Abstract
Malaria morbidity and mortality caused by both Plasmodium falciparum and Plasmodium vivax extend well beyond the African continent, and although P. vivax causes between 80 and 300 million severe cases each year, vivax transmission remains poorly understood. Plasmodium parasites are transmitted by Anopheles mosquitoes, and the critical site of interaction between parasite and host is at the mosquito's luminal midgut brush border. Although the genome of the “model” African P. falciparum vector, Anopheles gambiae, has been sequenced, evolutionary divergence limits its utility as a reference across anophelines, especially non-sequenced P. vivax vectors such as Anopheles albimanus. Clearly, technologies and platforms that bridge this substantial scientific gap are required in order to provide public health scientists with key transcriptomic and proteomic information that could spur the development of novel interventions to combat this disease. To our knowledge, no approaches have been published that address this issue. To bolster our understanding of P. vivax–An. albimanus midgut interactions, we developed an integrated bioinformatic-hybrid RNA-Seq-LC-MS/MS approach involving An. albimanus transcriptome (15,764 contigs) and luminal midgut subproteome (9,445 proteins) assembly, which, when used with our custom Diptera protein database (685,078 sequences), facilitated a comparative proteomic analysis of the midgut brush borders of two important malaria vectors, An. gambiae and An. albimanus.
Malaria transmission entails the obligatory development of Plasmodium in Anopheles mosquitoes (Fig. 1A). Although the majority of the 900,000 malaria deaths per year (caused primarily by Plasmodium falciparum) occur in Africa, malaria morbidity and mortality extend to other continents. Outside of Africa, malaria is caused by both P. falciparum and P. vivax. In fact, P. vivax has the widest geographic distribution among human malaria parasites and is responsible for between 80 and 300 million severe clinical cases every year (1). Despite this substantial burden of disease, P. vivax has received less attention, in terms of research efforts and resources, than P. falciparum (1, 2).
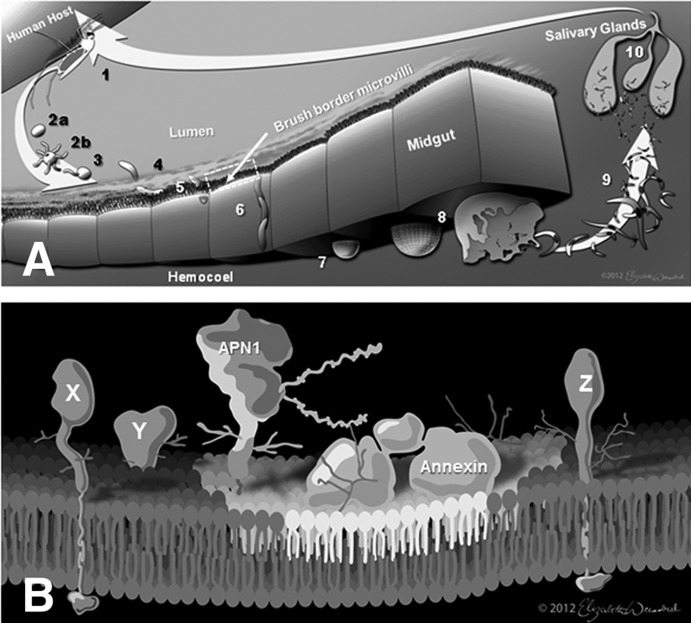
Fig. 1.
Role and biochemical characterization of the mosquito midgut brush border microvilli. A, Plasmodium developmental stages in the Anopheles mosquito. Following ingestion of infected blood (1), female macrogametes (2a) fuse with male microgametes (2b) to form zygotes (3) that then develop into the motile ookinetes (4). Ookinetes recognize and adhere to the mosquito midgut brush border microvilli (5) surface before midgut cell invasion (6). Once ookinetes make their way to the basal lamina, they develop into oocysts (7), which rupture upon maturation (8), releasing sporozoites into the abdominal hemocoel (9). The sporozoites that have invaded the salivary glands (10) are transmitted to a new human host when the mosquito takes another bloodmeal. Failure of the ookinete to attach to the midgut microvilli abrogates further development of the parasite in the mosquito. Boxed outlined area refers to a close-up of B. B, detergent resistant membranes (DRMs), often referred to as lipid rafts, are dynamic, mobile membrane microdomains (indicated by the light-colored area of the lipid bilayer) that have been hypothesized to be involved in clustering several transmission-blocking vaccine target proteins on the surface of the mosquito midgut apical membrane (see Ref. 9). The Anopheles gambiae Alanyl aminopeptidase N (APN1), the leading mosquito-based malaria transmission-blocking vaccine (TBV) target antigen, was previously shown to be enriched in DRMs (9). In a similar fashion, Annexin-like proteins (55, 56), which have also been shown to mediate parasite invasion of the midgut, were identified in the DRM proteome for An. gambiae. DRM-associated midgut surface proteins are regarded as a subset of the entire complement of surface proteins in the brush border midgut microvilli. To date, the identities of the microvillar proteins (indicated as proteins, X, Y, and Z) for any anopheline malaria vector species remains unknown, and this hinders functional characterization and verification of these molecules as potential ligands for Plasmodium ookinetes and as novel TBV targets.
Anopheles albimanus is one of the primary P. vivax mosquito vectors in the Americas. Specialized P. vivax–An. albimanus genotypic interactions have been shown to occur in Mexico (3), with a distinct genetic P. vivax population mirroring the geographic dispersal of An. albimanus (4). This degree of specificity underlying vector–parasite interactions suggest that genetic “compatibility” between parasite and mosquito, likely the outcome of co-evolutionary history, is an important factor in malaria epidemiology. An. albimanus is also a competent vector for P. falciparum (5, 6). Thus, despite evidence of P. vivax–An. albimanus co-evolution at the population level, certain interactions between parasites and mosquitoes may be conserved across different Anopheles/Plasmodium species combinations that sustain malaria transmission.
The prevailing notion that a subset of anophelines are “competent” malaria vectors (7, 8) reinforces the hypothesis that there is a conserved set of molecules on the midgut epithelial brush border microvilli (BBMV)1 surface of anopheline malaria vectors that act as binding-ligands for Plasmodium ookinetes (9) (Fig. 1A). Such mosquito ligands have been shown to be critical candidate targets for malaria transmission-blocking vaccines (TBVs) (10–13), and it has been hypothesized recently that a subset of the BBMV-associated glycoproteins can be clustered via detergent resistant membranes (DRMs) to form a receptor complex for the ookinete (9) (Fig. 1B). However, the current list of candidates is scant, and the remaining complement of TBV targets present on the BBMV, especially those that are not enriched in DRMs, remains unknown (Fig. 1B). Experiments that could validate these hypotheses are hampered by the dearth of molecular information available for non-model vectors.
Although the genome of the African malaria vector, Anopheles gambiae, has been sequenced, evolutionary divergence limits its utility as a reference across anophelines (14–17). Thus, our understanding of the similarities and differences among vivax–albimanus, falciparum–albimanus, and falciparum–gambiae interactions remains poor. Inspired by the renewed emphasis in the malaria community on advancing studies of P. vivax transmission biology, we developed a robust, hybrid sequencing workflow to produce high-quality, assembled transcriptomic data to drive comparative midgut proteomics analyses of the “model” P. falciparum mosquito vector, Anopheles gambiae, and a non-sequenced, predominantly P. vivax vector, An. albimanus. This workflow bridges the transcriptomic and proteomic gap between these anopheline vectors, thereby enabling studies aimed at providing new biological insight into the vivax–anopheles dyad that could translate into the development of novel interventions.
EXPERIMENTAL PROCEDURES
Mosquito Rearing and Midgut Dissection
Anopheles gambiae (Keele) (18) mosquitoes (5 to 6 days old, n = 1,000) at Johns Hopkins University and An. albimanus (3 to 5 days old, n = 2,000) white striped colony at the National Institute of Public Health, Mexico, were used and maintained following standard conditions [see “Methods in Anopheles Research,” available at the Malaria Research and Reference Reagent Resource Center (MR4) Web site]. Midguts from both species were dissected and stored frozen in PBS supplemented with protease inhibitor mixture (PIC).
Mosquito Midgut BBMV Preparation
BBMVs were prepared following established protocols (19), with modifications (20). Four hundred midguts were washed and resuspended in 200 μl in microvilli buffer (50 mm mannitol, 20 mm Tris-HCl pH 7.4, 1 mm PMSF, 3 mm imidazole-HCl) with PIC (Sigma, St. Louis, MO) and processed as described elsewhere (9).
SDS-PAGE, Immunoblot, and Aminopeptidase Activity Assay
SDS-PAGE and APN1 Western blots were carried out as described elsewhere (9). Aminopeptidase (APN) activity in An. gambiae BBMV was assayed with l-leucine p-nitroanilide as substrate in a 96-well plate in a final volume of 210 μl per well. The BBMV were diluted in APN buffer (10 mm Tris-HCl, pH 7.4, 150 mm NaCl) to a final concentration of 6 μg/ml, distributed in the wells, and incubated for 15 min at 37 °C. The APN substrate (2 mm in APN buffer) was then added, and the initial rate of free ρ-nitroanilide product formation at 405 nm (SpectraMax, Molecular Devices, Sunnyvale, CA, USA) was used to calculate the specific APN enzymatic activity.
Sample Preparation, Fractionation, and LC-MS/MS
Approximately 40 μg of BBMV proteins were resuspended in 8 m deionized urea and reduced and alkylated (in tris(2-carboxyethyl)phosphine and methyl methanethiosulfonate, respectively). Proteins were digested for 12 h at 30 °C using 1 μg LysC (Promega, Madison, WI). The resulting digests were diluted to 2 m urea/20 mm NH4HCO3 and digested overnight at 30 °C using 1 μg proteomics-grade trypsin (Promega). Digested, desalted, and dried peptides were separated using an Agilent 3100 OFFgel fractionator (Agilent, Santa Clara, CA). Samples were separated as described elsewhere (9), concentrated, and analyzed on an Agilent LC-MS system comprising a 1200 LC system coupled to a 6520 Q-TOF via an HPLC Chip (160 nL, 300 Å C18 150 mm column) Cube interface, using previously described parameters (9).
Library Preparation and Sequencing
Total RNA from 50 An. albimanus midguts was extracted using TRIzol (Invitrogen), DNase treated and cleaned with an RNeasy column (Qiagen, Valenci, CA, USA), and quality checked using a Bioanalyzer (Agilent Technologies). The mRNA libraries were constructed and sequenced, as described elsewhere (15, 21, 22), on a single lane of an Illumina HiSeq 2000, which generated ∼210 million 101 base pair paired end reads. The library preparation for Roche 454 sequencing data is described in greater detail elsewhere (23). However, transcriptome assembly for all transcriptome data followed the same analysis process described below.
Transcriptome Assembly
We assembled the ∼210 million 101 base pair paired end Illumina reads and ∼430,000 Roche 454 reads using Velvet (24) and Oases (25). To optimize our parameters, we first assembled a subset of the Illumina and 454 reads using 13 different k-mer values and found that k-mer values in the 40s gave the largest average and median-sized contigs and the largest cumulative assembly. To overcome memory constraints, we equally divided the complete set of paired end Illumina sequence reads into eight sets. We implemented the multiple k-mer assembly (26) independently for each read set, along with the 454 data, using three different k-mer values (43, 45, and 47). We merged and reassembled the resulting outputs of the 24 assemblies using Velvet and Oases using a k-mer value of 47, which produced the final transcriptome assembly. We filtered the assembly to retain genes that contained a single transcript, were longer than 300 base pairs, and had confidence scores of 1.0.
Transcriptome Mapping
As detailed in Ref. 23, briefly, An. albimanus midgut, abdominal cuticle, and dorsal vessel preparation was used to generate a set of 15,764 transcripts, ∼92% of which (15,441) mapped to the An. darlingi genome (11,430 predicted protein coding genes) and ∼57% of which (9,684) mapped to the An. gambiae genome (∼13,320 predicted protein coding genes). Given these data, we argue that the 16,699 proteins (our transcript set (15,764) plus the salivary gland Sanger contigs (935)) predicted from the current An. albimanus transcriptome should be virtually a complete transcriptome. We should emphasize here that because the transcriptome was assembled de novo, the data represent an independent sampling of the predicted protein sequences. With respect to proteome coverage, we find that of the 15,764 transcripts, 14,887 transcripts (∼94%) mapped to An. gambiae and 8,480 transcripts (∼54%) mapped to An. darlingi (NCBInr, August 11, 2011). The An. albimanus transcriptome dataset is available through VectorBase (23).
Peaklist Settings and Database Search for Proteomics and Transcriptomics
MS raw files for each sample were converted to mzXML format using Trapper 4.3.0 (Institute for Systems Biology, Seattle, WA). The peaklists from all OFFgel fractions for each replicate (12 to 24 fractions per replicate) were merged into a single mzXML data format file; three biological replicates from An. gambiae and three from An. albimanus, for a total of six mzXML files, contained ∼1,000,000 spectra. Data were uploaded and searched using the PepArML metasearch engine (27), which automatically conducts target and decoy searches using one or more of Mascot 2.2 (28), OMSSA 2.1.1 (29), and Tandem 2010.01.01.4 (30) with native, K-score 2010.01.01.4 (31), and S-score 2010.01.01.4 pluggable scoring modules, MyriMatch 1.5.8 (32), and Inspect 20110313 (33) with MS-GF spectral probability scoring (34). It also combines the search results using an unsupervised machine-learning strategy and estimates peptide identification false discovery rates using identifications from the reversed decoy searches (35).
The data were searched against an in-house Diptera FASTA database generated by extracting annotated sequences from the NCBInr (March 2011), with ∼ 685,078 entries from the Diptera order. The PepArML search was performed with variable modifications of methylmethanethiosulfonate and oxidized methionine, mass tolerances of 30 ppm and 20 ppm for precursor and fragment ions, respectively, and one missed cleavage. Search engines Mascot, OMSSA, and Tandem (native scoring) were used. The results were parsed into the MASPECTRAS 2 data analysis system with data filters of 5% peptide FDR and two peptides per protein minimum (36). The metasearch peptide identifications were combined and clustered together if peptide identifications were shared between them, as this indicates substantial sequence similarity and functional homology, and the leader of each group of proteins was considered a unique protein identification and a definite protein entry (36). Throughout this paper, we report only the unique identifications, specifically, the leader proteins of each protein group.
The resultant contigs from the assembled transcript data from Illumina and 454 pyrosequencing were translated in open reading frames using EMBOSS Transeq (http://www.ebi.ac.uk). The sequences were trimmed at the stop codon (TAA, TAG, and TGA) so as to (1) select the longest reading frame and (2) discard sequences that were less than 50 amino acids. All RNA-Seq transcripts that met these criteria from all six frames were selected to create a FASTA database (Anopheles protein database) with randomly generated unique identifiers, along with the contig name, assigned to each entry, providing a unique identifier for each entry. Because the transcriptome is un-annotated and transcript sequences are often less well characterized, resulting in unexpected cleavage sites, we considered the analysis as requiring greater search sensitivity, so the method was changed as follows: the individual MS raw files from An. albimanus and An. gambiae were searched against the transcript translated database with a semi-tryptic search. To maximize search sensitivity, all PepArML search engines were used: Mascot; OMSSA; X!Tandem with native, K-score, and S-score plugins; MyriMatch; and InsPecT +MS-GF. Amazon Web-Services Elastic Compute Cloud (EC2) resources were used to supplement the local PepArML computing resources to carry out this computationally intensive search on a total of 947,147 spectra. The remainder of the analysis was completed as described in a later section (Fig. 2). The data analysis system meets all standards regarding the Minimum Information About a Proteomics Experiment (MIAPE), and our data, including the Diptera FASTA database, have been deposited in the ProteomeExchange via the PRIDE partner repository with the dataset identifier PXD000062.
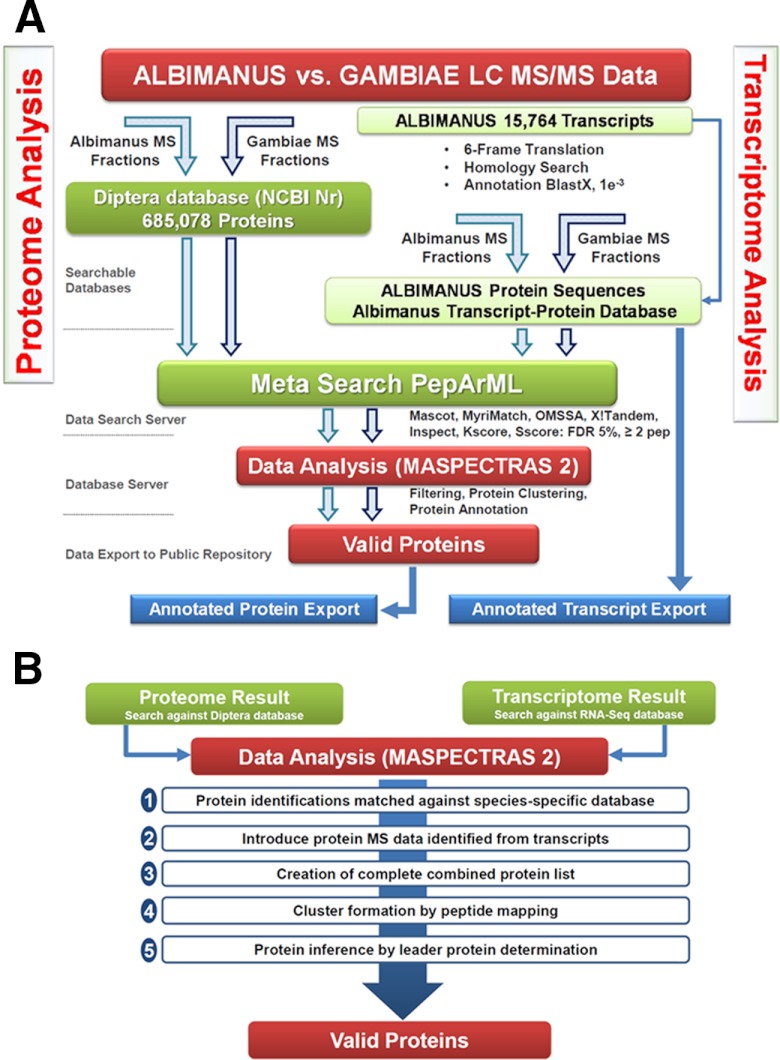
Fig. 2.
Transcriptomics-proteomics data analysis workflow. A, for proteome analysis (left), spectra from the vector species protein MS were searched against a custom-built Diptera database from NCBInr with search engines Mascot, OMSSA, and X!Tandem and combined with the PepArML unsupervised machine learning combiner. The results were then filtered according to statistical thresholds and stored in MASPECTRAS 2, allowing facilitated protein annotation, as well as grouping by shared peptides, removing isoforms, fragments, and proteins with functional commonality. To find proteins common to both An. albimanus and An. gambiae, we did direct sequence comparisons through the relational database contained in MASPECTRAS 2, for both total proteins and protein groups. For transcriptome analysis (right), 15,764 reads were sequenced by an Illumina and translated in silico in six frames to protein sequences; annotations were done by a BlastX homology search against NCBInr (October 2011). An. albimanus and An. gambiae protein MS spectra were searched against this constructed hypothetical protein database with multiple search engines. Proteins identified from transcriptomics analysis were annotated as to function by direct peptide sequence comparison against the identifications from the proteomics analysis. Finally, the annotated proteins were exported to a public repository. Proteins identified from the translated transcripts can be considered to have been identified by two orthogonal methods (RNA sequencing and MS); this provides further confirmation of their presence in the vector BBMV. Furthermore, most sequence annotations to species have been found by sequence homology to known proteins; this might leave out those proteins that have no homology to known proteins. With the method outlined here, by making no assumptions about the protein database, it is possible to detect entirely novel proteins from translated transcripts. B, extended peptide mapping process. The proteome and transcriptome data analysis step in MASPECTRAS 2. Proteome and transcriptome data are merged in MASPECTRAS 2, and a complete list of proteins is created. The clusters are then formed by mapping peptides to their respective proteins and checking for proteins that identify unique peptides. These proteins then are considered the leader sequences and are inferred.
Transcript Annotation and Analysis
Transcript sources were annotated by Blast2GO (37); specifically, the Gene Ontology (GO) database was searched by BLAST homology for annotations. NCBInr was used for the homology search with an E-value cutoff of 1.0 × 10−3. At the end of the ontology mapping process, unidentified sequences were searched again with InterPro (release 34.0), KEGG maps, and Enzyme codes.
RESULTS
A Hybrid Transcriptome Assembly Approach for the Development of Searchable Databases for Proteomics Analysis
Next-generation sequencing methods allow the acquisition of the entire transcriptome of non-model organisms that lack draft genomes (22, 23, 37) and represent a good option for quantitative transcriptomics in model organisms (21, 38), whose annotated genomes can be used to map the reads unambiguously. Recently, several studies have taken advantage of both short and long read RNA-Seq platforms to assemble the transcriptomes of non-model organisms (15, 22, 39), even in the absence of an appropriate reference genome (40). We combined Illumina and 454 reads to generate the hybrid reference transcriptome for downstream comparative proteomics analyses of the BBMV of An. gambiae and An. albimanus (supplemental Fig. S1).
A Quality-controlled Enriched Midgut BBMV Vesicle Preparation for Comparative Proteomics Analyses
To ensure a robust comparison, we characterized the An. gambiae and An. albimanus BBMV samples pre- and post-MS analyses. Protein banding patterns revealed both similarities and differences between the two species (supplemental Fig. S2A). Immunoblot staining of BBMV using antibodies against a known midgut TBV antigen, AgAPN1 (AGAP004809) (11, 12), suggests that this TBV antigen is conserved across the two species (supplemental Fig. S2B). We also demonstrated that the BBMV fractions were significantly enriched in APN activity, a quality indicator for apical membrane enrichment (41), and that the relative ratios among midgut homogenates, pellets, and BBMV are conserved between the two species (supplemental Fig. S2C). Therefore, these preparations represent the best available tool for studying the apical midgut surface. To generate a more complex picture of the BBMV proteome, we used in-solution isoelectric fractionation of peptides, which significantly improves the quantity, quality, and reproducibility of protein identifications (42) from three replicate samples for each species (supplemental Fig. S3). Because these are enrichments and not purifications, non-apically located proteins might be present and detected in subsequent analysis, but they can be accounted for in a post hoc manner following bioinformatics interrogation of the data.
Transcriptome-to-proteome Data Processing Platform
In a typical MS-based proteomics experiment, one relies on a sequence database containing all protein sequences of interest to match spectra against protein sequences, and in characterizing vaccine targets in the BBMV proteome we need to rely on available sequences from An. gambiae and other mosquitoes. However, complete annotation for non-model mosquito genomes is not available, and to our knowledge, no unified method is available for mining MS data by searching the transcriptome. We therefore developed an alternative data analysis method for transcript data, in which we search acquired MS data against a transcriptome assembly from the An. albimanus midgut, abdominal cuticle, and dorsal vessel, and we integrated this process into a complete data analysis system for proteomics data (Fig. 2A).
In general, the most common method for integrating RNA-Seq data into proteomics studies is to search MS data against sequences derived from transcript data directly (43–45). This direct method, however, has a high level of redundancy in the form of isoforms and splice-forms, and it might have poor coverage as a result of assembly error or sampling artifact. The situation is complicated by the protein inference problem (46), as results obtained by directly searching RNA-Seq data might be even more difficult to deconvolute than those from searches on protein databases. In order to determine which method was best suited to our experimental context, we compared the results of protein sequence MS searching to the results generated following a search against RNA-Seq sequences.
For the protein sequence MS search, rather than a broad protein sequence database search, we constructed a custom Diptera protein sequence database. Reducing the search space by limiting the sequences searched results in a lower search time, which relates to higher sensitivity and fewer false positives (47). However, an extensive decrease in database size might have a negative effect, causing either false negatives, if a protein of interest is excluded from the database, or false positives, if a too-small or too-large database breaks the assumptions on which a search algorithm is based (47). Initial search runs with MS data on a taxonomy-limited database with only the anopheline protein sequences available from NCBInr suggested a high false positive rate for identifications (data not shown), and we thus decided that a Diptera database provided a sufficiently limited search space.
The custom Diptera database contains 685,078 NCBInr protein sequences, including members of the family Culicidae (∼23% of sequences, including An. gambiae and 196 other anophelines) (Fig. 3A). We then assembled the output transcript reads from the two RNA-Seq platforms to generate the An. albimanus transcript-protein sequence database via a six-frame translation, with identifications from these being subsequently annotated via mapping against the GO database after a BlastX homology search (Fig. 2A, right). Because transcript data might have confounding factors such as unpredicted cleavage sites, for higher sensitivity we used more search engines than standard proteomics analyses (27). We also took into account the much shorter length of protein sequences derived from the hybrid assembly, as there will be many protein termini that are non-tryptic (48), and so the search was done including a semi-tryptic peptide parameter.
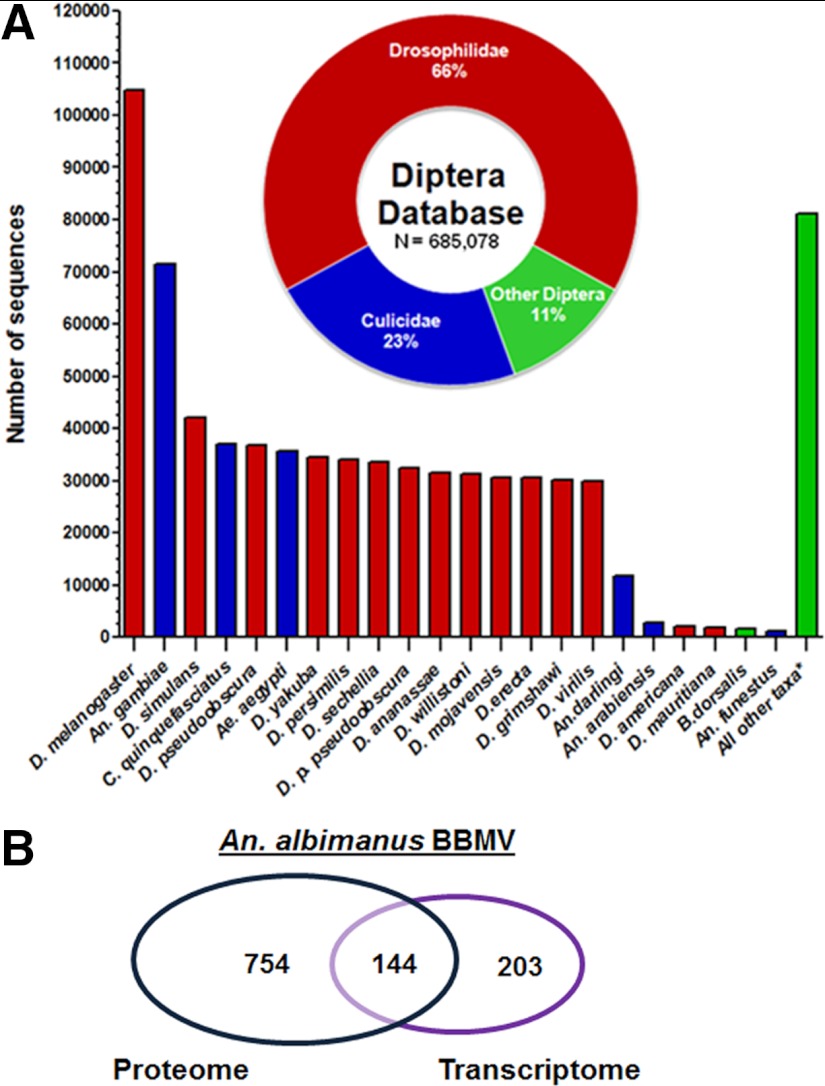
Fig. 3.
The Diptera database of translated protein sequences. A, the contribution of various species to the Diptera database of 685,078 translated protein sequences (cut-off of >1,000 sequences/species). A total of 299 mosquito species are included in the database. A total of 197 Anopheles mosquitoes are represented, of which 26 anophelines (blue columns) contributed 100 or more sequences. Inset: members of the Drosopholidae family constitute the majority of protein sequences in the Diptera database. The family Culicidae (Culicinae and Anophelinae sub-families) accounted for only 23% of the total sequences in the database, including non-blood feeding species (102 non-anopheline species). *Represents sequences contributed by all other taxa within Diptera (ranging from 2 to 999 sequences). B, the results from the BBMV proteomics and BBMV transcriptomics analyses are compared. The two approaches obtain different but overlapping sections of the midgut BBMV proteome, and we therefore use a combined method for our data analysis to maximize our coverage of the proteome (see text for details). To find proteins that are identified by RNA-Seq that are common to those proteins identified via a standard proteomics search method from both An. albimanus and An. gambiae, we performed a direct peptide mapping comparison through the relational database contained in MASPECTRAS 2, for both total proteins and protein groups.
Use of the PepArML machine learning combiner allowed a significant increase in peptide identifications. Furthermore, this boost in the number of peptide identifications could not be obtained solely through the use of a simple agreement heuristic, as the unsupervised machine-learning based combiner assigned about 30% to 50% more identifications at a 5% false discovery rate (FDR). The machine-learning combiner is able to weigh the contribution of each search engine according to its ability to discriminate true from false peptide identifications and to consider non-search-engine features computed directly from the spectra, the peptides, or properties of the peptide-spectrum match.
We then performed a comparison between the results of searching against this Diptera database and those from searching against the RNA-Seq sequences (Fig. 3B), and we found that the two approaches obtain different but overlapping sections of the proteome. The proteomic search gives a larger number of identifications. This might be due to the short sequence length from transcriptome data, which means many identifications (∼800) must be discarded because we limit protein identifications to two or more peptides. The Diptera database search in fact produces a larger number of identifications, but these results are from organisms different from, albeit related to, our target organism, and so these identifications cannot be immediately taken to be An. albimanus proteins. For this reason, we use the approach in Fig. 2A, which allows both proteomes identified by the two methods to be represented in the final dataset. Furthermore, in the method, the transcriptomic data are introduced before redundancy removal is carried out (36), allowing transcript data to inform the protein inference from the peptide data (Fig. 2B).
Thus, the transcriptome search results were mapped against the proteome result through a comparison of the detected peptides via a peptide mapping algorithm. Briefly, the mapping process utilized our protein grouping algorithm (36), which takes proteins to be grouped if they share peptides. In this case, the protein identifications from both the transcript-based searches and the Diptera search were pooled and then clustered according to shared peptides, and the leader sequence was taken as the protein with the most peptides thus matched. The process thereby allows both the transcript and Diptera searches to be equally weighted in inferring the final reported protein identification. The transcriptome mapping furthermore allows better annotation of protein identifications, and the transcriptome search allows the identification of novel proteins that might not be present in the Diptera database.
Analysis of BBMV Proteome via Traditional MS Proteomics and Transcriptome Sequence Database Searching
The An. gambiae proteome has been well studied for vaccine targets, so we decided to look for proteins that are common to the two mosquito vectors, because these would represent potentially conserved TBV antigens. From a preparation of An. gambiae BBMV, we found in total 676 proteins (supplemental Table S1, AngBBMV proteome). Of the 285 proteins identified in the An. albimanus preparation (AnaBBMV proteome) (supplemental Table S2, Figs. 4A–4C), 91% were shared by An. gambiae and An. albimanus (Fig. 4D, supplemental Table S3A); of a total of 961 proteins identified in both preparations, 28% were common between the two vectors. We used a protein grouping algorithm (see “Experimental Procedures”) to group our identifications and clustered the 2,190 common proteins to 278 functional protein groups. The analysis was carried out with a threshold value of 5% peptide FDR for all identifications and a minimum of two peptides identified per protein (supplemental Table S3B). The high rate of functional grouping of these identifications suggests that some of the overlap between proteomes might be due to redundancy in the database. Firstly, because the sequenced genomes of An. gambiae and other Diptera might contain many isoforms of the same proteins, many of the proteins identified might be similar in function. Also, because highly abundant proteins can “mask” less abundant proteins in MS experiments (49), much of the overlap between the proteomes detected here might be due to the repeated detection of isoforms of highly abundant proteins (e.g. actin). However, the 278 common functional groups provide interesting candidates for vaccine targets in An. albimanus, extending the work that has been done on An. gambiae.
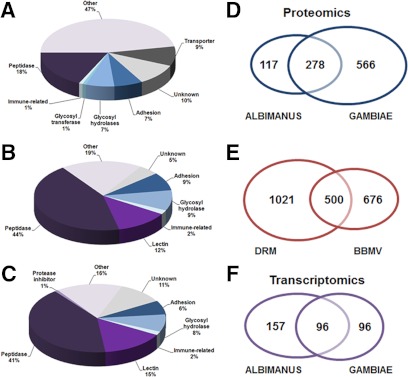
Fig. 4.
Molecular/functional classifications and comparative proteomics analyses of the anopheline mosquito midgut apical microvilli surface for An. albimanus and An. gambiae. A, functional classification of proteins that are unique to the An. albimanus BBMV (AnaBBMV) proteome (n = 117). B, functional classification of secreted/surface expressed proteins that are conserved between the AnaBBMV proteome (n = 105) and the An. gambiae surface expressed subset of the detergent resistant membrane (AngDRM-SE) subproteome (n = 191) (9). C, functional classification of secreted/surface expressed proteins that are conserved between the AngBBMV proteome (n = 121) and the AngDRM-SE. In A–C, “unknown” refers to proteins without corresponding GO annotation. D, we observed that midgut brush border midgut microvilli vesicle (BBMV) proteins are conserved between Anopheles gambiae and Anopheles albimanus. Out of 2,413 An. albimanus proteins, we noted 278 unique proteins in common between An. albimanus and An. gambiae at 5% FDR (minimum of two identifying peptides) for a protein, and (E) the An. gambiae midgut BBMV proteome overlaps with the An. gambiae midgut detergent resistant membrane (DRM) (n = 1,521) proteome. Approximately 32% of the DRM proteins were found in the BBMV proteome at 1% FDR (minimum of two identifying peptides). F, classification of search results with search against transcript sequences. We find a similar degree of overlap (27.5%, 96) between the proteome as identified from transcript sequences and that of the proteome as identified by the traditional proteomics method (28.9%, 278, as shown in D).
In general, very little is known about the full complement of An. albimanus proteins from various tissues, much less their respective functions and molecular interactions with malarial parasites. Prior to this work, there were only 315 An. albimanus proteins available in NCBInr, 290 of which are related in structure and are probably cytochrome proteins; the remaining 25 proteins constitute the genuine proteomics knowledge base for this vector to date. Our proteomic analysis of the BBMV alone generated 2,413 new An. albimanus proteins, which can be grouped functionally into 285 proteins (supplemental Table S2). Importantly, one result of the analysis is full annotations for all shared unique proteins (supplemental Tables S1 and S3).
The results of transcriptome mining are presented in Table I. In general, transcripts that could not be matched by a BLAST search could be proteins with no homology to known proteins, non-coding or regulatory RNA (ncRNA), or, possibly assembly artifacts. Although only 9,445 transcripts were matched by a BLAST homology search against the NCBInr database, the MS data were searched against all 15,764 translated transcripts. This was done to ensure that proteins with no known homology to a sequenced protein could be detected, as neither ncRNA nor assembly artifacts can match against protein-level MS data. These proteins are listed in supplemental Table S4A. We also searched the An. gambiae MS data against the An. albimanus transcriptome; this technique, analogous to the “reciprocal BLAST” technique in genomics (50), found 96 protein orthologs between the two species (supplemental Table S4B, Fig. 4F). This suggests that the remainder of the An. albimanus proteins identified, which were not orthologous to An. gambiae, might be entirely unique to An. albimanus.
Table I. In silico translation of assembled transcripts allows MS analysis of unsequenced genomes. Assembled transcript reads from 454 pyro sequencing and Illumina were annotated using BlastX. The homology search was done with the following criteria: E-value threshold of 1.0 × 10−3 and >20% similarity. This produces 9,445 BLAST identified proteins (60%), of which 3,442 (36%) have Gene Ontology annotation mappings. Searches against the RNA-Seq provisional “Anopheles protein database” using albimanus and gambiae MS data yielded 252 Anopheles albimanus proteins and 192 Anopheles gambiae proteins at 5% peptide false discovery rate with a minimum of two peptides per protein.
| Transcript reads | Oases/Velvet assembled contigs | BlastX predicted proteins | Proteins with ontology mapping | Provisional protein database search identifications |
|||||
|---|---|---|---|---|---|---|---|---|---|
| ALBIMANUS |
GAMBIAE |
||||||||
| Proteins identified | Peptide | Spectra | Proteins identified | Peptide | Spectra | ||||
| ∼210 million Illumina and ∼430,000 454 sequence reads | 15,764 | 9,445 | 3,442 | *252 | 1,624 | 22,484 | *192 | 1,183 | 15,601 |
* Indicates number of protein groups (see “Experimental Procedures”).
The Midgut BBMV Proteome Includes Apical Surface Proteins that Do Not Partition to DRMs (Lipid Rafts)
As expected, we found proteomic evidence of the presence of potential ookinete-interacting ligands (Table II) and immune-related proteins in the sugar-fed mosquito BBMV proteome, which was extensively described by Martinez-Barnetche et al. (23) (supplemental Tables S3B and S3C). Such information continues to improve our understanding of the concordance of midgut transcripts and the expression of their protein products before and after blood feeding (9). The An. gambiae midgut DRM (AgDRM) proteome, which includes proteins that preferentially reside in cholesterol-rich lipid microdomains on the brush border, has been recently published (9). We hypothesize that the BBMV proteome would encompass the majority of the DRM-resident proteins. Given that DRM isolation is contingent on Triton-X-100 insolubility at 4 °C, we would also expect that not all BBMV proteins partition to DRMs. In comparing the AgDRM sub-proteome (9) with the current BBMV proteome, we observed that specific proteins/protein families are present on the BBMV but do not reside in DRMs. Only ∼42% of the BBMV-associated proteins are found in DRMs (Fig. 4E). For example, three immune-related proteins that have been shown to mediate parasite development in An. gambiae, AGAP003656 (51), AGAP005471 (51), and AGAP000550 (9), are identified in the An. gambiae DRM sub-proteome and in the BBMV proteomes for both species. For An. gambiae, we observed that only 124 of the 191 DRM surface expressed (SE) proteins (proteins containing a canonical signal peptide) are also found in the BBMV (supplemental Tables S3B and S3C). In contrast, only 105 An. albimanus BBMV proteins were conserved in the AgDRM-SE protein subset (supplemental Table S3C). These data suggest that like the An. albimanus ortholog of the immune-related protein AGAP000550, which was found only in the SE protein subset of the AnaBBMV proteome, other immune genes might partition differentially across the midgut surface.
Table II. A proteomic and transcriptomic comparison of known/predicted ookinete-interacting proteins that reside in the BBMV of Anopheles albimanus and Anopheles gambiae. These six proteins are commonly considered as leading TBV targets. We find all six proteins in our transcriptome analysis of An. albimanus. All but one of these are identified in the An. gambiae transcriptome. At the protein level, all six proteins were identified in albimanus, some by homology to gambiae. Finally, proteomics was able to identified only three of six proteins in gambiae itself. It should be noted that AGAP003790, AGAP003721, and AGAP003722 were all identified by the same contigs, at the transcript level, and by the same peptides, at the protein level, suggesting that they are isoforms of the same protein.
| Accession number | Annotation | Proteomics |
Spectra | Transcriptomics |
Spectra | ||
|---|---|---|---|---|---|---|---|
| GAMBIAE | ALBIMANUS | GAMBIAE | ALBIMANUS | ||||
| AGAP004809 | AgAPN1 (12) | + | + | 1,200 | + | + | 1,077 |
| AGAP003790 | Annexin-like (55) | + | +a | 54 | + | + | 9 |
| AGAP003721 | Annexin-like (55) | − | +a | 54 | + | + | 9 |
| AGAP003722 | Annexin-like (55, 56) | − | +a | 54 | + | + | 9 |
| AGAP006209 | Carboxypeptidase B (56) | + | +a | 66 | + | + | 12 |
| AGAP010133 | Scavenger receptor, Croquemort homolog (54) | − | +a | 61 | − | + | 135 |
a Percent amino acid identity between GAMBIAE and ALBIMANUS orthologs as determined by BLAST for each NCBInr accession number. Accession numbers were mapped by sequence identity: AGAP003790 is 52% sequence identical to gi 312373765, AGAP003721 is 92% to gi 312373765, AGAP003722 is 84% to gi 312373765, AGAP006209 is 33% to gi312381583, and AGAP010133 is 65% to gi312374586.
Sequence Comparisons Reveal a High Degree of Conservation for Known and Novel Mosquito-based Malaria TBVs
Mosquito-based TBVs have received widespread attention as a critical intervention in the current era of malaria eradication. Candidate antigens for this type of TBV are molecules present on the luminal surface of the mosquito midgut and are known to mediate parasite invasion of the midgut tissue (13). To date, there are six leading candidates (Table II), and we found that all are conserved between An. gambiae and An. albimanus. Interestingly, although all six TBV candidates were identified in the AgDRM subproteome, three—AGAP004809, AGAP003790, and AGAP006209—were also identified in the AngBBMV proteome at 5% peptide FDR, suggesting that these proteins are abundant and do not partition preferentially to DRMs (9). AGAP004809, a female midgut specific alanyl APN N (APN1), is the leading mosquito-based TBV candidate (11, 12). As a direct result of this study, we were able to compare the protein sequences of the An. gambiae and An. albimanus orthologs of APN1 and observed a high degree of conservation, especially for the N-terminal fragment of APN1, which is the target of the malaria transmission-blocking antibodies (supplemental Fig. S2D). AGAP003721, AGAP003722, and AGAP010133 were detected in the AgDRM proteome because they are enriched in DRMs in An. gambiae, but they appear to be abundant in other domains of the An. albimanus midgut, and thus they are detected in AnaBBMV. It has been demonstrated that the An. albimanus midgut surface expresses calreticulin-like protein, a binding ligand for the P. vivax ookinete surface protein Pvs25, one of the leading parasite-based TBV antigens (52, 53). As expected, we identified the same SE calreticulin (gi 76797617) in our AnaBBMV dataset with three peptides, covering 10.76% of the protein sequence with 27 spectra.
DISCUSSION
Searching against sequence databases derived from the translation of newly generated mRNA transcripts into protein sequences allows for rapid identifications of proteins using mass spectra derived from unsequenced malaria mosquito vectors. We assembled both short and long reads from two independent RNA-Seq platforms to generate the An. albimanus transcript-protein database. To search the An. gambiae proteome, we produced a combined subset sequence database consisting of all available anopheline sequences, as well as all sequences corresponding to other members of the order Diptera. These two databases facilitated matches between experimentally and theoretically derived peptides. The use of the PepArML meta-search engine generally resulted in a 2- to 3-fold increase in the number of peptide identifications at 5% FDR relative to Mascot, OMSSA, and X!Tandem alone. We found that spectral features derived from the precursor mass-delta and theoretical isotope-cluster, isoelectric point, and retention-time modeling2 showed good discriminating power for the AngBBMV and AnaBBMV datasets. However, the use of metasearch is optional. A single search engine can be used, although fewer protein identifications should be expected (supplemental Table S5). Users should in particular be aware of the computing power required for a metasearch, which in our case was met by an Amazon EC2 computer cluster. Finally, to annotate novel proteins, we map them to known protein identifications via direct comparison of their peptide sequences. Thus, the overall analysis shows that we were able to obtain a 23% (203) increase in protein identifications by mining the transcriptome. We obtained 2- to 3-fold more single-peptide identifications, with multiple spectra, from transcriptome mining (supplemental Table S4A), which could eventually be manually validated and confirmed via immunoblot, although this is beyond the scope of the current work.
Importantly, proteins identified from the transcriptome-generated protein database were in fact identified by two orthogonal methods, RNA-Seq and protein MS. This provides stronger confirmation of the protein identification than MS alone. This method might identify not only unknown proteins, but also proteins that have no homology to any known protein. This might be a useful way of accessing the unique biology of Diptera, as well as that of other uncharacterized organisms. Importantly, through the implementation of this strategy, we were able to generate additional insight into the conservation of mosquito-based TBV candidate antigens and gain some understanding about the differences between anopheline vector species that might contribute to the unique genotypic interaction between the vector host and the malarial parasite at the midgut interface. Taken together, our platform might help to pave the way for proteogenomic analyses of the entire host range of anopheline malaria vectors and the panoply of Diptera that transmit veterinary and agricultural diseases.
Supplementary Material
Acknowledgments
We thank Travis Clark and Chelsea Baker of the Vanderbilt Genome Technology Core for help with Illumina sequencing. We thank Paul Eggleston and Hilary Hurd for providing the An. gambiae Keele University strain.
Footnotes
* This investigation received financial support from the Bloomberg Family Foundation (R.R.D.), the Johns Hopkins Malaria Research Institute (JHMRI) (R.R.D., J.A.S.), Grant No. HHSN268201000032C (N01-HV-00240), NHLBI, NIH (D.R.G), the Calvin A. and Helen L. Lang Fellowship (D.K.M.), the Advanced Computing Center for Research and Education at Vanderbilt University, the Austrian Ministry of Science and Research, and GEN-AU project BIN (FFG Grant No. 820962) (G.G.T). J.G.G. is funded by the Graduate Program in Biological Sciences at Vanderbilt University and NIAID, NIH, Grant No. F31AI091343–01.
 This article contains supplemental material.
This article contains supplemental material.
2 Gubbala, P., and Edward, N. (2010) Boosting Peptide Identification Performance by Combining Many Search Engines, Spectral Matching, and Proteotypic and Physicochemical Peptide Properties. Poster presented at the 2010 USHUPO Annual Conference, Denver, CO.
1 The abbreviations used are:
- BBMV
- brush border microvilli
- DRM
- detergent resistant membrane
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- MS
- mass spectrometry
- RNA-Seq
- next-generation RNA sequencing
- TBV
- transmission-blocking vaccine.
REFERENCES
- 1. Mueller I., Galinski M. R., Baird J. K., Carlton J. M., Kochar D. K., Alonso P. L., del Portillo H. A. (2009) Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect. Dis. 9, 555–566 [DOI] [PubMed] [Google Scholar]
- 2. Alonso P. L., Brown G., Arevalo-Herrera M., Binka F., Chitnis C., Collins F., Doumbo O. K., Greenwood B., Hall B. F., Levine M. M., Mendis K., Newman R. D., Plowe C. V., Rodriguez M. H., Sinden R., Slutsker L., Tanner M. (2011) A research agenda to underpin malaria eradication. PLoS Med. 8, e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodriguez M. H., Gonzalez-Ceron L., Hernandez J. E., Nettel J. A., Villarreal C., Kain K. C., Wirtz R. A. (2000) Different prevalences of Plasmodium vivax phenotypes VK210 and VK247 associated with the distribution of Anopheles albimanus and Anopheles pseudopunctipennis in Mexico. Am. J. Trop. Med. Hyg. 62, 122–127 [DOI] [PubMed] [Google Scholar]
- 4. Joy D. A., Gonzalez-Ceron L., Carlton J. M., Gueye A., Fay M., McCutchan T. F., Su X. Z. (2008) Local adaptation and vector-mediated population structure in Plasmodium vivax malaria. Mol. Biol. Evol. 25, 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collins W. E., Warren M., Skinner J. C., Richardson B. B., Kearse T. S. (1977) Infectivity of the Santa Lucia (El Salvador) strain of Plasmodium falciparum to different anophelines. J. Parasit. 63, 57–61 [PubMed] [Google Scholar]
- 6. Olano V. A., Carrillo M. P., Delavega P., Espinal C. A. (1985) Vector competence of Cartagena strain of Anopheles albimanus for Plasmodium falciparum and Plasmodium vivax. Trans. R. Soc. Trop. Med. Hyg. 79, 685–686 [DOI] [PubMed] [Google Scholar]
- 7. Collins W. E., McClure H., Strobert E., Skinner J. C., Richardson B. B., Roberts J. M., Galland G. G., Sullivan J., Morris C. L., Adams S. R. (1993) Experimental infection of Anopheles gambiae s.s., Anopheles freeborni and Anopheles stephensi with Plasmodium malariae and Plasmodium brasilianum. J. Am. Mosq. Control Assoc. 9, 68–71 [PubMed] [Google Scholar]
- 8. Kiszewski A., Mellinger A., Spielman A., Malaney P., Sachs S. E., Sachs J. (2004) A global index representing the stability of malaria transmission. Am. J. Trop. Med. Hyg. 70, 486–498 [PubMed] [Google Scholar]
- 9. Parish L. A., Colquhoun D. R., Mohien C. U., Lyashkov A. E., Graham D. R., Dinglasan R. R. (2011) Ookinete-interacting proteins on the microvillar surface are partitioned into detergent resistant membranes of Anopheles gambiae midguts. J. Proteome Res. 10, 5150–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lavazec C., Boudin C., Lacroix R., Bonnet S., Diop A., Thiberge S., Boisson B., Tahar R., Bourgouin C. (2007) Carboxypeptidases B of Anopheles gambiae as targets for a Plasmodium falciparum transmission-blocking vaccine. Infect. Immun. 75, 1635–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mathias D. K., Plieskatt J. L., Armistead J. S., Bethony J. M., Abdul-Majid K. B., McMillan A., Angov E., Aryee M. J., Zhan B., Gillespie P., Keegan B., Jariwala A. R., Rezende W., Bottazzi M. E., Scorpio D. G., Hotez P. J., Dinglasan R. R. (2012) Expression, immunogenicity, histopathology, and potency of a mosquito-based malaria transmission-blocking recombinant vaccine. Infect. Immun. 80, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dinglasan R. R., Kalume D. E., Kanzok S. M., Ghosh A. K., Muratova O., Pandey A., Jacobs-Lorena M. (2007) Disruption of Plasmodium falciparum development by antibodies against a conserved mosquito midgut antigen. Proc. Natl. Acad. Sci. U.S.A. 104, 13461–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dinglasan R. R., Jacobs-Lorena M. (2008) Flipping the paradigm on malaria transmission-blocking vaccines. Trends Parasitol. 24, 364–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holt R. A., Subramanian G. M., Halpern A., Sutton G. G., Charlab R., Nusskern D. R., Wincker P., Clark A. G., Ribeiro J. M., Wides R., Salzberg S. L., Loftus B., Yandell M., Majoros W. H., Rusch D. B., Lai Z., Kraft C. L., Abril J. F., Anthouard V., Arensburger P., Atkinson P. W., Baden H., de Berardinis V., Baldwin D., Benes V., Biedler J., Blass C., Bolanos R., Boscus D., Barnstead M., Cai S., Center A., Chaturverdi K., Christophides G. K., Chrystal M. A., Clamp M., Cravchik A., Curwen V., Dana A., Delcher A., Dew I., Evans C. A., Flanigan M., Grundschober-Freimoser A., Friedli L., Gu Z., Guan P., Guigo R., Hillenmeyer M. E., Hladun S. L., Hogan J. R., Hong Y. S., Hoover J., Jaillon O., Ke Z., Kodira C., Kokoza E., Koutsos A., Letunic I., Levitsky A., Liang Y., Lin J. J., Lobo N. F., Lopez J. R., Malek J. A., McIntosh T. C., Meister S., Miller J., Mobarry C., Mongin E., Murphy S. D., O'Brochta D. A., Pfannkoch C., Qi R., Regier M. A., Remington K., Shao H., Sharakhova M. V., Sitter C. D., Shetty J., Smith T. J., Strong R., Sun J., Thomasova D., Ton L. Q., Topalis P., Tu Z., Unger M. F., Walenz B., Wang A., Wang J., Wang M., Wang X., Woodford K. J., Wortman J. R., Wu M., Yao A., Zdobnov E. M., Zhang H., Zhao Q., Zhao S., Zhu S. C., Zhimulev I., Coluzzi M., della Torre A., Roth C. W., Louis C., Kalush F., Mural R. J., Myers E. W., Adams M. D., Smith H. O., Broder S., Gardner M. J., Fraser C. M., Birney E., Bork P., Brey P. T., Venter J. C., Weissenbach J., Kafatos F. C., Collins F. H., Hoffman S. L. (2002) The genome sequence of the malaria mosquito Anopheles gambiae. Science 298, 129–149 [DOI] [PubMed] [Google Scholar]
- 15. Hittinger C. T., Johnston M., Tossberg J. T., Rokas A. (2010) Leveraging skewed transcript abundance by RNA-Seq to increase the genomic depth of the tree of life. Proc. Natl. Acad. Sci. U.S.A. 107, 1476–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krzywinski J., Besansky N. J. (2003) Molecular systematics of Anopheles: from subgenera to subpopulations. Annu. Rev. Entomol. 48, 111–139 [DOI] [PubMed] [Google Scholar]
- 17. Zdobnov E. M., von Mering C., Letunic I., Torrents D., Suyama M., Copley R. R., Christophides G. K., Thomasova D., Holt R. A., Subramanian G. M., Mueller H. M., Dimopoulos G., Law J. H., Wells M. A., Birney E., Charlab R., Halpern A. L., Kokoza E., Kraft C. L., Lai Z., Lewis S., Louis C., Barillas-Mury C., Nusskern D., Rubin G. M., Salzberg S. L., Sutton G. G., Topalis P., Wides R., Wincker P., Yandell M., Collins F. H., Ribeiro J., Gelbart W. M., Kafatos F. C., Bork P. (2002) Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298, 149–159 [DOI] [PubMed] [Google Scholar]
- 18. Hurd H., Taylor P., Adams D., Underhill A., Eggleston P. (2005) Measuring the costs of mosquito resistance to malaria infection. Evolution 12, 2560–2572 [PMC free article] [PubMed] [Google Scholar]
- 19. Abdul-Rauf M., Ellar D. J. (1999) Isolation and characterization of brush border membrane vesicles from whole Aedes aegypti larvae. J. Invertebr. Pathol. 73, 45–51 [DOI] [PubMed] [Google Scholar]
- 20. Hauser H., Howell K., Dawson R. M. C., Bowyer D. E. (1980) Rabbit small intestinal brush-border membrane preparation and lipid-composition. Biochim. Biophys. Acta 602, 567–577 [DOI] [PubMed] [Google Scholar]
- 21. Gibbons J. G., Beauvais A., Beau R., McGary K. L., Latge J. P., Rokas A. (2012) Global transcriptome changes underlying colony growth in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell 11, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibbons J. G., Janson E. M., Hittinger C. T., Johnston M., Abbot P., Rokas A. (2009) Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol. Biol. Evol. 26, 2731–2744 [DOI] [PubMed] [Google Scholar]
- 23. Martinez-Barnetche J., Gomez-Barreto R. E., Ovilla-Munoz M., Tellez-Sosa J., Garcia-Lopez D. E., Dinglasan R. R., Ubaida Mohien C., Maccallum R. M., Redmond S. N., Gibbons J. G., Rokas A., Machado C. M., Cazares-Raga F., Gonzalez-Ceron L., Hernandez-Martinez S., Rodriguez-Lopez M. H. (2012) Transcriptome of the adult female malaria mosquito vector Anopheles albimanus. BMC Genomics 13, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zerbino D. R., Birney E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schulz M. H., Zerbino D. R., Vingron M., Birney E. (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28, 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Surget-Groba Y., Montoya-Burgos J. I. (2010) Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 20, 1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edwards N., Wu X., Tseng C. W. (2009) An unsupervised, model-free, machine-learning combiner for peptide identifications from tandem mass spectra. Clin. Proteomics 5, 23–36 [Google Scholar]
- 28. Perkins D. N., Pappin D. J. C., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 29. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X. Y., Shi W. Y., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 30. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 31. MacLean B., Eng J. K., Beavis R. C., McIntosh M. (2006) General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics 22, 2830–2832 [DOI] [PubMed] [Google Scholar]
- 32. Tabb D. L., Fernando C. G., Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 34. Kim S., Gupta N., Pevzner P. A. (2008) Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J. Proteome Res. 7, 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 36. Mohien C. U., Hartler J., Breitwieser F., Rix U., Rix L. R., Winter G. E., Thallinger G. G., Bennett K. L., Superti-Furga G., Trajanoski Z., Colinge J. (2010) MASPECTRAS 2: an integration and analysis platform for proteomic data. Proteomics 10, 2719–2722 [DOI] [PubMed] [Google Scholar]
- 37. Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 [DOI] [PubMed] [Google Scholar]
- 38. Pitts R. J., Rinker D. C., Jones P. L., Rokas A., Zwiebel L. J. (2011) Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics 12, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collins L. J., Biggs P. J., Voelckel C., Joly S. (2008) An approach to transcriptome analysis of non-model organisms using short-read sequences. Genome Inform. 21, 3–14 [PubMed] [Google Scholar]
- 40. DiGuistini S., Liao N. Y., Platt D., Robertson G., Seidel M., Chan S. K., Docking T. R., Birol I., Holt R. A., Hirst M., Mardis E., Marra M. A., Hamelin R. C., Bohlmann J., Breuil C., Jones S. J. M. (2009) De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data. Genome Biol. 10, R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eisen N. S., Fernandes V. F., Harvey W. R., Spaeth D. D., Wolfersberger M. G. (1988) Comparison of brush border membrane vesicles prepared by three methods from larval Manduca sexta midgut. Insect Biochem. 9, 337–342 [Google Scholar]
- 42. Horth P., Miller C. A., Preckel T., Wenz C. (2006) Efficient fractionation and improved protein identification by peptide OFFGEL electrophoresis. Mol. Cell. Proteomics 5, 1968–1974 [DOI] [PubMed] [Google Scholar]
- 43. Wang X., Slebos R. J. C., Wang D., Halvey P. J., Tabb D. L., Liebler D. C., Zhang B. (2012) Protein identification using customized protein sequence databases derived from RNA-Seq data. J. Proteome Res. 11, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Desgagne-Penix I., Khan M. F., Schriemer D. C., Cram D., Nowak J., Facchini P. J. (2010) Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol. 10, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adamidi C., Wang Y., Gruen D., Mastrobuoni G., You X., Tolle D., Dodt M., Mackowiak S. D., Gogol-Doering A., Oenal P., Rybak A., Ross E., Sanchez Alvarado A., Kempa S., Dieterich C., Rajewsky N., Chen W. (2011) De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res. 21, 1193–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nesvizhskii A. I., Aebersold R. (2005) Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 47. Tharakan R., Edwards N., Graham D. R. (2010) Data maximization by multipass analysis of protein mass spectra. Proteomics 10, 1160–1171 [DOI] [PubMed] [Google Scholar]
- 48. Wang H., Tang H. Y., Tan G. C., Speicher D. W. (2011) Data analysis strategy for maximizing high-confidence protein identifications in complex proteomes such as human tumor secretomes and human serum. J. Proteome Res. 10, 4993–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tabb D. L., Vega-Montoto L., Rudnick P. A., Variyath A. M., Ham A. J. L., Bunk D. M., Kilpatrick L. E., Billheimer D. D., Blackman R. K., Cardasis H. L., Carr S. A., Clauser K. R., Jaffe J. D., Kowalski K. A., Neubert T. A., Regnier F. E., Schilling B., Tegeler T. J., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Fisher S. J., Gibson B. W., Kinsinger C. R., Mesri M., Rodriguez H., Stein S. E., Tempst P., Paulovich A. G., Liebler D. C., Spiegelman C. (2010) Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 9, 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Salichos L., Rokas A. (2011) Evaluating ortholog prediction algorithms in a yeast model clade. PLoS One 6, e18755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garver L. S., Xi Z. Y., Dimopoulos G. (2008) Immunoglobulin superfamily members play an important role in the mosquito immune system. Dev. Comp. Immunol. 32, 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rodriguez M. D., Martinez-Barnetche J., Alvarado-Delgado A., Batista C., Argotte-Ramos R. S., Hernandez-Martinez S., Ceron L. G., Torres J. A., Margos G., Rodriguez M. H. (2007) The surface protein Pvs25 of Plasmodium vivax ookinetes interacts with calreticulin on the midgut apical surface of the malaria vector Anopheles albimanus. Mol. Biochem. Parasitol. 153, 167–177 [DOI] [PubMed] [Google Scholar]
- 53. Sattabongkot J., Tsuboi T., Hisaeda H., Tachibana M., Suwanabun N., Rungruang T., Cao Y. M., Stowers A. W., Sirichaisinthop J., Coleman R. E., Torii M. (2003) Blocking of transmission to mosquitoes by antibody to Plasmodium vivax malaria vaccine candidates Pvs25 and Pvs28 despite antigenic polymorphism in field isolates. Am. J. Trop. Med. Hyg. 69, 536–541 [PubMed] [Google Scholar]
- 54. Gonzalez-Lazaro M., Dinglasan R. R., Hernandez-Hernandez Fde L., Rodriguez M. H., Laclaustra M., Jacobs-Lorena M., Flores-Romo L. (2009) Anopheles gambiae Croquemort SCRBQ2, expression profile in the mosquito and its potential interaction with the malaria parasite Plasmodium berghei. Insect Biochem. Mol. Biol. 39, 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kotsyfakis M., Ehret-Sabatier L., Siden-Kiamos I., Mendoza J., Sinden R. E., Louis C. (2005) Plasmodium berghei ookinetes bind to Anopheles gambiae and Drosophila melanogaster annexins. Mol. Microbiol. 57, 171–179 [DOI] [PubMed] [Google Scholar]
- 56. Lavazec C., Bonnet S., Thiery I., Boisson B., Bourgouin C. (2005) cpbAg1 encodes an active carboxypeptidase B expressed in the midgut of Anopheles gambiae. Insect Mol. Biol. 14, 163–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.