Abstract
Cells infected with hepatitis C virus (HCV) become refractory to further infection by HCV (T. Schaller et al., J. Virol. 81:4591–4603, 2007; D. M. Tscherne et al., J. Virol. 81:3693–3703, 2007). This process, termed superinfection exclusion, does not involve downregulation of surface viral receptors but instead occurs inside the cell at the level of RNA replication. The originally infecting virus may occupy replication niches or sequester host factors necessary for viral growth, preventing effective growth of viruses that enter the cell later. However, there appears to be an additional level of intracellular competition between viral genomes that occurs at or shortly following mitosis. In the setting of cellular division, when two viral replicons of equivalent fitness are present within a cell, each has an equal opportunity to exclude the other. In a population of dividing cells, the competition between viral genomes proceeds apace, randomly clearing one or the other genome from cells in the span of 9 to 12 days. These findings demonstrate a new mechanism of intracellular competition between HCV strains, which may act to further limit HCV's genetic diversity and ability to recombine in vivo.
INTRODUCTION
Hepatitis C virus (HCV) is a positive-stranded, enveloped single-stranded RNA (ssRNA) virus in the Flavivirus family. Currently, HCV infects more than 180 million people worldwide, and the associated morbidity and mortality are second only to those caused by HIV among emerging infections (1). HCV is primarily transmitted parenterally, but vertical and sexual transmission may also occur. After acute infection, approximately 25% of patients spontaneously clear the virus. The remaining patients are chronically infected and may go on to develop hepatic steatosis, cirrhosis, and hepatocellular carcinoma (2).
Complete in vitro replication of an HCV molecular clone was first demonstrated in 2005, using the genotype 2a virus JFH-1 (3–5). This clone, isolated from a Japanese male patient with fulminant hepatitis (6), replicated robustly in Huh7 cells and produced infectious virions in the absence of cell culture adaptive mutations. The availability of this infectious molecular clone provided a powerful experimental model with many advantages over the previously described HCV replicons (7). More recently, other groups have constructed highly infectious intergenotypic chimeras of JFH-1 and other HCV strains by making substitutions in the region from core to a portion of NS2 (8–10). The genotype 2a/2a chimera Jc1 is especially infectious (10).
HCV blocks infection by other incoming HCV virions through a process known as superinfection exclusion (11, 12). This process appears to occur after the virus enters the cell, which is different from the superinfection exclusion mechanism found in many other viral systems in which downregulation of cell surface viral receptors is involved. The intracellular superinfection block during HCV infection might result from competition between the primary and secondary viruses involving sequestration of key host factor(s) needed for viral replication or through occupancy of replicative niches on the endoplasmic reticulum (ER) membrane.
Superinfection exclusion has clear implications for treating HCV infection. If HCV could successfully superinfect cells, the evolution of drug and/or vaccine resistance, especially in a virus that is already hypervariant, would be greatly enhanced. Superinfection exclusion during HCV replication likely reduces the prevalence of viral recombination, which left unchecked could result in an even greater degree of immune escape variants and drug-resistant strains within this already variable virus.
In this study, we have explored whether mechanisms beyond classical superinfection exclusion contribute to limiting the possibility of HCV recombination. We now define an additional mechanism that limits the degree of HCV coinfection. We specifically show that cells replicating two or more HCV viral genomes convert into cells replicating only one viral genome due to genetic bottlenecking occurring during, or shortly after, mitosis. Furthermore, this process is biased toward replicons that have accumulated higher levels of viral RNA in host cells. We postulate that this bottleneck involves disruption of the viral replication niches in mitotic cells.
MATERIALS AND METHODS
Cells and culture conditions.
Huh7.5 cells, a kind gift from C. M. Rice (The Rockefeller University) (13) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin (Mediatech). Cells were passaged when they became confluent. In all cases in which replicon-positive cells were grown using the Jc1/ΔE1E2NS5A-XFP-BSD replicons, blasticidin (Invitrogen) selection was begun 2 days posttransfection at 10 μg/ml. Continuous selection over a period of 45 days following transfection was used to obtain Jc1/ΔE1E2NS5A-GFP-BSD and Jc1/ΔE1E2NS5A-mKO2-BSD replicon cell lines (data not shown).
Plasmid construction.
The construction of the various HCV replicon constructs and lentiviral constructs is described in detail in the supplemental material.
RNA synthesis and transfection.
In vitro transcription of viral RNA and electroporation were carried out as previously described (4, 14), with minor modifications. Viral RNA was transcribed using a Megascript T7 kit (Ambion) and purified by LiCl precipitation. A total of 7.5 × 106 Huh7.5 cells were electroporated with 10 μg of viral RNA when transfecting a single strain of HCV or with 5 μg each when multiple strains were used.
Production of Huh7.5 cell lines by lentiviral transduction.
Lentiviral particles were produced using an HIV-1-based vesicular stomatitis virus (VSV)-G pseudotyped lentiviral system, as described previously (15). The transfer plasmid pSicoR H2B-EBFP2 or pSicoR hGem-GFP was cotransfected into 293T cells along with the HIV-1-based packaging construct pCMVΔR8.91 and the VSV-G envelope vector pMD.G. Supernatants were collected 48 h posttransfection and concentrated by ultracentrifugation for 2 h at 53,000 × g in an SW28 rotor. A total of 2.7 × 105 Huh7.5 cells were spinoculated with 700 μl of concentrated viral supernatant at 1,000 × g for 2 h at room temperature and expanded in culture. Cells were sorted by flow cytometry for enhanced blue fluorescent protein 2 (EBFP2) or green fluorescent protein (GFP) fluorescence to ensure full transduction of the resulting 7.5-H2B-EBFP2 and 7.5-GFP-hGem cell lines.
Flow cytometry.
In experiments where single-, dual-, or triple-replicon-containing cells were isolated by sorting, the indicated cell lines were transfected with 5 μg each of the HCV RNAs. The exception was the triply transfected cells used in the modeling analysis (see Fig. S2 in the supplemental material), where 3.33 μg each of the HCV RNAs was used for consistent comparison with dually transfected cells. Two days after electroporation, cells were sorted using a FACSAria III fluorescence-activated cell sorter (FACS) (Becton, Dickinson), plated on 24-well plates at 1 × 105 (Jc1/ΔE1E2NS5A-XFP replicons) or 5 × 104 (Jc1/ΔE1E2NS5A-XFP-BSD replicons) cells/well, and analyzed on an LSRII flow cytometer (Becton, Dickinson) at various times afterward. In experiments to analyze the loss of dual/triple-replicon cells, the cells were washed with medium every 2 days to remove dead/apoptotic cells and then fixed in 4% paraformaldehyde (Sigma) before flow cytometric analysis.
In apoptotic cell analysis, the cells were washed every 24 h so that the measured apoptosis rate would reflect only the preceding 24 h. Cells were trypsinized and combined with cells from the medium, washed once in annexin-V binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2), and stained with 7-aminoactinomycin-D (7-AAD) (2.5 ng/μl; eBioscience) and annexin-V allophycocyanin (APC) (1:20 dilution; BD Biosciences) in annexin-V binding buffer. Cells were immediately analyzed on an LSRII. To generate a positive apoptotic control, naïve Huh7.5 cells were treated for 24 h with 100 to 1,600 nM staurosporine (Sigma-Aldrich).
For cell cycle analysis, the cells were fixed in ice-cold 70% ethyl alcohol (EtOH) for 30 min, washed twice in phosphate-citrate buffer (200 mM Na2HPO4, 4 mM citric acid), treated with 100 μg/ml RNase A (Qiagen) for 30 min, and stained with 50 μg/ml propidium iodide (PI) (Invitrogen). Cells were immediately analyzed on an LSRII.
To analyze the loss of viral RNA in the decay of dual-replicon cultures, Huh7.5 cells were transfected with 5 μg each of Jc1/ΔE1E2NS5A-GFP-BSD and Jc1/ΔE1E2NS5A-mKO2-BSD RNA. Two days posttransfection, dual- and single-replicon cells were isolated by FACS (Sort 1). The dual-replicon cells were cultured in the presence of 10 μg/ml blasticidin (Invitrogen) for 9 to 12 days following Sort 1 to allow decay to occur (medium was changed every third day), and single-replicon and replicon-negative cells were isolated by FACS (Sort 2).
CFSE dilution and growth arrest studies.
For carboxyfluorescein diacetate-succinimidyl ester (CFSE) dilution analyses, cells were electroporated with 5 μg of Jc1/ΔE1E2NS5A-mCherry and Jc1/ΔE1E2NS5A-EBFP2 RNA and plated at 37.5 × 103 cells/well in 24-well plates or at 7.5 × 106 cells in 140-mm plates. One day later, cells were treated with 5 or 10 μM CFSE (Invitrogen) for 30 min, washed twice with medium, and analyzed for fluorophore expression and CFSE median fluorescence intensity on an LSRII at various times after the CFSE treatment. To generate growth-arrested Huh7.5 cells, cells were grown for 15 days on 140-mm plates coated with 50 μg/ml rat tail collagen type I (Becton, Dickinson) in the presence of 1% (vol/vol) dimethyl sulfoxide (DMSO) (Sigma-Aldrich), as described previously (16). After transfection of growth-arrested cells, cells were replated on collagen-coated culture vessels in the presence of 1% DMSO.
RNA isolation and quantitative PCR analysis.
Total RNA was extracted from each population with TRIzol (Invitrogen). Reverse transcription-PCR (RT-PCR) and subsequent real-time PCR were performed in two steps using the Superscript III first strand cDNA synthesis system (Invitrogen), followed by the Quantitect probe PCR system, or in one step using the Quantitect probe RT-PCR system (Qiagen). Real-time PCR was performed on a 7900HT fast real-time PCR system (Applied Biosystems), according to the manufacturer's protocol. The probes and primers were purchased from Applied Biosystems and are given in Table S2 in the supplemental material. The GFP probe/primer set was as described previously (17).
Live-cell epifluorescence microscopy.
Huh7.5 or 7.5-H2B-EBFP2 cells were electroporated with Jc1/ΔE1E2NS5A-GFP, Jc1/ΔE1E2NS5A-mCherry, or Jc1/ΔE1E2NS5A-EBFP2 RNA. The cells were plated at 5 × 104 cells/well in LabTek 8-well coverslip chambers (Nunc) and incubated for 2 days in a 37°C incubator. For time-lapse microscopy, cells were imaged on the UCSF Nikon Imaging Center Eclipse TI-E epifluorescence microscope (Nikon) at 37°C, 5% CO2. Images were colorized for clarity and ease of viewing using Photoshop (Adobe) in the publication figures. Videos in the supplemental material were assembled using ImageJ software (National Institutes of Health).
Statistical analysis.
Statistical analysis was carried out using Prism 5 software (Graphpad). For CFSE dilution curve analysis, semi-log regression and the extra sum of squares F test were used to determine whether the slopes of the dilution curves were significantly different. For data that were not amenable to regression analysis (analysis of transfection efficiency, RNA accumulation, and cell proliferation by cell count over time), the integrated area under the curve (AUC) was obtained, followed by one-way analysis of variance (ANOVA) with Bonferroni's multiple-comparison correction to ascertain statistical differences in the AUC. Regression curves and AUC data are not shown. The comparison of cell cycle progression in dual- and single-replicon-containing cells was assessed by Student's t test, as only one comparison was being made. For all other tests, one- or two-way ANOVAs with Bonferroni's multiple-comparison corrections were used, as appropriate.
RESULTS
Replication competence of the replicons used in the study.
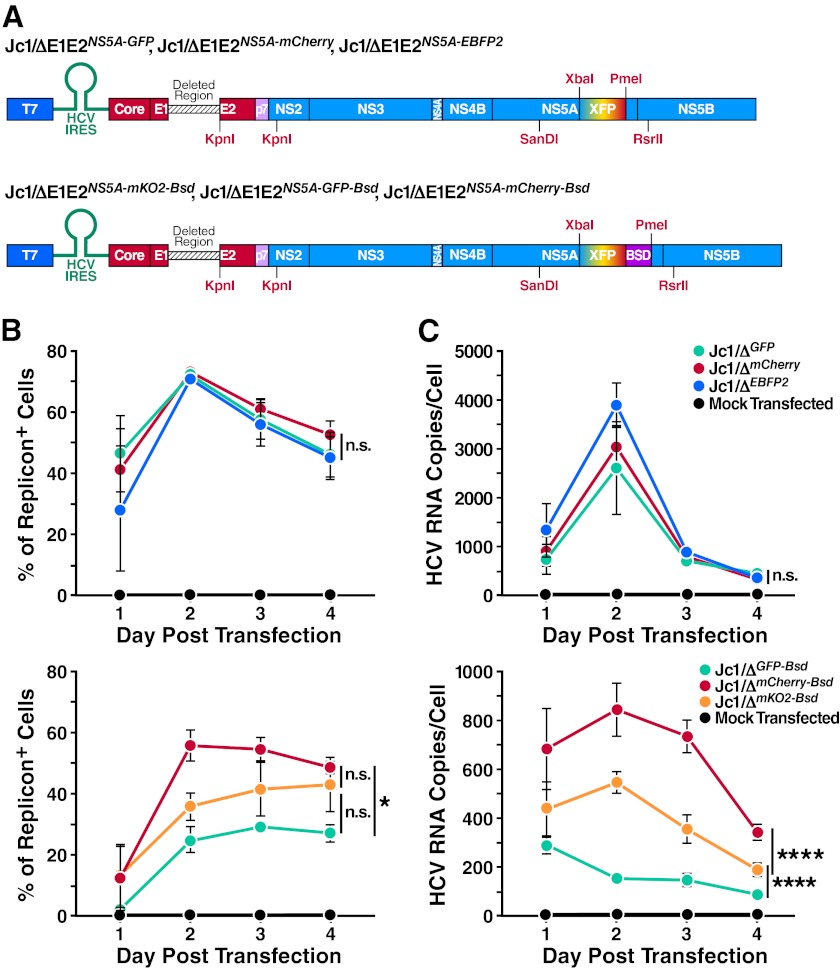
As we were interested in analyzing the replication of multiple HCV strains, we created fluorescently tagged monocistronic Jc1 replicon constructs similar to those described by Schaller et al. (11). By fusing fluorescent reporters to the viral NS5A protein, we could measure the replication potential of multiple replicon strains in one cell (Fig. 1A). By additionally adding a blasticidin resistance gene (BSD) in the context of NS5A domain III, we were able to select specifically for replicon-containing cells. To determine the relative fitness of the viral replicons, highly permissive Huh7.5 cells were singly transfected with 10 μg of each replicon. The efficiency of establishment of replication in host cells was measured by flow cytometry as well as viral replicon RNA accumulation by quantitative RT-PCR (Fig. 1B and C). As demonstrated by both the percentage of replicon-positive cells and RNA accumulation, the peak of replication occurred 2 days after transfection.
Fig 1.
Genomic HCV constructs employed in this study and relative efficiencies of each. (A) Schematic diagram of constructs used; XFP represents the fluorophore tag inserted into NS5A, and BSD represents the blasticidin resistance gene. (B, C) Comparison of replicon fitness by transfection efficiency and viral RNA accumulation in host cells. Huh7.5 cells were mock transfected or transfected with 10 μg of Jc1/ΔGFP, Jc1/ΔEBFP2, Jc1/ΔmCherry, Jc1/ΔGFP-BSD, Jc1/ΔmKO2-BSD, and Jc1/ΔmCherry-BSD replicon RNAs. (B) Transfection efficiency of each replicon strain. Flow cytometric analysis was carried out at the given time points to determine the percentage of replicon-positive cells. (C) RNA accumulation in host cells transfected with each replicon strain. RNA was extracted from transfected cells at the indicated time points and subjected to quantitative RT-PCR analysis using a core-specific HCV probe. A GAPDH probe was used for normalization of RNA samples, using mock-transfected cells as a calibrator. Error bars indicate standard errors of the means (±SEM). Differences in transfection efficiencies or RNA accumulation were assessed by obtaining the integrated area under the curve (AUC) for each independent experiment, followed by one-way ANOVA with Bonferroni's multiple-comparison correction to assess statistically significant differences (*, P < 0.05; ****, P < 0.0001; n.s., nonsignificant).
In agreement with prior studies (11), we did not observe differences in the fitness of different Jc1/ΔE1E2NS5A-XFP constructs (here referred to as Jc1/ΔXFP for simplicity). The Jc1/ΔE1E2NS5A-XFP-BSD replicons, however, did appear to differ in fitness, although differences in RNA accumulation were statistically greater than differences in transfection efficiency (Fig. 1C). It is known that GFP fusion to domain III of NS5A negatively affects the RNA replication efficiency of replicons (18); different inserted protein sequences (e.g., fluorescent proteins) may differentially affect the overall fitness. The addition of the BSD sequence reduced the replication efficiency of the Jc1/ΔXFP-BSD replicons overall, potentially unmasking changes that different fluorescent proteins exert on replicon fitness. The natively high replication efficiency of JFH-1-based replicons may have masked any subtle differences in replication of the Jc1/ΔXFP strains.
More than one genomic strain are unstable in HCV replicon-containing cells.
Superinfection exclusion has been demonstrated to occur at a postentry step in the viral life cycle (11, 12). Prior studies have also demonstrated a degree of compartmentalization of viral replication complexes (19, 20). We therefore hypothesized that intracellular competition for either limiting host proviral factors or replicative niches could occur even in cells productively replicating two strains of HCV. Intracellular competition would then lead to the progressive loss, or decay, of viral genomes until only one strain remained in a particular cell.
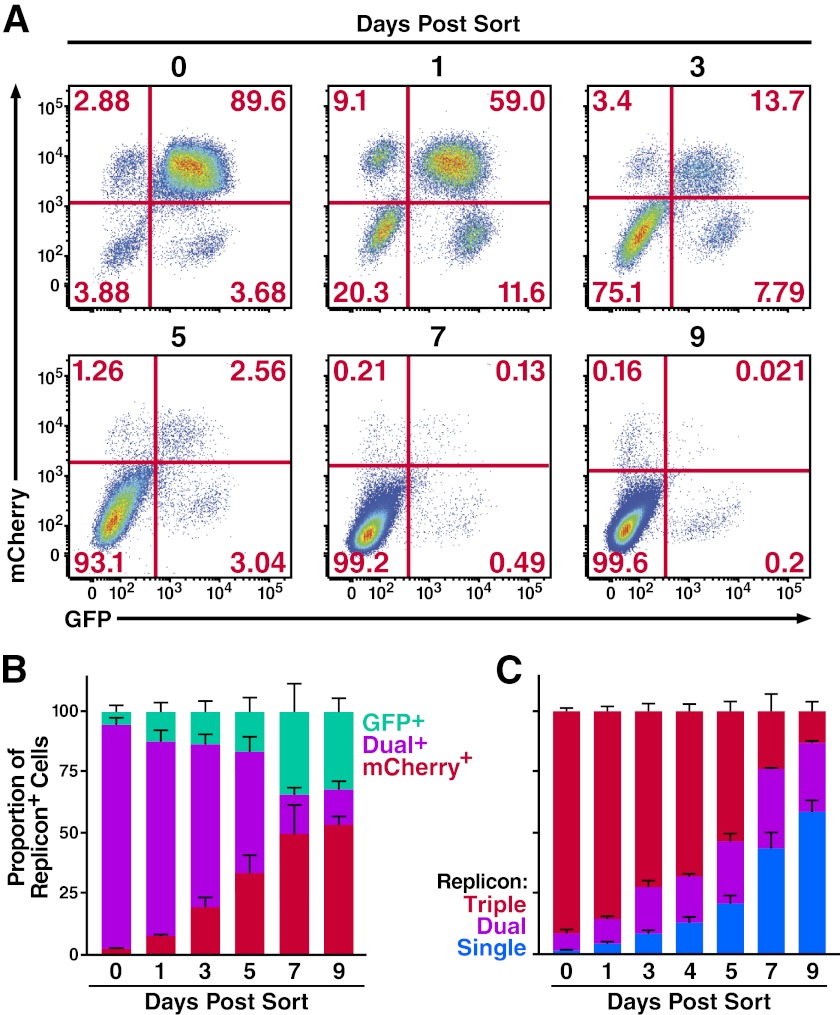
To address this possibility, studies were performed to test whether host cells could stably replicate two HCV genomic strains using the equally fit Jc1/ΔXFP replicons. These replicons lack the E1E2 proteins, preventing the confounding effects of spreading infection following the initial transfection. Huh7.5 cells were transfected simultaneously with Jc1/ΔE1E2NS5A-GFP and Jc1/ΔE1E2NS5A-mCherry. Dual-replicon (GFP+/mCherry+) cells were isolated by fluorescence-activated cell sorting 2 days later. These cells were then monitored at various time points postsorting by flow cytometry to determine the stability of the dual-replicon state. We found that dual-replicon cells were progressively lost over time with near-complete disappearance within 9 days (Fig. 2A). The replicon-negative cell population expanded in these cultures likely because HCV infection is associated with both apoptosis (21, 22) and cell cycle abnormalities (23, 24). However, when only the replicon-positive cells were evaluated (Fig. 2B), the loss of dual-replicon cells and a concomitant rise in single-replicon cells were readily apparent. Neither the Jc1/ΔGFP nor the Jc1/ΔmCherry genome seemed to have an advantage in this decay process, as there was no significant difference in the proportion of single-replicon-containing cells of each type (Table 1). However, in these experiments we could not rule out the possibility that single-replicon cells might exhibit a selective growth advantage over dual-replicon cells (less cell death and higher growth rate) culminating in more single-replicon cells after 7 to 9 days. To demonstrate the sample sizes of the analyzed cells, the absolute cell counts of all experiments analyzing the decay phenomena are provided in Fig. S1 in the supplemental material.
Fig 2.
Decay of cells multiply transfected with congenic Jc1 replicon strains. (A) The dual-replicon state is unstable. Huh7.5 cells were transfected with Jc1/ΔGFP and Jc1/ΔmCherry RNA, followed 48 h later by FACS isolation of dual-replicon (mCherry+/GFP+) cells. Cells were then analyzed by flow cytometry for mCherry/GFP expression on days 0, 1, 3, 5, 7, and 9 postsorting (data are representative of 5 independent experiments). (B) Flow cytometric analysis of replicon decay occurring within dual-replicon cells. The total number of replicon-positive cells was set to 100%. Jc1/ΔGFP-only replicon-containing cells are in green, Jc1/ΔmCherry-only replicon-containing cells are in red, and dual-replicon cells are in orange. Error bars indicate +SEM (n = 5). (C) Triple-replicon cells decay through a dual-replicon state to single-replicon cells. The total number of replicon-positive cells was set to 100%; blue, single-replicon cells (GFP+ only, mCherry+ only, or EBFP2+ only); green, dual-replicon cells (GFP+/mCherry+ only, mCherry+/EBFP2+ only, or GFP+/EBFP2+ only); red, triple-replicon cells (GFP+/mCherry+/EBFP2+). Error bars indicate +SEM (n = 4).
Table 1.
Statistical significance of the differences in the proportions of replicon-positive cells in dual-replicon decay experimentsa
| Transfection: Jc1/ΔGFP and Jc1/ΔmCherry | |||
|---|---|---|---|
| No. of days postsorting | Dual+ vs. GFP+ | Dual+ vs. mCherry+ | mCherry+ vs. GFP+ |
| 0 | P < 0.0001 | P < 0.0001 | NS |
| 1 | P < 0.0001 | P < 0.0001 | NS |
| 3 | P < 0.0001 | P < 0.0001 | NS |
| 5 | P < 0.001 | NS | NS |
| 7 | NS | P < 0.05 | NS |
| 9 | NS | P < 0.01 | NS |
Two-way ANOVA with Bonferroni's multiple-comparison correction was used to analyze the difference in proportions of each replicon-positive cell population. NS, nonsignificant; vs., versus.
We hypothesized that triple-replicon cells would decay in an ordered manner transitioning through a dual-replicon state and on to a single-replicon state reflecting the sequential loss of viral genomes. Conversely, if single-replicon cells simply displayed an increased growth advantage, such single-replicon cells would increase more quickly than dual-replicon cells. To distinguish between these possibilities, cells were transfected with Jc1/ΔGFP, Jc1/ΔmCherry, and Jc1/ΔEBFP2, and triple-replicon (GFP+/mCherry+/EBFP2+) cells were isolated by FACS 2 days later. At various times afterward, the cells were analyzed for the presence of GFP/mCherry/EBFP2 fluorescence. We observed an ordered progression of viral genome decay from triple- to dual- to single-replicon cells (Fig. 2C). Specifically, dual-replicon cells increased until day 5, when single-replicon cells became predominant (see Fig. S2 in the supplemental material).
Both dual- and single-replicon cells display a consistent exponential decay compared to replicon-negative cells (see Fig. S2 in the supplemental material). As isolated triple-replicon cells always contain a small proportion of contaminating dual- and single-replicon cells, we cannot completely rule out the null hypothesis that the increase in the number of dual- and single-replicon cells is a result of a selective growth advantage of these small numbers of contaminating cells. We have used the exponential decay rates of isolated dual- and single-replicon cells to model the expected numbers of dual- and single-replicon cells over time in isolated triple-replicon cell cultures. Interestingly, there are far more dual- and single-replicon cells over time in isolated triple-replicon cultures than would be expected from the simple expansion of contaminating cells (see Fig. S2 in the supplemental material). This fact further reinforces the conclusion that multiple-replicon cells sequentially eliminate viral genomes over time.
Dual-replicon cells do not exhibit higher rates of cell death.
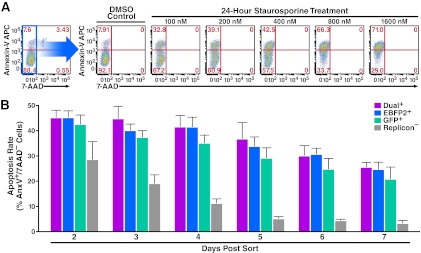
To further test a potential selective survival advantage for cells containing fewer HCV replicons, we measured the apoptosis rate in single- and dual-replicon cells. Huh7.5 cells were transfected with Jc1/ΔGFP and Jc1/ΔEBFP2 and then separated by FACS into dual-replicon (GFP+/EBFP2+), single-replicon (GFP+/EBFP2− or GFP−/EBFP2+), and replicon-negative (GFP−/EBFP2−) populations (see Fig. S3A in the supplemental material). Beginning 48 h postisolation, cells were stained with 7-AAD and annexin-V APC. We analyzed these cells by flow cytometry for early apoptotic cells in the standard manner, using staurosporine as a positive control (Fig. 3A). The 7-AAD−:annexin-V+ cells represent apoptotic cells, and 7-AAD+ cells represent dead cells. With adherent cells, dead cells are often excluded from the analysis (25), since they could potentially have died due to membrane rupture during trypsinization. However, we observed similar patterns in all apoptosis assays when dead cells were either included or excluded (data not shown).
Fig 3.
The apoptosis rates in Jc1/ΔXFP dual- and single-replicon-containing cells are similar. (A) Demonstration of the gating strategy used to analyze apoptotic cells. Huh7.5 cells were treated for 24 h with various concentrations of staurosporine to induce apoptosis or with DMSO vehicle (control). The cells were then stained with annexin-V APC and 7-AAD. Apoptotic cells are defined as 7-AAD−:annexinV+ cells. (B) No difference in apoptosis rates in dual- or single-replicon cells. Huh7.5 cells were transfected with Jc1/ΔGFP and Jc1/ΔEBFP2 RNA, followed 48 h later by FACS isolation of Jc1/ΔGFP-only replicon-containing cells, Jc1/ΔEBFP2-only replicon-containing cells, replicon-negative cells, and dual-replicon cells. Cells were stained with annexin-V APC and 7-AAD and analyzed at the given time points postisolation by flow cytometry. Cells were washed with medium 24 h postisolation and every 24 h thereafter, so the apoptosis rate reflects the preceding 24 h. Error bars indicate +SEM.
The apoptosis rates of dual-replicon and single-replicon cells were similar at all time points analyzed (P > 0.05) (Fig. 3B; Table 2). However, at each time point from 3 to 7 days postisolation, the apoptosis rate was significantly greater in each of the replicon-positive populations than in the replicon-negative population (P < 0.05). The initially high apoptosis rate of replicon-negative cells likely reflects the stresses of electroporation followed by FACS isolation. These findings highlight a cytopathic effect of HCV within these cells but further show that apoptosis rates do not differ between dual- and single-replicon cells.
Table 2.
Statistical significance of differences in apoptosis rates of dual-, single-, or replicon-negative cellsa
| Transfection: Jc1/ΔGFP and Jc1/ΔEBFP2 | ||||||
|---|---|---|---|---|---|---|
| No. of days postsorting | Dual+ vs. GFP+ | Dual+ vs. EBFP2+ | Dual+ vs. replicon− | GFP+ vs. EBFP2+ | GFP+ vs. replicon− | EBFP2+ vs. replicon− |
| 2 | NS | NS | P < 0.05 | NS | Ns | P < 0.05 |
| 3 | NS | NS | P < 0.0001 | NS | P < 0.05 | P < 0.01 |
| 4 | NS | NS | P < 0.0001 | NS | P < 0.001 | P < 0.0001 |
| 5 | NS | NS | P < 0.001 | NS | P < 0.05 | P < 0.01 |
| 6 | NS | NS | P < 0.001 | NS | P < 0.01 | P < 0.0001 |
| 7 | NS | NS | P < 0.05 | NS | NS | NS |
Two-way ANOVA with Bonferroni's multiple-comparison correction was used to analyze the difference in apoptosis rates of each replicon-positive cell population. NS, nonsignificant; vs., versus.
Isolated dual- and single-replicon cells exhibit no difference in proliferation as assessed by CFSE dilution.
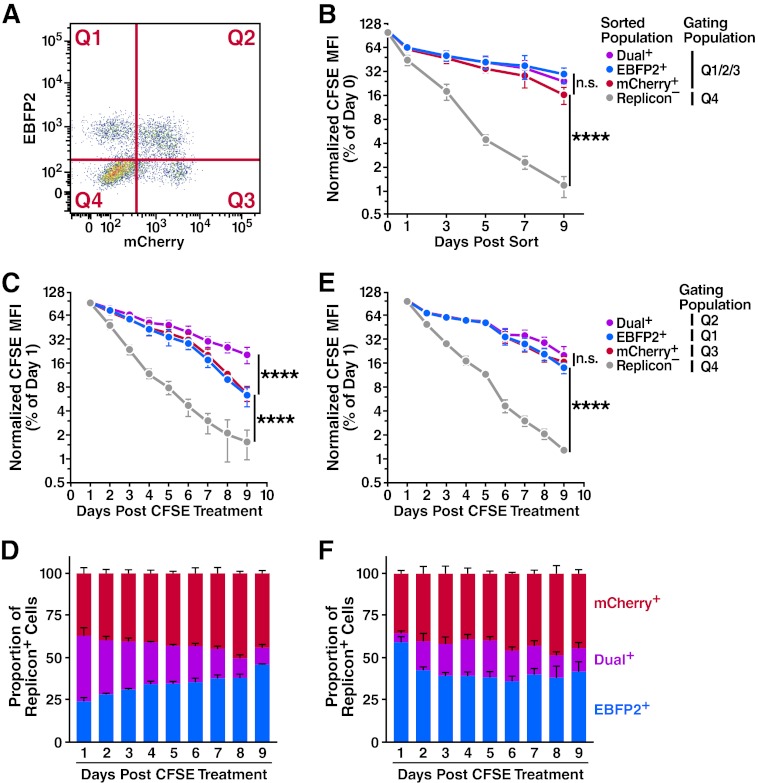
Single-replicon cells might have a selective advantage over dual-replicon cells by dividing more quickly. To test this possibility, we examined Huh7.5 cells transfected with Jc1/ΔmCherry and Jc1/ΔEBFP2 for CFSE dilution (22). As CFSE is equally partitioned between daughter cells following division, the dilution of CFSE fluorescence loss is an excellent marker for cellular proliferation. One day after transfection, we treated cells with 10 μM CFSE, and 24 h later dual-replicon (mCherry+/EBFP2+), single-replicon (mCherry−/EBFP2+ or mCherry+/EBFP2−), and replicon-negative cells (mCherry−/EBFP2−) were isolated by FACS (see Fig. S3B in the supplemental material). When analyzing CFSE dilution in replicon-positive cell samples, the replicon-negative cells were gated out to simplify the analyses (Fig. 4A).
Fig 4.
Progression through the cell cycle is important in the dual- to single-replicon decay process. Huh7.5 cells were transfected with Jc1/ΔEBFP2 and Jc1/ΔmCherry RNA and treated with 5 or 10 μM CFSE 24 h later. (A) Representative flow cytometry plot demonstrating gating quadrants. (B) No difference in proliferation of isolated dual- and single-replicon cells assessed by CFSE dilution. Two days after transfection, dual-replicon, single-replicon, and replicon-negative cells were isolated by FACS. Cells were analyzed for CFSE/EBFP2/mCherry by flow cytometry; the gating quadrants represent the cells used in the analysis. Values were normalized to the CFSE mean fluorescence intensity (MFI) at day 0 postsorting (n = 4). (C) In mixed-replicon cultures, dual-replicon cells dilute CFSE more slowly than single-replicon cells. Cells were not isolated by cell sorting; instead, each population was separated by the given quadrant gates (n = 3). (D) Decay of dual-replicon cells in mixed cultures. The chart indicates the proportion of single- and dual-replicon cells in the samples in panel C. (E) No significant difference in CFSE dilution of dual- or single-replicon cells in mixed-replicon DMSO-treated cultures. Huh7.5 cells were treated for 15 days with 1% DMSO to induce a differentiated, growth-arrested phenotype before transfection and were otherwise treated as in panel C (n = 3). (F) DMSO treatment of cells slows the dual-replicon decay process. The chart indicates the proportion of single- and dual-replicon cells in the samples in panel E. Error bars indicate SEM (****, P < 0.001; n.s., nonsignificant by extra sum of squares F test using semi-log regression).
In agreement with previous studies (21, 23, 24, 26), we did observe a significant proliferative defect in replicon-positive compared to replicon-negative cells (Fig. 4B). However, no significant difference in the CFSE dilution rate was observed with any of the replicon-positive samples (assessed by extra sum of squares F test, P > 0.05). If a proliferative advantage for single-replicon over dual-replicon cells existed, the slope of the CFSE dilution curves for dual- and single-replicon cells would have been significantly different. We conclude that Jc1/ΔXFP dual-replicon cells exhibit no apparent differences in either proliferation or apoptosis rates compared to single replicon cells. Therefore, the decay of dual-replicon cells is not an artifact caused by a selective growth or survival advantage of single-replicon cells that happen to be contaminating the dual-replicon population.
Mitosis is a key event promoting the decay of dual-replicon cells.
During mitosis, the endoplasmic reticulum (ER) membrane is disrupted (27, 28), which may cause the viral replication niches to break down. These events could produce a genetic bottleneck that would be responsible for the observed decay of dual-replicon cells. CFSE dilution analysis can be used to determine whether mitosis is indeed a key part of the decay of dual-replicon cells; if so, division of dual-replicon cells will result in both CFSE dilution and decay into single-replicon cells. To test this possibility, cells were transfected with Jc1/ΔmCherry and Jc1/ΔEBFP2 and treated with CFSE as in the previous experiment (Fig. 4C). However, dual- and single-replicon cells were not isolated from each other by flow-based sorting as in the prior experiment. In that experiment, using pure populations of single-replicon cells, decay of dual-replicon cells did not occur; thus, the CFSE dilution curves reflected the division of the single-replicon cells only (Fig. 4B). In this experiment, the decay of dual-replicon cells contributed to the overall population of single-replicon cells. Thus, in this experiment, the CFSE dilution rate of single-replicon cells would be expected to additionally reflect the division and attendant decay of dual-replicon cells. Hence, if mitosis is a key part of this decay, this should manifest as a difference in the slope of the CFSE dilution curve in dual- and single-replicon cells. In contrast to the similar rates of CFSE dilution of isolated, pure populations of single- and dual-replicon cells (Fig. 4B), there is indeed a significant difference in CFSE dilution rates of dual- and single-replicon cells in mixed cultures (P < 0.0001) (Fig. 4C).
If mitosis indeed forms the transition point in the decay from dual- to single-replicon cells, we should be able to slow the decay process by slowing cellular division rates. To test this possibility, the same CFSE dilution experiment was performed with cells treated with 1% DMSO for 15 days before transfection. This treatment is known to slow and ultimately arrest the growth of hepatoma cells (16). After 15 days, the confluent cells were indeed growth arrested as assessed by serial cell counts (data not shown). These cells were then transfected with the replicon RNAs. When the transfected cells were replated in the continued presence of 1% DMSO, they began to divide again, albeit more slowly. We observed that the DMSO-induced cell cycle retardation was sufficient to cause dual- and single-replicon cells to dilute CFSE at similar rates (Fig. 4E). Concomitantly, the DMSO treatment also slowed (or halted) the conversion of dual-replicon into single-replicon cells (Fig. 4D and F), indicating that mitosis may play an important role in the decay process.
Dual- and single-replicon cells are equally competent for proliferation.
To conclusively ensure that dual-replicon cells are not defective in proliferation relative to single-replicon cells, we examined the cell cycle profiles by DNA content analysis of Huh7.5 cells transfected with Jc1/ΔGFP and Jc1/ΔmCherry RNA. We compared the proportions of dual- and single-replicon cells in each phase of the cell cycle and found that there was no significant cell cycle arrest specific to dual-replicon cells (data not shown).
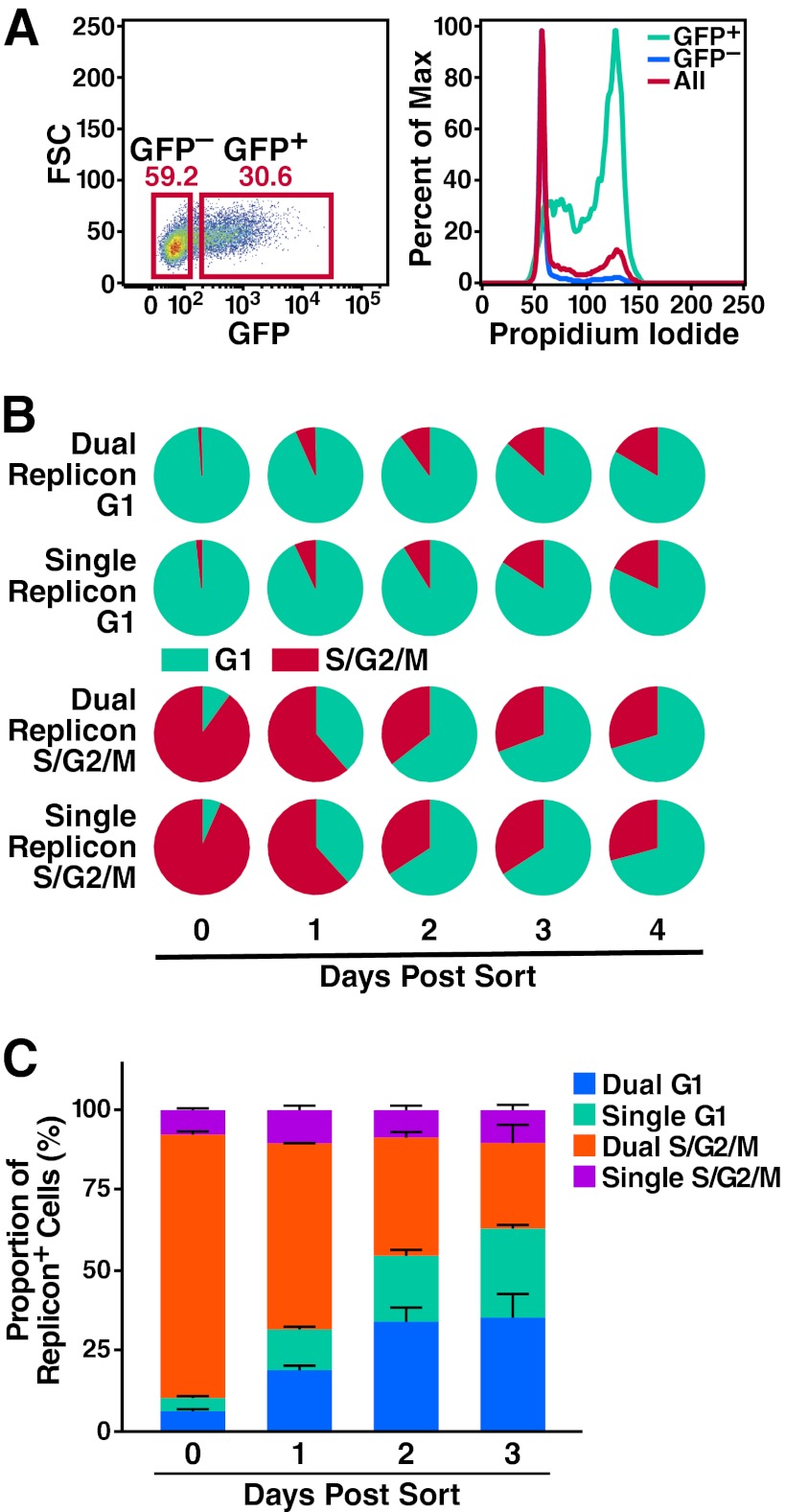
Since cell cycle profiles represent a relatively insensitive method for measuring proliferation competence, we elected to use a modified version of the Fucci system (29) to isolate dual- and single-replicon-containing premitotic and mitotic (S/G2/M-phase) cells or cells in the G1 phase. We then assessed the ability of these cells to progress into the next phase(s) of the cell cycle. The hCdt1 (human Cdt1) and hGem (human geminin) proteins are expressed during different phases of the cell cycle. Cdt1 is ubiquitinated by the SCFSkp2 complex during S and G2 phases, while Gem is ubiquitinated by the APC/C E3 ligase complex during late M and G1 phases. Both are rapidly degraded by the proteasome (30). Probes containing the region of hCdt1 and hGem necessary for ubiquitination/degradation fused to a fluorophore are selectively present during G1 and S/G2/M phases, respectively.
In our hands, a GFP-hGem (Fig. 5A) or an EBFP2-hGem (see Video S1 in the supplemental material) probe was sufficient to distinguish between S/G2/M- and G1-phase cells in stable Huh7.5 cell lines transduced with lentiviral constructs encoding these probes. Therefore, we used the hGem probe alone to reduce fluorophore overlap. 7.5-GFP-hGem cells were transfected with Jc1/ΔEBFP2 and Jc1/ΔmCherry RNA, and 2 days later, the following populations were isolated by FACS: G1-phase (GFP−) single-replicon cells (EBFP2+/mCherry−), G1-phase (GFP−) dual-replicon cells (EBFP2+/mCherry+), G2/S/M-phase (GFP+) single-replicon cells (EBFP2+/mCherry+), and G2/S/M-phase (GFP+) dual-replicon cells (EBFP2+/mCherry+).
Fig 5.
Mitosis is associated with the decay process in dual-replicon cells. (A) Validation of the fluorescent hGem probe for cells in S/G2/M phases of the cell cycle. 7.5-GFP-hGem cells were stained with PI and analyzed for GFP and PI by flow cytometry. As shown, the GFP+ cells are highly enriched in the S/G2/M phases of the cell cycle, while the GFP− cells are enriched for the G1 phase of the cell cycle. (B) Dual-replicon cells are equally competent as single-replicon cells for cell cycle progression. 7.5-GFP-hGem cells were transfected with Jc1/ΔGFP and Jc1/ΔmCherry RNA. Dual-replicon (EBFP2+/mCherry+) and single-replicon (mCherry+ only) cells were isolated in G1 (GFP−) and S/G2/M (GFP+) phases of the cell cycle by FACS. No difference was observed by Student's t test in the cell cycle phase of single- and dual-replicon cells at any time point (P > 0.2, n = 3). (C) As dual-replicon cells progress through mitosis, single-replicon cells are enriched. 7.5-GFP-hGem cells were transfected with Jc1/ΔEBFP2 and Jc1/ΔmCherry. Dual-replicon (EBFP2+/mCherry+) S/G2/M-phase (GFP+) cells were isolated by FACS and analyzed by flow cytometry for the dual- to single-replicon transition, as well as cell cycle progression. Error bars indicate +SEM (n = 3).
Beginning at 24 h postisolation, we used flow cytometry to assess whether cells had progressed through the G1-S transition (gain of GFP fluorescence) or through the M-G1 transition (loss of GFP fluorescence). Of note, we observed that dual- and single-replicon cells progressed through the cell cycle at equivalent rates (Fig. 5B), based on the fact that a delay in cell cycle progression in dual-replicon cells should cause them either to lose (M→G1) or gain (G1→S) GFP fluorescence more slowly than single-replicon cells. These findings further indicate that dual- and single-replicon cells proliferate at quite similar rates.
Dual-replicon cell transition through mitosis is associated with enrichment in single-replicon cells.
Slowing the cell cycle in replicon-transfected cells by DMSO treatment also slowed the decay of dual-replicon cells (Fig. 4). Accordingly, we investigated the possibility that the decay from dual- to single-replicon cells is linked to mitosis. 7.5-GFP-hGem cells were transfected with Jc1/ΔEBFP2 and Jc1/ΔmCherry RNA. Two days later, we isolated dual-replicon S/G2/M-phase cells by FACS (EBFP2+/mCherry+, dual-replicon; GFP+, S/G2/M phase). We next analyzed the cells beginning at day 1 postisolation for the transition into G1 phase (loss of GFP fluorescence) and the decay from dual-replicon cells into single-replicon cells. Completion of mitosis in dual-replicon cells (the transition from S/G2/M to G1 phase) was associated with a marked increase in the proportion of single-replicon cells relative to replicon-positive cells remaining in S/G2/M phase that had not completed cell division (Fig. 5C). At day 2 postsorting, when the proportions of post- and predivision cells were similar (54.8% and 45.2%, respectively), there were significantly more postdivision single-replicon cells than predivision single-replicon cells (P < 0.01). However, there was no difference in the proportion of post- versus predivision dual-replicon cells at this time point (P > 0.6).
These findings indicate that, following mitosis, dual-replicon cells tend to decay into a single-replicon state, although the persistence of postmitotic dual-replicon cells implies that decay does not necessarily occur in conjunction with every mitotic division. Nevertheless, these findings underscore the importance of mitosis in the viral genome decay process. We hypothesize that this decay results from a bottleneck in HCV genome diversity occurring during or shortly after mitosis. Of note, this bottleneck acts without regard to viral fitness, as we observed this decay phenomenon in congenic Jc1 viral strains differing only by their fluorophore tag.
Mitosis is associated with a loss of fluorescence from one of the NS5A fluorophore-tagged genomes in dual-replicon cells.
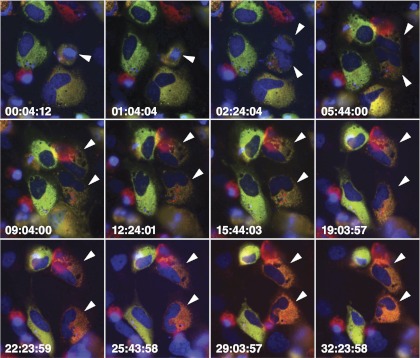
To further demonstrate that decay from the dual- to single-replicon state occurs during, or shortly after, mitosis, 7.5-H2B-EBFP2 cells were transfected with Jc1/ΔGFP & Jc1/ΔmCherry or Jc1/ΔGFP-BSD & Jc1/ΔmKO2-BSD RNA and, 2 days later, imaged via time-lapse microscopy for ∼40 h. Dual-replicon cells entering mitosis were identified by GFP/mCherry or GFP/mKO2 fluorescence and chromatin condensation of the H2B-EBFP2 tag. The relative fluorescence of the GFP/mCherry or GFP/mKO2 tags markedly shifts after mitosis (Fig. 6; see Videos S2 to S6 in the supplemental material). Although we never observed complete loss of fluorescence of one of the NS5A tags, a marked shift in fluorescence favoring one of the two replicons was observed in nearly every postmitotic daughter cell that was successfully imaged for more than 24 h following division. Near-complete loss of fluorescence of one of the NS5A tags was observed (see Videos S3 and S4 in the supplemental material) only when one of the two replicons already had an advantage. We hypothesize that one mitotic bottleneck may randomly lead to a bias in the number of genomes of one of the replicons; an additional round of mitosis and thus another bottleneck may be necessary to fully purge the other replicon.
Fig 6.
Loss of fluorophore-NS5A epifluorescence in dual-replicon cells after mitosis. 7.5-H2B-EBFP2 (blue) cells were transfected with Jc1/ΔGFP (green) and Jc1/ΔmCherry (red) RNA and imaged by time-lapse epifluorescence microscopy, with images taken every 20 min. Arrowheads indicate parental and daughter cells. Times are indicated as hours:minutes:seconds. Note the drastic shift in fluorescence favoring the Jc1/ΔmCherry replicon in both daughter cells.
Interestingly, in 9 of 12 observations displaying a shift in fluorescence, one replicon became dominant in both daughter cells; Video S6 in the supplemental material depicts one of the three experiments in which the replicons diverged in dominance in the daughter cells. This tendency to favor the same genome in both daughter cells suggests that the genetic bottleneck does not typically partition the genomes between daughter cells. Furthermore, Video S5 in the supplemental material demonstrates a shift in replicon fluorescence in a dual-replicon cell that initiates mitosis but fails to undergo cytokinesis, resulting in a multinucleate or multilobed nucleated cell. Thus, cytokinesis and partitioning of viral RNA to the daughter cells does not appear to be necessary for the genetic bottleneck; instead, the structural changes that occur in the host cell during mitosis may be the cause of the genetic bottleneck. In summary, mitosis of dual-replicon cells appears to restrict HCV genome recombination by promoting the elimination of one of the two viral genomes. This elimination depends on compartmentalization of the HCV genomes.
Decay of dual-replicon cells is biased toward the more-fit replicon.
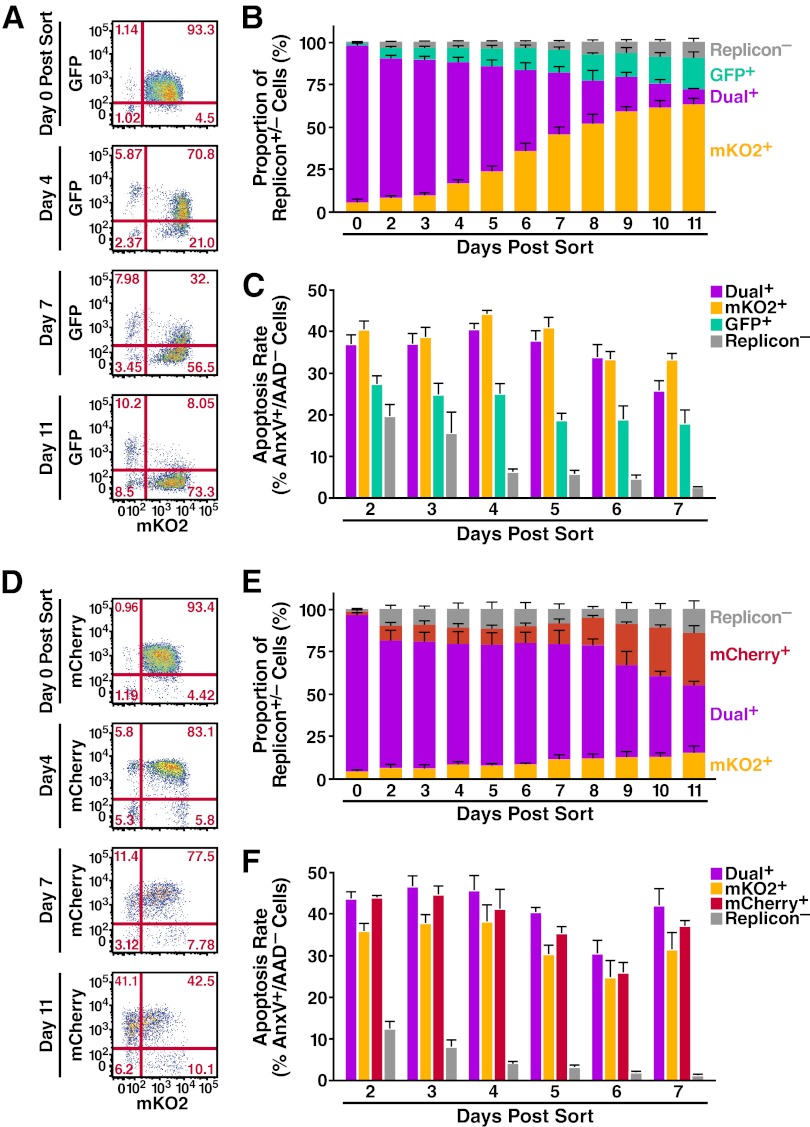
The previous analysis of the decay phenomenon in dual-replicon cells was performed using only replicons that were essentially equally replication competent. Although unexpected, the fitness differences of the Jc1/ΔXFP-BSD replicons permitted determination of whether the decay of dual-replicon cells is biased toward the replicon that accumulates higher levels of viral RNA. We predicted that the replicon with higher RNA levels would be more likely to survive the bottleneck. Huh7.5 cells were transfected with Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD or with Jc1/ΔmCherry-BSD and Jc1/ΔmKO2-BSD and then separated by FACS into dual-replicon, single-replicon, and replicon-negative populations (see Fig. S4 and S5 in the supplemental material). After isolation, blasticidin selection was used to prevent the confounding outgrowth of replicon-negative cells. Beginning 48 h postisolation, cells were stained with 7-AAD and annexin-V APC and analyzed by flow cytometry for GFP/mKO2/mCherry/7-AAD/annexin-V APC fluorescence.
Replicons that accumulate higher levels of RNA have an advantage in the decay process (Fig. 1 and 7A, B, D, E). The replicons, in order of increasing RNA accumulation, are Jc1/ΔGFP-BSD < Jc1/ΔmKO2-BSD < Jc1/ΔmCherry-BSD. In the decay process, the same order is observed: Jc1/ΔmKO2-BSD has the advantage over Jc1/ΔGFP-BSD, and Jc1/ΔmCherry-BSD has the advantage over Jc1/ΔmKO2-BSD. While the advantage of Jc1/ΔmKO2-BSD over Jc1/ΔGFP-BSD was quite marked, the advantage of Jc1/ΔmCherry-BSD over Jc1/ΔmKO2-BSD was not as pronounced (Tables 3 and 4). This may reflect the smaller difference in Jc1/ΔmCherry-BSD and Jc1/ΔmKO2-BSD RNA levels (Fig. 1C). Further, the decay process can apparently be slowed when the overall load of viral RNA is higher, as more dual-replicon cells are observed by day 11 when Jc1/ΔmCherry-BSD and Jc1/ΔmKO2-BSD are used than when Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD are used.
Fig 7.
Bias in decay of Jc1/ΔXFP-BSD dual-replicon-containing cells. Huh7.5 cells were transfected with the following RNAs: Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD (n = 5 independent experiments) (A, B, C) and Jc1/ΔmCherry-BSD and Jc1/ΔmKO2-BSD (n = 3 independent experiments) (D, E, F). Forty-eight hours later, the following populations were isolated by FACS: single-replicon-containing cells, dual-replicon cells, and replicon-negative cells. Replicon-containing cells were kept under selection with 10 μg/ml blasticidin after FACS isolation, with medium changed for all cells every 24 h. Cells were collected and stained with 7-AAD and annexin-V APC at the given time points. (A, D) Representative flow cytometry plots demonstrating the decay of each dual-replicon population. (B, E) Graphic representation of decay of dual-replicon cells. (C, F) Apoptosis rates in each isolated population of cells. Apoptosis rates were assessed as described for Fig. 4. Note that in each case, the bias in the decay of the dual-replicon cells is toward the replicon with higher RNA levels (Fig. 1). Error bars indicate +SEM.
Table 3.
Statistical significance of the differences in the proportions of replicon-positive cells in dual-replicon decay experimentsa
| Transfection: Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD | |||
|---|---|---|---|
| No. of days postsorting | Dual+ vs. GFP+ | Dual+ vs. mKO2+ | mKO2+ vs. GFP+ |
| 0 | P < 0.0001 | P < 0.0001 | NS |
| 2 | P < 0.0001 | P < 0.0001 | NS |
| 3 | P < 0.0001 | P < 0.0001 | NS |
| 4 | P < 0.0001 | P < 0.0001 | NS |
| 5 | P < 0.0001 | P < 0.0001 | P < 0.05 |
| 6 | P < 0.0001 | NS | P < 0.0001 |
| 7 | P < 0.0001 | NS | P < 0.0001 |
| 8 | NS | P < 0.0001 | P < 0.0001 |
| 9 | NS | P < 0.0001 | P < 0.0001 |
| 10 | NS | P < 0.0001 | P < 0.0001 |
| 11 | NS | P < 0.0001 | P < 0.0001 |
Two-way ANOVA with Bonferroni's multiple-comparison correction was used to analyze the difference in proportions of each replicon-positive cell population. NS, nonsignificant; vs., versus.
Table 4.
Statistical significance of the differences in the proportions of replicon-positive cells in dual-replicon decay experimentsa
| Transfection: Jc1/ΔmKO2-BSD and Jc1/ΔmCherry-BSD | |||
|---|---|---|---|
| No. of days postsorting | Dual+ vs. mKO2+ | Dual+ vs. mCherry+ | mKO2+ vs. mCherry+ |
| 0 | P < 0.0001 | P < 0.0001 | NS |
| 2 | P < 0.0001 | P < 0.0001 | NS |
| 3 | P < 0.0001 | P < 0.0001 | NS |
| 4 | P < 0.0001 | P < 0.0001 | NS |
| 5 | P < 0.0001 | P < 0.0001 | NS |
| 6 | P < 0.0001 | P < 0.0001 | NS |
| 7 | P < 0.0001 | P < 0.0001 | NS |
| 8 | P < 0.0001 | P < 0.0001 | NS |
| 9 | P < 0.0001 | P < 0.0001 | NS |
| 10 | P < 0.0001 | P < 0.001 | P < 0.01 |
| 11 | P < 0.0001 | NS | P < 0.05 |
Two-way ANOVA with Bonferroni's multiple-comparison correction was used to analyze the difference in proportions of each replicon-positive cell population. NS, nonsignificant; vs., versus.
Consistent replication of each replicon over the time course is demonstrated by the fact that isolated single-replicon populations remained >75% HCV positive over the period of study (see Fig. S4 and S5 in the supplemental material). To ensure that the blasticidin selection pressure was not too high for the less-fit replicons, live cell counts of isolated single- and dual-replicon cultures are shown in Fig. S6 in the supplemental material. Indeed, the less-fit replicon-containing cells actually survived/proliferated to a higher level.
Importantly, in both comparisons made between alternately fit replicons, the bias in decay of dual-replicon cells was toward the replicon that induced higher apoptosis rates (Fig. 7C and F) and lower proliferation rates in host cells (see Fig. S6 in the supplemental material). Again, when comparing the Jc1/ΔmCherry-BSD and Jc1/ΔmKO2-BSD replicons, the differences in apoptosis rates and proliferation were not significant (Tables 5 and 6). Apart from demonstrating that the level of RNA of each replicon is important in determining which replicon is lost following mitosis, these results conclusively demonstrate that contaminating single-replicon cells play no role in the observed decay. Otherwise, the replicon inducing more apoptosis and less proliferation in host cells would be expected to be at a disadvantage. In conclusion, the decay process is biased in cases where one replicon has the advantage over the other replicon in terms of a greater number of viral RNAs per cell. Additionally, the kinetics of decay are faster when the overall numbers of viral RNA per cell are lower.
Table 5.
Statistical significance of differences in apoptosis rates of dual-, single-, or replicon-negative cellsa
| Transfection: Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD | ||||||
|---|---|---|---|---|---|---|
| No. of days postsorting | Dual+ vs. GFP+ | Dual+ vs. mKO2+ | Dual+ vs. replicon− | GFP+ vs. mKO2+ | GFP+ vs. replicon− | mKO2+ vs. replicon− |
| 2 | ns | NS | P < 0.0001 | P < 0.01 | NS | P < 0.0001 |
| 3 | P < 0.05 | NS | P < 0.0001 | P < 0.01 | NS | P < 0.0001 |
| 4 | P < 0.001 | NS | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| 5 | P < 0.0001 | NS | P < 0.0001 | P < 0.0001 | P < 0.05 | P < 0.0001 |
| 6 | P < 0.001 | NS | P < 0.0001 | P < 0.01 | P < 0.01 | P < 0.0001 |
| 7 | ns | NS | P < 0.0001 | P < 0.001 | P < 0.01 | P < 0.0001 |
Two-way ANOVA with Bonferroni's multiple-comparison correction was used to analyze the difference in apoptosis rates of each replicon-positive cell population. NS, nonsignificant; vs., versus.
Table 6.
Statistical significance of differences in apoptosis rates of dual-, single-, or replicon-negative cellsa
| Transfection: Jc1/ΔmKO2-BSD and Jc1/ΔmCherry-BSD | ||||||
|---|---|---|---|---|---|---|
| No. of days postsorting | Dual+ vs. mKO2+ | Dual+ vs. mCherry+ | Dual+ vs. replicon− | mKO2+ vs. mCherry+ | mKO2+ vs. replicon− | mCherry+ vs. replicon− |
| 2 | NS | NS | P < 0.0001 | NS | P < 0.0001 | P < 0.0001 |
| 3 | NS | NS | P < 0.0001 | NS | P < 0.0001 | P < 0.0001 |
| 4 | NS | NS | P < 0.0001 | NS | P < 0.0001 | P < 0.0001 |
| 5 | NS | NS | P < 0.0001 | NS | P < 0.0001 | P < 0.0001 |
| 6 | NS | NS | P < 0.0001 | NS | P < 0.0001 | P < 0.0001 |
| 7 | NS | NS | P < 0.0001 | NS | P < 0.0001 | P < 0.0001 |
Two-way ANOVA with Bonferroni's multiple-comparison correction was used to analyze the difference in apoptosis rates of each replicon-positive cell population. NS, nonsignificant; vs., versus.
Transition of dual-replicon cells leads to explicit loss of viral genomes.
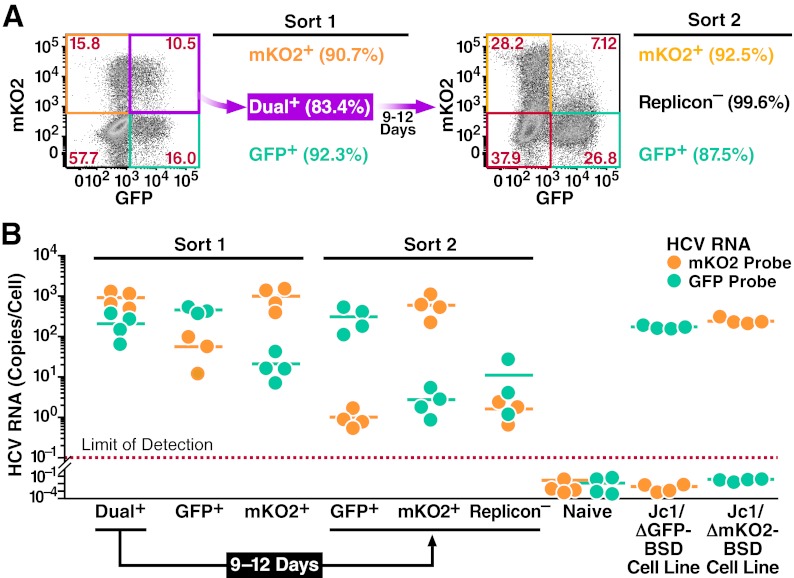
The decay of dual-replicon into single-replicon cells was previously measured by loss of fluorescence conferred by an NS5A-fluorophore fusion. We elected to further study this transition using an independent method since cryptic replication can occur below the limits of fluorescent detection. First, dual-replicon cells were isolated and allowed to transition into single-replicon cells, followed by isolation of the two single-replicon populations using flow cytometry. The relative level of each viral genome was then measured by quantitative RT-PCR. As replicon-negative cells quickly dominate in cultures of Jc1/ΔXFP-transfected cells, we used the Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD replicons, where blasticidin selection can minimize the numbers of replicon-negative cells. For these cultures, the selection pressure was less stringent, as new blasticidin-containing medium was added every 3 days, compared to every day in the earlier experiments. This allowed us to maximize the number of cells obtained at the end of the culture. However, this was not sufficient to keep replicon-negative cells from continuing to proliferate and apparently erased the advantage of the Jc1/ΔmKO2-BSD over the Jc1/ΔGFP-BSD replicon in the decay process (Fig. 8A).
Fig 8.
Explicit loss of viral RNA during the decay process. Huh7.5 cells were transfected with Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD RNA. Forty-eight hours later, the following populations were isolated by FACS (Sort 1): Jc1/ΔGFP-BSD-only replicon-containing cells, Jc1/ΔmKO2-BSD-only replicon-containing cells, and dual-replicon cells. The dual-replicon cells were further cultured for 9 days to allow decay to occur, and replicon-negative and single-replicon cells were isolated as before (Sort 2). The purity of each sorted population is indicated in parentheses. (A) Flow cytometric analysis of cells used for Sorts 1 and 2. Gates shown represent those used for isolation of each population. (B) Loss of viral RNA of one species during the decay process. RNA was extracted from each isolated population as well as naïve Huh7.5, stable Jc1/ΔGFP-BSD, and stable Jc1/ΔmKO2-BSD replicon-containing cell lines. Quantitative RT-PCR was used to determine the level of Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD RNA (mKO2/GFP probes). A GAPDH probe was used for normalization of the RNA samples, using the stable Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD cell lines as calibrators. Each data point is shown, with lines indicating the mean (n = 4 independent experiments).
After separation of the indicated populations in Sorts 1 and 2 (Fig. 8A; see Fig. S3C in the supplemental material), RNA was isolated from cells, reverse transcribed, and subjected to quantitative PCR using mKO2/GFP probes specific for each viral genome and a GAPDH probe to normalize for RNA quantities. Negative and positive control RNAs were isolated from untransfected Huh7.5 cells as well as Jc1/ΔmKO2-BSD and Jc1/ΔGFP-BSD stable-replicon cell lines (expanded for >45 days posttransfection under blasticidin selection; data not shown). As shown in Fig. 8B, following the decay of dual-replicon cells into single-replicon populations, Jc1/ΔGFP-BSD RNAs were >100-fold more abundant in GFP+ cells than in mKO2+ cells, and vice versa for Jc1/ΔmKO2-BSD RNA in mKO2+ versus GFP+ cells. Indeed, the level of Jc1/ΔGFP-BSD RNA in mKO2+ cells and the level of Jc1/ΔmKO2-BSD RNA in GFP+ cells approximated that found in the “replicon-negative” population, and this likely represents the level of contaminating cells in the culture following FACS isolation. The attempt was made in Sort 2 to reisolate dual-replicon cells following the decay period; due to the small numbers of these cells, we successfully reisolated dual-replicon cells only once for RNA analysis. In this case, the levels of Jc1/ΔmKO2-BSD and Jc1/ΔGFP-BSD RNA were comparable to the levels in the Sort 2 isolation of mKO2+ and GFP+ cells, respectively. These studies indicate that the decay of dual-replicon into single-replicon cells is associated with at least a 100-fold reduction in the RNA of one of the two replicon genomes. Hence, the decay phenomenon results in an explicit loss of replicon RNA.
DISCUSSION
In this study, we identify a new mechanism for intracellular competition between different HCV strains. We demonstrate that cells replicating two or more congenic HCV replicons decay over time into a single-replicon state and that this process is propelled by cellular mitosis. Importantly, the decay of isolated dual-replicon cells to a single-replicon state is not explained by a selective proliferation of a few contaminating single-replicon cells. Although HCV infection can induce cytotoxic effects and cell cycle abnormalities, these effects were not more prominent in dual-replicon than in single-replicon cells. The decay of dual-replicon cells seems instead to be due to a genetic bottleneck limiting the diversity of HCV strains. On a population level, when congenic, equally fit dual-replicon cells decay into a single-replicon state, neither strain dominates, indicating that this is generally a random phenomenon. However, we observe a clear bias in the dual-replicon decay when using replicons that accumulate different amounts of viral RNA, indicating that this process is influenced by viral fitness. Of note, when examining isolated dual-replicon cells during mitosis, the decay phenomenon tends to favor the same strain in both daughter cells, indicating that the reduction in genetic diversity is similar in both daughters. This decay phenomenon likely explains previous findings that competition can occur between HCV replicons, whereby the presence of one replicon reduces the replication levels of another replicon (20).
As the genetic bottleneck appears to occur every time the host cell divides, this represents a sequential bottleneck for HCV diversity in a population of dividing host cells. Sequential bottlenecks are well known in population genetics to lead to the loss of even beneficial mutations (31, 32). A population of two equally fit HCV replicon strains (e.g., Jc1/ΔGFP and Jc1/ΔmCherry) within one cell can be considered to be a haploid asexual population with a neutral difference at one locus. In this type of population, genetic drift will eventually lead to fixation of Jc1/ΔGFP or Jc1/ΔmCherry in a host cell, even in the absence of a bottleneck. The average number of generations, t̄1(p), required for fixation is given by the equation t̄1(p) = −(1/p)[2Ne(1 − p) ln (1 − p)] (33, 34). A new generation occurs each time the host cell divides (∼1.3 days in naïve Huh7.5 cells), and the frequency of one replicon “allele” initially is p = 0.5, since both have equal amounts of RNA. The “effective breeding population size,” Ne, represents the number of viral RNAs that populate the new generation of daughter cells. In the absence of a bottleneck, Ne will be ∼1,000 viral RNAs (Fig. 1), meaning that fixation will occur in about 5 years! However, according to our data, we can very conservatively estimate the average fixation at ∼9 days, meaning that one-half of the dual-replicon cells remain. This gives a value for Ne of ∼5, or the population bottleneck that survives mitosis at approximately five viral RNAs.
Of note, the probability that a neutral allele goes to fixation is simply the initial prevalence, p. For equally fit replicons, where p is equal to 0.5, this suggests equal numbers of single-replicon cells resulting from decay. However, for replicons that accumulate different amounts of RNA, the RNA levels can be used to calculate the expected numbers of single-replicon cells. The peak viral RNA accumulations 2 days after transfection for Jc1/ΔGFP-BSD, Jc1/ΔmKO2-BSD, and Jc1/ΔmCherry-BSD are 155, 549, and 845 copies/cell, respectively. This gives p = 0.220 for Jc1/ΔGFP-BSD in Jc1/ΔGFP-BSD and Jc1/ΔmKO2-BSD dual-replicon cells [155/(155 + 549)], meaning that 22.0% of the single-replicon cells resulting from the decay should be Jc1/ΔGFP-BSD positive. The actual measured proportion is 22.5% at day 11 (Fig. 7). In the case of Jc1/ΔmKO2-BSD in Jc1/ΔmKO2-BSD and Jc1/ΔmCherry-BSD dual-replicon cells, p = 0.394. The actual measured proportion of Jc1/ΔmKO2-BSD single-replicon cells at day 11 is in close agreement at 38.7%.
An important caveat to the hypothesis that a genetic bottleneck can occur during the HCV life cycle is that genomes of different viral strains must be compartmentalized separately. HCV RNA replication occurs in specialized replication complexes (RCs) termed the “membranous web,” which are thought to be formed by invaginations of the ER membrane (35–37). If viral genomes from multiple strains were fully mixed in the RCs, it would be difficult to understand how any process could specifically lead to loss of the genomes of one strain. However, of the RC components NS3, NS4B, NS5A, and NS5B, only the highly diffusible NS5A protein is capable of high levels of transcomplementation between defective replicons (19, 20), similar to what is observed for the related pestivirus bovine diarrhea virus (38). However, a lack of transcomplementation with specific NS5A mutants has demonstrated that even NS5A has some activity that is exerted only in cis (39). These studies thus demonstrate that viral genomes must be sequestered separately; otherwise, functional RC components could replicate defective genomes in trans. A more recent study using NS4B-defective replicons demonstrated low levels of transcomplementation; however, the authors postulate that this occurs only when two viral RNAs are incorporated into a newly formed RC (40).
The mechanism underlying this viral winnowing during mitosis is unclear. We suspect that structural changes within the ER or cytoskeletal network occurring during mitosis disrupt the niches normally occupied by HCV RCs. Major structural changes occur in the ER during mitosis, including the stripping of ribosomes and, controversially, a loss of either ER cisternae (28) or ER tubules (27). Interestingly, HCV nonstructural proteins preferentially localize to ER cisternae near mitochondria (41), and HCV RCs preferentially reside within cholesterol-rich lipid rafts (42–45). Since HCV RNA replication occurs on specialized regions of the ER membrane, many of the RCs in a productively infected cell may be disrupted during the major ER structural changes associated with mitosis. Alternatively, the rearrangement of the cytoskeletal network during mitosis (46) might be involved in the genetic bottleneck. The actin and microtubule networks are both necessary for HCV RNA replication (47), NS3 and NS5A specifically associate with tubulin and actin, and depolymerization of either leads to loss of the normal punctate pattern of RC localization (48). The loss of normal connections between HCV RCs or the ER with the cytoskeletal network during mitosis may indeed be responsible for this mitotic effect. It will be interesting in the future to examine the ultrastructure of HCV RCs by electron microscopy, to determine if there is a visible disruption during mitosis.
Superinfection exclusion in HCV infection has been characterized as host cells productively infected with one HCV strain becoming refractory to further HCV infection within 48 h after infection (11, 12). Superinfection exclusion was originally postulated to occur at the level of RNA replication or translation and not at the level of viral entry, since HIV particles pseudotyped with an HCV envelope showed no defect in viral fusion with HCV-infected cells (11). However, other work has called into question whether viral entry does play a role in superinfection exclusion. Some studies have found that viral receptors (claudin-1, NPC1L1, and CD81) are downregulated upon infection (12, 49, 50). However, another study reported upregulation of claudin-1 (51), and another found mixed effects (occludin and LDL-R downregulation; claudin-1 upregulation) (52). Regardless of the effects HCV infection has on viral receptor expression, a postentry block to further viral RNA replication has been well demonstrated in cells replicating HCV RNA (11, 12, 20, 53). This postentry block is thought to involve sequestration of one or more limiting host factor(s) or occupancy of replication sites, although the nature of these factor(s) or replicative niches remains to be characterized.
The genetic bottleneck in HCV genomic diversity that we observe may have some additional mechanistic parallels to the previously described postentry superinfection block. The genetic bottleneck occurring during mitosis may similarly result in one strain dominating because it is able to appropriate key host factors or fill replication sites emptied during the process of cell division. Importantly, both the genetic bottleneck and superinfection exclusion act even when viral strains are equally fit. Superinfection exclusion occurs between congenic equally fit strains of HCV, and the genetic bottleneck causes decay of cells dually infected with congenic equally fit strains of HCV. In the case of the genetic bottleneck, however, coinfection with alternately fit strains will demonstrate a bias over time toward the strain that accumulates higher levels of viral RNA. Importantly, the genetic bottleneck will act to eliminate one or more viral strains even if multiple infections occur within the 48-hour window when superinfection is permissible.
The existence of multiple tiers of intracellular competition between HCV strains has important implications for HCV biology. Each of these competition mechanisms serves to limit the time in which a cell is productively coinfected. Limiting productive coinfection reduces the chances of recombination between HCV strains and the transfer of drug resistance or adaptive mutations between viral strains. Indeed, intra- and intergenotypic recombinant strains of HCV are rare even among patients with multiple infections (54–56), although a few examples have been detected (57–61). Interestingly, it has been suggested that coinfection with multiple strains of HCV leads to exacerbation of chronic HCV disease correlates (62). Multiple levels of competition may help to explain the recent finding that when HCV-infected patients are transplanted with HCV-infected liver grafts, one strain (either donor or recipient) exhibits a dominance within as little as 1 day posttransplantation (63). Naturally, in a patient setting, differences in viral fitness come into play, instituting an additional level of competition between HCV strains. The relative dearth of natural recombinants of HCV strains may be a result of the interplay of the various levels of competition between strains in multitypic infections.
The in vivo relevance of a mitotic genetic bottleneck is as yet unclear, as most hepatocytes are not actively cycling. We further demonstrate (Fig. 4; see Fig. S6 in the supplemental material) that cellular division is retarded in host cells replicating HCV genomes, in line with previous findings (21, 24). Cell cycle arrest and decreased proliferation would act to slow the decay of multiply infected cells by this genetic bottleneck in vivo. Notably, one study found that HCV-infected cells in vitro are arrested specifically at the G2/M boundary (26), suggesting that HCV may have evolved a means of avoiding this bottleneck. On the other hand, some studies have indicated that HCV genome replication is enhanced in proliferating cells (64, 65) and hepatocyte proliferation increases in chronic HCV infection (66, 67). Additionally, the pathway leading from chronic HCV infection to fibrosis, cirrhosis, and hepatocellular carcinoma is intimately linked with an increase in hepatocyte proliferation and turnover (68, 69). A number of studies have suggested that this increase in hepatocyte proliferation is linked to the increased inflammation present in chronic HCV infection (70–73), raising the possibility that this bottleneck may have a greater role in vivo during the latter stages of HCV infection. Interestingly, alpha interferon (IFN-α), part of the present standard of care for chronic HCV treatment, has been demonstrated to induce G0/G1-phase cell cycle arrest (74, 75). In the context of the mitosis-associated bottleneck, IFN-α treatment would potentially act to retain a higher level of HCV diversity or facilitate viral recombination in treated patients.
A singular strength of HCV is its genomic diversity, leading to the emergence of immune escape variants and drug-resistant strains. Our results, aside from their potential role in elucidating the natural biology of HCV infection, suggest that targeting the stability of the HCV replication complex or mimicking the state of the cell in mitosis, aside from any antiviral effects, may act to nonspecifically reduce the quasispecies diversity of HCV. Hopefully, future studies will demonstrate to what extent these multiple levels of competition act on viral quasispecies diversity and recombination in patients. Understanding this process and learning how to exploit it may have important implications for future drug and vaccine development (55).
Supplementary Material
ACKNOWLEDGMENTS
We thank Ralf Bartenschlager (University of Heidelberg) for the Jc1 construct, Charles Rice (Rockefeller University) for Huh7.5 cells, Takaji Wakita (National Institute of Infectious Disease, Japan) for the JFH1 construct, and Matthew Spindler for the pSicoR construct. We thank Kurt Thorn and the Nikon Imaging Center at UCSF for assistance with microscopic analysis. We are grateful to Jane Gordon, Tara Rambaldo, and Sarah Elmes from the UCSF Laboratory for Cell Analysis, as well as Marielle Cavrois and the Gladstone Flow Cytometry Core, for assistance with flow cytometric assays. We thank Jill Dunham for critical reading of the manuscript, John Carroll for help in preparation of the figures, and members of the Greene laboratory for helpful discussions and support.
This work was supported by the Gladstone Institutes and funds from the U.S. Public Health Service (T32: A1060537-05 [UCSF Program in Microbial Pathogenesis and Host Defense]).
Footnotes
Published ahead of print 24 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01047-12.
REFERENCES
- 1. Lauer GM, Walker BD. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41–52 [DOI] [PubMed] [Google Scholar]
- 2. Flamm SL. 2003. Chronic hepatitis C virus infection. JAMA 289:2413–2417 [DOI] [PubMed] [Google Scholar]
- 3. Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 4. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64:334–339 [DOI] [PubMed] [Google Scholar]
- 7. Bartenschlager R, Sparacio S. 2007. Hepatitis C virus molecular clones and their replication capacity in vivo and in cell culture. Virus Res. 127:195–207 [DOI] [PubMed] [Google Scholar]
- 8. Dentzer TG, Lorenz IC, Evans MJ, Rice CM. 2009. Determinants of the hepatitis C virus nonstructural protein 2 protease domain required for production of infectious virus. J. Virol. 83:12702–12713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, Hoegh AM, Bukh J. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364–377 [DOI] [PubMed] [Google Scholar]
- 10. Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, Bartenschlager R. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schaller T, Appel N, Koutsoudakis G, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81:4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tscherne DM, Evans MJ, von Hahn T, Jones CT, Stamataki Z, McKeating JA, Lindenbach BD, Rice CM. 2007. Superinfection exclusion in cells infected with hepatitis C virus. J. Virol. 81:3693–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kato T, Date T, Murayama A, Morikawa K, Akazawa D, Wakita T. 2006. Cell culture and infection system for hepatitis C virus. Nat. Protoc. 1:2334–2339 [DOI] [PubMed] [Google Scholar]
- 15. Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 16. Sainz B, Jr, Chisari FV. 2006. Production of infectious hepatitis C virus by well-differentiated, growth-arrested human hepatoma-derived cells. J. Virol. 80:10253–10257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parsley TB, Chen B, Geletka LM, Nuss DL. 2002. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell 1:401–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Appel N, Herian U, Bartenschlager R. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79:896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Evans MJ, Rice CM, Goff SP. 2004. Genetic interactions between hepatitis C virus replicons. J. Virol. 78:12085–12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deng L, Adachi T, Kitayama K, Bungyoku Y, Kitazawa S, Ishido S, Shoji I, Hotta H. 2008. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J. Virol. 82:10375–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mateu G, Donis RO, Wakita T, Bukh J, Grakoui A. 2008. Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death. Virology 376:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blackham S, Baillie A, Al-Hababi F, Remlinger K, You S, Hamatake R, McGarvey MJ. 2010. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J. Virol. 84:5404–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, Katze MG. 2009. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 5:e1000269 doi:10.1371/journal.ppat.1000269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Engeland M, Ramaekers FCS, Schutte B, Reutelingsperger CPM. 1996. A novel assay to measure loss of plasma membrane asymmetry during apoptosis of adherent cells in culture. Cytometry 24:131–139 [DOI] [PubMed] [Google Scholar]
- 26. Kannan RP, Hensley LL, Evers LE, Lemon SM, McGivern DR. 2011. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J. Virol. 85:7989–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu L, Ladinsky MS, Kirchhausen T. 2009. Cisternal organization of the endoplasmic reticulum during mitosis. Mol. Biol. Cell 20:3471–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Puhka M, Vihinen H, Joensuu M, Jokitalo E. 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179:895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132:487–498 [DOI] [PubMed] [Google Scholar]
- 30. Tada S. 2007. Cdt1 and geminin: role during cell cycle progression and DNA damage in higher eukaryotes. Front. Biosci. 12:1629–1641 [DOI] [PubMed] [Google Scholar]
- 31. Edwards CT, Holmes EC, Wilson DJ, Viscidi RP, Abrams EJ, Phillips RE, Drummond AJ. 2006. Population genetic estimation of the loss of genetic diversity during horizontal transmission of HIV-1. BMC Evol. Biol. 6:28 doi:10.1186/1471-2148-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wahl LM, Gerrish PJ. 2001. The probability that beneficial mutations are lost in populations with periodic bottlenecks. Evolution 55:2606–2610 [DOI] [PubMed] [Google Scholar]
- 33. Hartl DL, Clark AG. 1997. Principles of population genetics, 3rd ed Sinauer Associates, Sunderland, MA [Google Scholar]
- 34. Kimura M, Ohta T. 1969. The average number of generations until fixation of a mutant gene in a finite population. Genetics 61:763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lindenbach BD. 2011. Understanding how hepatitis C virus builds its unctuous home. Cell Host Microbe 9:1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sklan EH, Glenn JS. 2006. HCV NS4B: from obscurity to central stage, p 245–266 In Tan SL. (ed), Hepatitis C viruses: genomes and molecular biology. Horizon Bioscience, Norfolk, United Kingdom: [PubMed] [Google Scholar]
- 38. Grassmann CW, Isken O, Tautz N, Behrens SE. 2001. Genetic analysis of the pestivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J. Virol. 75:7791–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fridell RA, Qiu D, Valera L, Wang C, Rose RE, Gao M. 2011. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J. Virol. 85:7312–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones DM, Patel AH, Targett-Adams P, McLauchlan J. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 83:2163–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mottola G, Cardinali G, Ceccacci A, Trozzi C, Bartholomew L, Torrisi MR, Pedrazzini E, Bonatti S, Migliaccio G. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31–43 [DOI] [PubMed] [Google Scholar]
- 42. Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461 [DOI] [PubMed] [Google Scholar]
- 43. Gao L, Aizaki H, He JW, Lai MM. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mannova P, Fang R, Wang H, Deng B, McIntosh MW, Hanash SM, Beretta L. 2006. Modification of host lipid raft proteome upon hepatitis C virus replication. Mol. Cell. Proteomics 5:2319–2325 [DOI] [PubMed] [Google Scholar]
- 45. Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heng YW, Koh CG. 2010. Actin cytoskeleton dynamics and the cell division cycle. Int. J. Biochem. Cell Biol. 42:1622–1633 [DOI] [PubMed] [Google Scholar]
- 47. Bost AG, Venable D, Liu L, Heinz BA. 2003. Cytoskeletal requirements for hepatitis C virus (HCV) RNA synthesis in the HCV replicon cell culture system. J. Virol. 77:4401–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lai CK, Jeng KS, Machida K, Lai MM. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J. Virol. 82:8838–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. 2009. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 83:2011–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sainz B, Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. 2012. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 18:281–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reynolds GM, Harris HJ, Jennings A, Hu K, Grove J, Lalor PF, Adams DH, Balfe P, Hubscher SG, McKeating JA. 2008. Hepatitis C virus receptor expression in normal and diseased liver tissue. Hepatology 47:418–427 [DOI] [PubMed] [Google Scholar]
- 52. Nakamuta M, Fujino T, Yada R, Aoyagi Y, Yasutake K, Kohjima M, Fukuizumi K, Yoshimoto T, Harada N, Yada M, Kato M, Kotoh K, Taketomi A, Maehara Y, Nakashima M, Enjoji M. 2011. Expression profiles of genes associated with viral entry in HCV-infected human liver. J. Med. Virol. 83:921–927 [DOI] [PubMed] [Google Scholar]
- 53. Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bernardin F, Herring B, Page-Shafer K, Kuiken C, Delwart E. 2006. Absence of HCV viral recombination following superinfection. J. Viral Hepat. 13:532–537 [DOI] [PubMed] [Google Scholar]
- 55. Blackard JT, Sherman KE. 2007. Hepatitis C virus coinfection and superinfection. J. Infect. Dis. 195:519–524 [DOI] [PubMed] [Google Scholar]
- 56. Zhou Y, Wang X, Hong G, Tan Z, Zhu Y, Lan L, Mao Q. 2010. Natural intragenotypic and intergenotypic HCV recombinants are rare in southwest China even among patients with multiple exposures. J. Clin. Virol. 49:272–276 [DOI] [PubMed] [Google Scholar]
- 57. Colina R, Casane D, Vasquez S, Garcia-Aguirre L, Chunga A, Romero H, Khan B, Cristina J. 2004. Evidence of intratypic recombination in natural populations of hepatitis C virus. J. Gen. Virol. 85:31–37 [DOI] [PubMed] [Google Scholar]
- 58. Kageyama S, Agdamag DM, Alesna ET, Leano PS, Heredia AM, Abellanosa-Tac-An IP, Jereza LD, Tanimoto T, Yamamura J, Ichimura H. 2006. A natural inter-genotypic (2b/1b) recombinant of hepatitis C virus in the Philippines. J. Med. Virol. 78:1423–1428 [DOI] [PubMed] [Google Scholar]
- 59. Kurbanov F, Tanaka Y, Avazova D, Khan A, Sugauchi F, Kan N, Kurbanova-Khudayberganova D, Khikmatullaeva A, Musabaev E, Mizokami M. 2008. Detection of hepatitis C virus natural recombinant RF1_2k/1b strain among intravenous drug users in Uzbekistan. Hepatol. Res. 38:457–464 [DOI] [PubMed] [Google Scholar]
- 60. Legrand-Abravanel F, Claudinon J, Nicot F, Dubois M, Chapuy-Regaud S, Sandres-Saune K, Pasquier C, Izopet J. 2007. New natural intergenotypic (2/5) recombinant of hepatitis C virus. J. Virol. 81:4357–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moreno P, Alvarez M, Lopez L, Moratorio G, Casane D, Castells M, Castro S, Cristina J, Colina R. 2009. Evidence of recombination in Hepatitis C Virus populations infecting a hemophiliac patient. Virol. J. 6:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kao JH, Chen PJ, Lai MY, Yang PM, Sheu JC, Wang TH, Chen DS. 1994. Mixed infections of hepatitis C virus as a factor in acute exacerbations of chronic type C hepatitis. J. Infect. Dis. 170:1128–1133 [DOI] [PubMed] [Google Scholar]
- 63. Ramirez S, Perez-del-Pulgar S, Carrion JA, Coto-Llerena M, Mensa L, Dragun J, Garcia-Valdecasas JC, Navasa M, Forns X. 2010. Hepatitis C virus superinfection of liver grafts: a detailed analysis of early exclusion of non-dominant virus strains. J. Gen. Virol. 91:1183–1188 [DOI] [PubMed] [Google Scholar]
- 64. Nelson HB, Tang H. 2006. Effect of cell growth on hepatitis C virus (HCV) replication and a mechanism of cell confluence-based inhibition of HCV RNA and protein expression. J. Virol. 80:1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Scholle F, Li K, Bodola F, Ikeda M, Luxon BA, Lemon SM. 2004. Virus-host cell interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J. Virol. 78:1513–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kronenberger B, Ruster B, Lee JH, Sarrazin C, Roth WK, Herrmann G, Zeuzem S. 2000. Hepatocellular proliferation in patients with chronic hepatitis C and persistently normal or abnormal aminotransferase levels. J. Hepatol. 33:640–647 [DOI] [PubMed] [Google Scholar]
- 67. Wilfredo Canchis P, Gonzalez SA, Isabel Fiel M, Chiriboga L, Yee H, Edlin BR, Jacobson IM, Talal AH. 2004. Hepatocyte proliferation in chronic hepatitis C: correlation with degree of liver disease and serum alpha-fetoprotein. Liver Int. 24:198–203 [DOI] [PubMed] [Google Scholar]
- 68. Kung JW, Currie IS, Forbes SJ, Ross JA. 2010. Liver development, regeneration, and carcinogenesis. J. Biomed. Biotechnol. 2010:984248 doi:10.1155/2010/984248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Levrero M. 2006. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene 25:3834–3847 [DOI] [PubMed] [Google Scholar]
- 70. Farinati F, Cardin R, D'Errico A, De Maria N, Naccarato R, Cecchetto A, Grigioni W. 1996. Hepatocyte proliferative activity in chronic liver damage as assessed by the monoclonal antibody MIB1 Ki67 in archival material: the role of etiology, disease activity, iron, and lipid peroxidation. Hepatology 23:1468–1475 [DOI] [PubMed] [Google Scholar]
- 71. Freeman A, Hamid S, Morris L, Vowler S, Rushbrook S, Wight DG, Coleman N, Alexander GJ. 2003. Improved detection of hepatocyte proliferation using antibody to the pre-replication complex: an association with hepatic fibrosis and viral replication in chronic hepatitis C virus infection. J. Viral Hepat. 10:345–350 [DOI] [PubMed] [Google Scholar]
- 72. Lake-Bakaar G, Mazzoccoli V, Ruffini L. 2002. Digital image analysis of the distribution of proliferating cell nuclear antigen in hepatitis C virus-related chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Dig. Dis. Sci. 47:1644–1648 [DOI] [PubMed] [Google Scholar]
- 73. Sarfraz S, Hamid S, Ali S, Jafri W, Siddiqui AA. 2009. Modulations of cell cycle checkpoints during HCV associated disease. BMC Infect. Dis. 9:125 doi:10.1186/1471-2334-9-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Matsuoka M, Tani K, Asano S. 1998. Interferon-alpha-induced G1 phase arrest through up-regulated expression of CDK inhibitors, p19Ink4D and p21Cip1 in mouse macrophages. Oncogene 16:2075–2086 [DOI] [PubMed] [Google Scholar]
- 75. Sangfelt O, Erickson S, Castro J, Heiden T, Gustafsson A, Einhorn S, Grander D. 1999. Molecular mechanisms underlying interferon-alpha-induced G0/G1 arrest: CKI-mediated regulation of G1 Cdk-complexes and activation of pocket proteins. Oncogene 18:2798–2810 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.