Abstract
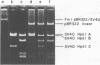
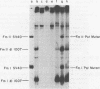
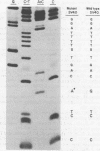
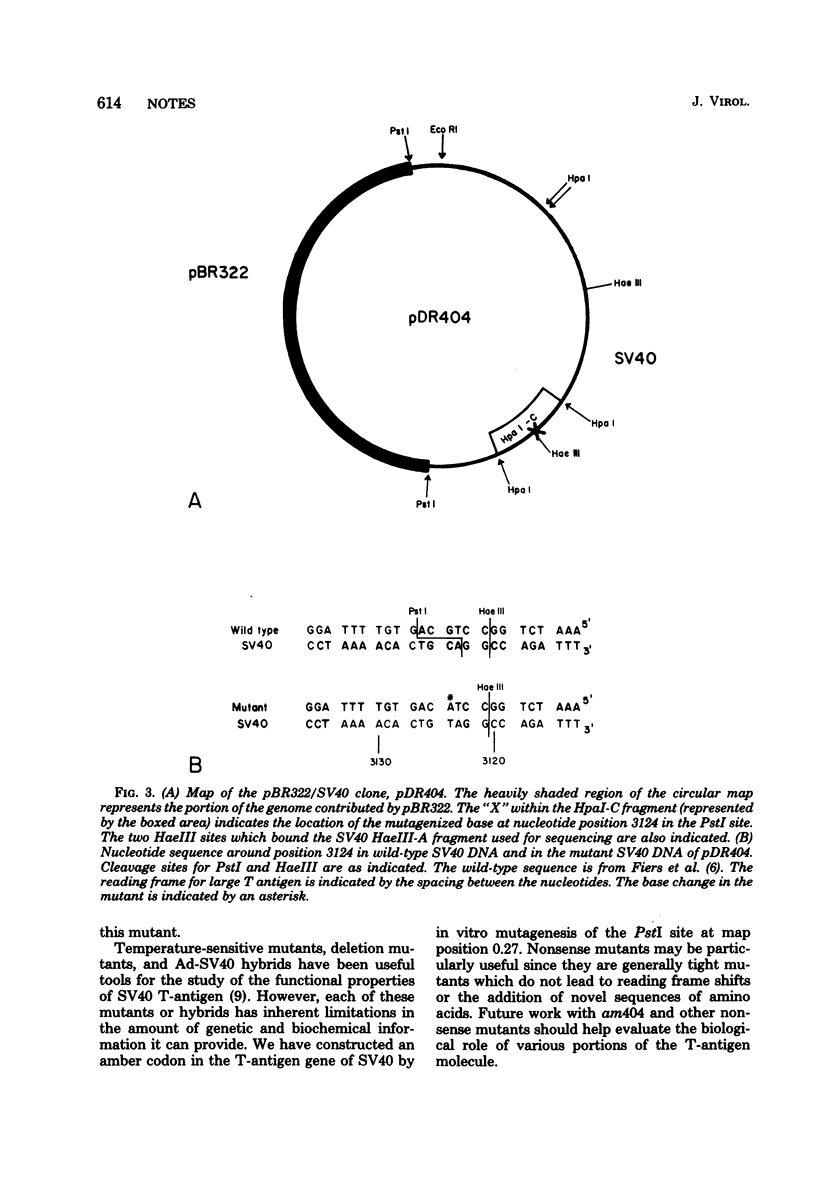
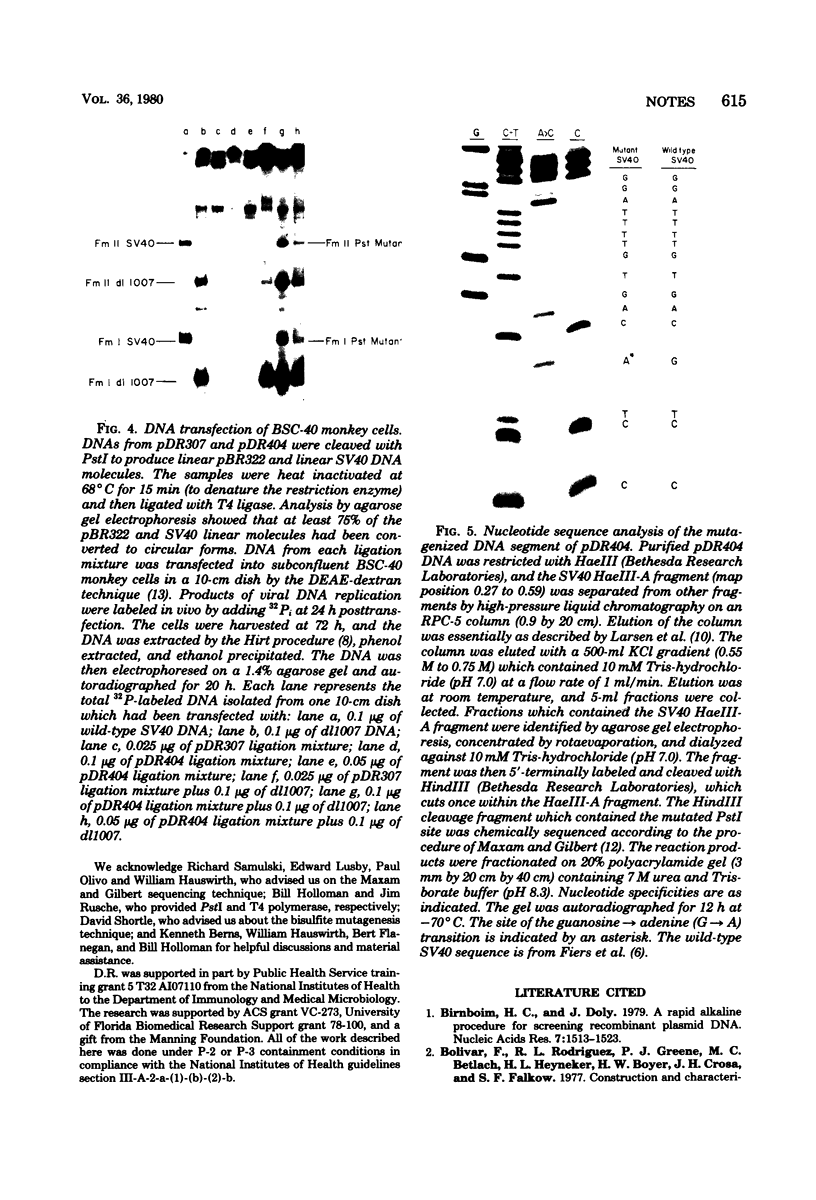
The site-directed bisulfite mutagenesis technique has been used to construct a specific mutation, am404, at nucleotide position 3124 in the simian virus 40 genome. The mutation was contained within a PstI restriction site (map position 0.27) and prevented cleavage by PstI at that position. Nucleotide sequence analysis of the mutagenized region indicated that only a single base pair change had occurred: a guanosine x cytosine leads to adenine x thymine transition. Comparison of the nucleotide sequence of am404 with the known DNA sequence of simian virus 40 indicted that the mutation in am404 resulted in the conversion of a glutamine codon to an amber codon. am404 could not replicate autonomously when transfected into monkey cells (BSC-40) but did replicate when it was cotransfected with the late deletion helper virus dl1007. On the basis of its position in the T-antigen, gene am404 should produce a T-antigen 24% shorter than the wild-type protein.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T., Crawford L. V. Simian virus 40 T-antigen: identification of tryptic peptides in the C-terminal region and definition of the reading frame. J Virol. 1980 May;34(2):315–329. doi: 10.1128/jvi.34.2.315-329.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund P. T. Analysis of nucleotide sequences at 3' termini of duplex deoxyribonucleic acid with the use of the T4 deoxyribonucleic acid polymerase. J Biol Chem. 1971 May 25;246(10):3269–3276. [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Hayatsu H., Miura A. The mutagenic action of sodium bisulfite. Biochem Biophys Res Commun. 1970 Apr 8;39(1):156–160. doi: 10.1016/0006-291x(70)90771-0. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Nathans D. The genome of simian virus 40. Adv Virus Res. 1977;21:85–173. doi: 10.1016/s0065-3527(08)60762-9. [DOI] [PubMed] [Google Scholar]
- Larson J. E., Hardies S. C., Patient R. K., Wells R. D. Factors influencing fractionation of DNA restriction fragments by RPC-5 column chromatography. J Biol Chem. 1979 Jun 25;254(12):5535–5541. [PubMed] [Google Scholar]
- Lindahl T. New class of enzymes acting on damaged DNA. Nature. 1976 Jan 1;259(5538):64–66. doi: 10.1038/259064a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- Muzyczka N. Persistence of phage lambda DNA in genomes of mouse cells transformed by lambda-carrying SV40 vectors. Gene. 1979 Jun;6(2):107–122. doi: 10.1016/0378-1119(79)90066-0. [DOI] [PubMed] [Google Scholar]
- Muzyczka N., Poland R. L., Bessman M. J. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J Biol Chem. 1972 Nov 25;247(22):7116–7122. [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Hirose T., Itakura K., Riggs A. D. Efficient correction of a mutation by use of chemically synthesized DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4268–4270. doi: 10.1073/pnas.75.9.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Brockman W. W., Nathans D. Biological activities of deletion mutants of simian virus 40. Virology. 1976 Dec;75(2):319–334. doi: 10.1016/0042-6822(76)90031-3. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H., Taniguchi T., Müller W., Meyer F., Weissmann C. Application of site-directed mutagenesis to RNA and DNA genomes. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):669–677. doi: 10.1101/sqb.1979.043.01.075. [DOI] [PubMed] [Google Scholar]