane 3). Among the mouse proteins bound to the complex, WDR33 was readily detectable, as were the two forms of mouse symplekin. Again, with the exception of CstF77, the binding of all of the proteins was greatly reduced when the processing-deficient FΔ87N FLASH mutant was used instead of FΔ51N (Fig. 2D, lane 4). Altogether, our results demonstrate that in mammalian nuclear extracts the FΔ51N/Lsm11 complex interacts with a specific combination of cleavage and polyadenylation factors: symplekin, all CPSF subunits, and CstF64 as the only CstF component.
The LDLY sequence in FLASH is essential for the efficient recruitment of polyadenylation factors.
The LDLY motif is absolutely essential for the activity of FLASH in processing (37), and the above results suggest that this motif might also be critical for binding the multitude of polyadenylation factors to the FLASH/Lsm11 complex. To test this assumption, we used the processing-deficient LDLY-4A mutant in which the LDLY sequence was replaced with 4 alanines (Fig. 1C). The mutation was made in the context of the longer FLASH FΔ28N (amino acids 29 to 139) fused N terminally to GST. No detectable amounts of polyadenylation factors were bound to glutathione beads in the absence of any recombinant protein or in the presence of either Lsm11 (amino acids 1 to 168) or FΔ28N alone (see lanes 2, 5, and 6, respectively, in Fig. S3B in the supplemental material). The complex of FΔ28N and Lsm11 efficiently interacted with all the polyadenylation factors previously identified for the Lsm11/FΔ51N complex: symplekin, CPSF160, CPSF100, CPSF73, and CPSF30 (Fig. 3A and B; see Fig. S3B). Most strikingly, the complex of Lsm11 and LDLY-4A did not interact with any of these proteins (Fig. 3A and B; see Fig. S3B). We conclude that the essential function of the LDLY motif in processing is to recruit a subset of polyadenylation factors to histone pre-mRNA.
To confirm the identity of polyadenylation factors associated with FLASH and Lsm11 and to potentially identify other interacting proteins, we used mass spectrometry. We assembled the complex of Lsm11 (amino acids 1 to 168) and either the FΔ28N FLASH (amino acids 29 to 139) or its LDLY-4A mutant and incubated each complex with a HeLa nuclear extract. The wild-type complex interacted with a number of proteins readily detectable in a polyacrylamide gel by silver staining as individual bands (Fig. 3C, lane 2). These bands and the remaining parts of the same lane and the corresponding regions in the lane containing the mutant complex (Fig. 3C, lane 3) were excised and analyzed by mass spectrometry. This analysis fully confirmed the results obtained by Western blotting, with the slowest-migrating silver-stained band in the lane containing the wild-type complex (Fig. 3C, lane 2) yielding a large number of peptides for CPSF160 and WDR33 (band 1). The faster-migrating bands contained symplekin, CPSF100, CPSF73, Fip1, and CstF64 (bands 2 to 7). All of these proteins were virtually undetectable in the precipitate bound to the complex of Lsm11 and LDLY-4A (Fig. 3D). Consistent with the results of Western blotting, proteomic analysis of the material bound to the wild-type complex did not detect any peptides from CstF50 and CF Im68 and only one peptide from CstF77. In addition, no traces of other polyadenylation factors, including the Pcf11 subunit of CF IIm, were detected. Finally, the complex of the FΔ28N FLASH and Lsm11 does not bind any of previously identified proteins specifically involved in 3′-end processing of histone pre-mRNAs, including ZFP100 and SLBP (not shown) (see Fig. S3B in the supplemental material).
Both the wild-type and mutant FLASH/Lsm11 complexes bound readily detectable amounts of DDB1 (damaged DNA binding protein 1) and its binding partner VprBP (viral protein R binding protein) (41) that comigrates with CPSF160 and WDR33 (Fig. 3C, lanes 2 and 3). Our subsequent studies revealed that VprBP and DDB1 bind directly to Lsm11, and this interaction is not linked to 3′-end processing of histone pre-mRNA in vitro (data not shown).
Independent proteomic analyses were conducted using other HeLa nuclear extract preparations and a nuclear extract prepared from mouse myeloma cells (not shown). The same polyadenylation factors invariably bound to the wild-type FLASH/Lsm11 complex, regardless of whether the extract was very active or poorly active in processing and whether the EDTA was omitted or included at the final concentration of 20 mM (not shown), as used in the in vitro processing reaction. The unique combination of polyadenylation factors bound to the complex of FLASH and Lsm11 was also not affected by pretreatment of the nuclear extract with RNase A (not shown).
We carried out preliminary experiments to determine which polyadenylation proteins make direct contacts and/or the strongest contacts with the FLASH/Lsm11 complex. We incubated the two recombinant proteins with a HeLa nuclear extract in the presence of 0.01 and 0.02% SDS. These low detergent concentrations do not interfere with either the interaction between FΔ28N variant and Lsm11 or the binding of their complex to glutathione beads (Fig. 3E, lanes 2 and 3). Interestingly, only two polyadenylation factors bound the complex in the presence of 0.01% SDS: symplekin and CstF64 (Fig. 3F, lane 4). The other tested proteins, CPSF160, CPSF100, CPSF73, and Fip1, no longer bound the complex. Symplekin and CstF64 are therefore among the proteins that likely directly contact the FLASH/Lsm11 complex (see Discussion).
Regions of Lsm11 required for recruiting polyadenylation factors.
Our previous analysis indicated that deletion of the first 40 amino acids of Lsm11 (LΔ40N) almost completely eliminated its interaction with FLASH (26). We used the GST-binding assay and various 35S-labeled deletion mutants of Lsm11 expressed using the TnT system (Fig. 4A and B) to define the minimal region of Lsm11 required to form a complex with FLASH. A short fragment encompassing the first 40 amino acids of Lsm11 (L40N) is not sufficient to interact with FΔ28N fused to GST (Fig. 4C, top panel, lane 3). However, extending this region to 65 amino acids resulted in a minimal Lsm11 (L65N) that interacted with FΔ28N as strongly as the entire N-terminal Lsm11 encompassing amino acids 1 to 168 (Fig. 4C, top panels, lanes 6 and 9).
Fig 4.
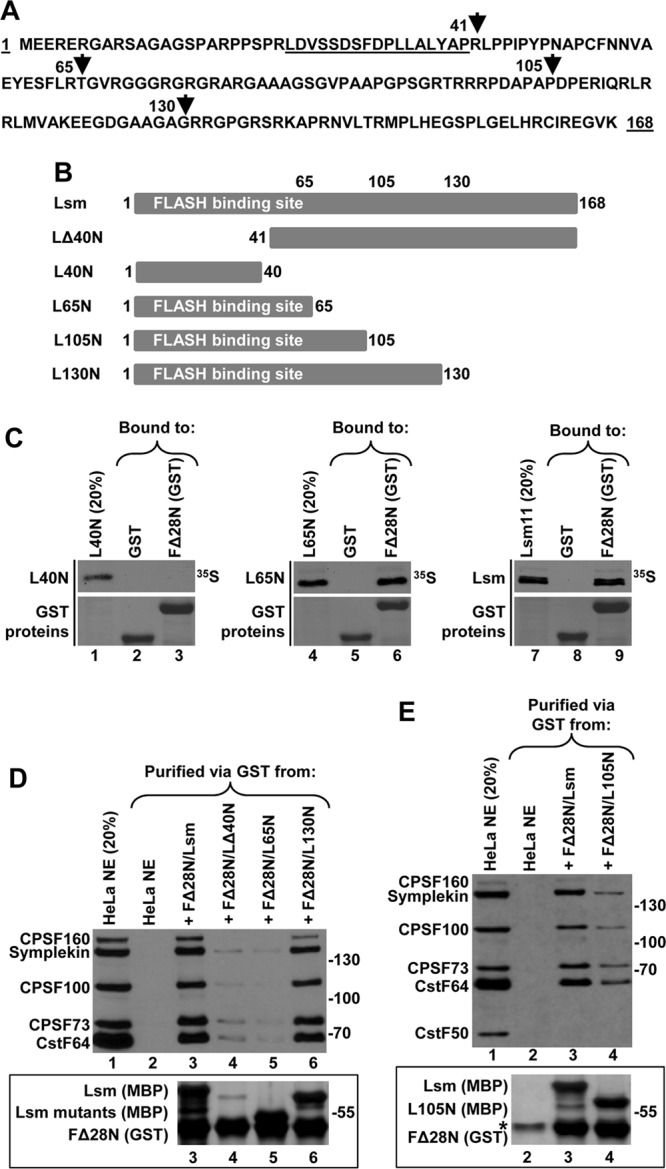
Sequences in Lsm11 required for binding the polyadenylation complex by the FLASH/Lsm11 complex. (A) Amino acid sequence of the N-terminal region of human Lsm11 (amino acids 1 to 168). The first or last amino acids of various truncations are indicated with arrowheads, and the FLASH-interacting region is underlined. (B) Diagrams of Lsm11 truncations used in the study. The MBP tag is fused N terminally to Lsm11 and is not indicated. (C) Binding of 35S-labeled Lsm11 truncations by either GST or FΔ28N tagged with GST, as indicated. GST-tagged proteins collected on GSH beads were stained with Coomassie blue (bottom panels). (D and E) Polyadenylation factors bound to GSH beads in the presence of the indicated FLASH and Lsm11 variants (lanes 3 to 6 in panel D and lanes 3 and 4 in panel E) were identified by Western blotting using specific antibodies. The recombinant proteins were stained with silver to monitor their recovery on GSH beads (bottom panels). The asterisk in panel E indicates a HeLa nuclear protein that nonspecifically binds to GSH beads and comigrates with FΔ28N (bottom panel, lane 2).
We bacterially expressed the same fragments of Lsm11 as fusions with MBP and tested their ability to interact with FΔ28N fused to GST and bind polyadenylation factors. As expected, only residual amounts of LΔ40N and polyadenylation factors were collected on glutathione beads (Fig. 4D, both panels, lane 4), consistent with the virtual inability of this Lsm11 mutant to form a complex with FLASH. Importantly, FΔ28N and the L65N mutant, while forming a very tight complex, were even less efficient in binding polyadenylation factors (Fig. 4D, both panels, lane 5). When the length of Lsm11 was increased from 65 to 130 N-terminal amino acids, the corresponding complex was nearly as efficient in binding symplekin, CstF64, and CPSF subunits as the wild-type complex containing the entire 168-amino-acid N-terminal Lsm11 (Fig. 4D, both panels, compare lanes 3 and 6). Shortening Lsm11 to the first 105 amino acids (L105N in Fig. 4B) resulted in a significant loss of binding activity by the FLASH/Lsm11 complex (Fig. 4E, both panels, lanes 3 and 4). Thus, a complex containing the first 130 amino acids of Lsm11 is sufficient for binding the polyadenylation factors.
Polyadenylation factors are present on the endogenous U7 snRNP.
We demonstrated that large amounts of the recombinant FLASH/Lsm11 complex bind a unique combination of polyadenylation factors in mammalian nuclear extracts. Does FLASH associate with the endogenous Lsm11 to recruit the same subset of polyadenylation factors to the U7 snRNP? To address this question, we purified the U7 snRNP on streptavidin beads using a 2′-O-methyl oligonucleotide that contains a biotin tag at the 3′ end and a sequence complementary to the first 15 nucleotides of the U7 snRNA (anti-U7). We also used a 3′-biotinylated 2′-O-methyl oligonucleotide with an unrelated sequence that served as a negative control (anti-Mock). The experiment was carried out with a nuclear extract from mouse myeloma cells, which contains about a 10-fold higher concentration of the U7 snRNP than extracts from HeLa cells (our unpublished observations).
As judged by the high enrichment of Lsm11 in the material bound to the anti-U7 oligonucleotide, this antisense oligonucleotide is very efficient in purifying the U7 snRNP (Fig. 5A, lane 2). The anti-Mock oligonucleotide did not result in any background of Lsm11, indicating that it has no affinity for the U7 snRNA (Fig. 5A, lane 3). Most importantly, readily detectable amounts of polyadenylation factors, including several CPSF subunits and CstF64, were present in the anti-U7 precipitate but not in the anti-Mock precipitate. Compared with the input, the amounts of these proteins were small and this reflects the extremely low concentration of endogenous U7 snRNP and FLASH and the large abundance of the polyadenylation factors in nuclear extracts. A specific antibody against the N-terminal region of FLASH encompassing amino acids 1 to 139 detected a band migrating at about 35 kDa in the anti-U7 precipitate that was not present in the control precipitate (Fig. 5A, lanes 2 and 3). This band most likely represents a FLASH degradation product encompassing the first ∼300 amino acids of the protein, i.e., the region that is sufficient for the interaction with Lsm11 and which contains the region used to generate the anti-FLASH antibody (26).
Only background amounts of CstF77 were detected in both the anti-U7 and anti-Mock precipitates, indicating that CstF77 is not part of a complex with CstF64 in the U7 snRNP (Fig. 5A, lanes 2 and 3). In agreement with the data obtained for the recombinant FLASH/Lsm11 complex, the U7 snRNP-bound proteins also did not contain detectable amounts of CstF50 or CF Im68. We conclude that in mammalian cells at least a fraction of the U7 snRNP exists as a preassembled complex containing the same set of polyadenylation factors that interact with the recombinant FLASH/Lsm11 complex.
Addition of recombinant FΔ28N but not the LDLY-4A mutant increased the amount of polyadenylation complex bound to the U7 snRNP but had no effect on the amount of CstF77 detected (Fig. 5B, lanes 2 and 3). Note that lane 3 compared to lane 2 was loaded with a smaller amount of U7 snRNP, as indicated by a weaker signal for Lsm11, yet symplekin, CPSF160, CPSF73, CPSF30, and CstF64 were clearly more abundant. These results confirm that a fraction of the endogenous U7 snRNP is associated with a subset of polyadenylation factors and support the notion that these factors are recruited to the U7 snRNP by FLASH interacting with Lsm11.
Composition of the processing complex assembled on histone pre-mRNA.
We next investigated whether the same cleavage and polyadenylation factors, including the endonuclease CPSF73, can be detected in vitro in a complex assembled on histone pre-mRNA. An obstacle in achieving this goal is that the cleavage reaction proceeds very rapidly in nuclear extracts incubated at room temperature, causing disruption of the processing complex. Previously, we used low concentrations of NP-40 (0.05 to 0.1%) to block the cleavage reaction and to arrest processing complexes during the assembly process (36). This strategy allowed us to capture SLBP and the U7 snRNP in association with histone pre-mRNA, but we failed to detect any polyadenylation factors. One possibility was that while NP-40 blocks processing, it does so by destabilizing the interaction of polyadenylation factors with FLASH bound to the U7 snRNP.
To avoid this potential limitation, we designed a new histone pre-mRNA substrate termed H2a/5m in which 5 nucleotides surrounding the major and three minor cleavage sites were modified with a 2′-O-methyl group (Fig. 5C). Our previous studies demonstrated that 2′-O-methyl nucleotides are resistant to hydrolysis by CPSF73 when it acts as a 5′ exonuclease (42). The H2a/5m substrate also contained two point mutations within the HDE that improved its interaction with the U7 snRNA and biotin at the 5′ end for subsequent purification of bound nuclear components on streptavidin beads. When incubated in a mouse nuclear extract, the H2a/5m pre-mRNA assembled into a stable complex containing highly enriched amounts of SLBP and, as judged by the presence of Lsm11, the U7 snRNP. This result indicates that the cleavage reaction was at least partially blocked (Fig. 5D, lane 2).
As determined using specific antibodies, the complex assembled on the H2a/5m pre-mRNA contained the same polyadenylation factors that associate with the recombinant FLASH/Lsm11 complex or the U7 snRNP, including symplekin, CPSF subunits, and CstF64 but lacking CstF50 and CF Im68 (Fig. 5D, lane 2). Addition of recombinant FLASH (amino acids 29 to 139) only slightly increased the amount of these factors in the processing complex, although the recombinant FLASH was efficiently delivered to the H2a/5m pre-mRNA through the interaction with Lsm11 (see Discussion). Importantly, blocking the U7 snRNA by a 2′-O-methyl oligonucleotide identical to the anti-U7 but lacking the biotin tag prevented the association of the U7 snRNP, recombinant FΔ28N, and all of the polyadenylation factors with the pre-mRNA substrate without affecting the binding of SLBP (Fig. 5D, lane 4). Again, only very small amounts of CstF77 were detected in both the presence and the absence of the anti-U7 oligonucleotide (Fig. 5D, compare lanes 2 to 4), indicating that this protein nonspecifically associates with the substrate and/or streptavidin beads and is not part of the complex with other polyadenylation factors. We conclude that a preassembled U7 snRNP containing the CPSF73 3′ endonuclease and multiple polyadenylation factors is delivered to histone pre-mRNA through the interaction between the HDE and the U7 snRNA for 3′-end processing (Fig. 5E).
DISCUSSION
Cleavage of animal replication-dependent histone pre-mRNAs critically depends on the U7 snRNP that interacts with the histone downstream element (HDE) located several nucleotides 3′ of the cleavage site. Histone pre-mRNAs do not share any sequence elements with canonical pre-mRNAs, and their cleavage is not followed by the polyadenylation step. Quite unexpectedly, previous studies showed that 3′-end processing of histone pre-mRNAs requires at least three components of the cleavage/polyadenylation machinery: the endonuclease CPSF73, CPSF100, and symplekin (31, 32, 34). For canonical pre-mRNAs, the CPSF73 endonuclease is brought to the vicinity of the cleavage site primarily by CPSF160, which recognizes the upstream AAUAAA element (2). However, it has not been determined how CPSF73 and the two other common subunits are recruited to the processing machinery assembled on histone pre-mRNAs.
A subset of polyadenylation factors interacts with the FLASH/Lsm11 complex.
Of the known components of the U7 snRNP-based processing machinery, Lsm11 was the most likely candidate for a protein that either directly or indirectly recruits CPSF73 to histone pre-mRNA. First, Lsm11 is the largest component of the U7-specific Sm ring and contains an extended N-terminal region of about 170 amino acids that is essential for processing (25). Furthermore, this extended region interacts with FLASH (26), a recently identified factor required for endonucleolytic cleavage of histone pre-mRNAs by CPSF73 (37). Finally, the base pair interaction between the HDE and the 5′ end of the U7 snRNA brings Lsm11 near the cleavage site (36).
In this study, we bacterially expressed the N-terminal fragments of FLASH and Lsm11 and showed that these two recombinant proteins interact with each other. Our initial gel filtration experiments demonstrated that the FLASH/Lsm11 complex is significantly larger than a simple heterodimer of the two proteins and suggest that it may consist of four molecules of FLASH and one molecule of Lsm11. This conclusion is consistent with the mechanism of 3′-end processing in which a single particle of the U7 snRNP interacts with histone pre-mRNA and with the observation that FLASH can self-associate through the N-terminal region (43).
Remarkably, the complex of recombinant FLASH and Lsm11 tightly interacts with a unique combination of polyadenylation factors present in mammalian nuclear extracts, whereas FLASH and Lsm11 individually are virtually incompetent in binding the same factors. Among nuclear proteins associated with the FLASH/Lsm11 complex were symplekin, CstF64, and all subunits of CPSF, including the recently discovered component WDR33 (6) and the endonuclease CPSF73. This is the first biochemical demonstration of an interaction between proteins specifically devoted to processing of histone pre-mRNAs (FLASH and Lsm11) with subunits of the cleavage and polyadenylation machinery. The two other subunits of CstF, CstF77 and CstF50, as well as the cleavage factors I and II, were absent, indicating that FLASH and Lsm11 interact with a very specific subset of polyadenylation factors.
The interaction of the FLASH/Lsm11 complex with CPSF subunits, CstF64, and symplekin was abolished by mutating the LDLY sequence in FLASH, a highly conserved cluster of amino acids found in FLASH orthologues of both vertebrates and invertebrates. Thus, all of these polyadenylation factors likely exist in the cell as a single complex that is specifically crafted to directly interact with the FLASH/Lsm11 complex. Due to the presence of the CPSF73 endonuclease, we refer to this complex as the histone pre-mRNA cleavage complex (HCC). The LDLY motif is absolutely essential for the activity of FLASH in processing of histone pre-mRNAs in mammalian nuclear extracts (37), and deletion of the corresponding LDIY sequence in Drosophila FLASH disrupts 3′-end processing in cultured S2 cells (38). Our current results demonstrate that the role of this motif in processing is to recruit the CPSF73 endonuclease and other components of the HCC to the FLASH/Lsm11 complex.
The details of the interaction between the FLASH/Lsm11 complex and the HCC are not known. In human Lsm11, amino acids 1 to 65 interact with FLASH and an additional region(s) located between amino acids 66 and 130 is required for binding the HCC by the FLASH/Lsm11 complex. In the simplest model, the LDLY motif in FLASH and amino acids 66 to 130 in Lsm11 have individually a weak affinity for one or more components of the HCC. When these two sequence elements are juxtaposed together in the FLASH/Lsm11 complex, they function in a cooperative manner, resulting in a very tight binding of the HCC. An important role in binding the HCC may also be played by the interface of the interacting Lsm11 and FLASH and/or potential structural rearrangements in each protein triggered by the formation of the Lsm11 and FLASH complex.
Intriguingly, the HCC is very similar in its composition to the heat labile factor (HLF) that was identified by Kolev and Steitz in HeLa cell nuclear fractions capable of restoring the ability of heat-treated nuclear extracts to process histone pre-mRNAs (32). The HLF contains symplekin as the most heat-sensitive component and cofractionated with all of the known CPSF subunits and CstF64. Similar to the complex recruited to histone pre-mRNA, the HLF lacks CstF50 and subunits of CF Im and CF IIm but was reported to contain CstF77, which we detect as a nonspecific contaminant. An attractive possibility is that the HCC and the HLF are identical and CstF77 is not a genuine part of the HLF but simply copurifies as a component of a different complex sharing similar chromatographic properties to the HCC. Taken together, our findings and the previous results of Kolev and Steitz demonstrate that the functional overlap between the canonical cleavage/polyadenylation and 3′-end processing of histone pre-mRNAs is much more extensive than previously anticipated, strongly suggesting that these two 3′-end processing reactions are evolutionarily related (44).
U7 snRNP as a preformed 3′-end processing unit.
The HCC is also associated with the endogenous U7 snRNP in the absence of a pre-mRNA substrate. Based on our experiments with recombinant Lsm11 and FLASH, the assembly of this “active” U7 snRNP is dependent on FLASH, which is limiting in many nuclear extracts. Indeed, addition of recombinant wild-type FLASH, but not the LDLY-4A mutant, to a nuclear extract stimulated the association of the polyadenylation complex with the U7 snRNP. Overall, our results reveal an unexpected complexity of the U7 snRNP, previously believed to consist solely of the U7 snRNA and the Sm ring.
Previous studies of U7 snRNP purified by several chromatographic steps did not reveal novel components of this snRNP other than Lsm10 and Lsm11, suggesting that the entire HCC dissociated from U7 snRNP during purification, perhaps as a result of FLASH proteolysis (45, 46). More recent proteomic studies identified CF Im68 as the only polyadenylation factor that associates with the U7 snRNP (47). We found no evidence that this protein is recruited by the FLASH/Lsm11 complex or is part of the endogenous U7 snRNP purified by the anti-U7 oligonucleotide. Further studies are required to determine the source of this contradiction.
We also isolated a processing complex assembled on a histone pre-mRNA that was resistant to cleavage as a result of the placement of five 2′-O-methyl nucleotides around the cleavage site. This processing complex contained SLBP, U7 snRNP, and the same combination of polyadenylation factors that bind the FLASH/Lsm11 complex, suggesting that the “active” form of U7 snRNP carrying the HCC is directly delivered to histone pre-mRNA through a single-step base-pairing interaction between the HDE and U7 snRNA. Addition of recombinant FLASH only slightly stimulated recruitment of the HCC to the modified histone pre-mRNA, suggesting that the interaction between the HCC and the U7 snRNP in the context of the entire processing machinery assembled on histone pre-mRNA is intrinsically less stable and/or the modified substrate was undergoing limited cleavage in neighboring sites that lack the 2′-O-methyl modification.
Potential roles of components of the HCC in 3′-end processing of histone pre-mRNAs.
Significant amounts of symplekin and CstF64 remain bound to the FLASH/Lsm11 complex in the absence of the CPSF subunits. Thus, symplekin in association with CstF64 may function as an adapter that bridges CPSF with the U7 snRNP/FLASH complex. Symplekin is a large and versatile protein that plays a scaffolding role in cleavage/polyadenylation (8, 11, 12, 48) and other processes (13, 49), and it may directly contact the LDLY motif in FLASH during 3′-end processing of histone pre-mRNAs.
CstF64 was previously shown to interact through overlapping regions with either CstF77 or symplekin, but it is unable to interact with these two proteins simultaneously (8, 40). The presence of CstF64 and symplekin in the complex bound to the FLASH/Lsm11 complex likely explains the concomitant lack of CstF77 and CstF50, as CstF50 interacts with CstF77 (8, 50). Thus, our data suggest that the previously puzzling mutually exclusive interaction of CstF64 with either symplekin or CstF77 allows formation of two alternative complexes in animal cells: one containing all three CstF subunits that functions in cleavage and polyadenylation and the other composed of CstF64 and symplekin that operates on histone pre-mRNAs (Fig. 5E). This interpretation is strongly supported by the recent in vivo studies of Schumperli and coworkers who in an elegant set of experiments addressed the role of the CstF64-symplekin interaction in 3′-end processing of canonical and histone pre-mRNAs. These authors designed a symplekin mutant that does not bind CstF64 and tested its ability to substitute for the endogenous symplekin depleted from HeLa cells by RNA interference (RNAi) (40). Importantly, this mutant protein could efficiently rescue cleavage/polyadenylation, but formation of mature histone mRNAs remained impaired, demonstrating that in vivo the symplekin-CstF64 interaction is required for 3′-end processing of histone pre-mRNA but not canonical pre-mRNAs. Intriguingly, CstF64 is the only known component of the cleavage and polyadenylation machinery that is upregulated severalfold as cells transit from G0 to S phase (51). In addition, depletion of CstF64 causes cell cycle arrest and apoptosis-like cell death (52), and this phenotype could be directly related to its role in generating mature histone mRNAs. Overall, our results suggest that the biochemical function of the CstF64/symplekin heterodimer is to interact with FLASH and Lsm11 and to bring other polyadenylation factors, including CPSF73 and CPSF100, to the U7 snRNP. This “active” U7 snRNP is ultimately recruited to histone pre-mRNA for 3′-end processing.
We tested the possibility that symplekin and CstF64 directly bind to the FLASH/Lsm11 complex by expressing symplekin and CstF64 using an in vitro transcription and translation system (TnT). However, we observed only weak interaction of these two polyadenylation factors with the FLASH/Lsm11 complex and the interaction was not dependent on the LDLY motif (not shown). Perhaps symplekin and/or CstF64 requires a posttranslational modification to stably interact with the FLASH/Lsm11 complex or effective concentrations of these two proteins generated by the TnT expression system are not sufficient to allow detectable interaction. Alternatively, there is an additional factor that bridges symplekin and CstF64 to the FLASH/Lsm11 complex that escaped our detection by mass spectrometry. We would not have detected proteins that migrated close to either the GST-FLASH or MBPl-Lsm11 used as bait.
With the exception of CPSF73, which contacts the cleavage site in histone pre-mRNA and functions as the 3′ endonuclease (31, 53), the role of the remaining CPSF subunits in the U7-dependent processing is largely unknown. CPSF100 is a noncatalytic homologue, binding partner, and perhaps a regulator of CPSF73 (33, 54). CPSF160, CPSF30, Fip1, and WDR33 may be essential for maintaining the integrity of the polyadenylation complex or play regulatory roles in vivo by connecting 3′-end processing of histone pre-mRNAs with other cellular processes. However, it is also possible that at least some of these subunits are simply passive bystanders that play no role in the U7-dependent processing.
Supplementary Material
ACKNOWLEDGMENTS
We thank Z. Mikinis and A. Wlodawer (NCI, Frederick, MD) for the plasmid expressing the fusion protein of the N-terminal Lsm11 and MBP and Y. Xiong (UNC at Chapel Hill) for antibodies against VprBP, DDB1, and Cul4A. We also thank J. Oledzki and A. Fabijanska for help with mass spectrometry analysis.
This work was supported by NIH grant GM29832 to Z.D. and W.F.M. J.D., M.K.-D., and M.D. were supported by grants POIG.02.02.00-00-025/09, CEPT, and TEAM/2011-7/1.
Footnotes
Published ahead of print 15 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00653-12.
REFERENCES
- 1. Colgan DF, Manley JL. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- 2. Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre-mRNA 3′-end processing. Cell Mol. Life Sci. 65: 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev. 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, III, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takagaki Y, Manley JL. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20: 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruegsegger U, Blank D, Keller W. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1: 243–253 [DOI] [PubMed] [Google Scholar]
- 10. De Vries H, Rüegsegger U, Hübner W, Friedlein A, Langen H, Keller W. 2000. Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 19: 5895–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghazy MA, He X, Singh BN, Hampsey M, Moore C. 2009. The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensable for pre-mRNA 3′-end processing. Mol. Cell. Biol. 29: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kyburz A, Sadowski M, Dichtl B, Keller W. 2003. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res. 31: 3936–3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barnard DC, Ryan K, Manley JL, Richter JD. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651 [DOI] [PubMed] [Google Scholar]
- 14. Dominski Z, Marzluff WF. 2007. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene 396: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muller B, Schumperli D. 1997. The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin. Cell Dev. Biol. 8: 567–576 [DOI] [PubMed] [Google Scholar]
- 16. Galli G, Hofstetter H, Stunnenberg HG, Birnstiel ML. 1983. Biochemical complementation with RNA in the Xenopus oocyte: a small RNA is required for the generation of 3′ histone mRNA termini. Cell 34: 823–828 [DOI] [PubMed] [Google Scholar]
- 17. Mowry KL, Steitz JA. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 238: 1682–1687 [DOI] [PubMed] [Google Scholar]
- 18. Scharl EC, Steitz JA. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 13: 2432–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schumperli D, Pillai RS. 2004. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell. Mol. Life Sci. 61: 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cotten M, Gick O, Vasserot A, Schaffner G, Birnstiel ML. 1988. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro RNA processing reaction. EMBO J. 7: 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin F, Schaller A, Eglite S, Schümperli D, Müller B. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16: 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Z-F, Whitfield ML, Ingledue TI, Dominski Z, Marzluff WF. 1996. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10: 3028–3040 [DOI] [PubMed] [Google Scholar]
- 23. Dominski Z, Zheng LX, Sanchez R, Marzluff WF. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19: 3561–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Streit A, Koning TW, Soldati D, Melin L, Schümperli D. 1993. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 21: 1569–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Azzouz TN, Gruber A, Schumperli D. 2005. U7 snRNP-specific Lsm11 protein: dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucleic Acids Res. 33: 2106–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell 36: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De Laurenzi V. 2006. FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. U. S. A. 103: 14808–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Cola A, Bongiorno-Borbone L, Bianchi E, Barcaroli D, Carletti E, Knight RA, Di Ilio C, Melino G, Sette C, De Laurenzi V. 2012. FLASH is essential during early embryogenesis and cooperates with p73 to regulate histone gene transcription. Oncogene 31: 573–582 [DOI] [PubMed] [Google Scholar]
- 29. Alm-Kristiansen AH, Saether T, Matre V, Gilfillan S, Dahle O, Gabrielsen OS. 2008. FLASH acts as a co-activator of the transcription factor c-Myb and localizes to active RNA polymerase II foci. Oncogene 27: 4644–4656 [DOI] [PubMed] [Google Scholar]
- 30. Imai Y, Kimura T, Murakami A, Yajima N, Sakamaki K, Yonehara S. 1999. The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature 398: 777–785 [DOI] [PubMed] [Google Scholar]
- 31. Dominski Z, Yang XC, Marzluff WF. 2005. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123: 37–48 [DOI] [PubMed] [Google Scholar]
- 32. Kolev NG, Steitz JA. 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 19: 2583–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kolev NG, Yario TA, Benson E, Steitz JA. 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 9: 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol. Cell 28: 692–699 [DOI] [PubMed] [Google Scholar]
- 35. Dominski Z, Sumerel J, Hanson RJ, Marzluff WF. 1995. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA 1: 915–923 [PMC free article] [PubMed] [Google Scholar]
- 36. Yang XC, Torres MP, Marzluff WF, Dominski Z. 2009. Three proteins of the U7-specific Sm ring function as the molecular ruler to determine the site of 3′-end processing in mammalian histone pre-mRNA. Mol. Cell. Biol. 29: 4045–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang XC, Xu B, Sabath I, Kunduru L, Burch BD, Marzluff WF, Dominski Z. 2011. FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol. Cell. Biol. 31: 1492–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff WF, Dominski Z. 2011. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA 17: 1132–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei N, Serino G, Deng XW. 2008. The COP9 signalosome: more than a protease. Trends Biochem. Sci. 33: 592–600 [DOI] [PubMed] [Google Scholar]
- 40. Ruepp MD, Schweingruber C, Kleinschmidt N, Schumperli D. 2011. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol. Biol. Cell 22: 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCall CM, Miliani de Marval PL, Chastain PD, Jackson SC, He YJ, Kotake Y, Cook JG, Xiong Y. 2008. Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1-CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol. Cell. Biol. 28: 5621–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang XC, Sullivan KD, Marzluff WF, Dominski Z. 2009. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol. Cell. Biol. 29: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kiriyama M, Kobayashi Y, Saito M, Ishikawa F, Yonehara S. 2009. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol. Cell. Biol. 29: 4729–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gilmartin GM. 2005. Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev. 19: 2517–2521 [DOI] [PubMed] [Google Scholar]
- 45. Pillai RS, Will CL, Lührmann R, Schümperli D, Müller B. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20: 5470–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith HO, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel ML. 1991. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc. Natl. Acad. Sci. U. S. A. 88: 9784–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ruepp MD, Vivarelli S, Pillai RS, Kleinschmidt N, Azzouz TN, Barabino SM, Schumperli D. 2010. The 68 kDa subunit of mammalian cleavage factor I interacts with the U7 small nuclear ribonucleoprotein and participates in 3′-end processing of animal histone mRNAs. Nucleic Acids Res. 38: 7637–7650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao J, Kessler M, Helmling S, O'Connor JP, Moore C. 1999. Pta1, a component of yeast CFII, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol. 19: 7733–7740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Keon BH, Schafer S, Kuhn C, Grund C, Franke WW. 1996. Symplekin, a novel type of tight junction plaque protein. J. Cell Biol. 134: 1003–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bai Y, Auperin TC, Chou CY, Chang CG, Manley JL, Tong L. 2007. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell 25: 863–875 [DOI] [PubMed] [Google Scholar]
- 51. Martincic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze MT, Milcarek C. 1998. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc. Natl. Acad. Sci. U. S. A. 95: 11095–11100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takagaki Y, Manley JL. 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2: 761–771 [DOI] [PubMed] [Google Scholar]
- 53. Dominski Z. 2010. The hunt for the 3′ endonuclease. Wiley Interdiscip. Rev. RNA 1: 325–340 [DOI] [PubMed] [Google Scholar]
- 54. Dominski Z. 2007. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 42: 67–93 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


