Abstract
Piccolo NuA4 is an essential yeast histone acetyltransferase (HAT) complex that targets histones H4 and H2A in nucleosome substrates. While Piccolo NuA4's catalytic subunit Esa1 alone is unable to acetylate nucleosomal histones, its accessory subunits, Yng2 and Epl1, enable Esa1 to bind to and to act on nucleosomes. We previously determined that the Tudor domain of Esa1 and the EPcA homology domain of Epl1 play critical roles in Piccolo NuA4's ability to act on the nucleosome. In this work, we pinpoint a loop within the Esa1 Tudor domain and a short basic region at the N terminus of the Epl1 EPcA domain as necessary for this nucleosomal HAT activity. We also show that this Esa1 Tudor domain loop region is positioned close to nucleosomal DNA and that the Epl1 EPcA basic region is in proximity to the N-terminal histone H2A tail, the globular region of histone H4, and also to nucleosomal DNA when Piccolo NuA4 interacts with the nucleosome. Since neither region identified is required for Piccolo NuA4 to bind to nucleosomes and yet both are needed to acetylate nucleosomes, these regions may function after the enzyme binds nucleosomes to disengage substrate histone tails from nucleosomal DNA.
INTRODUCTION
The regulation of gene expression in eukaryotic cells involves a complex orchestration of chromatin enzymes that modify or remodel the chromatin assembly of histone proteins and DNA (1, 2). Much has been learned about the identity and activities of chromatin modification and remodeling enzymes, and we are also beginning to appreciate the different genetic pathways through which these enzymes work. However, despite these significant advances, the precise mechanism by which chromatin enzymes function is largely not understood. In particular, we have a relatively poor understanding of how chromatin enzymes recognize and interact with their nucleosome substrate.
The discovery that the yeast Gcn5 protein possessed histone acetyltransferase (HAT) activity and was also a subunit of the SAGA transcriptional coactivator complex provided a critical link between histone acetylation and gene regulation (3, 4). Biochemical and structural studies of the catalytic domains of HAT enzymes equip us with insights into how the catalytic domain provides the structural environment to allow the acetyl-coenzyme A cofactor to react with the appropriate histone tail peptide substrates (5–8) and the mechanistic basis for how one histone modification can enhance or prevent a different modification on the same histone peptide, also known as cross talk (9, 10). Nevertheless, major fundamental issues, including how the histone modification enzymes function on their nucleosome substrates, remain unresolved. The catalytic subunit of histone modification enzymes often possess little or no activity on their physiologically relevant substrates of histone assembled into nucleosomes (4, 11–13). Many such enzyme catalytic subunits are components of multisubunit protein complexes that act on nucleosomes. For example, while the Gcn5 subunit possesses very poor HAT activity when assayed on a nucleosome substrate, the megadalton SAGA complex is able to acetylate both histone and nucleosome substrates (4). Similarly, the yeast Esa1 HAT fails to acetylate nucleosomal histones, but its parent multisubunit NuA4 complex will acetylate nucleosomes (11). In our previous studies, we identified minimal catalytic subcomplexes of SAGA (Ada2/Ada3/Gcn5) and NuA4 (Epl1/Yng2/Esa1, also known as the Piccolo NuA4 complex) with activities on nucleosome substrates similar to or even elevated compared to those of their parent megadalton complexes (11, 14).
The ability of chromatin modification enzymes like SAGA or NuA4 to act on nucleosome and not just naked histone substrates has important implications for their molecular mechanisms. Although the histone tails are the targets of SAGA- and NuA4-directed acetylation, it is unlikely that the tails are sufficient to engage these enzymes. The fact that the Gcn5 and Esa1 catalytic subunits require accessory subunits to modify nucleosomal histones suggests that these accessory proteins assist the catalytic subunit in interacting with the nucleosome and in acting on the histone tails. Parallels can be drawn for other chromatin modification enzymes, including the human histone demethylase LSD1, which requires CoREST to act on the nucleosome, and the yeast Set1 and human MLL1 histone methyltransferases, which require other components of the COMPASS or MLL complex to methylate nucleosomal histones (13, 15, 16). Some of the key unresolved issues include precisely how the surface of the nucleosome protein-DNA complex is recognized by these chromatin modification enzymes and the role of the accessory subunits in nucleosomal binding or catalytic activity.
In our previous studies of the Piccolo NuA4 catalytic subcomplex of the NuA4 complex, we identified regions of the component Epl1, Yng2, and Esa1 subunits necessary for the three proteins to form a complex and for nucleosomal HAT activity of the complex (17). We found that the Esa1 Tudor/chromo barrel domain found N-terminal to the catalytic HAT domain and a 20-residue region at the N terminus of the conserved EPcA domain of the Epl1 subunit were necessary for Piccolo NuA4 to acetylate nucleosomes. The Tudor domain found in Esa1 is part of the Tudor domain family, which includes the chromo, PWWP, MBT, and Tudor domains (18). Unlike the case of the chromodomain, which contains a peptide binding site often used to bind to methylated or unmodified histone tails, structural determination of the yeast Esa1 and related human MOF Tudor/chromo barrel domains shows that an N-terminal extension to the chromodomain occupies the chromodomain histone peptide binding site (19–21). Thus, the Tudor/chromo barrel domain is unlikely to bind to methylated histone peptides despite its structural similarity to the chromodomain. The exact function of the Tudor/chromo barrel domain in Esa1 and MOF is not clear, although RNA binding has been proposed as one possible function (20–22). In contrast to the Tudor/chromo barrel domain, no experimental structural information is available for the Epl1 EPcA domain.
In this study, we identify key residues within the Esa1 Tudor/chromo barrel domain and the Epl1 EPcA domains necessary for the Piccolo NuA4 complex to acetylate nucleosomal histones. The Esa1 Tudor/chromo barrel domain residues highlight a particular loop region that we show is in close proximity to nucleosomal DNA. We also provide evidence that a positively charged region of the Epl1 EPcA domain is close both to the N-terminal tail of histone H2A and to nucleosomal DNA. These results suggest new insights into how the Piccolo NuA4 enzyme interacts with its nucleosome substrate and catalyzes acetylation of nucleosomal histones.
MATERIALS AND METHODS
Protein expression and purification.
Wild-type Esa1, Yng2(2–18), and hexahistidine-tagged Epl1(51–380) subunits of Piccolo NuA4 and appropriate mutational variants were expressed using the pST44 polycistronic expression vector in BL21(DE3)pLysS cells as described previously (17, 23). The coexpressed complexes were purified by Talon (Clontech) cobalt affinity and SourceQ anion-exchange chromatography (24), and the engineered mutations did not adversely affect Piccolo NuA4 complex stability.
Recombinant Xenopus core histones and nucleosome core particles were prepared as described previously (25). Briefly, the four core histones were expressed individually in Escherichia coli and purified from inclusion bodies under denaturing conditions. The histone octamer was refolded from the individual core histones and then purified over a Superdex 200 16/60 size exclusion column. Nucleosome core particles were reconstituted from the purified histone octamer and recombinant Widom 601 DNA positioning sequence by gradient dialysis before purification by SourceQ anion-exchange high-performance liquid chromatography (HPLC).
HAT assay.
HAT assays were performed at least three times each on both recombinant histones and nucleosome core particles in 50 mM Tris-HC1 at pH 8.0, 50 mM KC1, 5% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 10 mM sodium butyrate using procedures described previously (4, 17). The HAT activity nucleosome preference was calculated by dividing the HAT activity on nucleosomes substrates by the HAT activity on free histone substrates.
Strep-Tactin pulldown assay.
One hundred picomoles of recombinant nucleosome core particles were immobilized on Strep-Tactin resin via histone H2B tagged with Strep-tag in H50 pulldown buffer (20 mM HEPES [pH 7.5], 50 mM NaCl, 100 μg/ml BSA [bovine serum albumin], 1% Triton X-100, and 0.1% sodium deoxycholate). After washing with H50 pulldown buffer twice, the same buffer or equivalent buffer containing 70 or 90 mM NaCl was used to wash the resin twice. After the resins were resuspended in 300 μl of desired binding buffer, 120 pmol of Piccolo NuA4 complexes were added. Proteins were eluted by heating the resin in 40 μl SDS-PAGE sample loading buffer and separating on an 18% acrylamide SDS-PAGE gel.
Preparation of Piccolo NuA4 containing Bpa and post-32P labeling assay.
Piccolo NuA4 complexes with p-benzoyl-l-phenylalanine (Bpa) incorporated at specific Esa1 positions were expressed using the pSup-BpaRS-6TRN suppression plasmid, kindly provided by Peter Schultz (26, 27), and pST44-yEsa1-Yng2(1-218)-HISNyEpl1(51-380) coexpression plasmids with the TAG amber codon introduced at appropriate Esa1 positions by site-directed mutagenesis. Expression of Piccolo NuA4 complexes containing Bpa were performed in 2× tryptone-yeast extract (2 × TY) growth medium supplemented with 1 mM Bpa (Bachem). Additional SourceISO hydrophobic chromatography was used to further purify Piccolo NuA4 complexes containing Bpa.
In the post-32P labeling assay, 32 pmol of Piccolo NuA4 incorporating Bpa at Esa1 K61 was incubated with 63 pmol recombinant nucleosome core particles in 40 μl cross-linking buffer (20 mM HEPES [pH 7.5], 50 mM KCl, 0.1 mM EDTA, and 5% glycerol) for 10 min at room temperature in the dark. Samples were irradiated with UV light at a distance of 5 cm using a Spectroline XX-15A lamp (365 nm) in a cold room for 20 min. After adding 40 μl of T2400 (20 mM Tris-Cl [pH 8.0] and 2.4 M NaCl) and 20 μl of Talon metal affinity resin in 200 μl T1200 (20 mM Tris-Cl [pH 8.0] and 1.2 M NaCl), the mixture was mixed with rotation at room temperature for 20 min before washing twice with T1200 and once with DNase I buffer (10 mM Tris-HCl [pH 7.6], 2.5 mM MgCl2, and 0.5 mM CaCl2). The sample was then digested using 3 U DNase I (Roche) in 40 μl DNase I buffer at 37°C for 1 h. After washing with T1200 twice and NEB buffer 3 (50 mM Tris-HCl [pH 7.9], 100 mM NaCl, and 10 mM MgCl2) once, the sample was treated with 10 U calf intestinal phosphatase (NEB) in 40 μl NEB buffer 3 at 37°C for 1 h. The resin was then washed twice with T1200 and once with T4 PNK buffer (70 mM Tris-HCl [pH 7.6] and 10 mM MgCl2) before incubation with 5 U T4 polynucleotide kinase (NEB) and 0.5 μl [γ-32P]ATP (3,000 Ci/mmol, 10 Ci/ml; MP Biomedical) in 40 μl T4 PNK buffer for 1 h. After this labeling step, the resin was washed 6 times with T4 PNK buffer plus 0.1 mM ATP. The Piccolo NuA4 complexes were eluted from the resin by adding 20 μl of SDS-PAGE sample loading buffer and heating the resin at 95°C for 5 min. The eluted proteins were separated by SDS-PAGE and visualized first by staining the gel with Coomassie blue and then by phosphorimaging after drying down the gel.
Yeast strains and viability tests.
Hemagglutinin (HA)-tagged ESA1 was subcloned into the yeast integrative vector pRS403 HIS marker plasmid. Site-directed mutagenesis was performed to create point mutations. pRS403 plasmids with HA-tagged wild-type or mutant ESA1 were linearized with PstI and transformed into QY118 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 esa1Δ::KanMX pLP795 (ESA1 ARS/CEN URA3) using standard methods (28). Desired clones were selected on synthetic complete (SC) plates lacking histidine and uracil. Viability tests were performed as follows. Yeast strains integrated with HA-tagged ESA1 constructs as well as wild-type ESA1 on a URA3 plasmid were grown overnight in yeast extract, peptone, adenine, and dextrose (YPAD) medium at 30°C and then diluted to an optical density of 1.5. Tenfold serial dilutions in water were spotted on both YPAD plates with or without 0.1% 5′-fluoroorotic acid (5-FOA). The plates were grown at 30°C for 2 to 5 days. Yeast strains integrated with HA-tagged esa1-D64A were selected from the 5-FOA plates.
Chromatin immunoprecipitation and qPCR.
One hundred milliliters of yeast culture (optical density at 600 nm [OD600] = 0.8 to 1.0) was cross-linked with formaldehyde (1% [vol/vol]) for 15 min at room temperature and quenched by adding glycine to 125 mM for 5 min at room temperature (29). Whole-cell extracts were prepared by zirconia bead disruption, and chromatin was sheared into fragments averaging 150 to 500 bp in size by using a Bioruptor instrument (Diagenode, Philadelphia PA). One hundred microliters of whole-cell extract was incubated with 2 to 4 μl of antibody overnight (anti-hyperacetylated histone H4 [06-866; Upstate] and anti-histone H3 [ab1791; Abcam). The immunoprecipitated DNA and input DNA were analyzed by quantitative PCR (qPCR) using primers covering the following regions: PYK1 promoter, −275 to −161; PYK1 ORF, +580 to +661; PMA1 promoter, −299 to −121; PMA1 open reading frame (ORF), +1138 to +1219 [numbers refer to distance in bases from the transcription start site]. Levels of histone H4 acetylation were normalized by measuring changes in nucleosome density in parallel using anti-H3 antibody. The results are presented as the means and standard deviations from at least three independent experiments.
Photo-cross-linking label transfer assay.
Piccolo NuA4 complexes with cysteines introduced into the Epl1 EPcA region were reduced in 10 mM DTT at room temperature for 30 min, before dialyzing twice against H200 buffer (20 mM HEPES [pH 7.5] and 200 mM NaCl) to remove the DTT. Forty-eight nanomoles of reduced Piccolo NuA4 complex was mixed with 48 nmol Mts-Atf-Biotin (dissolved in dimethyl sulfoxide [DMSO]) in a total reaction volume of 800 μl for 1 h at room temperature in the dark and removal of the nonreacted Mts-Atf-biotin by dialysis against H200 buffer. Sixty-four picomoles Mts-Atf-biotin-labeled Piccolo NuA4 complex was mixed with 80 pmol nucleosome core particles in 40 μl H50 buffer, incubated at room temperature in the dark for 10 min, irradiated with UV light for 5 min at a distance of 5 cm using a Spectroline XX-15B lamp (300 nm), and then reduced in 50 mM DTT at 70°C for 5 min. To detect protein-protein interactions, SDS-PAGE sample loading buffer was added to the samples. Samples were separated on an 18% acrylamide SDS-PAGE gel and blotted to nitrocellulose membranes. To ensure efficient transfer of histone proteins, Towbin buffer with 0.8% SDS was used. Horseradish peroxidase-conjugated streptavidin was used with chemiluminescent reagents to detect the proteins labeled with biotin. To detect protein-DNA interactions, samples were extracted four times with phenol-chloroform and once with chloroform. The DNA was then ethanol precipitated and spotted onto a Hybond-N+ blotting membrane (GE Healthcare). The DNA was then cross-linked to the membrane via UV 265-nm irradiation in a Stratalinker UV cross-linker (Stratagene), and the biotin-labeled DNA was detected using horseradish peroxidase-conjugated streptavidin and chemiluminescent reagents.
RESULTS
A conserved Esa1 Tudor loop region is critical for Piccolo NuA4 to acetylate nucleosomes.
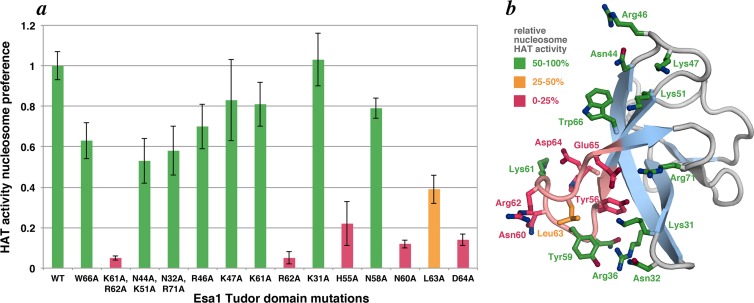
Our previous studies of the Piccolo NuA4 complex demonstrated that the Tudor domain of the catalytic Esa1 subunit is critical for Piccolo NuA4 to acetylate nucleosomes but not for Esa1 or Piccolo NuA4 to acetylate histones (17). To better understand the role of the Esa1 Tudor domain in nucleosome acetylation, we analyzed the effect of mutating residues on the surface of the Tudor domain. Given the structural similarity of the Esa1 Tudor domain and MOF chromo barrel domain to the archaeal Sso7 chromosomal protein, which binds to DNA (30), we designed alanine mutations to probe the Esa1 surface equivalent to the Sso7 DNA-binding surface. We had previously established that the His-tagged Epl1(51–380)/Yng2(1–218)/Esa1 Piccolo NuA4 complex v55 possesses robust nucleosomal HAT activity (17). To produce recombinant Piccolo NuA4 complexes, we coexpressed these full-length Esa1 proteins with the Yng2(1–218) and His-tagged Epl1(51–380) truncations using polycistronic expression in E. coli. As expected, the Esa1 Tudor domain mutations, which are outside the HAT domain sufficient to act on histones, did not significantly affect the ability of Piccolo NuA4 to acetylate histone substrates. Since none of the mutations examined in this work had a major effect on histone HAT activity (Table 1), comparing the nucleosome preference (the ratio of nucleosome HAT activity to histone HAT activity) allows us to correct for minor concentration differences between Piccolo NuA4 preparations (17). The Piccolo NuA4 complexes bearing the Esa1 W66A, N44A/K51A, or N32A/K71A mutations produced only relatively minor effects on nucleosomal HAT activity, but the K61A/R62A double mutation was severely compromised in its ability to acetylate a nucleosome substrate (Fig. 1a and Table 1). Specifically, the K61A/R62A mutant's nucleosomal preference was reduced to 4% relative to the unmutated v55 complex.
Table 1.
HAT activities of Piccolo NuA4 complexes containing point mutations in Esa1 Tudor domaina
| Esa1 Tudor mutation(s) | HAT activity (cpm) |
Preference for nucleosomes relative to wild type | |
|---|---|---|---|
| Histone | Nucleosome | ||
| None | 918 ± 53 | 5,351 ± 206 | 1.00 ± 0.07 |
| W66A | 746 ± 47 | 2,690 ± 135 | 0.63 ± 0.09 |
| K61A, R62A | 551 ± 85 | 215 ± 49 | 0.04 ± 0.01 |
| N44A, K51A | 793 ± 70 | 2,441 ± 349 | 0.53 ± 0.11 |
| N32A, R71A | 834 ± 59 | 2,829 ± 431 | 0.58 ± 0.12 |
| R46A | 1,106 ± 102 | 4,878 ± 865 | 0.70 ± 0.11 |
| K47A | 1,043 ± 36 | 5,324 ± 850 | 0.83 ± 0.20 |
| K61A | 1,071 ± 66 | 5,273 ± 811 | 0.81 ± 0.11 |
| R62A | 699 ± 58 | 330 ± 70 | 0.05 ± 0.03 |
| K31A | 843 ± 175 | 5,550 ± 691 | 1.03 ± 0.13 |
| H55A | 884 ± 57 | 1,134 ± 326 | 0.21 ± 0.11 |
| N58A | 879 ± 33 | 3,863 ± 284 | 0.79 ± 0.05 |
| N60A | 650 ± 20 | 549 ± 11 | 0.12 ± 0.02 |
| L63A | 776 ± 59 | 1,750 ± 212 | 0.39 ± 0.07 |
| D64A | 782 ± 56 | 558 ± 45 | 0.14 ± 0.03 |
Means ± SD are given.
Fig 1.
Mutations in an Esa1 Tudor domain loop region affect Piccolo NuA4's ability to acetylate nucleosomal histones. (a) HAT activity nucleosome preference (ratio of HAT activity on nucleosome substrates to HAT activity on histone substrates) for Piccolo NuA4 wild-type and Esa1 Tudor domain loop mutant enzyme complexes based on at least three measurements. Results are color coded as follows: green for 50 to 100%, orange for 25 to 50%, and red for 0 to 25% of wild-type nucleosome preference. (b) HAT activity results mapped onto Esa1 Tudor domain NMR structure (21). The loop region identified by the mutational studies is highlighted in pink. Molecular graphics were produced using the PyMOL software program.
We next designed single point mutations in the vicinity of this double mutation, including the individual K61A and R62A mutations. Piccolo NuA4 complex bearing the K61A mutation possessed a near-wild-type ability to acetylate nucleosomes, but the R62A mutation alone was sufficient to reduce the nucleosome preference to 5%, similar to the case of the K61A/R62A double mutation. Examination of the remaining mutants identified H55A, N60A, and D64A as reducing the nucleosome preference to less than 25% of that of the reference unmutated complex (Fig. 1a and Table 1). The L63A mutation had a modest effect (39% activity), and the K31A and N58A mutants possessed a near-wild-type nucleosomal preference. When mapped onto the nuclear magnetic resonance (NMR) structure of the Esa1 Tudor domain, the most severe mutations identified here (H55A, N60A, R62A, and D64A) and from our previous study (Y56A and E65L) (17) identify a loop between Esa1 Tudor domain strands S3 and S4 (Fig. 1b). The sequence of this loop is particularly well conserved among Esa1 from different species and is also conserved in the MOF protein, another member of the MYST family of histone acetyltransferases (20, 21). Removing positive charges on the surface of the Tudor domain is apparently not sufficient to reduce nucleosomal HAT activity, since the R46, K47, K51, K61, and R71 residues outside the loop region can be mutated to alanine without severely affecting the nucleosomal preference, suggesting specific electrostatic contacts with a defined orientation (Fig. 1a and b and Table 1).
The Esa1 Tudor loop region can cross-link to nucleosomal DNA.
A possible mechanism by which the Esa1 Tudor loop could affect acetylation of nucleosomal tails is if the loop region mediates interactions with the nucleosome. To examine if the Esa1 Tudor loop might interact with the nucleosome, we first analyzed if this loop region is proximal to the nucleosome in the Piccolo NuA4/nucleosome complex. We employed unnatural amino acid incorporation via amber STOP codon suppression (27) to site-specifically introduce a UV-dependent cross-linker amino acid analog into recombinant Piccolo NuA4 complex coexpressed in E. coli. In particular, the Bpa (p-benzoyl-l-phenylalanine) amino acid analog was introduced into several positions within or near the Esa1 Tudor loop region. Expression of Esa1 containing the amber TAG codon engineered into the Tudor loop was dependent on the presence of Bpa provided in the expression growth medium (data not shown). We prepared Piccolo NuA4 containing Bpa incorporated at Esa1 Tudor residues K31, Y56, Y59, and K61. Attempts to detect UV-dependent cross-linking of Piccolo NuA4 to the histone components of the nucleosome were unsuccessful: we did not observe any new bands corresponding to Esa1 cross-linked to histone proteins on Coomassie or silver-stained SDS-PAGE gels or in anti-Esa1 Western blots (data not shown).
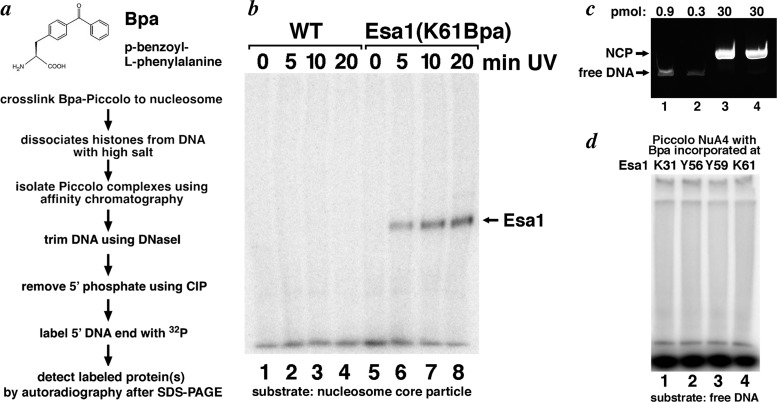
To investigate whether the Esa1 Tudor loop region is close to nucleosomal DNA, we developed a post-32P labeling procedure to detect cross-linking of Esa1 to DNA (Fig. 2a). Piccolo NuA4 complexes containing Bpa were incubated with recombinant nucleosome core particles and exposed to UV light. The nucleosomes but not the Piccolo NuA4 complex were then dissociated by high-salt treatment, and Piccolo NuA4 complexes were isolated by metal affinity chromatography via the His-tagged Epl1 subunit. DNase I was then used to trim nucleosomal DNA cross-linked to the Esa1 subunit. The small DNA fragment remaining was labeled by treatment with calf intestinal phosphatase to provide a free 5′-hydroxyl end and then T4 polynucleotide kinase and [32P]ATP to phosphorylate this 5′ end. No cross-linking to DNA was observed when Piccolo NuA4 complexes containing Bpa incorporated at Esa1 Tudor residue K31, Y56, or Y59 were incubated with nucleosomes. However, a band comigrating with Esa1 was observed when Piccolo NuA4 complexes with Bpa incorporated at Tudor residue K61 was bound to nucleosomes (Fig. 2b, lanes 5 to 8). This cross-linking was specific since it was UV dose dependent and was not present when wild-type Piccolo NuA4 enzyme without Bpa incorporated was used (Fig. 2b, lanes 1 to 4). The cross-linking was not due to direct labeling of Esa1 by polynucleotide kinase, since the band was absent if either the DNase I or phosphatase treatment was omitted, suggesting a requirement for a free DNA 5′ hydroxyl group (data not shown). We also considered the possibility that the signal resulted from Piccolo NuA4 binding to free DNA that might have dissociated from or contaminated our recombinant nucleosome preparation. We determined that the HPLC-purified recombinant nucleosome core particles used in these assays contained less than 1% free DNA (Fig. 2c) and thus performed the cross-linking/postlabeling procedure using free DNA present at 1% of the nucleosome concentration used in the nucleosome cross-linking experiment. In contrast to the case when Piccolo NuA4 is incubated with nucleosomes, we observed no cross-linking signal when free DNA was used (Fig. 2d), indicating that the cross-link to DNA results from Piccolo NuA4 interacting with nucleosomes and not free DNA. Thus, we conclude that the Esa1 Tudor loop residue K61 can be cross-linked to nucleosomal DNA in the Piccolo NuA4/nucleosome complex.
Fig 2.
Esa1 Tudor domain loop region cross-links to nucleosomal DNA. (a) Chemical structure of p-benzoyl-l-phenylalanine (Bpa) and experimental scheme for Bpa-based cross-linking to nucleosomal DNA. (b) UV dose-dependent Bpa-based cross-linking to nucleosomal DNA for wild-type and Esa1(K61Bpa) Piccolo NuA4 (the latter was produced by unnatural amino acid incorporation via amber codon suppression). The band was identified as Esa1 by comparing its mobility to the mobility of Piccolo NuA4 components Epl1, Yng2, and Esa1. (c) Nucleosome core particles used in cross-linking studies contain less than 1% free DNA. Ethidium bromide-stained native polyacrylamide gel of 0.9 pmol free DNA (same DNA fragment used to reconstitute nucleosome core particles) (lane 1), 0.3 pmol free DNA (lane 2), and 30 pmol recombinant nucleosome core particles (2 separate preparations, lanes 3 and 4) are shown. (d) Piccolo NuA4 with Bpa incorporated at Esa1 residue K31, Y56, Y59, or K61 (lanes 1 to 4, respectively) fails to cross-link to free DNA (1% of the nucleosome concentration used in the experiment shown in panel b) upon 20 min of UV exposure.
The Esa1 Tudor domain contributes to nucleosome binding by Piccolo NuA4.
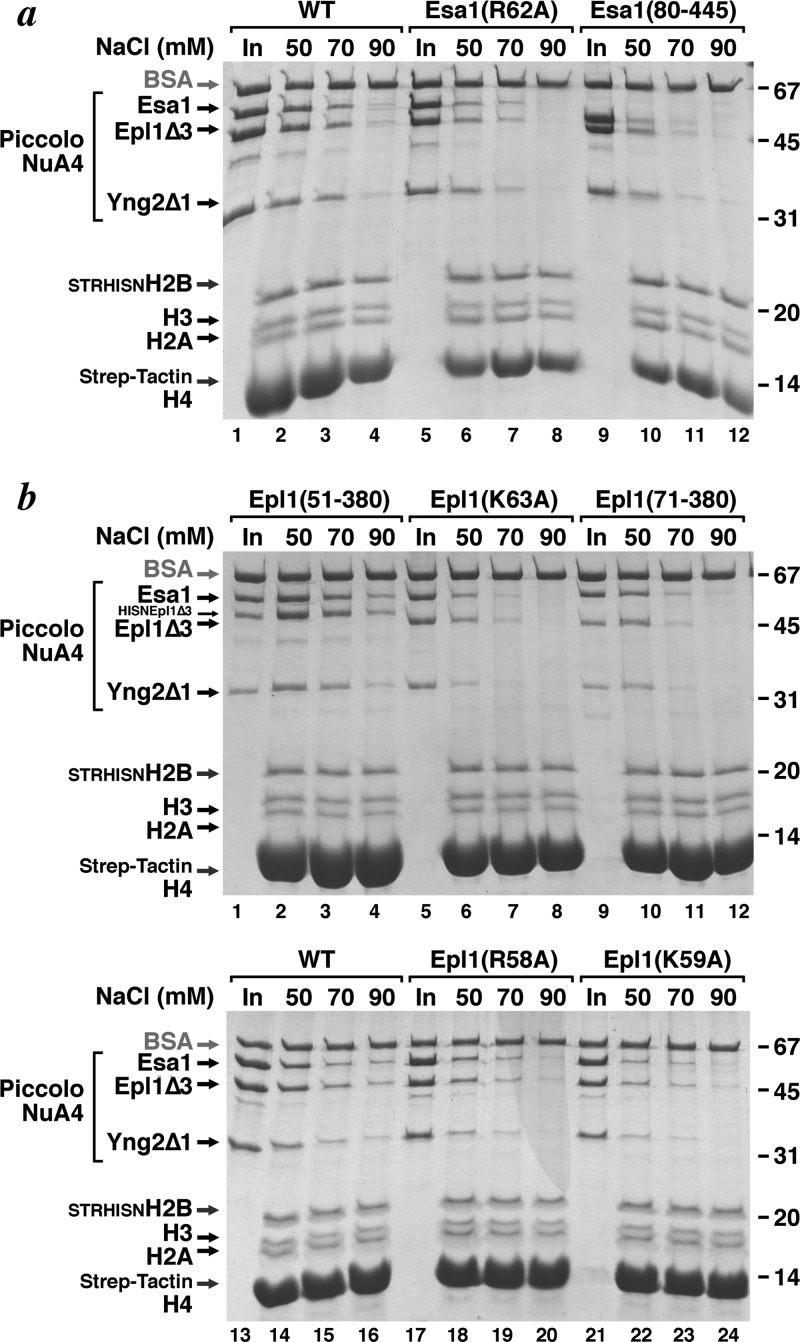
We have previously demonstrated through size exclusion chromatography and pulldown assays that the Esa1 Tudor domain was not necessary for Piccolo NuA4 to bind to the nucleosome (17). However, the assays used would not necessarily detect weaker binding due to reducing or eliminating one of the multiple contacts between the Piccolo NuA4 enzyme and the nucleosome. To analyze the role of the Esa1 Tudor domain in Piccolo NuA4 binding to the nucleosome, we used salt titration in a pulldown assay to examine interactions under increasingly stringent binding conditions (Fig. 3a). We immobilized nucleosomes via an N-terminal Strep-tag on histone H2B to Strep-Tactin resin and incubated Piccolo NuA4 with these immobilized nucleosomes in different salt concentrations. The components bound to the resin after extensive washing were released by heating the resin in SDS-PAGE sample loading buffer and fractionated by SDS-PAGE. We found that significantly less Piccolo NuA4 complex containing Esa1 Tudor R62A (which severely reduced nucleosome acetylation) bound to nucleosomes even at 50 mM NaCl than was the case with wild-type Piccolo NuA4 (Fig. 3a, lanes 2 and 6). Similar reductions were observed when binding was performed in 70 and 90 mM NaCl (Fig. 3a, lanes 3, 4, 7, and 8). Interestingly, comparable effects on binding were observed when the entire Esa1 Tudor domain was deleted from the Piccolo NuA4 complex (Fig. 3a, lanes 9 to 12). Thus, although the Esa1 Tudor domain is not necessary for Piccolo NuA4 to bind to the nucleosome, consistent with our prior size exclusion chromatography-based result, this domain and in particular the Tudor domain R62 residue appear to mediate some interaction with the nucleosome, presumably through nucleosomal DNA.
Fig 3.
Effect of Esa1 or Epl1 deletion or mutations on nucleosome binding by Piccolo NuA4. (a) Esa1 Tudor domain deletion or loop region mutations. The binding of Piccolo NuA4 to recombinant nucleosomes tagged on histone H2B with Strep-tag and immobilized on Strep-Tactin resin was examined in a pulldown assay (Coomassie blue-stained SDS-PAGE gel). Piccolo NuA4 input (30%) and contents of resin after washing with buffer containing 50, 70, and 90 mM NaCl are shown in lanes 1 to 4 for the wild-type enzyme, lanes 5 to 8 for the Piccolo NuA4 Esa1(R62A) point mutation variant, and lanes 9 to 12 for the Piccolo NuA4 enzyme with the Esa1 Tudor domain deleted. Duplicate experiments produced similar results, but the larger amount of Strep-Tactin in lanes 2 to 4 is not reproducible (not shown). Mobility of molecular weight markers is shown on the right. (b) Epl1 EPcA basic region deletion or point mutations. Piccolo NuA4 input (30%) and contents of resin after washing with buffer containing 50, 70, and 90 mM NaCl are shown in lanes 1 to 4 for wild-type enzyme (with a hexahistidine and TEV protease site [HISN] left on the Epl1Δ3 subunit) and in lanes 13 to 16 for wild-type enzyme (with a hexahistidine and TEV protease site [HISN] removed on the Epl1Δ3 subunit), lanes 5 to 8 for the Piccolo NuA4 Epl1(K63A) point mutation variant, lanes 9 to 12 for the Piccolo NuA4 enzyme with the Epl1 EPcA basic region residues 51 to 70 deleted, lanes 17 to 20 for the Piccolo NuA4 Epl1(R58A) point mutation variant, and lanes 21 to 24 for the Piccolo NuA4 Epl1(K59A) point mutation variant. The HISN tag on the N terminus of Epl1(51–380) does not appear to affect nucleosome interactions by Piccolo NuA4 (lanes 1 to 4 versus 13 to 16).
Esa1 Tudor domain mutations affect viability, growth rate, and global acetylation in yeast.
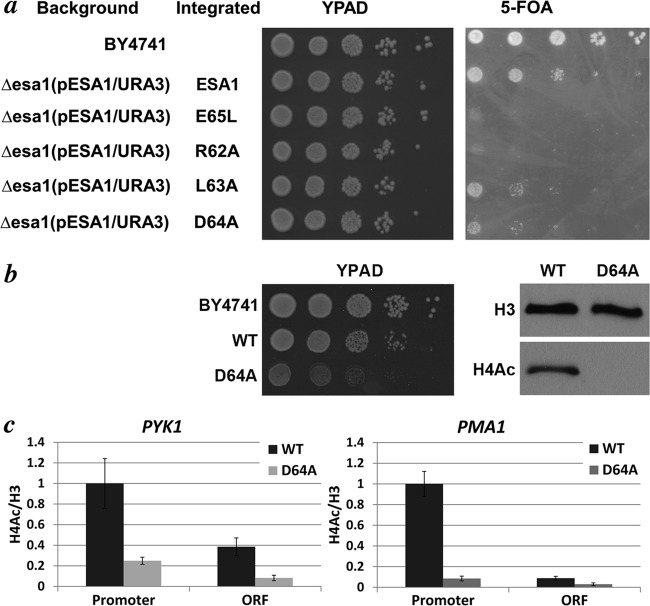
The effects of the Tudor domain mutations in yeast were examined using a URA3-based plasmid shuffle assay. Yeast strains bearing esa1-R62A on a URA3 plasmid were essentially unable to grow on 5-FOA plates, whereas strains expressing Esa1(L63A) or Esa1(D64A) displayed significant growth defects (Fig. 4a). The magnitude of these growth defects was largely consistent with the decreased nucleosome HAT activity of corresponding Piccolo NuA4 complexes in vitro. Yeast strains containing chromosomal integrations of esa1-D64A were analyzed further by growing them on plates and in liquid cultures and were found to grow at half the rate of the corresponding wild-type strain (4-h doubling time for the esa1-D64A strain versus 2 h for the wild-type strain). The global acetylation level of the esa1-D64A strain was then analyzed by Western blotting using an anti-hyperacetylated histone H4 antibody that recognizes acetylation of multiple histone H4 tail residues. Surprisingly, acetylated histone H4 not only was reduced but was actually not detectable in the esa1-D64A strain (Fig. 4b). The effect of the Esa1(D64A) mutation at the constitutively expressed PYK1 and PMA1 genes was also analyzed by chromatin immunoprecipitation (Fig. 4c). We found that histone H4 acetylation was substantially reduced at the promoters and within the open reading frames of both genes in the esa1-D64A strain compared to results for the wild-type strain. These results are consistent with previous findings that Esa1 Tudor domain loop mutations adversely affect yeast viability and growth rate (17, 21) and further show that global acetylation of histone H4 requires the Esa1 D64 side chain.
Fig 4.
The Esa1 Tudor domain loop region plays a critical role for yeast viability and for global H4 acetylation in yeast cells. (a) Serial dilution of yeast containing wild-type or Esa1 Tudor domain point mutants on YPAD or 5-FOA plates shows that the E65L and R62A mutants are more severely affected than the L63A and D64A mutants. (b) Slowed-growth phenotype of the Esa1(D64A) mutant on YPAD rich medium (left panel) and severe effect of the Esa1(D64A) point mutation on global H4 acetylation (right panel). (c) Chromatin immunoprecipitation analysis of histone H4 acetylation on the PYK1 and PMA1 promoter and open reading frame (ORF) in wild-type and Esa1(D64A) yeast cells (three experiments). Acetylated H4 occupancy at the promoter (normalized to H3) was set to 1.0.
Identification of residues in the Epl1 EPcA domain N-terminal basic region important for nucleosomal HAT activity.
Our previous studies had shown that the Epl1 residues 51 to 72 at the N terminus of its conserved EPcA domain are important for Piccolo NuA4 to acetylate nucleosomes and to bind to nucleosomes (17). To determine which residues within this relatively basic (positively charged) region were important for Piccolo NuA4's nucleosome acetylation activity, we constructed alanine mutations at individual residues, coexpressed these mutant Epl1(51–380) proteins with Yng2(1–218) and Esa1 in E. coli, purified the mutant Piccolo NuA4 complexes, and assayed HAT activity of these complexes using histone and nucleosome substrates (Fig. 5 and Table 2). Mutations at R58 and K59 had the largest effect, reducing nucleosome preference to 5% and 19% of wild-type activity, while the F55A and H65A mutants were reduced to approximately 45% wild-type activity. The other mutations examined, R54A, R56A, H57A, K63A, Q64A, and K67A, were associated with at least 60% of wild-type activity. Thus, just as with the Esa1 Tudor domain, mutations of specific Epl1 EPcA domain residues significantly compromise Piccolo NuA4's nucleosome HAT activity, but removing positive charges was not sufficient to affect nucleosome HAT activity.
Fig 5.
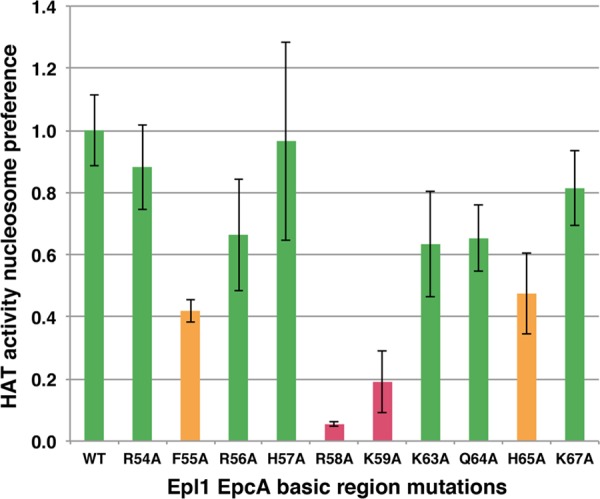
Specific mutations in the Epl1 EPcA N-terminal region affect nucleosomal HAT activity of Piccolo NuA4. HAT activity nucleosome preference for Piccolo NuA4 wild-type and Epl1 EPcA basic region mutant enzyme complexes based on at least three measurements. The same color code as in Fig. 1a is used.
Table 2.
HAT activities of Piccolo NuA4 complexes containing point mutations in Esa1 Tudor domaina
| Epl1 EPcA mutation | HAT activity (cpm) |
Preference for nucleosomes relative to wild type | |
|---|---|---|---|
| Histone | Nucleosome | ||
| None | 1,051 ± 134 | 6,206 ± 268 | 1.00 ± 0.11 |
| R54A | 569 ± 16 | 2,996 ± 430 | 0.88 ± 0.14 |
| F55A | 690 ± 65 | 1,581 ± 236 | 0.42 ± 0.04 |
| R56A | 763 ± 22 | 3,012 ± 650 | 0.66 ± 0.18 |
| H57A | 535 ± 56 | 2,877 ± 382 | 0.97 ± 0.32 |
| R58A | 800 ± 38 | 246 ± 12 | 0.05 ± 0.01 |
| K59A | 502 ± 107 | 509 ± 147 | 0.19 ± 0.10 |
| K63A | 618 ± 126 | 2,370 ± 679 | 0.63 ± 0.17 |
| Q64A | 556 ± 53 | 2,067 ± 225 | 0.65 ± 0.11 |
| H65A | 728 ± 81 | 2,071 ± 301 | 0.47 ± 0.13 |
| K67A | 909 ± 108 | 4,570 ± 100 | 0.81 ± 0.12 |
Means ± SD are given.
The Epl1 EPcA domain basic region can cross-link to the N-terminal tail of histone H2A and the globular domain of histone H4 in nucleosomes.
We were interested in utilizing a wider range of cross-linking or fluorescent agents than might be possible by unnatural amino acid incorporation via amber suppression. Such modifications are usually introduced into proteins by conjugating reactive side chains, such as cysteines, but the size and complexity of multisubunit complexes, such as Piccolo NuA4, often complicates conjugation to the desired specific residues. In particular, our reference Piccolo NuA4 v55 complex contains about 1,000 residues, including 13 cysteines. Fortunately, we discovered that these native cysteines in Piccolo NuA4 v55 are minimally chemically reactive, since relatively little conjugation of the fluorescent Alexa Fluor 647 maleimide dye to the native complex could be detected. However, a cysteine engineered into the Piccolo NuA4 Epl1 subunit could be specifically conjugated to the same dye (data not shown). Some background conjugation was detected even for the wild-type enzyme, possibly due to lower-efficiency reactivity between the maleimide and lysine residues (32, 33).
Having established that we could regioselectively label the Piccolo NuA4 complex with engineered cysteines, we pursued a label transfer cross-linking approach to determine what components of the nucleosome were proximal to the Epl1 EPcA domain basic region covering residues 51 to 72. We chose not to target residues in the basic region with the most severe effect on nucleosomal HAT activity, R58 and K59, to avoid possible complications if these residues were required for binding to the nucleosome. Instead, we selected the V62 and K63 residues, close to R58 and K59 (Piccolo NuA4 bearing the K63A mutation retained more than 60% of wild-type nucleosomal preference, suggesting that K63 was not required for interacting with the nucleosome or for HAT activity). We conjugated Piccolo NuA4 containing the Epl1 V62C and K63C point mutations with Mts-Atf-biotin, a trifunctional label transfer reagent (34–36). The sulfhydryl-reactive methanesulfonate (Mts) group allows conjugation to a reactive cysteine via a cleavable disulfide bond, the Atf (perfluorophenylazide) group allows UV-activated cross-linking within 11 Å to both protein and DNA, and the biotin moiety provides for sensitive detection. The Mts-Atf-biotin-conjugated Piccolo NuA4 complexes were incubated with recombinant nucleosome core particles, exposed to UV light, and treated with DTT to reduce the disulfide bond linking the cross-linking reagent to the Epl1 V62C or K63C residue (control experiments confirm that Mts-Atf-biotin-conjugated Piccolo NuA4 binds to nucleosome core particle similarly to binding of wild-type Piccolo NuA4 in a band shift gel; data not shown). The sample was then separated by SDS-PAGE and blotted, and the biotin group was probed using horseradish peroxidase-linked streptavidin (Fig. 6a and b). Our results show that a band corresponding to either histone H2A or H2B and a weaker band corresponding to histone H4 are detected when Piccolo NuA4 complex containing Epl1 K63C is used (Fig. 6b, lane 1). The absence of these bands when the Mts-Atf-biotin reagent is conjugated to the Piccolo NuA4 Epl1 V62C mutant indicates that the label transfer is specific, as does the finding that the bands are detected only in the presence of both nucleosomes and UV light (data not shown). The possibility that the Piccolo NuA4 cross-linked to dissociated histone dimer or free histones was examined by repeating the label transfer cross-linking using these substrates. Since we detected less than 1% of free DNA in our nucleosome preparation, we believe that less than 1% of our nucleosomes were dissociated into histones and DNA. Nevertheless, we performed the label transfer cross-linking assay using a 5% molar equivalence of core histones or histone H2A/H2B dimer, and we did not detect any biotin labeling of histones in these control experiments (data not shown). These results indicate that the cross-linking to the histone proteins we observed occurred between Piccolo NuA4 and nucleosomal histones.
Fig 6.
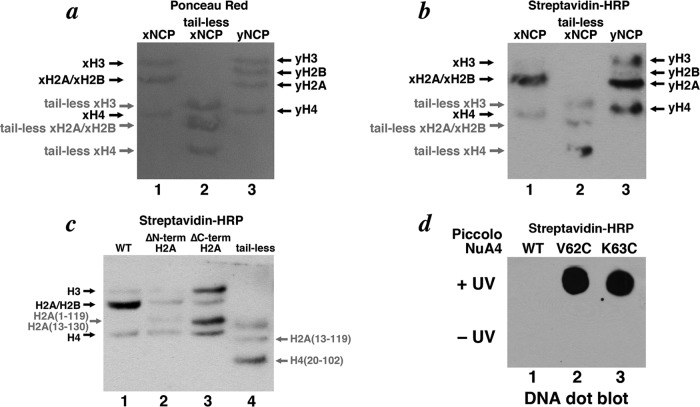
Label transfer experiments show that the Piccolo NuA4 Epl1 EPcA N-terminal region interacts with the N-terminal tail of histone H2A, the globular domain of histone H4, and nucleosomal DNA. (a and b) Western blots visualized using Ponceau red and horseradish peroxidase-linked streptavidin. Piccolo NuA4 conjugated with Mts-Atf-biotin on Epl1(K63C) was incubated with recombinant nucleosomes containing Xenopus core histones, tailless Xenopus core histones, and yeast core histones (lanes 1 to 3), cross-linking was activated by UV irradiation, and the biotin group was transferred to the nucleosome target protein. (c) Label transfer experiment using nucleosomes containing the wild type, histone H2A lacking its N-terminal tail, histone H2A lacking its C-terminal tail, or tailless core histones (lanes 1 to 4) indicate that the Epl1 EPcA basic region is proximal to the N-terminal histone H2A tail and the histone H4 globular region in the Piccolo NuA4/nucleosome complex. (d) Label transfer experiment samples for wild-type, Epl1(V62C) and Epl1(K63C) Piccolo NuA4 interacting with nucleosomes and analyzed on a DNA dot blot.
Because the Xenopus histones H2A and H2B present in our recombinant nucleosome core particles comigrate in an SDS-PAGE gel, our initial label transfer experiment was not able to identify which histone or whether possibly both histones were conjugated with biotin. To determine if Piccolo NuA4 K63C cross-linked to histone H2A or H2B, we prepared recombinant yeast nucleosome core particles and used these in the label transfer assay. Unlike Xenopus histones H2A and H2B, the corresponding yeast proteins were resolved from each other on SDS-PAGE. The results showed that only yeast H2A and not H2B was labeled, suggesting that histone H2A cross-linked to the Piccolo NuA4 (Fig. 6b, lane 3). A weaker labeling signal was again observed for histone H4, and a new fainter signal was seen for yeast histone H3. We have previously shown that Piccolo NuA4 binds to nucleosome core particles with each of the core histone tails removed, and so we also examined such tailless nucleosomes in the label transfer experiment. We found that much weaker labeling on H2A/H2B was present when tailless nucleosomes were used (Fig. 6b, lane 2 and c, lane 4), indicating that the cross-link was made to an H2A tail and not to the globular domain. In contrast, the signal for histone H4 was still present even when its N-terminal tail was removed, suggesting that the biotin label was transferred to the globular domain of histone H4.
Since histone H2A contains tail extensions on both the N- and C-terminal sides of its histone fold motif, we prepared recombinant nucleosome core particles containing histone H2A with either the N-terminal or C-terminal tail removed. When the H2A N-terminal tail was removed, the cross-linking signal to H2A was greatly reduced, indicating that the Piccolo NuA4 K63 cross-linked to the N-terminal tail of nucleosomal histone H2A (Fig. 6c, lane 2). In contrast, H2A lacking its C-terminal tail was still labeled with biotin (Fig. 6c, lane 3). We also observed significantly more labeling of H3 in nucleosomes lacking the C-terminal tail of H2A compared to results for wild-type or H2A N-terminally truncated nucleosomes.
The Epl1 EPcA domain basic region can cross-link to DNA in nucleosomes.
We also used Piccolo NuA4 Epl1 V62C and K63C variants in label transfer experiments to examine if the EPcA basic region could cross-link to nucleosomal DNA. After incubating Mts-Atf-biotin-conjugated Piccolo NuA4 with nucleosomes, UV-activated cross-linking, and reducing with DTT to transfer the biotin group to the cross-linked nucleosome subunit, the samples were phenol extracted to remove all proteins. Nucleic acids in the phenol extraction aqueous phase were then ethanol precipitated and detected on a dot blot using horseradish peroxidase-linked streptavidin. The results showed that nucleosomal DNA was labeled for Piccolo NuA4 conjugated at either V62C or K63C in a UV-dependent manner (Fig. 6d). This biotin label transfer to DNA likely occurs when Piccolo NuA4 binds to nucleosomes and not to free DNA, since control experiments using 1% equivalent concentrations of free DNA failed to detect any label transfer to DNA (data not shown). Thus, we conclude that the Epl1 EPcA basic region can also cross-link to nucleosomal DNA when Piccolo NuA4 interacts with nucleosomes.
We next examined the effect of point mutations in or deletion of the Epl1 EPcA basic region on the interaction between Piccolo NuA4 complexes and nucleosomes using the salt titration pulldown assay. We found that Piccolo NuA4 containing the Epl1 EPcA K63A point mutation or a deletion of the EPcA basic region was significantly compromised in its interaction with nucleosomes in 90 mM NaCl (Fig. 3b). Both Piccolo NuA4 variants still interacted with nucleosomes in 50 mM NaCl, albeit less strongly than wild-type Piccolo NuA4. The Epl1 EPcA R68A and K59A point mutations that had the strongest effects on Piccolo NuA4's ability to acetylate nucleosomes have apparently milder effects on nucleosome binding: both mutants show reduced binding in 50 mM NaCl compared to the wild-type enzyme, but weak interactions with the nucleosomes were detected even in 90 mM NaCl. These results suggest that, as with the Esa1 Tudor domain loop mutations, mutations in the EPcA basic region affect nucleosomal HAT activity more strongly than they appear to affect nucleosomal binding.
DISCUSSION
Our studies of the yeast Piccolo NuA4 histone acetyltransferase complex have pinpointed sequences within two conserved domains of a catalytic (Esa1) and noncatalytic (Epl1) subunit that, in addition to the HAT domain, play a critical role in the enzyme's ability to act on a nucleosomal substrate. We have identified a loop region of the Esa1 Tudor domain, which is required for nucleosomal HAT activity, and have shown that this region is in close proximity to nucleosomal DNA when Piccolo NuA4 binds to the nucleosome. This Esa1 Tudor domain loop region is also important for yeast viability and for global acetylation of histone H4, the major target of Piccolo NuA4's acetylation activity. Our studies also highlight residues in a basic region of the Epl1 conserved EPcA domain, which we find to be proximal to the N-terminal tail of histone H2A, the globular domain of histone H4 and nucleosomal DNA.
The Tudor/chromo barrel domain of Esa1 and related MYST family proteins has been proposed to bind to RNA (20–22). The isolated Esa1 Tudor domain was found to bind to poly(U) RNA in a pulldown assay and less tightly to double-stranded DNA (21). Mutations in the same Esa1 Tudor domain loop region that we independently identified decreased binding to RNA. In a separate study, the related MOF chromo barrel domain interacted with RNA or double-stranded DNA (dsDNA) on a native polyacrylamide gel, and this interaction was disrupted by point mutations in the corresponding chromo barrel loop region (37). Our experiments show that the Esa1 Tudor domain loop region plays a critical role in Piccolo NuA4's acetylation of nucleosomal histones in vitro and in vivo. It appears unlikely that RNA binding by the Esa1 Tudor domain is involved in nucleosome acetylation in our in vitro experiments, since RNA was not included in our HAT assays using recombinant Piccolo NuA4 purified through multiple chromatography steps. Furthermore, including RNase A in the histone acetyltransferase assays had no effect on the HAT activity of Piccolo NuA4 (data not shown).
Our experiments using unnatural amino acid incorporation to substitute the photoreactive Bpa amino acid at specific Esa1 positions indicate that the Esa1 Tudor domain loop region is within 7 Å of nucleosomal DNA in the Piccolo NuA4/nucleosome complex. Unfortunately, we are not able to specify the location along the nucleosomal DNA where this cross-link occurs, since our attempts to map the cross-link were not successful. Together with our finding that mutations in this loop region adversely affect binding to nucleosomes, the cross-linking results suggest that one role of the Esa1 Tudor domain may be to interact with nucleosomal DNA. It is possible that the major role of the Esa1 Tudor domain is to contribute incrementally to nucleosome binding by contacting nucleosomal DNA. However, it is worth noting that deleting the Esa1 Tudor domain or mutating the Tudor domain at position R62 (17) (Fig. 1) had more severe effects on nucleosomal HAT activity than might have been anticipated based on their modest effects on nucleosome binding (Fig. 3). This suggests that the Esa1 Tudor domain role in nucleosomal HAT activity extends beyond simply interacting with the nucleosome.
Consistent with the effect of the loop region mutations on nucleosomal HAT activity in vitro, the Esa1 Tudor domain plays a critical role in vivo, as our experiments examining the effect of Tudor domain point mutations, such as Esa1 D64A, show. A single mutation in the Esa1 Tudor domain loop region is sufficient to kill yeast cells. In contrast, mutations in the Esa1 HAT domain, which render the enzyme catalytically inactive in vitro, allow slow growth in vivo (38). Thus, for reasons that are not apparent, mutations in the Esa1 Tudor domain cause a more severe phenotype in yeast than mutations in the Esa1 catalytic domain.
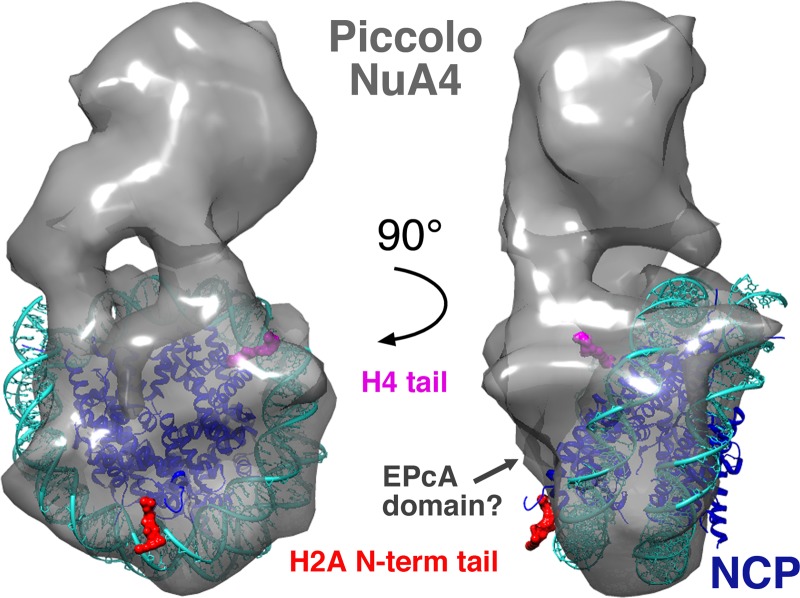
Our investigations of the N terminus of the Epl1 subunit EPcA domain suggest that particular residues in this approximately 20-residue region are important for Piccolo NuA4's nucleosomal HAT activity. Mutations of the Epl1 EPcA R58 and K59 residues substantially decreased Piccolo NuA4's ability to acetylate nucleosomes in vitro, but equivalent mutations of several other basic residues (R54, R56, K63, and K67) in this region had much milder effects. Our label transfer cross-linking studies indicate that this Epl1 EPcA basic region is in proximity (within 11 Å for the cross-linking agent used) to the N-terminal tail of histone H2A and to nucleosomal DNA. Since the histone H2A N-terminal tail exits from within the nucleosome core particle by making contact with DNA, it is perhaps not so surprising that cross-links to both this histone tail and nucleosomal DNA were detected in our experiments. These results provide new insight into where the Epl1 subunit and the N terminus of its EPcA domain in particular are located when bound to the nucleosome as part of the Piccolo NuA4 complex. The recent low-resolution structure of the Piccolo NuA4/nucleosome complex determined by cryo-electron microscopy (cryo-EM) offers a structural framework to interpret this finding (39). The structural model shows electron density on the surface of the histone octamer portion of the nucleosome and proximal to the H2A N-terminal tail, which could correspond to the Epl1 EPcA domain (Fig. 7). Furthermore, we detect cross-links between the Epl1 EPcA N-terminal region and the globular domain of histone H4. Interestingly, we had previously observed direct interaction between Esa1 and part of the histone fold globular region of histone H4 (40). The Epl1 cross-linking results could provide an explanation for why Esa1 requires Epl1 to interact with the nucleosome: significant contacts are contributed by the Epl1 EPcA domain. However, the Epl1 EPcA domain is unlikely to account for all the interactions between Piccolo NuA4 and the nucleosome, since the cryo-EM structural model for Piccolo NuA4/nucleosome complex shows extensive interactions across a broad surface of the nucleosome. Perhaps consistent with this multivalent interaction, we find that although the Piccolo NuA4 complexes containing the Epl1 EPcA R58A, R59A, and K63A mutations have reduced nucleosome binding activity in pulldown assays (Fig. 3b), these modest reductions do not appear to be sufficient to account for the significantly lowered nucleosomal HAT activity of the R58A or K63A mutant complex (Fig. 5).
Fig 7.
Low-resolution cryo-electron microscopy structure of the Piccolo NuA4/nucleosome complex (two orthogonal views) provides a structural model to interpret Piccolo NuA4 label transfer cross-linking results. The crystal structure of the nucleosome core particle (42) has been docked into the cryo-electron microscopy reconstruction of the Piccolo NuA4/nucleosome core particle complex (39). The N-terminal tails of histone H2A and H4 are highlighted in red and pink, respectively. A possible location for the EPcA domain based on our cross-linking data is indicated.
The cryo-electron microscopy structure of the Piccolo NuA4/nucleosome complex and our biochemical studies highlight two aspects that should be considered when assessing any mechanism for Piccolo NuA4's acetylation function. The first is that Piccolo NuA4 recognizes the histone tail residues in the structural context of the nucleosome. To do so, Piccolo NuA4 presumably either (i) interacts first with the body of the nucleosome and then binds to the histone H4 or H2A tail before catalyzing acetylation of these tail lysine residues, (ii) interacts first with the histone H4 or H2A tail before it contacts the body of the nucleosome, or (iii) engages histone tails and the nucleosome body simultaneously. Since Piccolo NuA4 can bind to the nucleosome in the absence of any histone tails (17) and since possibility iii seems unlikely, we favor possibility i. Given that neither the Esa1 Tudor domain loop nor the Epl1 EPcA basic region is required for binding to nucleosomes, this suggests either or both domains act post-nucleosome binding. We have proposed that the Esa1 Tudor domain in Piccolo NuA4 competes with the histone tails for nucleosomal DNA and in so doing releases histone tails to enter the Esa1 catalytic site (17). Our acetylation, cross-linking, and binding results provide additional data consistent with this hypothesis for the Epl1 EPcA domain in addition to the Esa1 Tudor domain. This hypothesis could explain why the Esa1 Tudor domain and EPcA basic region are not required for Piccolo NuA4 to bind nucleosomes and yet are important for the enzyme to acetylate histone tails in nucleosomes but not naked histones. Second, Piccolo NuA4 makes multiple interactions with the nucleosome that involve both protein-protein and protein-DNA contacts. It is perhaps not surprising that binding of a large and complicated macromolecular assembly like the nucleosome would involve multivalent contacts. Our crystallographic studies of the RCC1/nucleosome complex provide a precedent for how a single chromatin factor can make intricate protein-protein and protein-DNA contacts with different surfaces of the nucleosome (41). The interactions of the three-subunit Piccolo NuA4 complex with the nucleosome are likely to be at least as complicated.
ACKNOWLEDGMENTS
We thank Allen Minns, Justin Malloy, Bryan Thurston, Kristen Wiley, and Dylan Schlaich for technical support, Rob McGinty for comments on the manuscript, and members of the Tan laboratory and the Penn State gene regulation community for helpful discussion and advice.
This work was supported by Public Health Service grant GM-60489 from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print 29 October 2012
REFERENCES
- 1. Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res. 21: 381– 395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rando OJ, Winston F. 2012. Chromatin and transcription in yeast. Genetics 190: 351– 387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843– 851 [DOI] [PubMed] [Google Scholar]
- 4. Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11: 1640– 1650 [DOI] [PubMed] [Google Scholar]
- 5. Marmorstein R. 2001. Structure of histone acetyltransferases. J. Mol. Biol. 311: 433– 444 [DOI] [PubMed] [Google Scholar]
- 6. Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. 1999. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401: 93– 98 [DOI] [PubMed] [Google Scholar]
- 7. Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. 1999. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. U. S. A. 96: 8931– 8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. 2000. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell 6: 1195– 1205 [DOI] [PubMed] [Google Scholar]
- 9. Clements A, Poux AN, Lo Pillus W-SL, Berger SL, Marmorstein R. 2003. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol. Cell 12: 461– 473 [DOI] [PubMed] [Google Scholar]
- 10. Lee J-S, Smith E, Shilatifard A. 2010. The language of histone crosstalk. Cell 142: 682– 685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J. 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17: 1415– 1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197– 208 [DOI] [PubMed] [Google Scholar]
- 13. Patel A, Vought VE, Dharmarajan V, Cosgrove MS. 2011. A novel non-SET domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 286: 3359– 3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277: 7989– 7995 [DOI] [PubMed] [Google Scholar]
- 15. Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. 2006. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13: 713– 719 [DOI] [PubMed] [Google Scholar]
- 16. Lee MG, Wynder C, Cooch N, Shiekhattar R. 2005. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437: 432– 435 [DOI] [PubMed] [Google Scholar]
- 17. Selleck W, Fortin I, Sermwittayawong D, Côté J, Tan S. 2005. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 25: 5535– 5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. 2003. The Tudor domain “Royal Family”: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28: 69– 74 [DOI] [PubMed] [Google Scholar]
- 19. Eissenberg JC. 2012. Structural biology of the chromodomain: form and function. Gene 496: 69– 78 [DOI] [PubMed] [Google Scholar]
- 20. Nielsen PR, Nietlispach D, Buscaino A, Warner RJ, Akhtar A, Murzin AG, Murzina NV, Laue ED. 2005. Structure of the chromo barrel domain from the MOF acetyltransferase. J. Biol. Chem. 280: 32326– 32331 [DOI] [PubMed] [Google Scholar]
- 21. Shimojo H, Sano N, Moriwaki Y, Okuda M, Horikoshi M, Nishimura Y. 2008. Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain. J. Mol. Biol. 378: 987– 1001 [DOI] [PubMed] [Google Scholar]
- 22. Akhtar A, Zink D, Becker PB. 2000. Chromodomains are protein-RNA interaction modules. Nature 407: 405– 409 [DOI] [PubMed] [Google Scholar]
- 23. Tan S, Kern RC, Selleck W. 2005. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40: 385– 395 [DOI] [PubMed] [Google Scholar]
- 24. Barrios A, Selleck W, Hnatkovich B, Kramer R, Sermwittayawong D, Tan S. 2007. Expression and purification of recombinant yeast Ada2/Ada3/Gcn5 and Piccolo NuA4 histone acetyltransferase complexes. Methods 41: 271– 277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luger K, Rechsteiner TJ, Richmond TJ. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304: 3– 19 [DOI] [PubMed] [Google Scholar]
- 26. Chin JW, Martin AB, King DS, Wang L, Schultz PG. 2002. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99: 11020– 11024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ryu Y, Schultz PG. 2006. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods 3: 263– 265 [DOI] [PubMed] [Google Scholar]
- 28. Gietz D, St Jean A, Woods RA, Schiestl RH. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharma VM, Tomar RS, Dempsey AE, Reese JC. 2007. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 27: 3199– 3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao YG, Su SY, Robinson H, Padmanabhan S, Lim L, McCrary BS, Edmondson SP, Shriver JW, Wang AH. 1998. The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat. Struct. Biol. 5: 782– 786 [DOI] [PubMed] [Google Scholar]
- 31. Reference deleted.
- 32. Cobb CE, Beth AH. 1990. Identification of the eosinyl-5-maleimide reaction site on the human erythrocyte anion-exchange protein: overlap with the reaction sites of other chemical probes. Biochemistry 29: 8283– 8290 [DOI] [PubMed] [Google Scholar]
- 33. Smyth DG, Blumenfeld OO, Konigsberg W. 1964. Reactions of N-ethylmaleimide with peptides and amino acids. Biochem. J. 91: 589– 595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chahal HK, Dai Y, Saini A, Ayala-Castro C, Outten FW. 2009. The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry 48: 10644– 10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW. 2007. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 282: 13342– 13350 [DOI] [PubMed] [Google Scholar]
- 36. Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. 2011. Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. U. S. A. 108: E472– E479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Conrad T, Cavalli FMG, Holz H, Hallacli E, Kind J, Ilik I, Vaquerizas JM, Luscombe NM, Akhtar A. 2012. The MOF chromobarrel domain controls genome-wide H4K16 acetylation and spreading of the MSL complex. Dev. Cell 22: 610– 624 [DOI] [PubMed] [Google Scholar]
- 38. Decker PV, Yu DY, Iizuka M, Qiu Q, Smith MM. 2008. Catalytic-site mutations in the MYST family histone acetyltransferase Esa1. Genetics 178: 1209– 1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chittuluru JR, Chaban Y, Monnet-Saksouk J, Carrozza MJ, Sapountzi V, Selleck W, Huang J, Utley RT, Cramet M, Allard S, Cai G, Workman JL, Fried MG, Tan S, Côté J, Asturias FJ. 2011. Structure and nucleosome interaction of the yeast NuA4 and Piccolo-NuA4 histone acetyltransferase complexes. Nat. Struct. Mol. Biol. 18: 1196– 1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Berndsen CE, Selleck W, McBryant SJ, Hansen JC, Tan S, Denu JM. 2007. Nucleosome recognition by the Piccolo NuA4 histone acetyltransferase complex. Biochemistry 46: 2091– 2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Makde RD, England JR, Yennawar HP, Tan S. 2010. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467: 562– 566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. White CL, Suto RK, Luger K. 2001. Structure of the yeast nucleosome particle reveals fundamental changes in internucleosome interactions. EMBO J. 20: 5207– 5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reference deleted.