Abstract
HIV-1 R5 viruses vary extensively in their capacity to infect macrophages. R5 viruses that confer efficient infection of macrophages are able to exploit low levels of CD4 for infection and predominate in brain tissue, where macrophages are a major target for infection. HIV-1 R5 founder viruses that are transmitted were reported to be non-macrophage-tropic. Here, we investigated the sensitivities of macrophage-tropic and non-macrophage-tropic R5 envelopes to neutralizing antibodies. We observed striking differences in the sensitivities of Env+ pseudovirions to soluble CD4 (sCD4) and to neutralizing monoclonal antibodies (MAbs) that target the CD4 binding site. Macrophage-tropic R5 Envs were sensitive to sCD4, while non-macrophage-tropic Envs were significantly more resistant. In contrast, all Envs were sensitive to VRC01 regardless of tropism, while MAb b12 conferred an intermediate neutralization pattern where all the macrophage-tropic and about half of the non-macrophage-tropic Envs were sensitive. CD4, b12, and VRC01 share binding specificities on the outer domain of gp120. However, these antibodies differ in their ability to induce conformational changes on the trimeric envelope and in specificity for residues on the V1V2 loop stem and β20-21 junction that are targets for CD4 in recruiting the bridging sheet. These distinct specificities of CD4, b12, and VRC01 likely explain the observed differences in Env sensitivity to inhibition by these reagents and provide an insight into the envelope mechanisms that control macrophage tropism. We present a model where the efficiency of bridging-sheet recruitment by CD4 is a major determinant of HIV-1 R5 envelope sensitivity to soluble CD4 and macrophage tropism.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) entry into cells involves interactions with CD4 and coreceptor CCR5 or CXCR4 to trigger fusion of the virus and cell membranes. In vivo, HIV-1 infection is limited mainly to cells expressing CD4 and an appropriate coreceptor. These include T cells, macrophages, and dendritic cells as well as their progenitors (1–3). In the past, HIV-1 R5 viruses that use CCR5 were described as macrophage-tropic or M-tropic, reflecting a view that such viruses infect macrophages in addition to T cells (4–6). However, studies from our group and others have shown that R5 viruses vary extensively in their capacity to infect macrophages (7–14). This variation results mainly from differences in the ability of HIV-1 to exploit low levels of CD4 on macrophages for infection (10, 12, 14, 15). Recent studies reported that founder viruses transmitted either sexually (16, 17) or via mother to child (18) were non-macrophage-tropic and that they persist in immune tissue even in late disease (10, 12, 19). Nevertheless, highly macrophage-tropic R5 variants are increasingly detected in late disease (20–22) and are predominant in brain tissue of subjects with HIV-associated neurocognitive disorders (7, 10, 12, 14).
The HIV-1 envelope glycoprotein is a trimer comprising three copies each of gp120 and transmembrane gp41. Many high-resolution crystal structures of monomeric gp120 of HIV-1 have been reported (23–28). However, all lack the V1V2 loops and carry deletions and mutated glycosylation sites, and they frequently lack the V3 loop. In addition, many represent the gp120 structure that forms after gp120 has bound CD4 (29), with the bridging sheet (usually recruited by CD4) fully assembled. The only unliganded, high-resolution structure of gp120 that is distinct from the CD4-bound forms was reported for simian immunodeficiency virus (SIV) (30). This structure shows an unformed bridging sheet and a more extended CD4 binding loop.
The different crystal structures of the gp120 monomers have been augmented by less-resolved cryo-electron tomographic images of native Env trimers on virions (31–36). These structures show that the V1V2 loops are positioned at the apex of the unliganded trimer and proximal to the V3 loops. They also reveal how the V1V2 loops move from the trimer apex when Env is activated by CD4 (31, 33). The movement of V1V2 exposes V3 and releases a determinant on the V1V2 stem for recruitment by CD4 and bridging-sheet formation, thus enabling the formation and exposure of the coreceptor binding site (26, 32). It remains less clear how variations in the structure and function of the trimer control a range of different Env properties, including R5 macrophage tropism and neutralization sensitivity.
Env determinants that control R5 macrophage tropism are complex. Dunfee et al. identified an asparagine at residue 283 which is prevalent in macrophage-tropic Envs from brain and in individuals with neurological complications (7). Residue 283 is a contact site for CD4, and an asparagine at that position confers a higher affinity for CD4 and an increased ability to infect macrophages via low CD4 (7). We confirmed the role of N283 in macrophage-tropic Env (12, 37). However, the identification of many macrophage-tropic Envs that lack N283 (and non-macrophage-tropic Envs with N283) indicates that other determinants play a significant role (13, 19). We identified complex determinants that included residues on the N-terminal flank of the CD4 binding loop as well as changes in the glycan shield that may affect the exposure of CD4 contact residues (37, 38). We also reported a single residue in the V1 loop that modulated macrophage infectivity (39) and identified determinants in the V3 loop that contribute to macrophage tropism (37). Together these observations implicate sites within or proximal to the CD4 binding site (CD4bs) and at the apex of the trimer as determinants of R5 macrophage tropism.
Here, we investigated HIV-1 R5 Envs that differ in their capacity to infect primary macrophages by measuring their sensitivities to newly described potent neutralizing monoclonal antibodies (MAbs) that target different Env sites, including the CD4bs (28, 40). In particular, we compared the potent CD4bs MAb VRC01 with b12 and soluble CD4 (sCD4). VRC01 and other highly potent CD4bs MAbs with similar specificities were reported to mimic binding of the host receptor CD4 (28, 41, 42). However, our data show that VRC01 potently neutralized pseudoviruses via all of the Envs tested, contrasting with sCD4, which was highly preferential for macrophage-tropic Envs, and with b12, which neutralized all macrophage-tropic but only some of the non-macrophage-tropic Envs that carried the b12 epitope. These distinct specificities reflect different abilities of sCD4, b12, and VRC01 to induce conformational changes and to bind determinants on the bridging sheet following attachment to the outer domain of gp120 on functional trimers (26, 28, 43–45). The different neutralization profiles and bridging-sheet specificities as well as distinct abilities to induce conformational changes in the trimer have led us to conclude that non-macrophage-tropic Envs are substantially more resistant to CD4-induced conformational changes than highly macrophage-tropic R5 variants. The ability of CD4 to recruit the bridging sheet is therefore a major determinant of HIV-1 R5 macrophage tropism.
MATERIALS AND METHODS
HIV-1 envelope glycoproteins.
We selected 48 HIV-1 R5 Envs, including many that were clearly macrophage-tropic or non-macrophage-tropic (Table 1). We included Envs that had been well characterized in our previous studies for tropism and other properties (10–12, 19, 37). Where possible, we included groups of Envs that had been amplified from single individuals and that included both macrophage-tropic and non-macrophage-tropic Envs. Mainly, we used a panel of Envs that we had previously studied (11) and included more recently amplified and characterized Envs (19).
Table 1.
HIV-1 envelope clones
| Patient | Macrophage-tropic Envs |
Non-macrophage-tropic Envs |
||
|---|---|---|---|---|
| Envelope | Origin | Envelope | Origin | |
| NA20 | B59 | Brain | LN3 | Lymph node |
| B76 | Brain | LN8 | Lymph node | |
| B501 | Brain | LN10 | Lymph node | |
| 23-14-2 | Lymph node | LN14 | Lymph node | |
| LN16 | Lymph node | |||
| NA420 | B13 | Brain | LN40 | Lymph node |
| B33 | Brain | LN85 | Lymph node | |
| B42 | Brain | |||
| NA118 | B12 | Brain | ||
| LN27 | Lymph node | |||
| LN33 | Lymph node | |||
| NA176 | B93 | Brain | ||
| NA353 | B27 | Brain | ||
| 7766 | FL1 (FL19-54-50) | Brain | SP1 (SP13-33-41) | Spleen |
| 6568 | FL1 (FL11-1-249) | Brain | SP1 (SP6-11-9) | Spleen |
| 10017 | FL1 (FL9-1-2) | Brain | SP2 (SP10-9-65) | Spleen |
| SP3 (SP9-8-57) | Spleen | |||
| CA110 | OC1 (OC58-11-9) | Brain | SP1 (SP52-13-34) | Spleen |
| P1114 | C98-15 | Plasma | C95-65 | Plasma |
| C98-18 | Plasma | C96-26 | Plasma | |
| C98-27 | Plasma | |||
| C98-28 | Plasma | |||
| C98-67 | Plasma | |||
| P43 | 380.1 | Semen | 378.2 | Blood |
| 380.4 | Semen | |||
| P3 | 164-1.4 | Blood | ||
| 180-6.4 | Blood | |||
| 196-10.1 | Semen | |||
| 197-9.3 | Semen | |||
| 199-8.5 | Semen | |||
| P31 | 350.1 | Blood | ||
| 351.6 | Blood | |||
| 308.2 | Semen | |||
| JR | JR-FL | Brain | JR-CSF | Cerebrospinal fluid |
Preparation and titration of Env+ pseudovirions.
env+ rev+ pSVIIIenv or pcDNA 3.1D/V5-His-TOPO were cotransfected into 293T cells with env− pNL43. Env+ pseudovirions were harvested after 48 h, clarified by low-speed centrifugation, and frozen as aliquots at −152°C. Env+ pseudovirions were titrated on HeLa TZM-BL cells, which carry β-galactosidase and luciferase reporter genes controlled by HIV long terminal repeat (LTR) promoters (46). Infected cells were visualized at 48 h after infection as focus-forming units (FFU) following staining for β-galactosidase activity. Since Env+ pseudovirions are capable of only a single round of replication, individual cells or small groups of divided cells were counted as foci.
Neutralization and inhibition assays.
Neutralization and inhibition assays were performed as described previously using HeLa TZM-BL cells and a luminescence readout (12).
Production of monomeric gp120.
Monomeric gp120 was produced in 293T cells. Briefly, pJW4303 carrying gp120 sequences was transfected into 293T cells using calcium phosphate. Supernatant was harvested after 48 h and clarified by low-speed centrifugation, protease inhibitor added, and aliquots frozen at −80°C.
Determination of ligand binding to monomeric gp120 by ELISA.
The amount of gp120 in 293T supernatants was estimated by titration using a capture enzyme-linked immunosorbent assay (ELISA) and by comparison to a standard concentration of IIIB gp120 (47). To estimate binding of CD4bs MAbs to gp120, serial 2-fold dilutions of each MAb were added to captured gp120. A dilution of gp120 that saturated the capture antibody was used throughout.
Statistical analyses.
Fifty percent inhibitory concentrations (IC50s) for the different MAbs, sCD4, T20, and maraviroc were estimated by GraphPad Prism 5 for Mac OS X and confirmed using a robust semiparametric regression model (48) implemented by R package drc (49). When no inhibition was observed, when inhibition failed to reach 50%, or when the model-fitting algorithms either did not converge or reported an extrapolating estimate out of the experimental ranges with a very wide confidence interval, we report the lower dose boundary (e.g., >50 μg/ml) or winsorized IC50 estimates obtained manually from Excel plotted graphs. Two-tailed, nonparametric Mann-Whitney tests were used to evaluate whether statistically significant differences existed between distributions of IC50s for the different ligands between macrophage-tropic and non-macrophage-tropic envelopes. Correlations were tested using nonparametric, two-tailed Spearman analyses.
RESULTS
Sensitivities of macrophage-tropic and non-macrophage-tropic R5 Env+ pseudovirions to neutralization by broadly active CD4 binding site monoclonal antibodies.
Highly macrophage-tropic R5 Envs are consistently detected in brain tissues of subjects with HIV-associated neurological issues (7, 9, 10, 12–14). The brain is protected by the blood-brain barrier, which limits the amount of immunoglobulin present (50–52). HIV variants replicating in the brain are thus not exposed to the high levels of neutralizing antibodies that bombard their counterparts in immune tissue. We expected that HIV variants in the brain might have evolved a more open structure so that CD4 contact residues are more efficiently exposed to enhance Env-CD4 interactions. Such Envs would then be able to confer infection of brain macrophages that express lower levels of CD4 than CD4+ T cells (53–55).
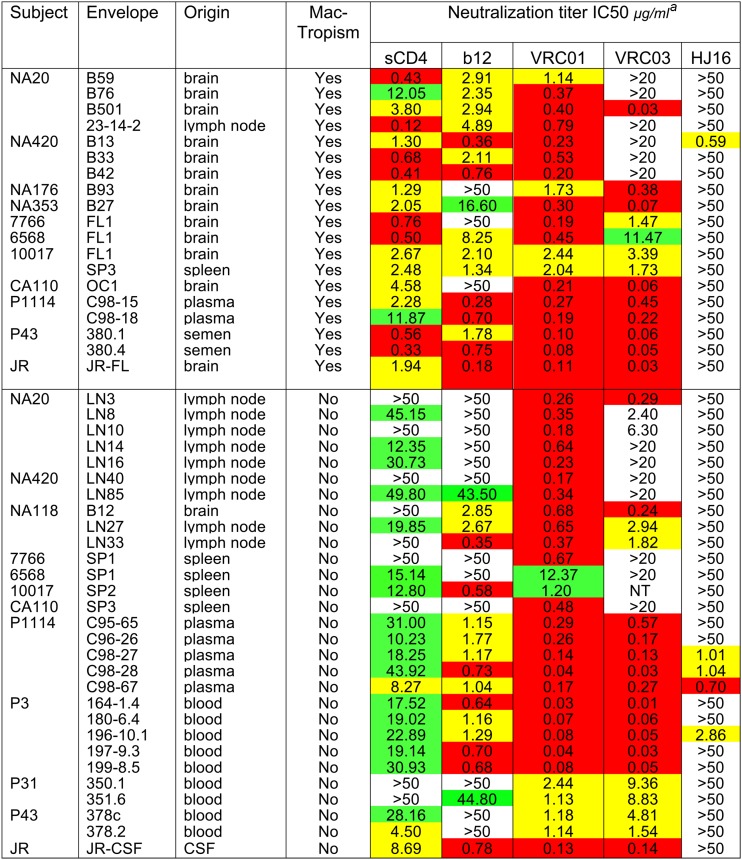
In an earlier study, we reported that R5 macrophage tropism correlated with sensitivity to sCD4 inhibition (11). Thus, pseudovirions carrying macrophage-tropic Envs were generally sensitive and non-macrophage-tropic Envs more resistant. Initially, we concluded that this observation supported a more exposed CD4bs on macrophage-tropic R5 Envs than on non-macrophage-tropic Envs. However, when we tested Env+ pseudovirions for their sensitivities to CD4bs MAbs b12 and b6, we did not detect a clear correlation with macrophage tropism (11). Here, we investigated the sensitivities of an extended panel of Env+ pseudovirions (Table 1) to newly described CD4bs MAbs, including VRC01 and VRC03 (28, 43) and HJ16 (56) and compared with b12 and sCD4 (Fig. 1 and Table 2). Our results show that the neutralization patterns for each of the different CD4bs MAbs varied considerably. They confirm a highly significant correlation between macrophage tropism and sensitivity to sCD4 but not b12. In addition, there was no correlation between macrophage infectivity and sensitivity to VRC01, VRC03, or HJ16. Moreover, VRC01 neutralized all Env+ pseudovirions tested regardless of macrophage tropism or tissue origin, while VRC03 inhibited about 70% of the pseudovirions, without a significant correlation with tropism.
Fig 1.
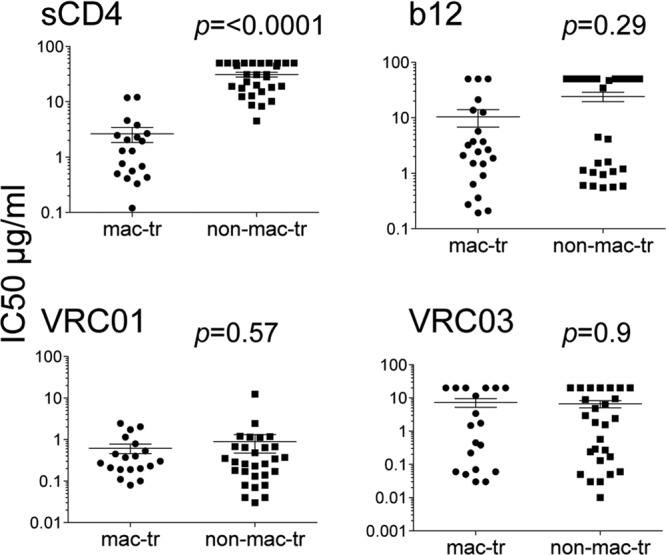
R5 macrophage-tropic Env+ pseudovirions are more sensitive to neutralization by sCD4 but not VRC01. Mann-Whitney analyses comparing macrophage-tropic and non-macrophage-tropic Env+ pseudovirions for sensitivity to sCD4 and CD4bs MAbs are shown. Only for sCD4 was there a significant difference in sensitivity between macrophage-tropic and non-macrophage-tropic Env+ pseudovirions. Nonparametric, two-tailed tests were carried out on IC50s for sCD4 and each CD4bs MAb. See also Table 2.
Table 2.
Sensitivities of HIV-1 Env+ pseudovirions to neutralization by sCD4 and CD4bs MAbs
aIC50s: red, <1.0 μg/ml; yellow, >1 μg/ml, <10 μg/ml; green, >10 μg/ml, <50 μg/ml. NT, not tested.
Env resistance to the broadly active CD4bs MAbs could be due to amino acid variation within the epitope that abrogates MAb recognition. Alternatively, it is possible that the target epitope is sterically blocked or in a different conformation in the context of the trimeric envelope spike even though it is present on the monomer. We investigated whether the b12 epitope could be detected on monomeric gp120s derived from resistant and sensitive Envs using ELISAs. Our results show that the three macrophage-tropic Envs (NA176 B93, 7766 FL1, and CA110 OC1), which resisted b12, reacted poorly in ELISAs, explaining their resistance (Fig. 2A). In contrast, monomeric gp120s from non-macrophage-tropic LN8 and LN40 bound b12 strongly, indicating that the epitope was present but either sterically protected or conformed differently on the trimer (Fig. 2A).
Fig 2.
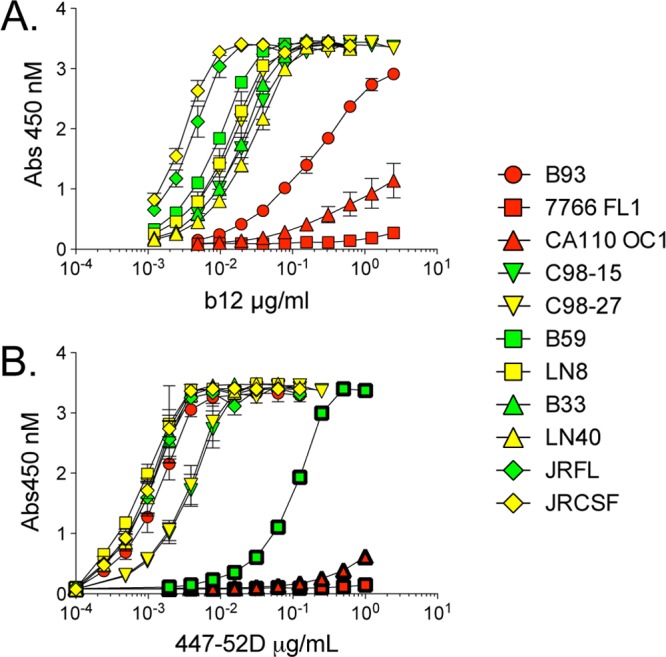
Presence and exposure of b12 and 447-52D epitopes on neutralization-sensitive and -resistant Envs. Results of ELISAs estimating CD4bs MAb b12 and V3 loop MAb 447-52D binding to monomeric gp120 derived from resistant and sensitive Envs are shown. (A) Monomeric gp120 from b12-resistant macrophage-tropic Envs (red symbols) (B93, 7766 FL1, and CA110 OC1) does not efficiently bind b12. In contrast, gp120 from b12-resistant non-macrophage-tropic Envs (yellow symbols) (C98-27, LN8, and LN40) bind as strongly as sensitive macrophage-tropic Envs (green symbols), indicating that the b12 epitope is present on the monomer but protected on the functional trimer. These data also indicate that all macrophage-tropic R5 Envs that carry the b12 epitope are sensitive to b12 (Table 2). (B) The 447-52D epitope was detected on monomeric gp120 from 447-52D neutralization-sensitive macrophage-tropic C98-15 and B33 as well as on gp120 from neutralization-resistant C98-27 and LN40 Envs derived from subjects P1114 and NA420, respectively. These data indicate that the 447-52D epitope is present on resistant envelopes C98-27 and LN40 but is hidden or in a different conformation on the trimer. Similarly, gp120s from NA20 LN8, NA176 B93, JR-FL, and JR-CSF each bound to 447-52D efficiently, even though Env+ pseudovirions were resistant to neutralization. The 447-52D epitopes of these Envs are therefore not presented on the trimer. In contrast, gp120s from NA176 B93, 7766 FL1, and NA20 B59 (symbols with thick borders) bound 447-52D inefficiently, which explained their resistance to neutralization by this MAb. These last three Envs lack the GPXR motif on the V3 crown that is targeted by 447-52D (Table 4).
These data show that macrophage-tropic Envs were all sensitive to sCD4, VRC01, and b12 (if the b12 epitope was present). In contrast, the non-macrophage-tropic Envs were much more resistant to sCD4, partially sensitive to b12, and universally sensitive to VRC01 (Fig. 1; Table 2). Since VRC01 shares overlapping binding sites with CD4 and b12 on gp120, these data are striking.
Sensitivities of macrophage-tropic and non-macrophage-tropic Envs to heterologous neutralizing antibodies in HIV-1+ sera.
If macrophage-tropic envelope trimers carry a more open structure, then they would be expected to be generally more sensitive to neutralizing antibodies in HIV-1-positive (HIV-1+) sera. To investigate this, we selected 5 HIV-1+ sera that conferred strong heterologous neutralizing activity. The results shown in Fig. 3 and Table 3 reveal that these sera neutralized both macrophage-tropic and non-macrophage-tropic Env+ pseudovirions without significant differences. The neutralization specificities of these HIV-1+ sera are not known. However, it is worth noting that there were highly significant correlations of IC50s for sera 8735-92, 30194-64, and Z01679 with those for VRC01 but not for sCD4 (P values of 0.0002. 0.0014, and 0.007, respectively, for correlations with VRC01 [Spearman analyses]).
Fig 3.
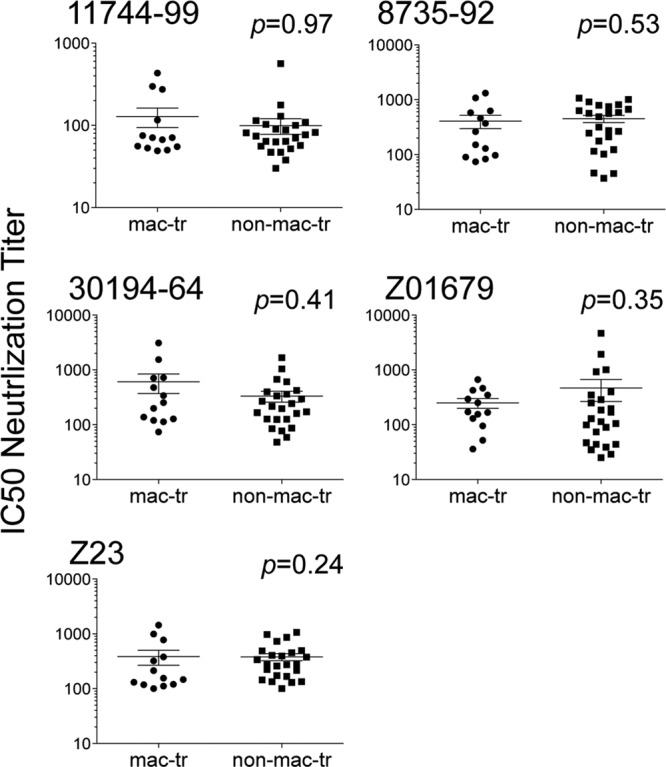
Macrophage-tropic R5 Env+ pseudovirions are not significantly more sensitive to neutralizing antibodies in HIV-1+ sera. Mann-Whitney analyses comparing macrophage-tropic and non-macrophage-tropic Env+ pseudovirions for sensitivity to HIV-1+ human sera are shown. There was no significant difference in the sensitivities of macrophage-tropic and non-macrophage-tropic Env+ pseudovirions to neutralization by 5 different HIV-1+ human sera. See also Table 3.
Table 3.
Sensitivities of HIV-1 Env+ pseudovirions to neutralization by heterologous neutralizing antibodies present in HIV-1+ sera
aIC50s: red, >1,000 reciprocal serum dilution; yellow, >100 reciprocal serum dilution, <1,000 reciprocal serum dilution; green, >10 reciprocal serum dilution, <100 reciprocal serum dilution.
Sensitivity of Env+ pseudovirions to 447-52D, PG9, and PG16 indicates that some macrophage-tropic R5 Envs have alterations in the V1, V2, and V3 loops at the apex of the trimer.
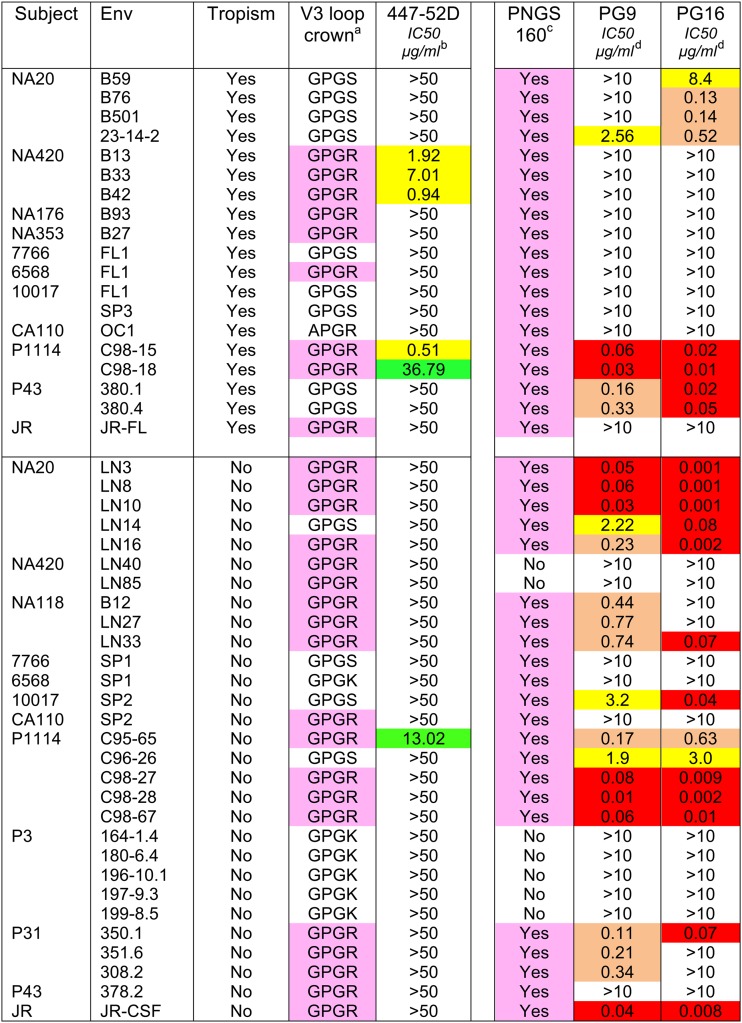
Several studies have identified residues within or proximal to the CD4bs as determinants of macrophage tropism (7, 12, 37). However, we also previously identified a conserved residue in the V1 loop that modulated macrophage infectivity for about 25% of Envs tested (39) and showed that residues within V3 contributed to macrophage tropism (37). In the unliganded Env, the V1V2 loops are located at the trimer apex close to the V3 loop. During entry, the V1V2 loops are repositioned following CD4 binding to expose V3 and allow bridging-sheet determinants on the V1V2 stem to be recruited. The identification of V1 and V3 loop residues that affect infection of macrophages via low CD4 is consistent with alterations at the apex of the trimer that facilitate V1V2 movement and bridging-sheet formation following CD4 binding. To investigate whether structural alterations at the trimer apex could be detected, we tested the sensitivities of macrophage-tropic and non-macrophage-tropic Envs to neutralizing antibodies that bind the V3 loop (447-52D binds the V3 loop crown [57]) or that recognize quaternary sites on V2 and V3 loops (PG9 and PG16 [58]).
Table 4 shows that several macrophage-tropic Envs derived from two subjects (P1114 and NA420) were sensitive to V3 loop MAb 447-52D, while all non-macrophage-tropic Envs (including related Envs from the same individuals) were resistant. Sensitive Envs included C98-15 (which we previously reported [39]) and three Envs from patient NA420 (B33, B13, and B42).
Table 4.
Sensitivities of HIV-1 Env+ pseudovirions to neutralization by the V3 loop-specific MAb 447-52D and to PG9 and PG16, which bind to a quaternary epitope that contains determinants in the V2 and V3 loops
a447-25D recognizes the GPGR motif on the crown of the V3 loop (57).
bIC50s for 447-52D: yellow, <10 μg/ml; green, >10 μg/ml, <50 μg/ml.
cThe PNG at asparagine 160 is a dominant target for PG9 and PG16 (58).
dIC50s for PG9 and PG16: red, <0.1 μg/ml; tan, >0.1 μg/ml, <1.0 μg/ml; yellow, >1.0 μg/ml, <10 μg/ml.
We next used ELISAs to assess whether resistant macrophage-tropic or non-macrophage-tropic Envs carried the 447-52D epitope on monomeric gp120 to establish the extent that the 447-52D epitope was protected from exposure on the trimer. Several gp120s derived from macrophage-tropic R5 Envs that were resistant to 447-52D failed to bind efficiently (e.g., CA110 OC1, 7766 FL1, and NA20 B59). However, for NA176 B93 and JR-FL, there was strong binding, indicating that the epitope was present but either protected within the trimer or in a conformation not recognized (Fig. 2B). Similarly, several non-macrophage-tropic gp120s tested (NA20 LN8, NA420 LN40, and P1114 C98-27) showed strong binding to 447-52D, confirming that the epitope was present on the monomer but not presented on the trimer. These binding data are also consistent with the presence or absence of the V3 loop sequence (GPXR) that represents the predominant target sequence for 447-52D (57) (Table 4).
Together, these data indicate that some of the macrophage-tropic Envs carried an exposed 447-52D epitope, consistent with increased exposure of the V3 loop crown. Nevertheless, for other highly macrophage-tropic Envs (e.g., NA176 B93) the 447-52D epitope was protected on the trimer even though it was efficiently recognized on the monomer. Exposure of the V3 loop crown is therefore apparent for some of the macrophage-tropic Envs but is not universal.
We also tested MAbs PG9 and PG16, which predominantly target a glycan (N160) at the N terminus of V2 but are strongly affected by determinants in the V3 loop (58). Our rationale was that these MAbs might also detect differences in the arrangement of V1V2 and V3 at the trimer apex. Our data show that macrophage-tropic and non-macrophage-tropic Envs from several individuals showed divergent sensitivities to PG9 or PG16 (Table 4). These sets included JR-CSF and JR-FL as well as Envs from subjects NA20 and 10017, where non-macrophage-tropic Envs were sensitive to both MAbs, while macrophage-tropic envelopes were resistant. Curiously, Envs from subject 43 showed reversed sensitivity and resistance to PG9 and PG16. Nevertheless, for Envs that carried the N160 glycan (which is critical for PG9 and PG16 binding [58]), a significant difference in sensitivity between macrophage-tropic and non-macrophage-tropic Envs was detected for PG9 (P = 0.002) but not quite for PG16 (P = 0.05) (Fig. 4). In summary, our data with 447-52D, PG9, and PG16 have identified sets of Envs from 6 of 11 individuals where there is evidence consistent with a different arrangement of V1V2 and V3 loops at the trimer apex for macrophage-tropic and non-macrophage-tropic Envs.
Fig 4.
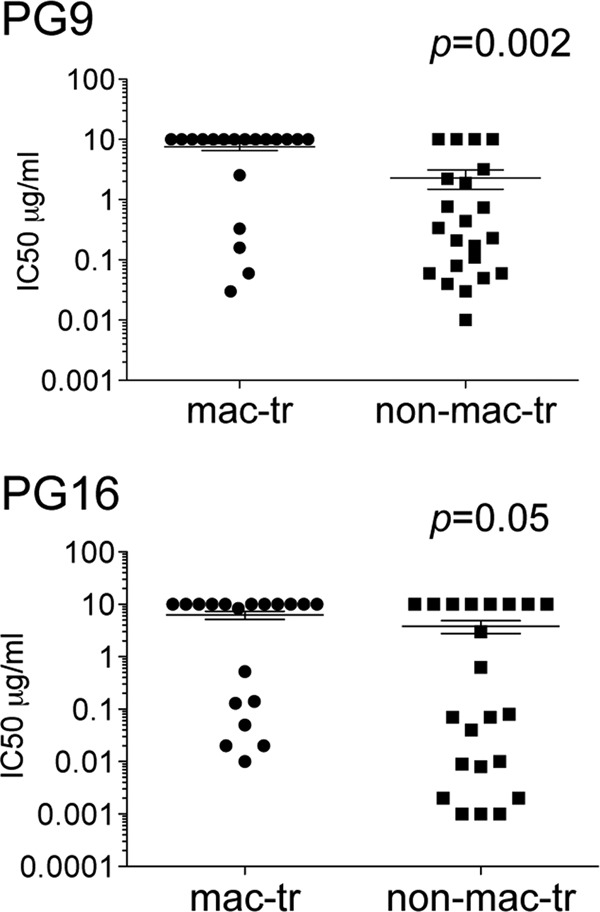
Macrophage-tropic R5 Env+ pseudovirions are significantly more resistant to PG9 neutralization but not to PG16. Mann-Whitney analyses comparing macrophage-tropic and non-macrophage-tropic Env+ pseudovirions for sensitivity to MAbs PG9 and PG16 (which recognize a quaternary epitope comprising determinants in V2 and V3), are shown.
Limited changes in the sensitivity of macrophage-tropic R5 Env+ pseudovirions to MAbs and reagents that target other Env sites and downstream events in fusion.
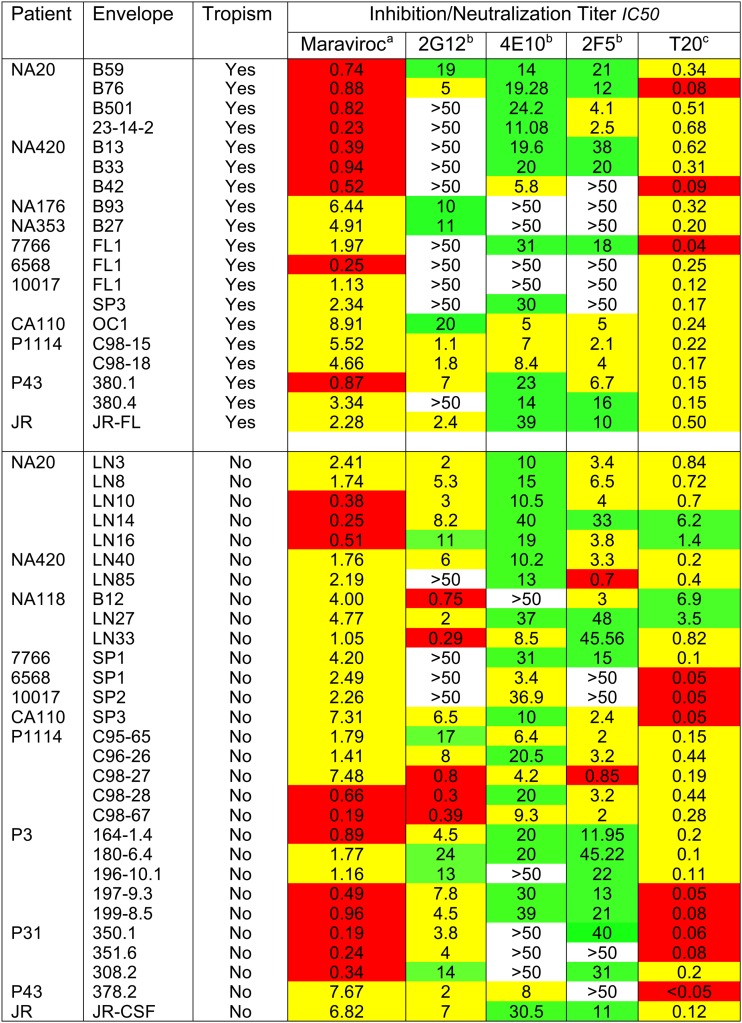
We next investigated the panel of Envs for their sensitivities to MAbs or reagents that target other Env sites or entry events subsequent to Env-CD4 binding. This approach was aimed at establishing whether Env changes that led to increased or decreased macrophage tropism were localized within or proximal to the CD4bs and at sites on the trimer apex or, alternatively, conferred more extensive effects on Env structure that would affect other functions, including coreceptor interactions and gp41 conformational changes that result in fusion. Non-macrophage-tropic Envs were significantly more sensitive to 2G12 than macrophage-tropic Envs (P = 0.006), confirming our previous observations on a smaller panel of Envs (11). 2G12 binds to a glycan complex on the outer domain of gp120. The decreased sensitivity to 2G12 for macrophage-tropic Envs is likely associated with the loss of a critical glycan or a change in the orientation of one or more glycans, as we previously reported (38). Such changes most likely influence exposure of CD4 contact residues on the outer domain of gp120, although effects on coreceptor interactions are also possible. Figure 5 and Table 5 show that there were no significant differences between macrophage-tropic and non-macrophage-tropic Envs in sensitivity to maraviroc or to each of the gp41 reagents, including MAbs 2F5 and 4E10, or to the fusion inhibitor, T20. Together, these data indicate that major or “global” structural or functional alterations between trimers of macrophage-tropic and non-macrophage-tropic Envs were not apparent and emphasize that the differences described above are localized to sites within or proximal to CD4 contact residues and to the trimer apex.
Fig 5.
Macrophage-tropic R5 Env+ pseudovirions do not significantly differ from non-macrophage-tropic Envs in sensitivity to maraviroc, T20, 2F5, and 4E10. However, macrophage-tropic R5 Envs are significantly more resistant to neutralization by the glycan-specific MAb 2G12. Mann-Whitney analyses comparing macrophage-tropic and non-macrophage-tropic Env+ pseudovirions for sensitivity to the CCR5 antagonist maraviroc, the 6-helix bundle inhibitor T20, gp41 MAbs 4E10 and 2F5, and the gp120 glycan-specific MAb 2G12 are shown.
Table 5.
Sensitivities of HIV-1 Env+ pseudovirions to neutralization or inhibition by reagents that bind envelope sites distal from the CD4bs and/or block entry events downstream from CD4 binding
aIC50s for maraviroc: red, <1 ng/ml; yellow, <10 ng/ml, >1 ng/ml.
bIC50s for MAbs 2G12, 4E10, and 2F5: red, <1 μg/ml; yellow, <10 μg/ml, >1 μg/ml; green, <50 μg/ml, >10 ng/ml.
cIC50s for T20: red, <0.1 μg/ml; yellow, <1 μg/ml, >0.1 μg/ml; green, <10 μg/ml, >1 μg/ml.
DISCUSSION
We report striking differences in the ability of sCD4 and different CD4bs ligands to neutralize macrophage-tropic and non-macrophage-tropic R5 Env+ pseudovirions. While sCD4 mainly neutralized macrophage-tropic Envs, VRC01 conferred efficient neutralization of all Env+ pseudovirions regardless of tropism. The CD4bs MAb b12 conferred an intermediate pattern where all macrophage-tropic Envs that carried the b12 epitope were neutralized, while some but not all of the non-macrophage-tropic Envs were sensitive. The new and potent CD4bs MAbs, e.g., VRC01, have been described as “highly active agonistic antibodies against the CD4 binding site (HAADs) that mimic binding of the host receptor CD4” (41). The binding sites for VRC01, b12, and sCD4 overlap on the outer domain of gp120 (26, 41, 43, 45). However, these reagents have different requirements for binding the bridging sheet and vary in their capacities to induce conformational changes in the trimer (43). CD4 recruits determinants on the stem of the V1V2 loops and at the junction of the β20-β21 strands to assemble the bridging sheet present on the gp120/sCD4 structure (26). However, the V1V2 stem determinant that binds CD4 is not critical for binding either of these MAbs (28, 45). Nevertheless, b12 does induce conformational changes in the trimer, although less extensively than CD4 (32). In contrast, VRC01, like other recently described potent CD4bs MAbs (PGV04, 3BNC117, and NIH45-46) (42), does not induce conformational changes on the trimer (43, 59). Finally, while mutation of bridging-sheet determinants was reported to confer profound effects on sCD4 and, to a lesser extent, b12 neutralization, they barely affect VRC01 (43). These distinct specificities of CD4, b12, and VRC01 for the bridging sheet along with their different abilities to neutralize via macrophage-tropic and non-macrophage-tropic Envs have led us to propose that R5 macrophage tropism is determined by the ability of CD4 to recruit contact residues on the V1V2 stem and β20-21 junction to assemble the bridging sheet (Fig. 6).
Fig 6.
Model showing how Env substitutions affect macrophage tropism of R5 Envs. Substitutions in gp120 residues that increase macrophage tropism occur in residues directly contacting CD4 or that affect their exposure. These substitutions confer an increased affinity for CD4 and may slow the “off rate,” making it more likely that the V1V2 stem on the inner domain will be recruited. Alternatively, a higher affinity for gp120 may more efficiently induce gp120 conformational changes so that the V1V2 stem is exposed for CD4 binding. Finally, substitutions in residues at sites on the trimer apex may facilitate conformational changes induced by CD4, which move the V1V2 loops to expose both the V1V2 stem and the V3 loop for binding to CCR5.
The ability of CD4 to recruit the bridging sheet may be influenced by changes within or proximal to CD4 contact residues, which directly affect the affinity of the Env for CD4. An increased Env-CD4 affinity potentially leads to a slower “off rate” (as reported for gp120 carrying N283 [7]), thus increasing the time frame for the V1V2 stem to be bound by CD4. Alternatively, an increased affinity for CD4 may enhance the induction of conformational changes that move V1V2 at the apex of the trimer, again increasing the likelihood that the V1V2 stem will be exposed and bound by CD4. An additional mechanism involves the ease with which conformational changes are elicited by CD4 binding, i.e., movement of the V1V2 loops away from V3 at the trimer apex (31). Our experiments with 447-52D, PG9, and PG16 have suggested that (at least for some Envs) the V3 loop may be more exposed or that the arrangement of V1V2 and V3 loops is altered at the trimer apex. For C98-15, a conserved V1 residue controls macrophage tropism as well as sCD4 and 447-52D sensitivity (39), unequivocally associating a single V1 residue at the trimer apex with V3 loop exposure, CD4 binding, and macrophage tropism. The data presented here incorporating VRC01 sensitivity now implicate accessibility of CD4 contact residues on the bridging-sheet segments as the major factor in determining low CD4 use and macrophage tropism.
Our data show that the Env determinants that confer HIV-1 R5 macrophage tropism are focused on the Env's interaction with CD4 and the immediate ensuing conformational changes and do not reflect major changes in envelope structure. This is evident from the lack of correlations between macrophage tropism and sensitivity to other neutralizing MAbs or inhibitors that target gp120-coreceptor interactions or gp41 conformational changes as well as by the consistent insensitivity of all Envs to the CD4i MAb 17b (data not shown) The absence of significant variation in sensitivities to these reagents further emphasize the marked difference in sensitivity to trimer apex reagents 447-52D, PG9, and PG16 discussed above. Perhaps most surprisingly, we were unable to detect differences in sensitivity to heterologous neutralizing antibodies in HIV-1+ human sera. Since most of the macrophage-tropic Envs used here were derived from the immune-privileged environment of brain tissue (50–52), we had expected them to have evolved a more open structure that would expose antibody epitopes and confer increased sensitivity to neutralizing antibodies. However, while Envs may evolve enhanced exposure of CD4 contact residues and subtle exposure of V3 residues at the trimer apex, they have not incurred more extensive alterations that would confer a more global neutralization sensitivity.
Previously, we had hypothesized that a more open structure of macrophage-tropic R5 Envs evolving in the brain would efficiently expose the CD4bs along with conserved neutralization epitopes and have relevance for vaccine development. Our data do not support a wide-open structure for such Envs. However, the identification of residues proximal to CD4 contact sites on the outer domain of gp120 that modulate macrophage tropism (37) and evidence for alterations in the glycan shield (as indicated by the increased resistance of macrophage-tropic Envs to 2G12) may indicate increased exposure of sites on the gp120 outer domain that are the initial contacts for CD4. Such Envs may more readily elicit antibodies targeting CD4 contact sites on the outer domain and have application for vaccine development.
In summary, our data point to a model where R5 macrophage tropism depends on the ability of CD4 to recruit bridging-sheet determinants on the V1V2 stem and the β20-21 junction. Different mechanisms can be envisaged to achieve this, which involve changes in (i) CD4 contact residues that directly affect affinity for CD4, (ii) proximal or distal residues that affect exposure of CD4 contact residues, including the loss or change in the orientation of glycans that protect CD4 contact residues, and (iii) residues at the trimer apex that facilitate conformational changes induced by CD4. HIV-1 Envs in the brain may evolve to be highly macrophage-tropic via all three mechanisms. Thus, their increased capacity to bind and respond to CD4 will enable the efficient formation and exposure of the bridging sheet and coreceptor binding site in the presence of low levels of CD4 on the surfaces of macrophages.
ACKNOWLEDGMENTS
Our study was supported by NIH R01 grants MH64408, AI089334, HD049273, and P01 AI082274.
We thank Pham Phung, Terri Wrin, and Yolande Lie (Monogram Biosciences Inc.) for technical assistance. We also thank John Mascola for providing VRC01 and VRC03 and for critical comments on the manuscript. Thanks also go to James Robinson (Tulane University) for providing 17b. We acknowledge and thank the University of Massachusetts Center for AIDS Research (P30-AI42845), the NIH AIDS Research and Reference Reagent Program and the Centre for AIDS Reagents, NIBSC, United Kingdom, for services and reagents.
Footnotes
Published ahead of print 10 October 2012
REFERENCES
- 1. Clapham PR, McKnight A. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 83:1809–1829 [DOI] [PubMed] [Google Scholar]
- 2. Gorry PR, Ancuta P. 2011. Coreceptors and HIV-1 pathogenesis. Curr. HIV/AIDS Rep. 8:45–53 [DOI] [PubMed] [Google Scholar]
- 3. McNamara LA, Collins KL. 2011. Hematopoietic stem/precursor cells as HIV reservoirs. Curr. Opin. HIV AIDS 6:43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asjo B, Morfeldt Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo EM. 1986. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet ii:660–662 [PubMed] [Google Scholar]
- 5. Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215–219 [DOI] [PubMed] [Google Scholar]
- 6. Tersmette M, de Goede RE, Al BJ, Winkel IN, Gruters RA, Cuypers HT, Huisman HG, Miedema F. 1988. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J. Virol. 62:2026–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, Wolinsky SM, Gabuzda D. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. U. S. A. 103:15160–15165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goodenow MM, Collman RG. 2006. HIV-1 coreceptor preference is distinct from target cell tropism: a dual-parameter nomenclature to define viral phenotypes. J. Leukoc. Biol. 80:965–972 [DOI] [PubMed] [Google Scholar]
- 9. Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, Bell JE, Bannert N, Crawford K, Wang H, Schols D, De Clercq E, Kunstman K, Wolinsky SM, Gabuzda D. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J. Virol. 78:6915–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peters PJ, Duenas-Decamp MJ, Sullivan WM, Brown R, Ankghuambom C, Luzuriaga K, Robinson J, Burton DR, Bell J, Simmonds P, Ball J, Clapham P. 2008. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peters PJ, Sullivan WM, Duenas-Decamp MJ, Bhattacharya J, Ankghuambom C, Brown R, Luzuriaga K, Bell J, Simmonds P, Ball J, Clapham PR. 2006. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J. Virol. 80:6324–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. 2011. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog. 7:e1002286 doi:10.1371/journal.ppat.1002286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, Bell JE, Wolinsky SM, Gabuzda D. 2007. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360:105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin-Garcia J, Cao W, Varela-Rohena A, Plassmeyer ML, Gonzalez-Scarano F. 2006. HIV-1 tropism for the central nervous system: brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology 346:169–179 [DOI] [PubMed] [Google Scholar]
- 16. Alexander M, Lynch R, Mulenga J, Allen S, Derdeyn CA, Hunter E. 2010. Donor and recipient envs from heterosexual human immunodeficiency virus subtype C transmission pairs require high receptor levels for entry. J. Virol. 84:4100–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kishko M, Somasundaran M, Brewster F, Sullivan JL, Clapham PR, Luzuriaga K. 2011. Genotypic and functional properties of early infant HIV-1 envelopes. Retrovirology 8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez-Perez MP, O'Connell O, Lin R, Sullivan WM, Bell J, Simmonds P, Clapham PR. 2012. Independent evolution of macrophage-tropism and increased charge between HIV-1 R5 envelopes present in brain and immune tissue. Retrovirology 9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gray L, Sterjovski J, Churchill M, Ellery P, Nasr N, Lewin SR, Crowe SM, Wesselingh SL, Cunningham AL, Gorry PR. 2005. Uncoupling coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from macrophage tropism reveals biological properties of CCR5-restricted HIV-1 isolates from patients with acquired immunodeficiency syndrome. Virology 337:384–398 [DOI] [PubMed] [Google Scholar]
- 21. Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, Naif HM. 1999. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J. Virol. 73:9741–9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tuttle DL, Anders CB, Aquino-De Jesus MJ, Poole PP, Lamers SL, Briggs DR, Pomeroy SM, Alexander L, Peden KW, Andiman WA, Sleasman JW, Goodenow MM. 2002. Increased replication of non-syncytium-inducing HIV type 1 isolates in monocyte-derived macrophages is linked to advanced disease in infected children. AIDS Res. Hum. Retroviruses 18:353–362 [DOI] [PubMed] [Google Scholar]
- 23. Diskin R, Marcovecchio PM, Bjorkman PJ. 2010. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat. Struct. Mol. Biol. 17:608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329–1339 [DOI] [PubMed] [Google Scholar]
- 26. Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose N, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid J, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U. S. A. 109:5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841 [DOI] [PubMed] [Google Scholar]
- 31. Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. U. S. A. 108:11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S. 2011. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J. Virol. 85:12114–12123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2:e83 doi:10.1371/journal.ppat.0020083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852 [DOI] [PubMed] [Google Scholar]
- 36. Zhu P, Winkler H, Chertova E, Taylor KA, Roux KH. 2008. Cryoelectron tomography of HIV-1 envelope spikes: further evidence for tripod-like legs. PLoS Pathog. 4:e1000203 doi:10.1371/journal.ppat.1000203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duenas-Decamp MJ, Peters PJ, Burton D, Clapham PR. 2009. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J. Virol. 83:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duenas-Decamp MJ, Clapham PR. 2010. HIV-1 gp120 determinants proximal to the CD4 binding site shift protective glycans that are targeted by monoclonal antibody, 2G12. J. Virol. 84:9608–9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Musich T, Peters PJ, Duenas-Decamp MJ, Gonzalez-Perez MP, Robinson J, Zolla-Pazner S, Ball JK, Luzuriaga K, Clapham PR. 2011. A conserved determinant in the V1 loop of HIV-1 modulates the V3 loop to prime low CD4 use and macrophage infection. J. Virol. 85:2397–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. 2011. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334:1289–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Olivera TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton D, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. 2011. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J. Virol. 85:8954–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moore JP. 1990. Simple methods for monitoring HIV-1 and HIV-2 gp120 binding to soluble CD4 by enzyme-linked immunosorbent assay: HIV-2 has a 25-fold lower affinity than HIV-1 for soluble CD4. AIDS 4:297–305 [DOI] [PubMed] [Google Scholar]
- 48. Nottingham QJ, Birch JB. 2000. A semiparametric approach to analysing dose-response data. Stat. Med. 19:389–404 [DOI] [PubMed] [Google Scholar]
- 49. Ritz C, Streibig JC. 2005. Bioassay analysis using R. J. Stat. Softw. 12:1–22 [Google Scholar]
- 50. Bullard DE, Bourdon M, Bigner DD. 1984. Comparison of various methods for delivering radiolabeled monoclonal antibody to normal rat brain. J. Neurosurg. 61:901–911 [DOI] [PubMed] [Google Scholar]
- 51. Kuang F, Wang BR, Zhang P, Fei LL, Jia Y, Duan XL, Wang X, Xu Z, Li GL, Jiao XY, Ju G. 2004. Extravasation of blood-borne immunoglobulin G through blood-brain barrier during adrenaline-induced transient hypertension in the rat. Int. J. Neurosci. 114:575–591 [DOI] [PubMed] [Google Scholar]
- 52. Triguero D, Buciak JB, Yang J, Pardridge WM. 1989. Blood-brain barrier transport of cationized immunoglobulin G: enhanced delivery compared to native protein. Proc. Natl. Acad. Sci. U. S. A. 86:4761–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bannert N, Schenten D, Craig S, Sodroski J. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophage-tropic human immunodeficiency viruses. J. Virol. 74:10984–10993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. U. S. A. 96:5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mori K, Rosenzweig M, Desrosiers RC. 2000. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J. Virol. 74:10852–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O'Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805 doi:10.1371/journal.pone.0008805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538–7542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Miiro G, Serwanga J, Pozniak A, McPhee D, Manigart O, Mwananyanda L, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Allen S, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Falkowska E, Ramos A, Feng Y, Zhou T, Moquin S, Walker LM, Wu X, Seaman MS, Wrin T, Kwong PD, Wyatt RT, Mascola JR, Poignard P, Burton DR. 2012. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J. Virol. 86:4394–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]