Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is a common muscle disease whose molecular pathogenesis remains largely unknown. Over-expression of FSHD region gene 1 (FRG1) in mice, frogs, and worms perturbs muscle development and causes FSHD–like phenotypes. FRG1 has been implicated in splicing, and we asked how splicing might be involved in FSHD by conducting a genome-wide analysis in FRG1 mice. We find that splicing perturbations parallel the responses of different muscles to FRG1 over-expression and disease progression. Interestingly, binding sites for the Rbfox family of splicing factors are over-represented in a subset of FRG1-affected splicing events. Rbfox1 knockdown, over-expression, and RNA-IP confirm that these are direct Rbfox1 targets. We find that FRG1 is associated to the Rbfox1 RNA and decreases its stability. Consistent with this, Rbfox1 expression is down-regulated in mice and cells over-expressing FRG1 as well as in FSHD patients. Among the genes affected is Calpain 3, which is mutated in limb girdle muscular dystrophy, a disease phenotypically similar to FSHD. In FRG1 mice and FSHD patients, the Calpain 3 isoform lacking exon 6 (Capn3 E6–) is increased. Finally, Rbfox1 knockdown and over-expression of Capn3 E6- inhibit muscle differentiation. Collectively, our results suggest that a component of FSHD pathogenesis may arise by over-expression of FRG1, reducing Rbfox1 levels and leading to aberrant expression of an altered Calpain 3 protein through dysregulated splicing.
Author Summary
Alternative splicing is a major contributor to the complexity of human cells, and its disruption can lead to a wide range of human disorders. FSHD is one of the most important muscle diseases. While muscle differentiation defects have been widely reported in the disease, the molecular mechanisms responsible are largely unknown. We found that expression of the alternative splicing factor Rbfox1 is a direct FRG1 target, and its expression decreased in the muscles of a mouse model of FSHD and FSHD patients. Moreover, alternative splicing of Calpain 3, encoding for a protease involved in muscle differentiation, is regulated by Rbfox1 and is altered in the muscles of the mouse model of FSHD and FSHD patients. Interestingly, we found that Rbfox1 is required for muscle differentiation and that this activity is likely mediated by Calpain 3 alternative splicing. Hence, our results suggest that decreased expression of Rbfox1 and aberrant Calpain 3 splicing contribute to the muscle differentiation defects of FSHD patients.
Introduction
Facioscapulohumeral Muscular Dystrophy (FSHD, OMIM 158900), the third most common myopathy with an incidence of 1 in 15,000 in the human population [1], [2], is characterized by progressive wasting of a specific subset of skeletal muscles [3], [4]. Myogenic defects in FSHD have been widely reported [5]–[11], but the molecular mechanism responsible for them is currently unknown. While FSHD is primarily a disease of skeletal muscles, epilepsy, mental retardation and autism have also been described in severely affected FSHD infants [12]–[15]. FSHD is inherited as an autosomal dominant disorder but is caused by a peculiar molecular mutation [1], [16] involving deletion of tandemly repeated 3.3 kbp sequences, called D4Z4 [17]–[20], in the subtelomeric region of chromosome 4 (4q35). The D4Z4 deletion causes a Polycomb/Trithorax epigenetic switch leading to increased expression of several 4q35 genes specifically in FSHD patients [21]–[23], offering an explanation for its dominant phenotype. Since expression of multiple genes is affected, the molecular pathogenesis of FSHD has been challenging to untangle, and as yet no therapy is available for FSHD patients.
Among the genes up-regulated in FSHD, FRG1 (FSHD region gene 1) is a likely contributor to FSHD pathogenesis since it is required for normal muscle development [24] and its over-expression in mice, Xenopus and C. elegans causes an FSHD-like phenotype [24]–[27]. The precise function of FRG1 is still unknown, but there is evidence for a role in RNA processing [25], [28]–[34]. For example, several studies reported association of FRG1 with the spliceosome [28], [30], [32], [33]. Moreover, FRG1 assumes a speckled nuclear distribution pattern characteristic of mammalian splicing factors [34]. Finally, altered splicing of the muscle-expressed genes MTMR1 and TNNT3 has been reported in FSHD [25].
Muscle tissues, like brain, are rich in their use of tissue-specific alternative splicing events to regulate gene expression and produce specialized protein isoforms. Many of these events display enrichment for putative binding sites for the evolutionary conserved, tissue-specific Rbfox family of alternative splicing regulators: Rbfox1 (Fox-1 or A2BP1), Rbfox2 (Fox-2 or Rbm9) and Rbfox3 (Fox-3 or NeuN) [35]. Rbfox1 is expressed in brain, skeletal muscle and heart [35]–[38], while Rbfox2 has a broader expression pattern, being detected in whole embryo, stem cells, hematopoietic cells and in adult brain, heart and ovary [35], [36], [39]–[42]. In contrast, Rbfox3 has been observed only in neurons [36], [43]. So far, few genes have been experimentally validated as Rbfox family targets in muscle. In this paper, we show that FRG1 over-expression in mouse muscle is associated with widespread alternative splicing perturbations that appear to delay or inhibit proper muscle development at a cellular level. We show that FRG1 over-expression decrease Rbfox1 RNA. In FRG1 mouse muscles, C2C12 myoblasts over-expressing FRG1, and in FSHD patients, down-regulation of Rbfox1 expression leads to altered splicing of Rbfox1-dependent muscle exons. We further show that Rbfox1 is required for myogenesis and part of this requirement may involve correct regulation of Calpain 3 alternative splicing. Our results provide a molecular mechanism for myogenic defects in FSHD and identify possible therapeutic targets.
Results
Widespread mRNA expression level and alternative splicing changes in muscles of FRG1 mice
A peculiar distribution of affected muscles and the progressive character of the disease are among the features shared between FSHD patients and FRG1 mice [25]. To investigate the molecular connections between FRG1 over-expression and the impairment of muscle development and function, we took a genome-wide approach. Since FRG1 is thought to function in pre-mRNA splicing [28], [30], [32], [33], we employed splicing-sensitive microarrays [44], [45]. To identify changes that occur differently in muscles differentially affected in FSHD, we compared RNA extracted from vastus lateralis (highly affected) and biceps brachii (mildly affected) muscles of three FRG1 mice and control littermates [25]. To follow disease progression, we analyzed 4-week-old animals (no signs of muscular dystrophy) and 13-week-old mice (fully developed muscular dystrophy) [25]. The full results for genes investigated at transcriptional and splicing levels are displayed in Tables S1 and S2, respectively.
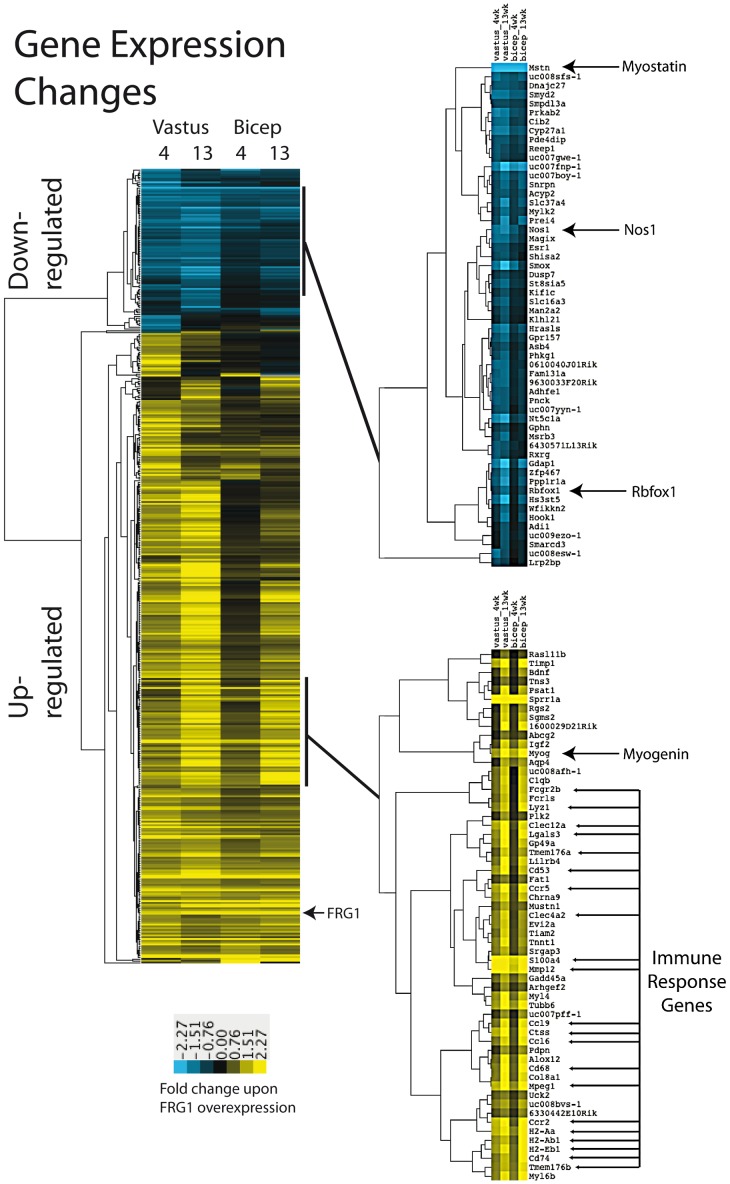
We identified 440 genes whose expression changes more than 2 fold (q<0.05) in at least one of the four experiments (Table S3a). The effect of FRG1 over-expression was very coherent among the four sets of samples. Indeed, all genes that changed did so in the same direction in every sample in which they changed. Upon hierarchical clustering (Figure 1), two groups of genes, those down-regulated and those up-regulated upon FRG1 over-expression were clearly defined. No clear functional enrichment was observed upon GO analysis of the down-regulated genes, although there were clear signs of altered muscle development (see below). In contrast, up-regulated genes showed strong enrichment for inflammation and immune response genes (e.g. GO:0002376 P = 3.40E-05), as well as enrichment for contractile proteins. To directly assess the presence of an inflammatory infiltrate, we performed a specific histological staining using antibodies against the pan-hematopoietic marker CD45 and a quantitative analysis of CD45 positive cells by FACS. We found no evidence of inflammation in 4-week-old FRG1 mice (Figure S1). Hence, these results suggest that the muscle fibers are the source of the up-regulation of inflammatory genes identified by our microarray analyses performed in 4-week-old FRG1 mice. Among the genes affected by FRG1 over-expression were several whose responses indicated developmental perturbations. The mRNA of Myostatin, an inhibitor of muscle development [46], was strongly down-regulated (7–10 fold). On the contrary, the mRNA of Myogenin, a promoter of muscle cell development [47], was strongly up regulated (3–7 fold). The mRNA of Nos1, a regulator of myogenic stem cells activation and differentiation frequently affected in muscular dystrophy [48], was down-regulated primarily in vastus (3 fold). We concluded that FRG1 overexpression has a profound effect on the muscle transcriptome, including induction of immune response and inflammatory genes, and alteration of proper muscle development.
Figure 1. Gene expression changes in muscles of mice over-expressing FRG1.
Average linkage clustering was applied to the fold change values of the 440 genes whose expression changes by at least 2 fold (SAM q<0.5) in at least one of the four comparison datasets. Selected nodes are expanded on the right. Genes with increased expression in FRG1 over-expressing muscles are shown in yellow in the heatmap, while those whose expression decreased are in blue.
We next searched the data for changes in alternative splicing. We observed 1735 splicing events whose include/skip isoform ratio changed significantly (|Sepscore|≥0.3; p = 0, see [49], [50]) between FRG1 and wild type mice in vastus lateralis at 4 weeks of ages (V4w), 1005 events in biceps brachii at 4 weeks of ages (B4w), 1895 events in vastus lateralis at 13 weeks of ages (V13w) and 1454 events in biceps brachii at 13 weeks of ages (B13w) (Table S3b). Based on this simple count, more events were altered in the more affected muscle (vastus), and 13-week-old mice were more affected than 4-week-old mice. Among the transcripts identified were Mtmr1 and Tnnt3, previously shown aberrantly spliced in FSHD [25]. We selected 48 genes for validation using semi-quantitative RT-PCR with RNA extracted from vastus lateralis muscle at 4 weeks of age from three new FRG1 mice and control littermates that were not used for the microarray analysis (Table S4). An example of the typical results obtained for the different RNA splicing modes is displayed in Figure 2a. For the 48 genes, the microarray results were validated by RT-PCR in 83% of the cases (Figure S2), confirming that more genes are affected, and are more severely affected in vastus lateralis than in biceps (Figure 2b and Figure S3, Table S3b).
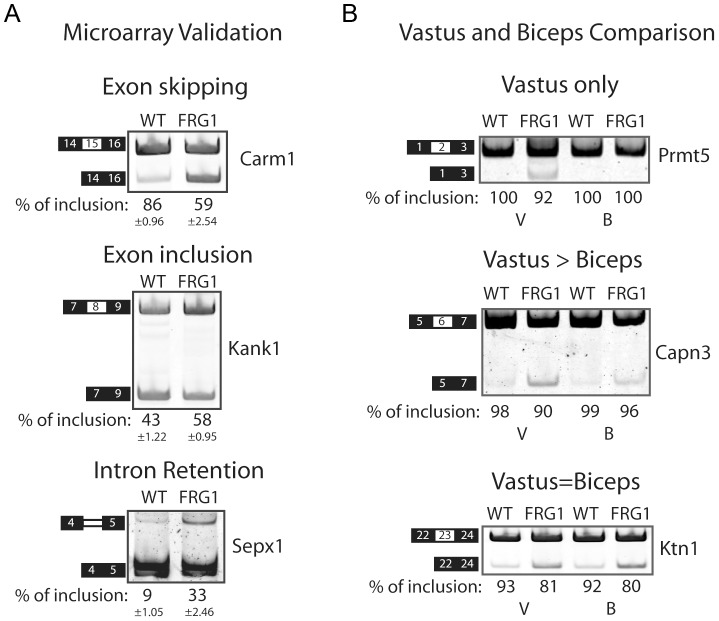
Figure 2. RT–PCR validation of splicing events belonging to different classes.
(a) RT-PCR performed using RNA extracted from vastus lateralis muscle of FRG1 and control littermates (n = 3 per group, 4 weeks old mice). Examples of increased exon skipping (Carm1), exon inclusion (Kank1) or intron retention (Sepx1) in FRG1 mice is shown. RT-PCR products from three individual FRG1 and control WT mice were quantified using the Typhoon and the skipping rates were calculated. Samples were judged as being different from WT if a t-test indicated that the sample was unlikely to be from the WT distribution with P<0.05. (b) Examples of alternative exons affected only (Prmt5), primarily (Capn3) or similarly (Itga7) in FRG1 mice vastus lateralis compared to biceps brachii muscles. Numbers below gel images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
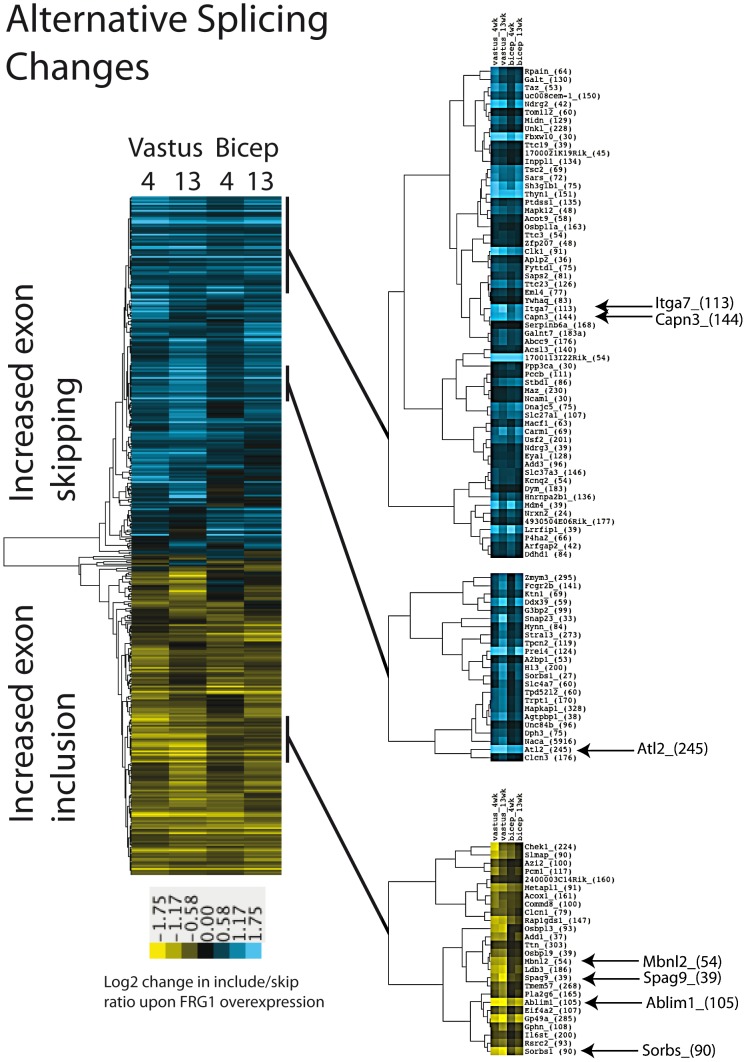
To study the pattern of splicing alterations in vastus and biceps muscles at 4- and 13- weeks, we clustered affected exons (q = 0, |Sepscore|≥0.3) by the log2 values of the ratio of inclusion/skipping ratios in FRG1 tissues relative to wild type (Figure 3). Like the transcript level defects, the splicing defects were coherent, especially among strongly affected exons. Affected exons belong to genes involved in calcium regulation, muscle cell development and alternative splicing in muscle. For example, we noted increased inclusion of a 54 nucleotide exon in the mRNA for splicing factor Mbnl2 (Figure 3), however the significance of this change for the activity of Mbnl2 is not known. Our current lack of knowledge in general for the functional implications of most alternative splicing modifications makes it difficult to interpret the global impact for the changes identified by the microarrays.
Figure 3. Alternative splicing changes in muscles of mice over-expressing FRG1.
Average linkage clustering was applied to alternative cassette exon splicing events with |Sepscore|≥0.3 in at least one of the four comparison datasets (q = 0). Selected nodes are expanded on the right. Exons which shown increased inclusion in FRG1 over-expressing muscles are shown in yellow in the heatmap, while exons with decreased inclusion are represented in blue.
To assess whether the observed splicing changes are associated with sequence motifs that might betray the splicing factor controlling them, we performed motif analysis.
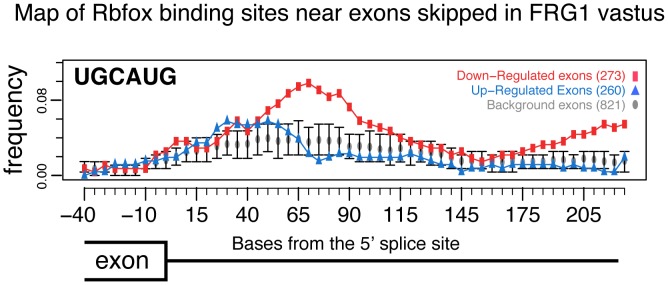
We counted the frequency of 6-mer “words” in the sequences within and surrounding exons whose inclusion was affected by FRG1 over-expression. We found enrichment of TGCATG (P = 0.003), which is the highest ranking word in our search recognizably associated with a known RNA binding protein family, in this case, Rbfox [35]. This enrichment is found downstream of exons whose skipping increases in the FRG1 muscles relative to wild type. To map the location of this motif in the exons that possess it, we measured the frequency of TGCATG in sliding windows across the exon set, as compared to the background frequency among exons which splicing did not change in the experiment (Figure 4). We observed a strong peak suggesting enrichment of the Rbfox binding site TGCATG in the region 70–90 nucleotides downstream from the 5′ splice site of exons whose inclusion decreased upon FRG1 overexpression in 4-week-old vastus muscle. This location implicated one or more Rbfox family members in the activation of a set of exons normally expressed in vastus at this time. Furthermore this suggested that FRG1 over-expression compromised function of one or more Rbfox family members. Consistent with this, we found that Rbfox1 gene expression is down-regulated in tissues over-expressing FRG1 (Figure 1).
Figure 4. RNA map of Rbfox motifs downstream of exons.
Each point represents the average frequency of UGCAUG in the 150 bp downstream of the 260 exons whose inclusion increases (blue triangles), the 273 exons whose inclusion decreases (red squares) or the 821 expressed alternative cassette exons whose splicing did not change in the comparison (gray circles, q>0.2 and |Sepscore|<0.3). Error bars indicate 95% confidence intervals of the mean frequency distribution for this population of background exons.
Splicing alterations are similar to those in FRG1 over-expressing cultured myoblasts, but distinct from those in mdx mice
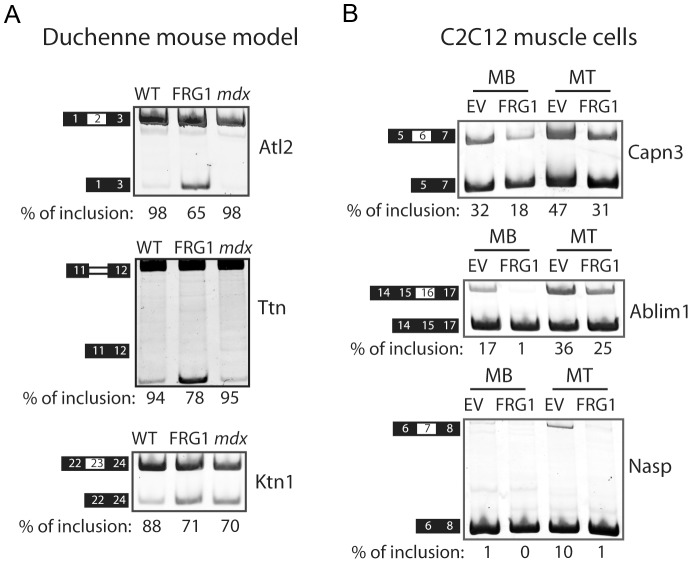
The altered splicing events identified here could be primary consequence of FRG1 over-expression, secondary consequence of FRG1 affecting expression of splicing factors for example, or far downstream consequences observable for any muscle disease. Most of the splicing changes were present in FRG1 mice at 4 weeks (Table S3b), when the animals did not show signs of muscular dystrophy at histological and ultra-structural levels [25]. This strongly suggested that the altered splicing events were primarily associated with ongoing FRG1 over-expression and related to early disease, before the appearance of changes due to muscle wasting. To assess this, we analyzed splicing in mdx mice, the mouse model of Duchenne muscular dystrophy [51], as an example of a muscle disease with a distinct genetic cause. This test showed that the vast majority (30 out of 40 events analyzed) of the events altered in FRG1 overexpressing mice were spliced normally in mdx mice (Figure 5a and Figure S4). This excluded the possibility that most of the splicing alterations identified in FRG1 mice were simply due to a common molecular phenotype found in all muscular dystrophies. To asses whether the phenotype observed in mouse tissues primarily concerns cell-independent functions or arises only in the presence of complex sets of cell types in developing tissues, we compared stable C2C12 muscle cells expressing either the empty vector (C2C12-EV) or FRG1 at low levels (C2C12-FRG1) ([25] and Figure S5). We analyzed both proliferating (myoblast, MB) and differentiating (myotubes, MT) cells. For the majority of the genes tested, we found that C2C12- FRG1 cells displayed splicing alterations similar to FRG1 mice (Figure 5b and Figure S5). Collectively, these findings indicated that FRG1 over-expression induced a characteristic set of splicing changes that can be observed in both cultured muscle cells and in tissues, and is distinct from that observed in a different model of muscular dystrophy.
Figure 5. Alternative splicing changes are a primary consequence of FRG1 overexpression.
(a) Examples of alternative exons (Atl2) or introns (Ttn) spliced normally in the mouse model of Duchenne muscular dystrophy, mdx mice, and example of an alternative exon similarly altered in FRG1 and mdx mice (Ktn1). (b) RT-PCR analysis of mRNA splicing variants from proliferating (MB) and differentiating (MT) C2C12 muscle cells over-expressing FRG1. Examples of alternative splicing changes present in both MB and MT (Capn3), only in MB (Ablim1), or only in MT (Nasp). Numbers are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
The alternative splicing factor Rbfox1 is selectively down-regulated in mice and cells over-expressing FRG1 and in FSHD patients
A recent report indicated that recombinant FRG1 binds RNA in vitro and is associated to FXR1 and FRG1 mRNAs [52]. If FRG1 is an RNA binding protein, affected splicing events could be directly controlled by FRG1. Using the same antibodies and experimental conditions to perform RNA immunoprecipitation (RIP) as reported [52], we could not detect specific FRG1 association with the set of RNAs whose splicing is altered in FRG1 over-expressing C2C12 cells (Figure S6a). FRG1 RIP was also performed using an alternative protocol that we have successfully applied to other RNA-binding proteins (see below), with the same result (Figure S6b).
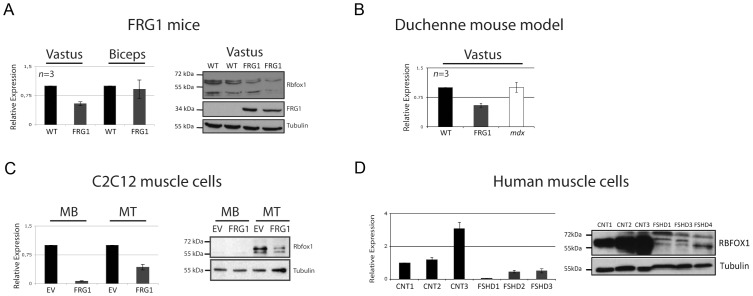
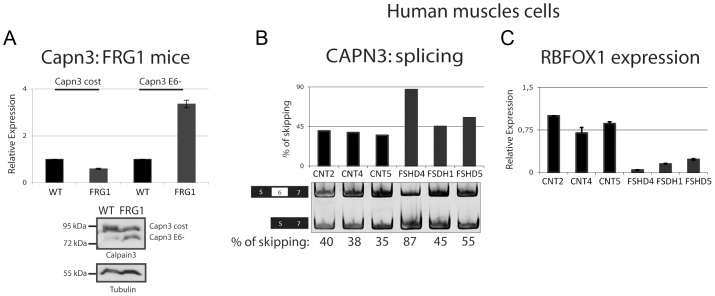
Since we did not find evidence of FRG1 binding to the RNAs whose splicing is altered in FRG1 mice, we investigated the hypothesis that FRG1 might alter splicing through its effect on the activity or the expression of known splicing factors. Since FRG1 over-expression appeared to down-regulate Rbfox1 mRNA and inhibited the activation of exons with Rbfox1 binding sites, we decided to examine more carefully the expression of Rbfox splicing factors upon FRG1 over-expression. While Rbfox2 and Rbfox3 expression was not significantly affected (data not shown), expression of Rbfox1 protein was significantly reduced in vastus lateralis of FRG1 mice (Figure 6a). Furthermore, the extent of down-regulation of Rbfox1 expression paralleled the severity of the disease in different muscles (Figure 6a). In addition, Rbfox1 was expressed at normal levels in mdx mice, indicating that its down-regulation in FRG1 mice is not generally associated with muscular dystrophy (Figure 6b). Rbfox1 mRNA down-regulation was also displayed by both proliferating (MB) and differentiating (MT) C2C12-FRG1 muscle cells, indicating that it is a cellular consequence of FRG1 over-expression (Figure 6c, left). Rbfox1 protein down-regulation was evident in differentiating (MT) C2C12-FRG1 muscle cells, while the expression level of Rbfox1 in proliferating (MB) C2C12-FRG1 muscle cells was below the sensitivity of our immunoblotting (Figure 6c, right). Importantly, RBFOX1 mRNA and RBFOX1 protein were down-regulated in primary human muscle cells derived from FSHD patients (Figure 6d).
Figure 6. Rbfox1 is selectively down-regulated in mice, in cells over-expressing FRG1, and in FSHD patients.
(a) Left panel: real-time RT-PCR analysis showing that in FRG1 mice Rbfox1 expression is preferentially downregulated in vastus lateralis compared to biceps brachii. Gapdh expression was used for sample normalization. Right panel: immunoblotting on vastus lateralis using anti-Rbfox1 antibody. Tubulin was used as loading control and FRG1 antibody as confirmation of the genotype. (b) Real-time RT-PCR analysis showing that Rbfox1 expression is normal in mdx mice. (c) Left: real-time RT-PCR analysis showing that Rbfox1 expression is down-regulated in proliferating (MB) or differentiating (MT) C2C12 muscle cells over-expressing FRG1. Right: immunoblotting using protein extracted from the same samples as in (c, left) using anti-Rbfox1 antibodies. Tubulin was used as loading control. Note that in proliferating C2C12 cells Rbfox1 protein is almost undetectable, while its levels significantly increase during myogenic differentiation. (d) Left panel: real-time RT-PCR analysis of RBFOX1 expression using RNA extracted from muscle cells derived from three different normal individuals and three different FSHD patients. Right panel: immunoblotting using protein extracted from the same samples as in (d, left) using anti-RBFOX1 antibody. Tubulin was used as loading control.
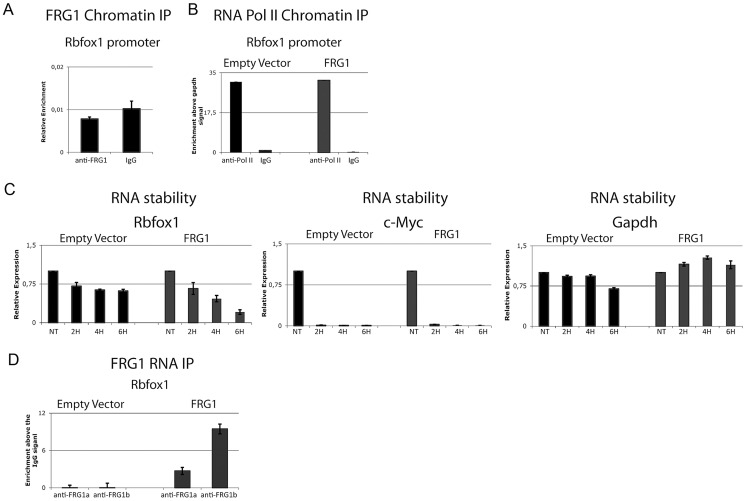
FRG1 over-expression could down-regulate Rbfox1 expression at transcriptional or post-transcriptional level. To investigate if FRG1 regulates Rbfox1 at the transcriptional level, we performed chromatin immunoprecipitation (ChIP) using anti-FRG1 antibodies. We found no evidence of FRG1 binding to the promoter of the Rbfox1 gene (Figure 7a). To further investigate this, we performed ChIP using anti-RNA pol II antibodies, as it is known that RNA pol II recruitment correlates with the transcriptional rate [53]. As shown in Figure 7b, we found no change in RNA pol II loading on the Rbfox1 promoter in FRG1 over-expressing cells compared to control. Collectively, these results indicate that FRG1 does not regulates Rbfox1 expression at the transcriptional level and suggest that it could act at post-transcriptional level. To test this, we monitored the stability of the Rbfox1 mRNA upon FRG1 over-expression. As shown in Figure 7c, the stability of the Rbfox1 mRNA was greatly reduced in FRG1 over-expressing cells compared to control. The result appears specific since we found no change in the stability of two other mRNAs: c-Myc and Gapdh. To investigate if Rbfox1 could be a direct FRG1 target, we performed RIP following UV-crosslinking. As shown in Figure 7d, using two different anti-FRG1 antibodies [52], we found an FRG1 association to the Rbfox1 mRNA selectively in cells over-expressing FRG1. Altogether, our results strongly suggest that FRG1 down-regulates Rbfox1 by decreasing the stability of its mRNA.
Figure 7. FRG1 regulates the stability of the Rbfox1 mRNA.
(a) ChIP of FRG1 in the promoter region of the Rbfox1 gene. (b) The distribution of RNA pol II on C2C12-EV and C2C12-FRG1 cells in the promoter region of the Rbfox1 gene. (c) Real-time RT-PCR of the kinetics of Rbfox1, c-Myc and Gapdh expression after 8 hours of ActD (Actinomycin D) treatment on C2C12-EV and C2C12-FRG1 cells. (d) RNA-IP experiment on samples from (c) using anti-FRG1 antibodies or control IgG antibodies. Immunoprecipitated material was analyzed by real-time RT-PCR, normalized versus the relative input and plotted as fold enrichment versus the IgG. RT-minus control experiments showed the absence of DNA contamination (data not shown).
Identification of direct Rbfox1 targets altered in FRG1 mice
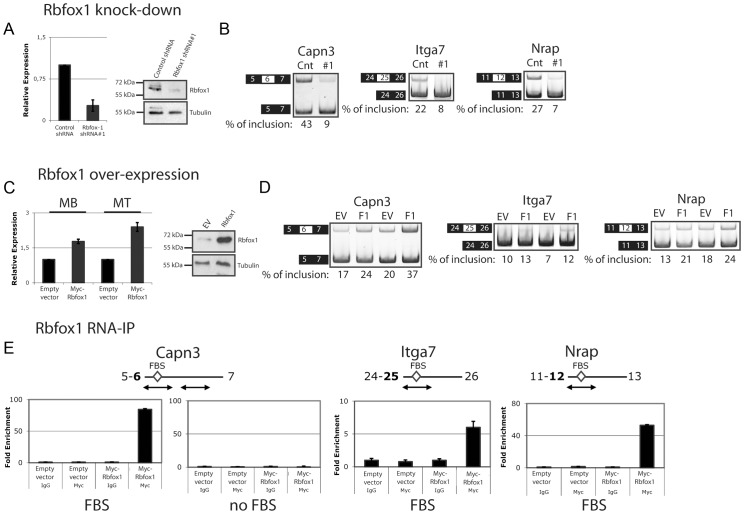
To test directly whether Rbfox1 regulated splicing of the exons we identified in FRG1 over-expressing cells and tissues, we performed Rbfox1 knockdown and overexpression studies in C2C12 cells expressing normal levels of FRG1. We selected two independent shRNAs (Rbfox1 shRNA #1 and #2) that significantly decreased Rbfox1 expression in C2C12 muscle cells at both its mRNA and protein levels (Figure 8a and Figure S7b). For all pre-mRNAs displaying putative Fox binding sites (FBS) analyzed (6/6), Rbfox1 knockdown caused alternative splicing changes similar to those detected in C2C12-FRG1 (Figure 8b and Figure S7a and S7c). None of the pre-mRNA lacking putative FBS analyzed (5/5) displayed altered splicing upon Rbfox1 knockdown (Figure S7a and S7c). The directions of the alternative splicing changes caused by Rbfox1 knockdown agrees with the positions of the putative FBS as previously noted [35], see Figure 8b, Figure S7a and S7c).
Figure 8. Rbfox1 down-regulation is responsible for significant portion of the splicing alterations in FRG1 mice.
(a) Specific Rbfox1 knockdown was confirmed by real-time RT-PCR and immunoblotting using RNAs and proteins isolated from C2C12 muscle cells expressing a control non-silencing shRNA or an shRNA specific for Rbfox1 (shRNA#1). (b) Examples of alternative splicing changes caused by Rbfox1 knockdown are showed. Numbers are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons. (c) Rbfox1 overexpression causes alternative splicing changes opposite to FRG1 over-expression. Specific Rbfox1 over-expression was confirmed by real-time RT-PCR and immunoblotting using RNAs and proteins isolated from C2C12 muscle cells expressing an empty vector (EV) or a Myc-tagged Rbfox1 (F1) either in proliferating or differentiating C2C12 muscle cells. (d) Examples of alternative splicing changes caused by Rbfox1 over-expression are showed. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons. (e) Selective in vivo association of Rbfox1 to target regions displaying putative Fox binding sites (FBS). RIP experiment on samples from (c) using anti-Myc or control IgG antibodies. Immunoprecipitated material was analyzed by RT-PCR, quantified using the Typhoon, normalized versus the relative input and plotted as fold enrichment versus the IgG. RT-minus control experiments showed the absence of DNA contamination (data not shown).
Next, we analyzed alternative splicing in C2C12 cells over-expressing Rbfox1 (Figure 8c). In this case, for all pre-mRNAs analyzed (6/6) Rbfox1 over-expression caused alternative splicing changes opposite to those detected in C2C12-FRG1 or Rbfox1 knockdown C2C12 cells, and these also agree with the position of the putative FBS (Figure 8d, Figure S7d). To determine if Rbfox1 regulated pre-mRNAs were bound by Rbfox1 in vivo we performed RIP experiments. As shown in Figure 8e and Figure S7e, Rbfox1 was associated with all pre-mRNAs displaying the putative FBS by RIP. No Rbfox1 RIP signal was found for a region of the Calpain 3 transcript lacking a putative FBS (Figure 8e) or to the control Gapdh transcript (Figure S7e) suggesting that the tested pre-mRNAs were direct Rbfox1 targets. Collectively, these results strongly suggest that Rbfox1 down-regulation is responsible for a subset of the alternative splicing changes observed in cells and muscles engaged in the FRG1 overexpression associated with FSHD.
Altered Calpain 3 splicing in FSHD patients
Among the pre-mRNAs displaying aberrant splicing as a result of FRG1 overexpression in muscles (Figure 3), or after Rbfox1 knockdown and Rbfox1 overexpression in cultured cells (Figure 8), we focused our attention on Calpain 3 (Capn3). Capn3 encodes for a muscle-specific Calcium-dependent protease [50] and its mutation is responsible for limb-girdle muscular dystrophy type 2A (LGMD2A) [49]. A Capn3 alternative splicing isoform lack exon 6 (Capn3 E6–) was increased in FRG1 mice (Figure 3). Increased Capn3 E6– upon FRG1 over-expression was confirmed by semi-quantitative RT-PCR (Figure 2b and Figure 5b), real-time RT-PCR and immunoblotting (Figure 9a). Notably, increased CAPN3 E6- expression was observed in FSHD muscle cells (Figure 9b) that also displayed Rbfox1 down-regulation (Figure 9c).
Figure 9. Alternative splicing isoform of Calpain 3 increased in FRG1 mice and in FSHD patients.
(a) Real-time RT-PCR and immunoblotting analysis confirming that the Capn3 alternative splicing isoform lacking exon 6 (Capn3 E6-) is increased in the vastus lateralis muscle from FRG1 mice. (b) RT-PCR analysis of CAPN3 splicing in human muscle cells derived from three different healthy subjects and three different FSHD patients indicates increased expression of CAPN3 E6- isoform in FSHD patients. Numbers below the image are the percentage of exon skipping. Black boxes illustrate constitutive exons, white boxes alternatively spliced exon. RT-PCR products were quantified using the Typhoon. (c) RBFOX1 expression analysis was performed on RNA extracted from the same samples as in (b) by real-time RT-PCR.
Transgenic mice with muscle-specific increased Capn3 E6– expression display some similarity to FRG1 mice [25], [54]. For example, both models display kyphosis, reduced muscle mass, reduced muscle fiber cross-sectional area and increased number of centrally nucleated muscle fibers [25], [54]. Moreover, in both models vastus lateralis is more affected than biceps brachii [25], [54]. Finally, Evans blue dye staining and creatine kinase levels are normal in Capn3 E6– and FRG1 mice indicating that muscle disease in both models, like in FSHD patients, do not compromise sarcolemma integrity [25], [54]. Based on this, it is tempting to speculate that an increase in Capn3 E6– isoform expression as a consequence of FRG1-mediated down regulation of Rbfox1 could contribute to FSHD.
FRG1 over-expression, Rbfox1 knockdown of Capn3 E6– all inhibit myogenic differentiation
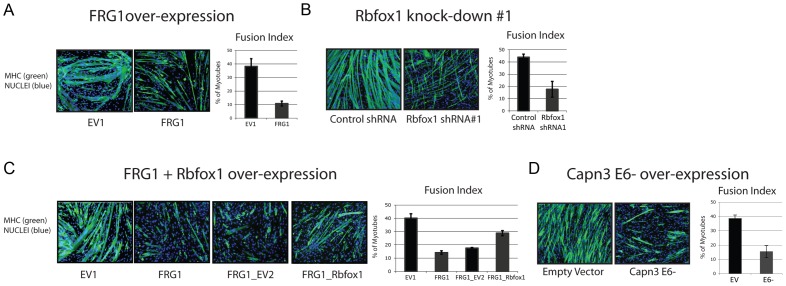
Several reports indicate a muscle differentiation defect in FSHD [5]–[7], [9]–[11]. To investigate whether this phenotype might be mediated by FRG1 over-expression, we compared the differentiation capability of C2C12 muscle cells over-expressing FRG1 (FRG1) or the empty vector (EV1). As shown in Figure 10a, FRG1 over-expression significantly reduced the differentiation capability of C2C12 cells. Quantification of the fusion index showed that FRG1 cells form only 25% of the myotubes formed by EV1 (Figure 10a). C2C12 cells expressing two independent shRNAs specific for Rbfox1 (shRNA#1 and #2) also displayed a reduction in myogenesis, although lower compared to FRG1 cells (compare Figure S7b, Figure 10a and Figure 10b).
Figure 10. Altered Calpain 3 splicing inhibits myogenesis.
(a) C2C12 muscle cells over-expressing FRG1, (b) Rbfox1 knockdown with shRNA#1 and (d) over-expressing Capn3 E6- display a reduced myogenic differentiation. (c) C2C12 muscle cells over-expressing FRG1 and Rbfox1 present increase myogenic differentiation compared to C2C12-FRG1. Left: immunofluorescence using antibodies specific for skeletal muscle myosin heavy chain (MHC), a typical marker of terminally differentiated muscle cells. Right: fusion index defined as the percentage of nuclei belonging to MHC-positive cells with three or more nuclei. The values reported in the graph are the means plus-minus standard deviations from two separate experiments performed in triplicate.
FRG1 and Rbfox1 might act independently on muscle differentiation. To establish a link between the two proteins, we performed rescue experiments by over-expressing Rbfox1 in FRG1 over-expressing cells. By performing differentiation experiments, we observed a partial amelioration of the phenotype in Rbfox1/FRG1 over-expressing cells compared to control cell lines (Figure 10c). This result strongly suggests that Rbfox1 plays an important role in the myogenic inhibition caused by FRG1 over-expression.
Importantly, a differentiation defect was also displayed in C2C12 muscle cells selectively over-expressing Capn3 E6– (Figure S8 and Figure 10d). Thus, as with the mouse models, the C2C12 cell system suggested that an increase in Capn3 E6– isoform expression as a consequence of FRG1-mediated down-regulation of Rbfox1 could contribute to FSHD.
Discussion
FSHD is a complex disease in which the severity, rate of progression and distribution of muscle weakness display great variability even among close family relatives. The unusual nature of the mutation that causes FSHD and its complex effect on chromatin surrounding the 4q35 region makes it highly unlikely that the root cause can be attributed to a single gene. Two 4q35 genes have been investigated as candidate FSHD genes, DUX4 and FRG1 [23], [55], [56]. Individually, each gene causes myopathic changes when over-expressed in muscle cells in vivo [25], [57]. For Dux4, a disruption of muscle development/differentiation through competition with Pax3/Pax7 myogenic regulators has been reported [58]. Our gene expression profiling of FRG1 mice (Figure 1) and the myogenic defects caused by FRG1 over-expression in C2C12 cells (Figure 10) indicate that FRG1 also disrupts muscle differentiation. Surprisingly, we found that a significant number of genes involved in inflammation and immune response are up-regulated in vastus muscles of FRG1 mice at 4 weeks of age (Figure 1). This is particularly intriguing since at this age FRG1 mice do not show any evidence of infiltrate/inflammation by histology, immunohistochemistry with anti-CD45 (expressed on all hematopoietic cells, except erythrocytes and plasma cells) antibodies or by FACS analysis of single cell preparations from the muscle (Figure S1). Moreover, at 4 weeks of age there is no sign of necrosis or muscle regeneration in FRG1 mice (i.e. no increase in centrally nucleated muscle fibers) (Xynos et al., submitted). Hence, it appears that the increased expression of inflammation and immune response genes originates directly from the muscle cells themselves. In classical muscular dystrophies, inflammation is secondary to the necrosis of muscle fibers. On the contrary, our results suggest that in FRG1 mice recruitment of hematopoietic cells is induced by the muscle directly expressing inflammation and immune response genes, before the appearance of muscle fiber necrosis and regeneration. This is in agreement with the fact that in FSHD, unlike classical muscular dystrophies, the inflammation is not simply an event secondary to muscle wasting but takes place early on in the disease and could triggers the disease process [59]–[62]. Based on this, it is tempting to speculate that a spurious activation of inflammation and immune response genes within muscle cells by FRG1 (and possibly DUX4, [63]) could play a role in FSHD onset and progression.
While the specific function of FRG1 is still unknown, an increasing body of evidence suggests a role in alternative splicing regulation [25], [28]–[30], [32], [33]. In this study, we investigated this by a genome-wide accounting of alternative splicing events affected by FRG1 over-expression. In both FRG1 mice and in FSHD patients, different muscles are affected differentially, the disease worsens with time [25], and the splicing profiles of differentially affected muscles of FRG1 mice at different ages correlates with the severity of the disease (Table S3b). Importantly, most of these splicing alterations are already evident in pre-symptomatic FRG1 mice, suggesting that they are related to disease onset and early progression (Figure 2). Moreover, comparisons with the splicing profile of the mouse model of Duchenne muscular dystrophy strongly indicate that the identified splicing changes are specific for the FSHD model rather than as a signature of diseased muscle (Figure 5a). Finally, most of the splicing alterations also occur in C2C12 muscle cells over-expressing FRG1 (Figure 5b), indicating a fundamental cellular response rather than a complex and secondary tissue effect. Collectively, these data identify a large set of splicing alterations that are a primary consequence of FRG1 over-expression and are involved in disease onset.
Our results suggest that the exons identified by the splicing-sensitive arrays are not direct FRG1 targets and that FRG1 function at least in part by regulating the expression of the splicing factor Rbfox1 at post-transcriptional level. We found that the Rbfox1 recognition sequence is over-represented in the alternative splicing events altered in FRG1 mice (Figure 4) and by Rbfox1 knockdown, over-expression and RIP experiments we confirmed that these are direct Rbfox1 targets (Figure 8 and Figure S7). Rbfox1 expression is down-regulated in FRG1 mice (Figure 6a), C2C12 cells over-expressing FRG1 (Figure 6c), and in FSHD patients (Figure 6d). We also found that the level of Rbfox1 down-regulation parallels the severity of the disease in different muscles of FRG1 mice, while it is expressed at normal levels in the mouse model of Duchenne muscular dystrophy (Figure 6b). Importantly, our results suggest that the FRG1 over-expression down-regulates the expression of Rbfox1 by decreasing the stability of its mRNA (Figure 7). Collectively, these results support a role for altered Rbfox1 expression in FSHD. While FSHD is primarily a disease of the skeletal muscle, epilepsy, mental retardation and autism have also been described in severely affected patients [12]–[15]. Since the same symptoms are caused by Rbfox1 mutation or down-regulation [64]–[73], it is tempting to speculate that Rbfox1 down-regulation could be involved in the neurological manifestations of FSHD. Among the transcripts displaying FRG1 and Rbfox1 dependent aberrant splicing, we focused on Capn3, as mutations in Capn3 cause LGMD2A [49], and FSHD and LGMD share clinical features [74], [75]. The clinical spectrum of FSHD can include prominent pelvic girdle weakness and, in some individuals, only minimal facial muscle involvement, leading to confusion in diagnosis [74]. Indeed, it has been reported that ∼8% of patients with a diagnosis of LGMD may in fact have FSHD [75].
We found that FRG1 over-expression is associated with increased expression of a Capn3 splicing isoform lacking exon 6 (Capn3 E6-) in both FRG1 mice and FSHD patients (Figure 2 and Figure 9b). Interestingly, transgenic mice over-expressing Capn3 E6- display phenotypic similarities to FRG1 mice [25], [54]. The fact that Capn3 E6- is already increased in pre-symptomatic FRG1 mice (Figure 2b and Figure 9a) and that its expression is normal in the mouse model of Duchenne muscular dystrophy (Figure S4) suggests that altered Capn3 splicing is a primary consequence of FRG1 overexpression and is not simply secondary to muscle wasting. This is confirmed by the fact that FRG1 over-expression in tissue culture is sufficient to drive increased Capn3E6- expression (Figure 5b). Intriguingly, we found that Capn3 E6- expression correlates with the degree of muscular dystrophy in different muscles and at different ages (Figure 2 and Figure 3) suggesting that altered Capn3 E6- could explain the differential susceptibility of different muscles to FRG1 over-expression and the progression of the disease over time.
Capn3 belongs to the Calpain family of non-lysosomal, soluble cytosolic calcium-dependent proteases [76]. Capn3 is highly expressed in the skeletal muscle and numerous data suggest a role for Capn3 in the maintenance of muscle integrity and function [76]. Capn3 presents three insertion sequences that differentiate it from all other calpains and might contribute to its muscle-specific functions. These regions, called NS, IS1, and IS2 are encoded by exons characterized by developmentally regulated alternative splicing [77]. Exon 6 encodes for IS1, a Capn3 domain involved in auto-inhibitory function through autolytic cleavage [76]. Capn3 E6-, encoding the isoform of Capn3 lacking IS1, is not expressed in healthy adult muscle and appears to be primarily important during development and muscle regeneration [76]. Accordingly, Capn3 E6- over-expression in adult muscle results in muscles that appear developmentally abnormal, underlining the importance of this alternative splicing event [54]. It has been suggested that alternative splicing isoforms of Capn3 could serve specific roles in regulating muscle differentiation [54]. Intriguingly, we found that Capn3 E6- over-expression in C2C12 muscle cells is sufficient to cause a differentiation defect similar to FRG1 over-expression or Rbfox1 down-regulation (Figure 10). Since myogenic defects have been reported in FSHD [5]–[11], it is tempting to speculate that altered Capn3 splicing could contribute to the pathogenesis of FSHD.
In conclusion, our studies have provided insight on the molecular pathogenesis of one of the most important muscle diseases. Moreover, our findings advance the understanding of the alternative splicing regulation of myogenic differentiation and identify possible therapeutic targets for the treatment of FSHD.
Materials and Methods
Ethics statement
All procedures involving human samples were approved by the Fondazione San Raffaele del Monte Tabor Ethical Committee. All animal procedures were approved by the Institutional Animal Care and Use Committee of the Fondazione San Raffaele del Monte Tabor and were communicated to the Ministry of Health and local authorities according to Italian law.
Human samples
Human primary myoblasts were obtained from the Telethon BioBank of the C. Besta Neurological Institute, Milano, Italy.
Animals and tissue preparation
All mice were maintained in Specific Pathogen Free conditions. FRG1-high transgenic mice were described previously [25]. mdx mice were purchased from the Jackson laboratory (Bar Harbor, ME, USA). Skeletal muscle from both vastus lateralis and biceps brachii were carefully dissected from individual age- and sex-matched mice and maintained in RNA later (Ambion). RNA was isolated using RNeasy Fibrous Tissue Midi or Mini Kit (Qiagen) according to the manufacturer's instructions. All RNA samples were treated with DNase I. Concentration, DNA-free RNA purity and integrity was determined by Nanodrop and by Agilent 2100 Bioanalyzer. Only samples with RNA integrity number ≥8 were used.
Splicing-sensitive microarrays
Ribosomal RNAs were removed from samples using the RiboMinus Transcriptome Isolation Kit (Invitrogen) according to the manufacturer's instructions. An amplified, biotinylated complementary DNA target was produced using the GeneChip Whole Transcript Sense Target Labeling and Control Reagents kit (Affymetrix) according to the manufacturer's instructions. Each sample target was hybridized overnight to a Mouse GeneSplice Array (Affymetrix PN 540092). Hybridized arrays were processed using the Affymetrix Fluidics Station 450 and scanned with an Affymetrix GeneChip scanner. Microarray data have been deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE32073. Sepscore-based alternative splicing analysis was performed as previously described [45]. Expression was estimated according to probe sets that measure constitutive features for each gene. Differentially expressed genes were identified using SAM (Significance Analysis of Microarrays) [78]. Hierarchical clustering was done using Cluster [79] and visualized using Java Treeview [80].
Sequence motif analysis
“Word counting” was performed by counting the number of cassette exons containing a specific 6mer within a window of sequence (e.g. 150 nucleotides of flanking downstream intron). The difference in counts of sequences containing the motif of interest from the test and background sets of exons are tested for significance by Fisher's exact test.
We identified areas enriched in the TGCATG motif by sliding a window of 50 nucleotides in 5-nucleotide steps over portions of the intron flanking the alternative exon. Specifically, we examined the area starting at the 5′ splice site and extending 150 nucleotides downstream. At each step, we contrasted the number of motifs observed for each of three groups of alternative splicing events: those exons repressed in 4 week vastus (Sepscore<0.3, q = 0), those induced at 4 weeks (Sepscore>0.3, q = 0), and background events that were from genes expressed in 4 week vastus but that showed no significant change in splicing (|Sepscore|<0.3, p>0.2). For the repressed and induced cassette exons, we plotted the motif frequency within the window: the total number of motifs observed divided by the number of events. We then randomly selected a set of background events equal in size to the smaller of the set of repressed or induced events and measured the motif frequency. From 100 such samplings, we estimated the average background frequency (plotted as points), and the 95% confidence intervals (plotted as error bars).
Cell culture, myogenic differentiation, and RNA extraction
C2C12 muscle cells and 293T cells were obtained from the ATCC - LGC Standards (Sesto San Giovanni, MI, Italy) were grown in DMEM (EuroClone) supplemented with 10% FBS (EuroClone) and 1% antibiotic at 37°C in 5% CO2 and 5% O2. To induce muscle differentiation, C2C12 muscle cells were growth to near confluence and then switched to DMEM 2% horse serum for three days (EuroClone). Fresh differentiation medium was changed every day. To determine the extent of myogenic differentiation, differentiating C2C12 cells were fixed with 4% of paraformaldehyde and stained with antibodies against the differentiation marker myosin heavy chain (MHC) (mouse monoclonal, 1∶2, #MF20, Developmental Studies Hybridoma Bank, University of Iowa) according to manufacturer's instructions. At least 1,500 nuclei from MHC-positive cells were counted from several random fields. The fusion index was calculated as follows: (MHC-stained myocytes containing >2 nuclei/total number of nuclei)×100. All experiments were performed in triplicate. RNA from C2C12 muscle cells was extracted using Trizol and PureLink RNA kit (Invitrogen) according to manufacturer's instructions.
RT–PCR validation of microarray results
cDNA was generated using SuperScript III First-strand synthesis supermix for qRTPCR (Invitrogen). 25 ng of cDNA were used as a template for PCR with each primer pair (designed with Primer3 software; Table S5). PCR reactions were carried out using Go Taq Polymerase (Promega), were separated on native TBE acrylamide gels and stained with SYBER Gold (Invitrogen). Gels were imaged and signals quantified with a Typhoon FLA 9000 Biomolecular Imager. We quantified the RT-PCR products from three individual mice for each genotype or muscle type and calculated skipping rates. We judged samples as being different from wild-type if a t-test indicated that the sample was unlikely to be from the wild-type distribution with P<0.05.
Quantitative real-time PCR (qPCR)
qPCR was performed with SYBR Green PCR Master Mix (Invitrogen) and a BioRad CFX96 machine using 10 ng of cDNA in 20 µl total volume per reaction with the primers describe in Table S6. Gapdh was used for normalization and relative amounts of mRNA were calculated using the comparative CT (threshold cycle) method.
Plasmids constructs, infection, and transfection
All constructs were fully sequenced before use. For RNA interference, shRNAs against Rbfox1 (sh#1 CCCAGACACAACCTTCTGAAA; sh#2 CCGACAAATGTTTGGTCAATT) and the control non-silencing shRNA (TCTCGCTTGGGCGAGAGTAAG) in pLKO.1 were purchased from Open Biosystems (Huntsville, AL). Viruses were packaged in 293T cells and used to infect WT C2C12 cells according to manufacturer's instructions. Infected cells were selected with relative antibiotics (puromycin 0.5 µg/ml final) and maintained as a polyclonal population. Generation of C2C12-EV and C2C12-FRG1 was described elsewhere [24]. For the experiments reported in Figure 10, FRG1 ORF was cloned into the lentiviral vector pCCL.sin.cPPT.SV40ployA.eGFP.minCMV.hPGK.deltaNGFR.WPRE (EV1) [81] and Rbfox1 ORF was cloned into the pBABE-puro retroviral vector (EV2). EV1 and FRG1 viruses were packaged in 293T cells using psPAX2 packaging plasmid and pMD2G envelope plasmid. The medium containing the lentiviral particles was harvested after 27 h, 34 h and 48 h, filtered by using a 0.22 mm filter unit and right after used to transduce WT C2C12 cells. After 48 h, transduced cells were 70 to 80% GFP positive. Then, EV2 and Rbfox1 viruses were packaged in 293T cells using VSV-G expression vector and gag/pol expression vector. The medium containing the retroviral particles was harvested after 27 h, 34 h and 48 h, filtered by using a 0.22 mm filter unit and right after used to transduce FRG1 cells. After 48 h, FRG1-EV2 and FRG1-Rbfox1 transduced cells were put under puromycin selection (puromycin 0.5 µg/ml final) for 5 days. The construct-containing mouse A713 Rbfox1 ORF fused to a myc-epitope [82] was a kind gift of Dr. S. Kawamoto and was subcloned in pIRES-Puro (Clontech). Calpain 3 lacking exon 6 ORF was obtained by RT-PCR from FRG1 mice muscles and was cloned in pIRES-Neo (Clontech). To generate stable cells, 10 µg of Fox1-IRES-Puro, Capn3 E6-IRES-Neo plasmids and the appropriate empty vector controls were used to transfect C2C12 cells with Lipofectamine LTX (Invitrogen) following manufacturer's instructions. 48 hours post-transfection, cells were selected with puromycin or neomycin (neomycin 1 µg/ml final). Resistant cells were maintained as polyclonal populations.
Protein extracts, immunoblotting, and antibodies
C2C12 and primary human muscle cells were lysed in SDS sample buffer at 95°C for 10 min. Muscle extracts were homogenized using buffer containing 0.125 M Tris-HCl pH6.4; 10% glycerol, 4% SDS, 4 M urea, 10% mercaptoethanol and 0.01% bromophenol blue (final pH of the buffer 6.8) according to the datasheet of CAPN-12A2 Novocastra antibody. Protein extracts were separated using a SDS-PAGE gel and transferred onto a nitrocellulose membrane (GE Healthcare). Immunoblots were incubated with primary antibodies against Rbfox1 (generous gifts from Dr. Douglas Black [83] and Dr. Thomas Cooper [84]), Calpain 3 (mouse monoclonal, 1∶250 #NCL-CAPN-12A2, Novocastra), HA (mouse monoclonal, 1∶500 #MMS-101, Covance) and anti-α tubulin antibody (mouse monoclonal, 1∶10,000, #049K4767, Sigma) as loading control. Subsequently, 1∶10,000 dilution of peroxidase-conjugated donkey anti-mouse IgG (#715-035-015. Jackson ImmunoResearch) was added. Detection was performed by super signal west pico chemioluminescence substrate (Thermo Scientific).
Chromatin immunoprecipitation (ChIP) and RNA immunoprecipation (RIP)
Antibodies against FRG1 were kindly provided by DR. PL Jones [52]. Anti-RNA pol II (Millipore #17-620), anti-Myc monoclonal antibody (MMS-164P, Covance) or control mouse or rabbit IgG (715-035-015 and 011-000-003, Jackson ImmunoResearch) were also used. ChIP was performed as described in [21]. RIP displayed in Figure S6a was performed as [52]. RIP displayed in Figure 7d was performed as UV-RIP described in [21]. RIP displayed in Figure S6b, Figure 8 and Figure S7e was performed as ChIP described in [21]. The primers used are described in Table S6.
Supporting Information
CD45 staining in FRG1 mice. (a) Left panel: representative images of vastus lateralis staining against pan-hematopoietic marker CD45. Right panel: representative images of vastus lateralis Gömöri trichrome staining using a ×20 objective analysis. (b) Quantitative analysis of CD45 positive cells.
(TIF)
RT-PCR validation of splicing-sensitive microarrays. RT-PCR analysis was performed on RNA extracted from three independent wild type and FRG1 mice at 4 weeks of age. For each gene, representative images as well as quantification of triplicate mice data with standard deviations are shown. For the quantification, RT-PCR products from three individual FRG1 and control WT mice were quantified using the Typhoon and the skipping rates were calculated. Samples were judged as being different from WT if a t-test indicated that the sample was unlikely to be from the WT distribution with P<0.05. Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
(TIF)
Alternative splicing changes correlate with disease severity in different muscles. RT–PCR analysis of alternative splicing using RNA extracted from vastus lateralis (severely affected) and biceps brachii (mildly affected) muscles from wild type and FRG1 mice at 4 weeks of age. Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
(TIF)
Alternative splicing changes in FRG1 mice are not secondary to muscular dystrophy. RT-PCR analysis of alternative splicing using RNA extracted from vastus lateralis of gender, age and background-matched WT, FRG1 and mdx mice. Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
(TIF)
Alternative splicing changes are a primary consequence of FRG1 over-expression in tissue culture. (a) Specific FRG1 over-expression was confirmed by real-time RT-PCR and immunoblotting using RNAs and proteins isolated from C2C12 muscle cells expressing a Flag-HA empty vector (EV) or a Flag-HA-tagged FRG1 (FRG1) either in proliferating or differentiating C2C12 muscle cells. (b) RT-PCR analysis of alternative splicing using the same samples as in (a). Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
(TIF)
Altered transcripts are not direct FRG1 targets. (a) RIP experiments on control and over-expressing FRG1 C2C12 muscle cells using anti-FRG1 (HS2) or control IgG antibodies. (b) RIP experiments on control and over-expressing FRG1 C2C12 muscle cells using anti-FRG1 (HS2), anti-FRG1 (L-07 sc-101050, SCBT) or control IgG antibodies. Anti-FRG1 immunoprecipitated material did not show any enrichment versus control IgG. RT-minus control experiments showed the absence of DNA contamination.
(TIF)
Rbfox1 knockdown causes alternative splicing changes similar to FRG1 over-expression, Rbfox1 over-expression causes opposite results. (a) Left: schematic representation of the regions analyzed showing the location of the putative Fox binding site (FBS) and RT-PCR analysis of alternative splicing in C2C12 muscle cells expressing Rbfox1 shRNA#1 for genes with FBS. Right: RT-PCR analysis of alternative splicing in C2C12 muscle cells expressing Rbfox1 shRNA#1 for genes without FBS. (b) Specific Rbfox1 knockdown using a second shRNA was confirmed by real-time RT-PCR and immunoblotting using RNAs and proteins isolated from C2C12 muscle cells expressing a control shRNA or an shRNA specific for Rbfox1 (shRNA#2). Rbfox1 knockdown with shRNA#2 display a reduced myogenic differentiation. (c) RT-PCR analysis of alternative splicing in C2C12 muscle cells expressing Rbfox1 shRNA#2 (#2) for genes containing putative FBS (left) or for genes without FBS (right). (d) RT-PCR analysis of alternative splicing in C2C12 muscle cells over-expressing Rbfox1 (F1) for genes with putative FBS. Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron. (e) Selective in vivo association of Rbfox1 to target regions displaying putative Fox binding sites (FBS).
(TIF)
Real-time PCR analysis in proliferating (MB) or differentiating (MT) C2C12 muscle cells confirming selective Capn3 E6- isoform over-expression. Real-time RT-PCR analysis was performed on RNA extracted from proliferating (MB) or differentiating (MT) C2C12 expressing the empty vector or Capn3 E6- using primers specific for the Capn3 isoform containing exon 6 (Capn3 cost) or the alternative splicing isoform lacking exon 6 (Capn3 E6-).
(TIF)
Genes investigated at transcriptional levels.
(XLS)
Genes investigated at splicing levels.
(XLS)
Distribution of alternative splicing events and gene expression changes. (a) Top: Gene expression microarray data were filtered for q = 0 and fold change >1.5. Bottom: Summary of the genes altered only in vastus lateralis, in vastus lateralis and biceps brachii or only in biceps brachii at 4 or 13 weeks of age. Summary of the genes altered only at 4 weeks, at 4 and 13 weeks or only at 13 weeks of age in vastus lateralis or in biceps brachii. (b) Top: Splicing microarray data were filtered for q = 0 and |Sepscore|≥0.3. Significant splicing events were categorized by RNA processing mode. Bottom: Summary of the total events altered only in vastus lateralis, in vastus lateralis and biceps brachii or only in biceps brachii at 4 or 13 weeks of age. Summary of the total events altered only at 4 weeks, at 4 and 13 weeks or only at 13 weeks of age in vastus lateralis or in biceps brachii.
(XLS)
Lists of transcripts identified by splicing-sensitive microarray that were validated by RT-PCR. Gene symbol and gene name is listed.
(PDF)
List of primers used for the RT-PCR validation of the splicing-sensitive microarrays.
(TIF)
List of primers used for real-time RT-PCR, RNA-IP, and ChIP.
(TIF)
Acknowledgments
We thank Chris W. Smith and Juan Valcarcel for critically reading the manuscript. Maria Victoria Neguembor conducted this study as partial fulfillment of her PhD in Molecular Medicine, Program in Cellular and Molecular Biology, San Raffaele University, Milan, Italy.
Funding Statement
Support for the Gabellini Laboratory came from the European Research Council (ERC StG 204279, ncRNAs-Splicing-FSHD, http://erc.europa.eu), the Italian Epigenomics Flagship Project (http://www.epigen.it), the Italian Ministry of Health (GRO8-21, http://www.salute.gov.it), and the FSHD Global Research Foundation (http://www.fshdglobal.org). DG is a Dulbecco Telethon Institute Assistant Scientist (TCP05001, http://dti.telethon.it). Research in MA's laboratory is supported by NIH grant GM084317. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cabianca DS, Gabellini D (2010) The cell biology of disease: FSHD: copy number variations on the theme of muscular dystrophy. J Cell Biol 191: 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flanigan KM, Coffeen CM, Sexton L, Stauffer D, Brunner S, et al. (2001) Genetic characterization of a large, historically significant Utah kindred with facioscapulohumeral dystrophy. Neuromuscul Disord 11: 525–529. [DOI] [PubMed] [Google Scholar]
- 3. Pandya S, King WM, Tawil R (2008) Facioscapulohumeral dystrophy. Phys Ther 88: 105–113. [DOI] [PubMed] [Google Scholar]
- 4. Shahrizaila N, Wills AJ (2005) Significance of Beevor's sign in facioscapulohumeral dystrophy and other neuromuscular diseases. J Neurol Neurosurg Psychiatry 76: 869–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barro M, Carnac G, Flavier S, Mercier J, Vassetzky Y, et al. (2008) Myoblasts from affected and non-affected FSHD muscles exhibit morphological differentiation defects. J Cell Mol Med 14: 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Celegato B, Capitanio D, Pescatori M, Romualdi C, Pacchioni B, et al. (2006) Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics 6: 5303–5321. [DOI] [PubMed] [Google Scholar]
- 7. Morosetti R, Mirabella M, Gliubizzi C, Broccolini A, Sancricca C, et al. (2007) Isolation and characterization of mesoangioblasts from facioscapulohumeral muscular dystrophy muscle biopsies. Stem Cells 25: 3173–3182. [DOI] [PubMed] [Google Scholar]
- 8. Stadler G, Chen JC, Wagner K, Robin JD, Shay JW, et al. (2011) Establishment of clonal myogenic cell lines from severely affected dystrophic muscles - CDK4 maintains the myogenic population. Skelet Muscle 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tupler R, Perini G, Pellegrino MA, Green MR (1999) Profound misregulation of muscle-specific gene expression in facioscapulohumeral muscular dystrophy. Proc Natl Acad Sci U S A 96: 12650–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winokur ST, Barrett K, Martin JH, Forrester JR, Simon M, et al. (2003) Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul Disord 13: 322–333. [DOI] [PubMed] [Google Scholar]
- 11. Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, et al. (2003) Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet 12: 2895–2907. [DOI] [PubMed] [Google Scholar]
- 12. Brouwer OF, Padberg GW, Bakker E, Wijmenga C, Frants RR (1995) Early onset facioscapulohumeral muscular dystrophy. Muscle Nerve 2: S67–72. [PubMed] [Google Scholar]
- 13. Funakoshi M, Goto K, Arahata K (1998) Epilepsy and mental retardation in a subset of early onset 4q35-facioscapulohumeral muscular dystrophy. Neurology 50: 1791–1794. [DOI] [PubMed] [Google Scholar]
- 14. Miura K, Kumagai T, Matsumoto A, Iriyama E, Watanabe K, et al. (1998) Two cases of chromosome 4q35-linked early onset facioscapulohumeral muscular dystrophy with mental retardation and epilepsy. Neuropediatrics 29: 239–241. [DOI] [PubMed] [Google Scholar]
- 15. Saito Y, Miyashita S, Yokoyama A, Komaki H, Seki A, et al. (2007) Facioscapulohumeral muscular dystrophy with severe mental retardation and epilepsy. Brain Dev 29: 231–233. [DOI] [PubMed] [Google Scholar]
- 16. Neguembor M, Gabellini D (2010) In Junk We Trust: Repetitive DNA, Epigenetics and Facioscapulohumeral Muscular Dystrophy. Epigenomics 2: 271–287. [DOI] [PubMed] [Google Scholar]
- 17. Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, et al. (1994) Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet 3: 1287–1295. [DOI] [PubMed] [Google Scholar]
- 18. van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, et al. (1993) FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 2: 2037–2042. [DOI] [PubMed] [Google Scholar]
- 19. Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, et al. (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 2: 26–30. [DOI] [PubMed] [Google Scholar]
- 20. Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, et al. (1994) The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res 2: 225–234. [DOI] [PubMed] [Google Scholar]
- 21. Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, et al. (2012) A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149: 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gabellini D, Green MR, Tupler R (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110: 339–348. [DOI] [PubMed] [Google Scholar]
- 23. Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, et al. (2010) A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329: 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanel ML, Wuebbles RD, Jones PL (2009) Muscular dystrophy candidate gene FRG1 is critical for muscle development. Dev Dyn 238: 1502–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gabellini D, D'Antona G, Moggio M, Prelle A, Zecca C, et al. (2006) Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature 439: 973–977. [DOI] [PubMed] [Google Scholar]
- 26. Liu Q, Jones TI, Tang VW, Brieher WM, Jones PL (2010) Facioscapulohumeral muscular dystrophy region gene-1 (FRG-1) is an actin-bundling protein associated with muscle-attachment sites. J Cell Sci 123: 1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wuebbles RD, Hanel ML, Jones PL (2009) FSHD region gene 1 (FRG1) is crucial for angiogenesis linking FRG1 to facioscapulohumeral muscular dystrophy-associated vasculopathy. Dis Model Mech 2: 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, et al. (2010) Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. Rna 16: 2384–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davidovic L, Sacconi S, Bechara EG, Delplace S, Allegra M, et al. (2008) Alteration of expression of muscle specific isoforms of the fragile X related protein 1 (FXR1P) in facioscapulohumeral muscular dystrophy patients. J Med Genet 45: 679–685. [DOI] [PubMed] [Google Scholar]
- 30. Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ (2002) Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. Rna 8: 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim SK, Lund J, Kiraly M, Duke K, Jiang M, et al. (2001) A gene expression map for Caenorhabditis elegans. Science 293: 2087–2092. [DOI] [PubMed] [Google Scholar]
- 32. Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, et al. (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- 33. Rappsilber J, Ryder U, Lamond AI, Mann M (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Koningsbruggen S, Dirks RW, Mommaas AM, Onderwater JJ, Deidda G, et al. (2004) FRG1P is localised in the nucleolus, Cajal bodies, and speckles. J Med Genet 41: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuroyanagi H (2009) Fox-1 family of RNA-binding proteins. Cell Mol Life Sci 66: 3895–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Damianov A, Black DL (2009) Autoregulation of Fox protein expression to produce dominant negative splicing factors. Rna 16: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McKee AE, Minet E, Stern C, Riahi S, Stiles CD, et al. (2005) A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang ZZ, Zheng S, Nikolic J, Black DL (2009) Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol 29: 4757–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baraniak AP, Chen JR, Garcia-Blanco MA (2006) Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol 26: 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, et al. (2006) Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem 281: 12468–12474. [DOI] [PubMed] [Google Scholar]
- 41. Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL (2005) Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol 25: 10005–10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, et al. (2009) An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 16: 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim KK, Adelstein RS, Kawamoto S (2009) Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem 284: 31052–31061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, et al. (2007) Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sugnet CW, Srinivasan K, Clark TA, O'Brien G, Cline MS, et al. (2006) Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol 2: e4 doi:10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walsh FS, Celeste AJ (2005) Myostatin: a modulator of skeletal-muscle stem cells. Biochem Soc Trans 33: 1513–1517. [DOI] [PubMed] [Google Scholar]
- 47. Berkes CA, Tapscott SJ (2005) MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 16: 585–595. [DOI] [PubMed] [Google Scholar]
- 48. Finanger Hedderick EL, Simmers JL, Soleimani A, Andres-Mateos E, Marx R, et al. (2011) Loss of sarcolemmal nNOS is common in acquired and inherited neuromuscular disorders. Neurology 76: 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, et al. (1995) Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 81: 27–40. [DOI] [PubMed] [Google Scholar]
- 50. Sorimachi H, Imajoh-Ohmi S, Emori Y, Kawasaki H, Ohno S, et al. (1989) Molecular cloning of a novel mammalian calcium-dependent protease distinct from both m- and mu-types. Specific expression of the mRNA in skeletal muscle. J Biol Chem 264: 20106–20111. [PubMed] [Google Scholar]
- 51. Bulfield G, Siller WG, Wight PA, Moore KJ (1984) X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci U S A 81: 1189–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sun CY, van Koningsbruggen S, Long SW, Straasheijm K, Klooster R, et al. (2011) Facioscapulohumeral muscular dystrophy region gene 1 is a dynamic RNA-associated and actin-bundling protein. J Mol Biol 411: 397–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamashita R, Sathira NP, Kanai A, Tanimoto K, Arauchi T, et al. (2011) Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res 21: 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Spencer MJ, Guyon JR, Sorimachi H, Potts A, Richard I, et al. (2002) Stable expression of calpain 3 from a muscle transgene in vivo: immature muscle in transgenic mice suggests a role for calpain 3 in muscle maturation. Proc Natl Acad Sci U S A 99: 8874–8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, et al. (2009) RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet 18: 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, et al. (2010) Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet 6: e1001181 doi:10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, et al. (2011) DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol 69: 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, et al. (2008) An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J 27: 2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arahata K, Ishihara T, Fukunaga H, Orimo S, Lee JH, et al. (1995) Inflammatory response in facioscapulohumeral muscular dystrophy (FSHD): immunocytochemical and genetic analyses. Muscle Nerve 2: S56–66. [PubMed] [Google Scholar]
- 60. Figarella-Branger D, Pellissier JF, Serratrice G, Pouget J, Bianco N (1989) [Immunocytochemical study of the inflammatory forms of facioscapulohumeral myopathies and correlation with other types of myositis]. Ann Pathol 9: 100–108. [PubMed] [Google Scholar]
- 61. Frisullo G, Frusciante R, Nociti V, Tasca G, Renna R, et al. (2011) CD8(+) T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J Clin Immunol 31: 155–166. [DOI] [PubMed] [Google Scholar]
- 62. Munsat TL, Piper D, Cancilla P, Mednick J (1972) Inflammatory myopathy with facioscapulohumeral distribution. Neurology 22: 335–347. [DOI] [PubMed] [Google Scholar]
- 63. Geng LN, Yao Z, Snider L, Fong AP, Cech JN, et al. (2012) DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell 22: 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bhalla K, Phillips HA, Crawford J, McKenzie OL, Mulley JC, et al. (2004) The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet 49: 308–311. [DOI] [PubMed] [Google Scholar]
- 65. Davis LK, Maltman N, Mosconi MW, Macmillan C, Schmitt L, et al. (2012) Rare inherited A2BP1 deletion in a proband with autism and developmental hemiparesis. Am J Med Genet A 158A: 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, et al. (2012) RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet 21: 4171–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gallant NM, Baldwin E, Salamon N, Dipple KM, Quintero-Rivera F (2011) Pontocerebellar hypoplasia in association with de novo 19p13.11p13.12 microdeletion. Am J Med Genet A 155A: 2871–2878. [DOI] [PubMed] [Google Scholar]
- 68. Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, et al. (2011) The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet 43: 706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kaynak B, von Heydebreck A, Mebus S, Seelow D, Hennig S, et al. (2003) Genome-wide array analysis of normal and malformed human hearts. Circulation 107: 2467–2474. [DOI] [PubMed] [Google Scholar]
- 70. Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, et al. (2007) Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet 144B: 869–876. [DOI] [PubMed] [Google Scholar]
- 71. Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, et al. (2011) Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A 155A: 2386–2396. [DOI] [PubMed] [Google Scholar]
- 72. Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. (2007) Strong association of de novo copy number mutations with autism. Science 316: 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, et al. (2011) Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bushby KM (1999) Making sense of the limb-girdle muscular dystrophies. Brain 122 (Pt 8) 1403–1420. [DOI] [PubMed] [Google Scholar]
- 75. van der Kooi AJ, de Visser M, Barth PG (1994) Limb girdle muscular dystrophy: reappraisal of a rejected entity. Clin Neurol Neurosurg 96: 209–218. [DOI] [PubMed] [Google Scholar]
- 76. Beckmann JS, Spencer M (2008) Calpain 3, the “gatekeeper” of proper sarcomere assembly, turnover and maintenance. Neuromuscul Disord 18: 913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fougerousse F, Bullen P, Herasse M, Lindsay S, Richard I, et al. (2000) Human-mouse differences in the embryonic expression patterns of developmental control genes and disease genes. Hum Mol Genet 9: 165–173. [DOI] [PubMed] [Google Scholar]
- 78. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Saldanha AJ (2004) Java Treeview–extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- 81. Amendola M, Venneri MA, Biffi A, Vigna E, Naldini L (2005) Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nat Biotechnol 23: 108–116. [DOI] [PubMed] [Google Scholar]
- 82. Nakahata S, Kawamoto S (2005) Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res 33: 2078–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee JA, Tang ZZ, Black DL (2009) An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev 23: 2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kalsotra A, Wang K, Li PF, Cooper TA (2010) MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev 24: 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CD45 staining in FRG1 mice. (a) Left panel: representative images of vastus lateralis staining against pan-hematopoietic marker CD45. Right panel: representative images of vastus lateralis Gömöri trichrome staining using a ×20 objective analysis. (b) Quantitative analysis of CD45 positive cells.
(TIF)
RT-PCR validation of splicing-sensitive microarrays. RT-PCR analysis was performed on RNA extracted from three independent wild type and FRG1 mice at 4 weeks of age. For each gene, representative images as well as quantification of triplicate mice data with standard deviations are shown. For the quantification, RT-PCR products from three individual FRG1 and control WT mice were quantified using the Typhoon and the skipping rates were calculated. Samples were judged as being different from WT if a t-test indicated that the sample was unlikely to be from the WT distribution with P<0.05. Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
(TIF)
Alternative splicing changes correlate with disease severity in different muscles. RT–PCR analysis of alternative splicing using RNA extracted from vastus lateralis (severely affected) and biceps brachii (mildly affected) muscles from wild type and FRG1 mice at 4 weeks of age. Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
(TIF)
Alternative splicing changes in FRG1 mice are not secondary to muscular dystrophy. RT-PCR analysis of alternative splicing using RNA extracted from vastus lateralis of gender, age and background-matched WT, FRG1 and mdx mice. Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
(TIF)
Alternative splicing changes are a primary consequence of FRG1 over-expression in tissue culture. (a) Specific FRG1 over-expression was confirmed by real-time RT-PCR and immunoblotting using RNAs and proteins isolated from C2C12 muscle cells expressing a Flag-HA empty vector (EV) or a Flag-HA-tagged FRG1 (FRG1) either in proliferating or differentiating C2C12 muscle cells. (b) RT-PCR analysis of alternative splicing using the same samples as in (a). Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron.
(TIF)
Altered transcripts are not direct FRG1 targets. (a) RIP experiments on control and over-expressing FRG1 C2C12 muscle cells using anti-FRG1 (HS2) or control IgG antibodies. (b) RIP experiments on control and over-expressing FRG1 C2C12 muscle cells using anti-FRG1 (HS2), anti-FRG1 (L-07 sc-101050, SCBT) or control IgG antibodies. Anti-FRG1 immunoprecipitated material did not show any enrichment versus control IgG. RT-minus control experiments showed the absence of DNA contamination.
(TIF)
Rbfox1 knockdown causes alternative splicing changes similar to FRG1 over-expression, Rbfox1 over-expression causes opposite results. (a) Left: schematic representation of the regions analyzed showing the location of the putative Fox binding site (FBS) and RT-PCR analysis of alternative splicing in C2C12 muscle cells expressing Rbfox1 shRNA#1 for genes with FBS. Right: RT-PCR analysis of alternative splicing in C2C12 muscle cells expressing Rbfox1 shRNA#1 for genes without FBS. (b) Specific Rbfox1 knockdown using a second shRNA was confirmed by real-time RT-PCR and immunoblotting using RNAs and proteins isolated from C2C12 muscle cells expressing a control shRNA or an shRNA specific for Rbfox1 (shRNA#2). Rbfox1 knockdown with shRNA#2 display a reduced myogenic differentiation. (c) RT-PCR analysis of alternative splicing in C2C12 muscle cells expressing Rbfox1 shRNA#2 (#2) for genes containing putative FBS (left) or for genes without FBS (right). (d) RT-PCR analysis of alternative splicing in C2C12 muscle cells over-expressing Rbfox1 (F1) for genes with putative FBS. Numbers below images are the percentage of exon inclusion. Black boxes illustrate constitutive exons, white boxes alternatively spliced exons and double lines represent the affected intron. (e) Selective in vivo association of Rbfox1 to target regions displaying putative Fox binding sites (FBS).
(TIF)
Real-time PCR analysis in proliferating (MB) or differentiating (MT) C2C12 muscle cells confirming selective Capn3 E6- isoform over-expression. Real-time RT-PCR analysis was performed on RNA extracted from proliferating (MB) or differentiating (MT) C2C12 expressing the empty vector or Capn3 E6- using primers specific for the Capn3 isoform containing exon 6 (Capn3 cost) or the alternative splicing isoform lacking exon 6 (Capn3 E6-).
(TIF)
Genes investigated at transcriptional levels.
(XLS)
Genes investigated at splicing levels.
(XLS)
Distribution of alternative splicing events and gene expression changes. (a) Top: Gene expression microarray data were filtered for q = 0 and fold change >1.5. Bottom: Summary of the genes altered only in vastus lateralis, in vastus lateralis and biceps brachii or only in biceps brachii at 4 or 13 weeks of age. Summary of the genes altered only at 4 weeks, at 4 and 13 weeks or only at 13 weeks of age in vastus lateralis or in biceps brachii. (b) Top: Splicing microarray data were filtered for q = 0 and |Sepscore|≥0.3. Significant splicing events were categorized by RNA processing mode. Bottom: Summary of the total events altered only in vastus lateralis, in vastus lateralis and biceps brachii or only in biceps brachii at 4 or 13 weeks of age. Summary of the total events altered only at 4 weeks, at 4 and 13 weeks or only at 13 weeks of age in vastus lateralis or in biceps brachii.
(XLS)
Lists of transcripts identified by splicing-sensitive microarray that were validated by RT-PCR. Gene symbol and gene name is listed.
(PDF)
List of primers used for the RT-PCR validation of the splicing-sensitive microarrays.
(TIF)
List of primers used for real-time RT-PCR, RNA-IP, and ChIP.
(TIF)