If the whole is more than the sum of its parts, a complete description of any system will require delineating the separate contributions of each individual part, as well as the contributions of interactions between these different components. The need to consider context-dependent interactions when describing a system also holds true in the molecular world. For instance, the BK voltage– and ligand-activated K+ channel, comprising a central pore (P), together with voltage-sensing (V) and substrate-binding (S) gating domains, represents a modular molecular system able to integrate inputs in the form of chemical and electrical potentials to produce a precise and tunable physiological response. The output of the system corresponds to the overall chemical free energy of channel gating (), reflecting the transition from the resting closed state, where both signals are off, to the final open state, where both voltage and ligand concentration are at saturating levels. This output is a reflection of the entire domain interaction network in which domain transitions are coupled to pore opening. A full description of in this system, therefore, requires delineating the energetic contributions of the individual P, V, and S domains of all possible PV, PS, and VS pairwise domain interactions and of the third-order PVS interaction involving the three domains. Defining these energetic contributions requires the formulation of a channel gating model that adequately describes the function of the polymodal ion channel of interest. Because such models are proposed only after painstaking steady-state and transient kinetics analyses have been performed, the identification of a shortcut for obtaining these first- and higher-order contributions to would be a welcome development. In this issue of The Journal of General Physiology, two papers, one by Chowdhury and Chanda and the other by Sigg, present rigorous thermodynamic formalisms for the analysis of polymodal voltage- and ligand-dependent ion channels that enable accurate determination of the net free energy of channel gating, along with all possible pairwise interaction energies between the P, V, and S domains in a model-free manner.
A context-dependent interaction formalism for energy parsing
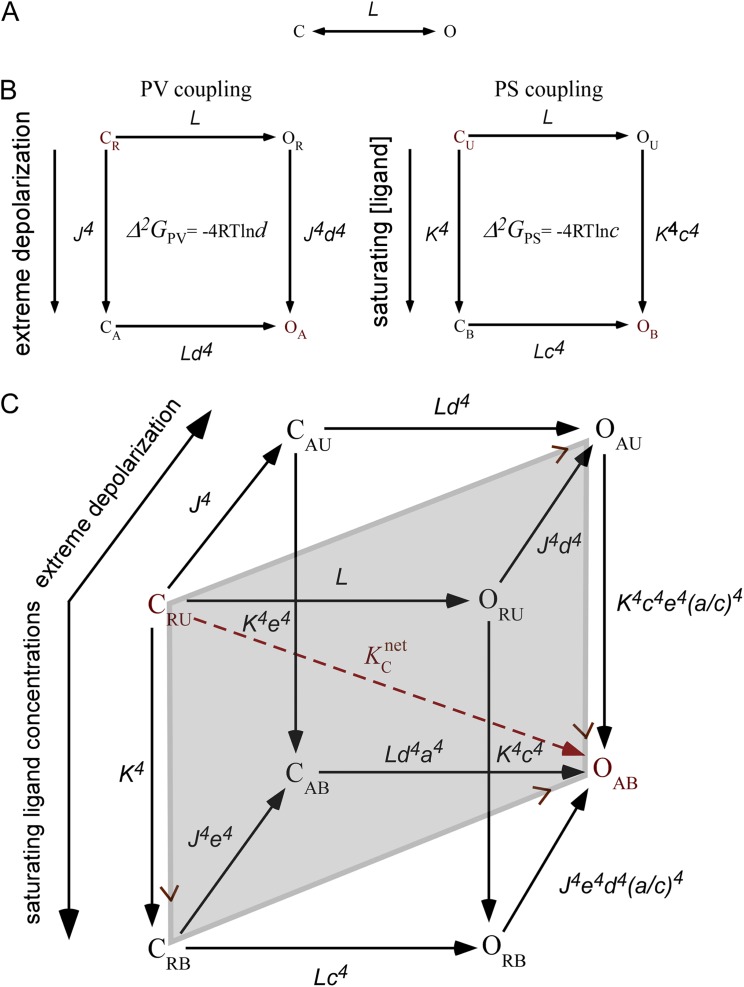
The problem of energy parsing in complex modular protein systems can, in principle, be addressed using a conceptual framework that considers the hierarchy of domain organization. Asking how one would design a voltage- and ligand-activated ion channel from scratch would help in formulating such formalism and would set the stage to understand the importance of Chowdhury and Chanda’s and Sigg’s findings. Consider first a simple channel comprising only a pore domain that undergoes a conformational transition between two states, one closed (C) and one open (O), as dictated by the equilibrium constant L (Fig. 1 A). If the pore contains a gating charge, then the voltage-dependent Boltzmann equation would adequately describe channel gating, where pore opening and pore gating charge movement occur concomitantly. In this simple channel, Lo, the chemical component of L at 0 mV (≡ ΔGP), determines . If one next adds either a voltage-sensing or substrate-binding domain to the pore, giving rise to a voltage-gated and ligand-gated ion channel, respectively, then would be parsed to the individual intrinsic contributions of the pore (P) domain and either the voltage-sensing (V) or substrate-binding (S) domains and of the PV or PS pairwise interaction free energy between them. The schemes depicted in Fig. 1 B suggest one possible model for how such coupling might be achieved. In these schemes, both the V and S domains undergo two-state transitions, with the V domain switching between the resting (R) and activated (A) states, and the S domain cycling between the unbound (U) and bound (B) states, as described by the respective J and K equilibrium constants.1 Voltage application or ligand binding would affect both the open and closed conformations of the channel, as in the Monod–Wyman–Changeux allosteric model (Monod et al., 1965). In both of these channel systems, is described by one additional intrinsic parameter, either the Ka or Jo equilibrium constant (ΔGS or ΔGV, respectively), aside from the contribution of the pore (Lo), and an additional coupling constant describing the interaction of the pore domain with either the ligand-binding domain (c) or the voltage-sensing domain (d), as appropriate. In energetic terms, these constants are written as Δ2GPS = −4RTlnc and Δ2GPV = −4RTlnd, where the dimension 2 in the ΔG notation is added to indicate the contribution of the second-order interaction between two domains, as opposed to the first-order ΔGP, ΔGV, and ΔGS contributions of the individual P, V, and S domains. The final stage in constructing a dually voltage- and ligand-activated channel involves the addition of a third voltage-sensing or ligand-binding domain, as required. Energy parsing for this three-component (P, V, and S) channel system must consider all possible interactions along the allosteric network.
Figure 1.
Constructing a voltage- and ligand-activated BK-like ion channel. (A) A two-state channel with only a pore (P) domain undergoing a conformational transition between closed (C) and open (O) states, as governed by the L equilibrium constant (L = Loexp(−zV/RT)). (B) The same channel, to which a voltage-sensing (V) or ligand-binding (S) gating domain has been added. Transitions of voltage sensors between rest (R) and activated (A) states are manifested by the domain equilibrium constant J (= Joexp(−zV/RT)), whereas K (= Ka[S], where Ka is the association constant) reflects the transition of the S domain between the unbound (U) and bound (B) states. The fourth power in the equilibrium or interaction constants reflects the tetrameric organization of the voltage- or ligand-gated channels undergoing independent and sequential gating charge and ligand-binding transitions, respectively (Monod et al., 1965). The d and c interdomain interaction constants reflect the ratio of the equilibrium constants for V activation in the open and closed states (d), or the ratio of ligand affinity to the open and closed conformations (c). Resting and final open states are colored red. (C) The same channel as in B to which a voltage-sensing or ligand-binding gating domains has been added (as required) to yield a dually voltage- and ligand-activated BK-like channel. The interaction constant e represents the coupling between the voltage and ligand-binding domains (left face of the cube), whereas the a coupling constant represents the coupling between the pore and the ligand-binding domain, where the V domain has already been activated (back face of the cube). The dashed red arrow indicates the overall channel equilibrium constant (), indicative of the resting to final open-state transition (CRU→OAB transition). The equilibrium constants of any transition along the thermodynamic cube are indicated next to the arrows. Arrows in these schemes are drawn to indicate possible paths that can be taken, although all represent equilibrium between the states connected.
The gating model of the BK voltage– and ligand-activated K+ channel (Rothberg and Magleby, 1999; Horrigan and Aldrich, 2002) provides an example of one possible description of three-domain gating (Fig. 1 C). In this thermodynamic cubic construct, the x, y, and z directions represent channel opening, Ca2+ binding, and voltage-sensor transitions, respectively. To fully describe this system, two additional interaction constants are needed, namely the e constant, reflecting the interaction between the V and S domains (left face of the cube; Fig. 1 C), and the a constant, reflecting the interaction between the P and S domains, much as constant c does, yet accounting for the voltage-sensor domain in the activated conformation (back face of the cube). Such context-dependent interaction allows the evaluation of the mutual coupling between the three P, V, and S domains of the polymodal channel (Δ3GPVS). This third-order interaction can be viewed as the effect of the third domain on the magnitude of coupling between the other two domains, as given by the a/c ratio obtained by comparing the interaction energies determined for any two opposing faces of the cube.2 The equilibrium constants of any transition along the 3-D construct of Fig. 1 C can be expressed using the appropriate L, J, and K domain equilibrium constants and the a, c, d, and e interdomain interaction constants, thus yielding a full description of the polymodal BK channel. The net chemical equilibrium constant for channel gating in this case, , represents the concentration ratio of the resting CRU state, where both V and S domains are not activated, and the fully open OAB end state, where both V and S signals are saturated (red dashed arrow in Fig. 1 C). The following expression is obtained for = LoJo4Ka4c4d4e4(a/c)4, which can also be written as:
| (1) |
It can be seen that is indeed parsed to the sum of the individual first-order contributions of the P, V, and S domains and to all possible second- and third-order interactions thereof. By using the energy parsing conceptual framework delineated above, a polymodal channel with up to n gating domains can be constructed. for such a channel is parsed to the sum of all n individual domain contributions and to all their possible higher-order contributions up to the nth-order contribution, reflecting the mutual interaction of the pore and the other n−1 gating domains. The context-dependent interaction formalism for energy parsing in a polymodal ion channel presented here is reminiscent of that previously used to evaluate the contribution of residue interaction networks to protein stability (Horovitz and Fersht, 1992) or channel gating (Sadovsky and Yifrach, 2007). Such formalism, applied here to domain-level organization, is based on thermodynamic considerations and requires knowing only the list of parts comprising the channel and is thus model independent. In principle, although the context-dependent energetic contributions to can be evaluated experimentally using high-order thermodynamic double mutant cycle analysis (Horovitz and Fersht, 1990), such an analysis is not straightforward to perform at the domain level. How to evaluate these contributions is what concerns Chowdhury, Chanda, and Sigg.
The Chowdhury and Chanda analysis for dually activated channels
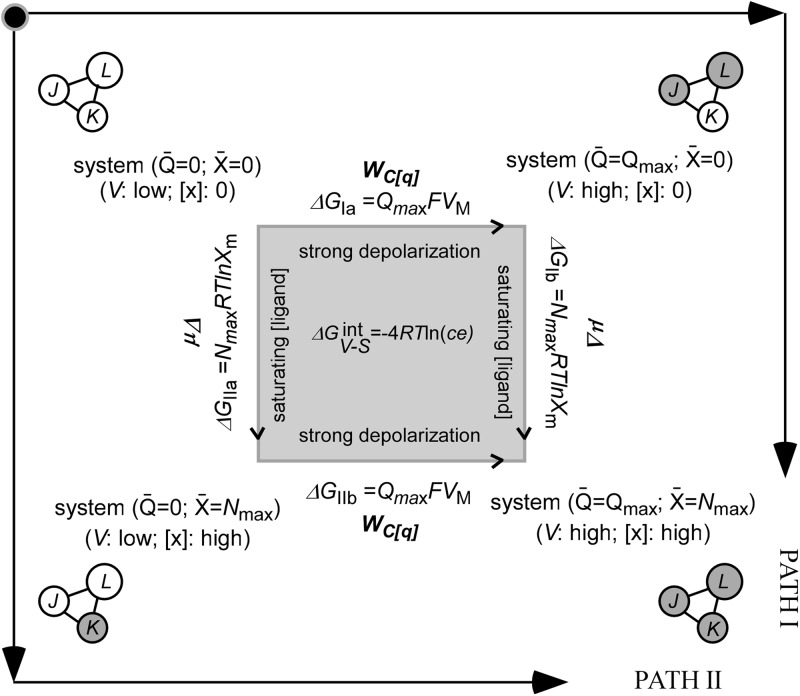
The authors’ papers offer powerful thermodynamic tools, based on Wyman’s seminal linkage analysis, to address energy parsing in polymodal ion channels, allowing for the model-free evaluation of the output of the whole system and of the contributions of the individual system components and their interactions. Linkage analysis is a thermodynamic formalism originally developed and applied to describe the effects of two different ligands of a macromolecule on each other’s binding, as in the Bohr effect in hemoglobin (Wyman, 1964; 1967). Chowdhury and Chanda (2013) extend this formalism to the case of electrochemical equilibria, as in the BK model allosteric channel. Initially considering whole system output (), the authors present a rigorous thermodynamic description of the free energy () for a voltage- and ligand-activated channel system. They show that is a thermodynamic state function with respect to voltage (V) and ligand concentration (S or X in the authors’ notation), and that the overall free energy for this system () can be parsed into two separate contributions corresponding to the voltage-dependent () and ligand-dependent () pathways. Assuming that the signals are turned on one after the other, there are two ways to switch from the resting closed state to the fully open end state, as described in the thermodynamic cycle in Fig. 2 (adopted from Fig. 2 of both the Chowdhury and Chanda, 2013, and Sigg, 2013, papers). Along Path I, all gating charges are first moved, and then ligands can bind. Along Path II, the channel is first saturated with ligands, and then gating charge movement occurs. Regardless of the path taken, these contributions can be accurately evaluated directly from the respective Q-V and -S (with being the fractional ligand saturation) curves using the median metrics; i.e., the median voltage (VM) and median ligand concentration (Sm) directly read off the appropriate curve, provided that the total number of channel gating charges (Qmax) and ligand-binding sites (Nmax) is known. No model is required for evaluating (= QmaxFVM) or (= NmaxRTlnSm) in this manner (Chowdhury and Chanda, 2012). In applying their strategy to the BK channel, the authors demonstrate that median metrics analysis works; essentially identical values were obtained when they were calculated according to the Horrigan and Aldrich (2002) model parameters or based on the sum of the appropriate and gating energies of the voltage and ligand pathways.
Figure 2.
A thermodynamic linkage cycle for assessing coupling between the voltage- and ligand-dependent pathways (adopted from Fig. 2 of both the Sigg, 2013, and the Chowdhury and Chanda, 2013, papers). The cycle presents the four extreme channel states populated in the absence of any signal (top left, the resting state), where either of the voltage or the ligand signals are saturating (top right and bottom left corners, respectively), or where both signals are turned on, giving rise to the fully open end state (bottom right). The states are either described schematically, as in the Sigg paper (with the P, S, and V domains designated by their respective L, K, and J equilibrium constants), or by text, as in the Chowdhury and Chanda paper. For each transition, the corresponding energies and work functions are indicated above the arrows.
Chowdhury and Chanda next evaluate the overall free energy of interaction between the voltage and ligand pathways, , again in a model-independent manner. As the authors show, is a manifestation of heterotropic linkage between the two signals. Its value can be obtained by comparing any two parallel transitions along the thermodynamic cycle presented in Fig. 2 with a given pair of parallel transitions addressing the context dependence of signal activation. For example, the upper and lower transitions reflect the switching on of the voltage signal once the ligand pathway is off (upper transition) and a second time when the ligand signal is on (lower transition). The free energy difference between these two energies is thus a measure of the degree of synergy or coupling between the two pathways. If the two pathways are uncoupled, then no context-dependent interaction exists and = 0. In practical terms, the magnitude of can be determined from the shift in the Q-V curve between zero and saturating ligand concentration, as evaluated by ΔVM ( = QmaxFΔVM).
The manner by which Chowdhury and Chanda calculate is model independent. However, understanding the true meaning of this parameter and defining what contributes to the coupling inherent therein require that a model be considered. In the context of the BK channel gating model (Fig. 1 C), the Chowdhury, Chanda, and Sigg model–independent cycle presented in Fig. 2 corresponds to the diagonal gray-shaded cycle drawn at the center of the thermodynamic cube in Fig. 1 C, offering a way to understand the connection between experiment and model. In this gray-shaded cycle, the two horizontal transitions (CRU→OAU and CRB→OAB) can be experimentally measured and compared to obtain (= QmaxFΔVM, the context dependence of gating charge translocation), whereas the two vertical legs are “model-rooted” and can as well be used to evaluate . In the framework of this model, and assuming no context-dependent interactions (a/c = 1), this pathway-dependent coupling is determined by −4RTln(ce), as obtained by the ratio of equilibrium constants of the two vertical transitions of the gray cycle. Thus, the overall pathway-dependent coupling is determined by the sum of second-order interactions of the ligand-binding domain with the pore (c) and voltage-sensing (e) domains.3 Using the ratio of ZOA/ZCR partition functions that sum all OA and CR states lying along the vertical transitions of the gray cycle in Fig. 1 C (all states with a different number of ligands bound), the authors rigorously show that the median metrics for evaluating is indeed a function of the ce product of interaction constants, thus linking experiment to model observables.
Using the median metrics analysis of Q-V curves in the presence or absence of saturating [Ca2+], generated using the Horrigan and Aldrich (2002) model parameters, Chowdhury and Chanda demonstrate that practically identical negative values are obtained for , as compared with the reported c and e interaction parameters of the BK channel (Horrigan and Aldrich, 2002). The minus sign of indicates a positive heterotropic linkage between the two signals, meaning that gating charge movement is facilitated by ligand binding, and the other way around. Thus, both voltage and ligand pathways act synergistically to open the BK channel pore.
Chowdhury and Chanda’s findings described thus far addressed the overall free energy of the system and the overall interaction energy between the component voltage and ligand pathways. From a thermodynamic point of view, these two overall quantities are obtained by considering the two resting and open-edge states of the system, which are experimentally accessible under extreme conditions. However, as indicated by Chowdhury and Chanda, a full description of the free energy landscape () of a dually activated BK-like channel requires measuring charge translocation Q-V curves at different ligand concentrations and ligand binding -S curves at different voltages. Although Q-V curves can be easily measured at different ligand concentrations, measuring -S curves at different voltages is not straightforward. To overcome this challenge, the authors use linkage principles and demonstrate that ligand-binding curves at any voltage can be calculated by measuring the Q-V curve at different ligand concentrations, provided that a ligand-binding curve at a reference voltage (usually at 0 mV) is available. This is another important tool that Chowdhury and Chanda provide, derived from their rigorous thermodynamic analysis. The power of the authors’ thermodynamic approach is further shown in their analysis of other channel systems, such as the HCN and CNG channels.
The Sigg analysis for polymodal channels
Chowdhury and Chanda restricted their analysis to dually activated channels, such as the BK channel. The paper by Sigg (2013) extends the linkage analysis used by Chowdhury and Chanda (2010, 2013) to the realm of polymodal ion channels in general and offers a complete thermodynamic formalism for studying allosterically regulated polymodal ion channels, with the dually voltage-and ligand-activated BK channel model allosteric protein being one example. In Sigg’s general formalism, pore or gating domains influence each other in a manner described by the basic linkage cycles presented in Fig. 1 B. Using such elementary building blocks, various different polymodal ion channels can be constructed, each with a unique interaction network. Sigg thus provides a thermodynamic answer to the question posed above regarding how one would construct a polymodal ion channel, focusing on a particular type of channel in which domain linkages assume the signature of the Wyman linkage cycle (Fig. 1 B). As such, Sigg offers a thermodynamic model–dependent language for constructing polymodal channels that parallels the context-dependent and model-independent interaction formalism for energy parsing discussed above.
To define the language of his model, Sigg has devised a linkage diagram tool that concisely illustrates the connectivity between the different domains, along with the entire pairwise interaction network of a polymodal channel. This linkage diagram tool, together with the accompanying set of rules, facilitates the identification of channel states that contribute to the interaction between any two domains, as delineated by the appropriate model sub-partition functions. The overall partition function (Z) of the channel gating model is a weighted sum of all channel states and is a polynomial function of the equilibrium constants of the various channel domains and the interdomain interaction constants. The Z partition function is directly related to the free energy of the system, (= −RTlnZ). Although the Sigg study offers analysis of complex gating models involving up to 20 domains, for the sake of simplicity, the discussion here is restricted to the BK channel, a nine-domain system composed of a pore and four voltage-sensing and four calcium-binding domains (Scheme 2 in Sigg, 2013). In the context of this model, a full description of energy parsing for such a system requires, in addition to defining core domain energies, that the PS, PV, and VS interaction energies (the respective c, d, and e constants) be determined (Fig. 1 C).
At the heart of Sigg’s analysis is a thermodynamic linkage cycle, similar to that depicted in Fig. 1 B, that addresses the context-dependent activation of a principal domain (termed a “work function”) in response to the full activation of a linked component (a process referred to as a “lever operation”). Such cycles reflect the heterotropic linkage or interaction between the two domains. As before, the basic idea in using such linkage cycles is to choose the right cycle, where the context-dependent activation of the principal domain is directly related to experimental measure, and the other pair of parallel transitions—the lever operation of activating the secondary domain—lies along the model framework.4 One such cycle is described in Fig. 2, as discussed previously, and also addressed by Sigg. Sigg considers gating charge movement, as described by the horizontal transitions in this cycle, as a global marker of the system activation, with the relevant experimentally oriented work function describing this transition defined as WC[q], the electrical capacitance energy. WC[q] is the intrinsic energy caused by the voltage-dependent pathway (by definition, WC[q] = −QmaxFVM ≡ −), as determined by Chowdhury and Chanda in their Q-V–based median metrics analysis. The parallel vertical transitions reflecting ligand binding at saturating concentrations correspond to the chemical potential lever, μΔ. Like Chowdhury and Chanda, Sigg shows that the context dependence of WC[q], in the presence or absence of saturating ligand concentrations (= −), determines the ce product of interaction constants.
The second work function introduced by Sigg is the Hill transform of the G-V steady-state measurement, WH(g) (WH(g) = kTln(G/(Gmax − G))).5 Sigg shows that WH(g) is a local marker of pore activation that determines WL, the negative value of the free energy of pore opening. Because WH(g) is a marker of pore activation, it must reflect the ratio of all the open and closed states of the channel (regardless of whether or not the V and S domains are activated), as described by all states lying at the respective right and left faces of the cube in Fig. 1 C. This is reflected by the ratio of the model open and closed sub-partition functions (ZO/ZC) that sums all of these states. Sigg shows that the ZO/ZC ratio determines the cd product of interaction constants, thus linking between the experimentally measured WH(g) and the specific model parameters.
The two experimentally oriented work functions described above assume that the model interaction energies can be determined from experiments, in particular as the context dependence of WC[q] determines the ce product and WH(g) itself determines the value of the cd product of interaction constants. To distinguish between the c and d interaction constants, the context dependence of WH(g) can be evaluated using the respective chemical (μΔ) and electrical (VΔ) potential levers. The relevant linkage cycles for achieving this correspond to the respective front and upper faces of the thermodynamic cube shown in Fig. 1 C.6 In the front face linkage cycle, the context dependency of WH(g) is evaluated by measuring its value in the presence or absence of saturating ligand concentration (μΔ), as given by the upper and lower pore-opening transitions. Using the parallel pair of model-rooted vertical transitions, this context dependence determines the c interaction constant, a value that can also be expressed as the ZOR/ZCR ratio of the partition functions that sum all OR and all CR states contributing to this coupling (all states along the right and left transitions of this cycle with their different number of bound ligands). According to Sigg’s notation, μΔWH(g) = −4WC (WC = −RTlnc). Similarly, using the upper face linkage cycle, the context dependence of WH(g) can be evaluated in the presence or absence of the voltage signal (VΔ). Aided by a parallel pair of vertical transitions, this dependence yields the d interaction constant, where VΔWH(g) = −4WD (WD = −RTlnd). It can be shown that d is determined by the ZOU/ZCU ratio of partition functions that sum all OU and CU states contributing to this coupling and having different numbers of activated voltage sensors.
Now that the WH(g) linkage cycles can be shown to correlate between experiments and model observables, as demonstrated by Sigg, it becomes a straightforward process to obtain the interaction parameters from WH(g) linkage plots. Such linkage plots describe the voltage dependence of WH(g) in the presence and absence of saturating Ca2+ concentrations. WH(g) plots, by definition the Hill transform of the G-V curve, resemble the traditional Hill plots (log(/1-) vs. log[S] plots) and exhibit two parallel “floor” and “ceiling” asymptotes, and a steeper rising phase in between. All the information we are after is hidden in these plots. As can be seen in the WH(g) linkage plot presented in Fig. 6 of Sigg (2013), WC and WD along with the pore gating charge (qL) can be directly read off these plots. The height difference between the floor and ceiling asymptotes of the WH(g) curve in the absence of ligand equals −4WD, whereas the height difference between the ceiling asymptotes of WH(g) in the presence and absence of saturating ligand concentrations corresponds to −4WC. Once determined in this manner, WC can be used to extract the WE interaction energy from the Q-V–based WC[q] linkage cycle (Fig. 2), which, as discussed earlier, determines the ce product of interaction constants. Now, all three possible interdomain interaction energies have been determined.
Sigg next tested the applicability of his linkage analysis on an expanded 17-domain BK-like channel model using simulated voltage-ramp–induced ionic and gating current data (based on decay time constants reported by Horrigan and Aldrich, 2002) to generate the corresponding G-V (or rather its Hill transform) and Q-V curves in the presence or absence of saturating calcium ions.7 Read directly from these graphs, such analysis yielded estimates for the c, d, and e coupling constants very close to the actual values determined by Horrigan and Aldrich (2002) in their detailed analysis. In addition, the Sigg paper addresses other aspects of linkage analysis, including different BK-like models having more than one type of voltage-sensor or Ca2+-binding domain, or where the pore is not assumed to be binary. Moreover, several issues related to the underlying assumptions of the model, like the assumption that the particles are uni-modal or that the allosteric constants are modality insensitive, are also considered, as are the advantages and limitations of the linkage analysis and its applicability to other channel types, such as TRP channels.
The two papers discussed here, together with two other papers from the Chanda group (Chowdhury and Chanda, 2010, 2012), enrich the channel biophysicist’s toolbox with powerful thermodynamic tools that can provide valuable information on almost everything we need to know to fully describe energy parsing in polymodal channels. Remarkably, this information is derived from conventional G-V and Q-V measurements in an essentially model-free manner and can be rationalized with only elementary knowledge of the allostery of the system, with no knowledge of the states of a system, their weights, or the rate constants dictating their life span. Together, these two studies open the way for a model-independent approach to studying voltage- (and ligand-) dependent ion channel gating. The domain interaction energies obtained from using such a model-independent approach may constrain the set of possible channel gating models, offering a shortcut to identifying the most complete model. Furthermore, as pointed out by Chowdhury and Chanda, the easily accessible interaction energies obtained from such an approach may be useful in scanning mutagenesis analysis aimed at identifying the network of residues involved in mediating interdomain communication.
With the wealth of information derived from Hill transformation of G-V curves, including the intrinsic pore-opening free energy and the overall system energy accurately evaluated from the Q-V curve, one might ponder the fate of the traditional Hill-like Boltzmann equation. Given the thermodynamics studies of Sigg and of Chowdhury and Chanda, it seems that we no longer need the poor energy estimates usually derived from the V1/2 metrics of the two-state channel model, making use of the Boltzmann equation unnecessary (Chowdhury and Chanda, 2012). However, despite its weaknesses, we can still find some use for the Boltzmann equation. First, this equation can be used for the fast screening of mutations in the search for those that affect cooperativity between channel subunits, as reflected by the z slope of the equation. Second, the Boltzmann equation still provides a framework definition to connect with a specific gating model, revealing the true meaning of the Hill coefficient for channel gating transitions (Yifrach, 2004). Lastly, the Hill equation is still with us, almost 50 years since Wyman offered his ideas on thermodynamic linkage that form the basis for all of the concepts considered here.
On a final note, the Sigg (2013) and Chowdhury and Chanda (2013) papers present complete thermodynamic analyses that when used to study complex molecular systems, like polymodal ion channels, yield valuable information from simple measurements and with no model in mind. Such thermodynamics spectacles allow one to examine the system at its extreme ends but ignore the details and properties associated with the numerous intermediate states, usually studied under the realm of kinetics analyses; but still, the advances considered can reveal much about the system as a whole, about its parts and about what is more than “the sum of its parts”—all of their synergistic interactions.
Acknowledgments
Work in my lab is supported by the Israel Science Foundation (grant u88/12).
Footnotes
In these and later schemes, a pore-centric approach is adopted. Model channel states are annotated as either closed (C) or open (O), with subscript indices representing the states of the other two V and S domains—whether R or A, or U or B—for the respective V and S domains. Such a pore-centric description directly relates to the final output of the channel evaluated in the G-V measurements.
For the BK channel, it is usually assumed that no context-dependent interaction exists, meaning that there is no third-order contribution to (a/c = 1). This is frequently explained by a lack of intersubunit interactions between the voltage-sensing and ligand-binding domains. Coupling between the V and S domains, as reflected by the e interaction constant, is thought to be mediated by the pore domain of the same subunit. However, coupling between these domains can principally be mediated through the pore in both the closed and open conformations (left and right faces of the cube in Fig. 1 C), giving rise to a state-dependent interaction of any two domains.
It is interesting to note that when context-dependent interactions are not neglected, = −4RTln(ce)(a/c); i.e., the overall pathway coupling is greater (or smaller depending on the value of the (a/c) ratio) than the sum of the coupling free energies because of the interaction of the S domain with the V (e) and P domains (c). This is a manifestation of non-additivity, a characteristic property of modular hierarchical systems.
In practical terms, levers constrain the possible configuration space of the channel so that only those states that comprise a sub-scheme of the general model are visited, a step that proved instrumental in understanding models of the BK channel (Rothberg and Magleby, 1999; Horrigan and Aldrich, 2002).
WH(g) is analogous to the lnε function defined by Chowdhury and Chanda (2010) in their χ-value analysis of mutational effects on cooperativity. Principally, WH(g)-like Hill transform functions can be defined for any domain, provided that a specific marker for activation is available.
The y and z directions in Fig. 1 C, respectively, describe saturating ligand concentrations and strong depolarization; they correspond, by definition, to the μΔ and VΔ levers.
In generating his simulated data, Sigg uses a slow voltage-ramp protocol instead of the conventional step protocol. The major advantage of using such a protocol is that the entire (quasi-) equilibrium G-V and Q-V curves can be obtained with a single sweep.
References
- Chowdhury S., Chanda B. 2010. Deconstructing thermodynamic parameters of a coupled system from site-specific observables. Proc. Natl. Acad. Sci. USA. 107:18856–18861 10.1073/pnas.1003609107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Chanda B. 2012. Estimating the voltage-dependent free energy change of ion channels using the median voltage for activation. J. Gen. Physiol. 139:3–17 10.1085/jgp.201110722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Chanda B. 2013. Free-energy relationships in ion channels dually activated by voltage and ligand. J. Gen. Physiol. 141:11–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz A., Fersht A.R. 1990. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J. Mol. Biol. 214:613–617 10.1016/0022-2836(90)90275-Q [DOI] [PubMed] [Google Scholar]
- Horovitz A., Fersht A.R. 1992. Co-operative interactions during protein folding. J. Mol. Biol. 224:733–740 10.1016/0022-2836(92)90557-Z [DOI] [PubMed] [Google Scholar]
- Horrigan F.T., Aldrich R.W. 2002. Coupling between voltage sensor activation, Ca2+ binding and channel opening in large conductance (BK) potassium channels. J. Gen. Physiol. 120:267–305 10.1085/jgp.20028605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J., Wyman J., Changeux J.P. 1965. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12:88–118 10.1016/S0022-2836(65)80285-6 [DOI] [PubMed] [Google Scholar]
- Rothberg B.S., Magleby K.L. 1999. Gating kinetics of single large-conductance Ca2+-activated K+ channels in high Ca2+ suggest a two-tiered allosteric gating mechanism. J. Gen. Physiol. 114:93–124 10.1085/jgp.114.1.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovsky E., Yifrach O. 2007. Principles underlying energetic coupling along an allosteric communication trajectory of a voltage-activated K+ channel. Proc. Natl. Acad. Sci. USA. 104:19813–19818 10.1073/pnas.0708120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigg D. 2013. A linkage analysis toolkit for studying allosteric networks on ion channels. J. Gen. Physiol. 141:29–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman J., Jr 1964. Linked functions and reciprocal effects in hemoglobin: a second look. Adv. Protein Chem. 19:223–286 10.1016/S0065-3233(08)60190-4 [DOI] [PubMed] [Google Scholar]
- Wyman J. 1967. Allosteric linkage. J. Am. Chem. Soc. 89:2202–2218 10.1021/ja00985a037 [DOI] [Google Scholar]
- Yifrach O. 2004. Hill coefficient for estimating the magnitude of cooperativity in gating transitions of voltage-dependent ion channels. Biophys. J. 87:822–830 10.1529/biophysj.104.040410 [DOI] [PMC free article] [PubMed] [Google Scholar]