Abstract
The human importin-β family consists of 21 nucleocytoplasmic transport carrier proteins that carry proteins and RNAs across the nuclear envelope through nuclear pores in specific directions. These transport carriers are responsible for the nucleocytoplasmic transport of thousands of proteins, but the cargo allocation of each carrier, which is necessary information if one wishes to understand the physiological context of transport, is poorly characterized. To address this issue, we developed a high-throughput method to identify the cargoes of transport carriers by applying stable isotope labeling by amino acids in cell culture to construct an in vitro transport system. Our method can be outlined in three steps. (1) Cells are cultured in a medium containing a stable isotope. (2) The cell membranes of the labeled cells are permeabilized, and proteins extracted from unlabeled cells are transported into the nuclei of the permeabilized cells. In this step, the reaction system is first depleted of all importin-β family carriers and then supplemented with a particular importin-β family carrier of interest. (3) Proteins in the nuclei are extracted and analyzed quantitatively via LC-MS/MS. As an important test case, we used this method to identify cargo proteins of transportin, a representative member of the importin-β family. As expected, the identified candidate cargo proteins included previously reported transportin cargoes as well as new potential cargoes, which we corroborated via in vitro binding assays. The identified cargoes are predominately RNA-interacting proteins, affirming that cargoes allotted to the same carrier share functional characteristics. Finally, we found that the transportin cargoes possessed at least two classes of signal sequences: the well characterized PY-nuclear localization signals specific for transportin, and Lys/Arg-rich segments capable of binding to both transportin and importin-β. Thus, our method will be useful for linking a carrier to features shared among its cargoes and to specific nuclear localization signals.
Most genetic processes, including chromosome replication and transcription, occur in the nucleus, and thus the selection of proteins that enter the nucleus and act there is crucial for cellular processes (1). During interphase, all nuclear proteins cross the nuclear envelope via nuclear pores, channels that constitute a selective permeability barrier for macromolecules. Only a fraction of proteins traverse these channels by means of free diffusion; most nuclear proteins are imported into or exported out of the nucleus with the assistance of importin-β (Imp-β)1 family proteins (Imp-βs) (2, 3). Imp-βs interact with the nuclear pore complex in a way that allows them to travel in and out of the nucleus. To drive cargo transport, they utilize the GTPase cycle of small GTPase Ran. Import carriers bind to their cargoes in the cytoplasm and then travel to the nucleus, where they release their cargoes upon binding to RanGTP. The RanGTP-bound carriers recycle back to the cytoplasm, where RanGTP is converted to RanGDP. Export carriers form trimeric complexes with their cargoes and RanGTP in the nucleus, and the export complexes disassemble in the cytoplasm (2, 3). The human genome encodes 21 Imp-βs, of which 12 are nuclear import carriers, 7 are export carriers, and 2 are bidirectional carriers (4, 5). Among the Imp-βs, Imp-β is special in that it often uses Imp-α family proteins as adapters to bind to cargoes, but the transport activity is still basically attributable to Imp-β (6).
Imp-βs import thousands of cargo proteins into the nucleus and export a large number. Although some cargoes are transported by multiple carriers, most cargoes are expected to be transported via specific interactions, and a group of cargoes transported by a carrier might share specific functions. Therefore, to reveal the role of each carrier in various cellular processes, it is imperative to define which carriers transport which cargoes. Unfortunately, only a limited number of cargo–carrier interactions have been characterized, and in some cases few or no cargoes have been linked to specific Imp-β family carriers.
Systematic searches for cargo proteins have been performed, especially for those dependent on Imp-α to associate with Imp-β, using Far Western-based cellular proteomics (7) and the screening of in vitro virus libraries (8) and oriented peptide libraries (9). The experiments using synthetic libraries helped to refine the classical nuclear localization signal (cNLS) sequences to which Imp-α binds, but they did not detect natural cargo proteins directly. Cargo proteins for Imp-β family carriers have mainly been identified using affinity methods (4, 10–14) or yeast two-hybrid screening (15). These methods were based on Imp-cargo protein binding. Recent studies, however, have reported that the functions of Imp-βs are not limited to nucleocytoplasmic transport and are also relevant to diverse cellular processes (16–18). In this study, we designed a transport-based method for cargo identification by combining stable isotope labeling by amino acids in cell culture (SILAC) (19), an in vitro transport system (20, 21), and MS. Our use of SILAC-based quantitative MS differs from the typical methods so far reported that compare cellular protein levels under different conditions. We used SILAC to prepare permeabilized recipient cells for an in vitro transport system and incubated them with unlabeled proteins, which are the source of import substrates, and thereby proteins imported into the nuclei could be distinguished from the proteins of the recipient cells.
We selected transportin (Trn) (importin-β2/karyopherin-β2) (22) as a representative example of the Imp-β family carriers because many of its cargo proteins have been identified (3, 14), its cargo recognition has been characterized structurally (23), and potential cargo proteins have been predicted (24).
Historically, the first Trn-binding site identified was the “M9 sequence” (25), which we now understand to be an instance of the PY-NLS (24), a Trn-recognition signal which almost all known Trn cargoes appear to contain. In terms of sequence constraints, the PY-NLS is a loosely defined motif usually ending with a proline-tyrosine (Pro-Tyr) dipeptide, which can be further subdivided into hydrophobic and basic types, depending on the nature of its N-terminal region (24). Previous work also suggests the possible existence of another NLS for Trn, distinct from the PY-NLS. In 1998, Jäkel and Görlich reported a basic region resembling a cNLS within residues 32–74 of RL23A (rpL23a) that can serve as an NLS for Trn as well as the import carriers Imp-β, importin 5 (RanBP5), and importin 7 (RanBP7) (10). RL23A is the only clear instance of this so-called β-like import receptor binding (BIB) domain, but circumstantial evidence hints that this might be a more general signal: (1) the ribosomal proteins rpS7 and rpL5 contain similar sequences and are imported by several Imp-βs, including Trn, although binding to Trn could not be confirmed in vitro (10); (2) the ribosomal proteins rpS6 (26) and rpL7a (27) contain basic regions that serve as NLSs, although their carriers are not yet identified; and (3) the C-terminal domain of the nucleoporin ELYS contains a basic segment and interacts with both Trn and Imp-β (28).
The present study was performed with two goals: (1) to establish our transport assay as a valuable new tool that can complement conventional techniques for import carrier cargo identification, and (2) to discover novel Trn cargoes and gain insight into the range of sequences recognized by Trn.
EXPERIMENTAL PROCEDURES
The buffers, ATP regeneration system, proteins, antibodies, cytosolic and nuclear extracts, evaluation of the transport system, and identification of Ran-regulated Trn-interacting proteins via a pull-down method are described in the supplemental data.
In Vitro Transport
A transport system modified from a previous study (20, 21) was used. Adherent HeLa-S3 cells were labeled with u-13C6 Lys and u-13C6 Arg via SILAC and seeded onto a 30-mm glass plate in a dish. In vitro transport was performed on the glass plate. (Materials were kept on ice thereafter unless otherwise stated.) The cells were then rinsed in transport buffer (TB) (20 mm HEPES-KOH (pH 7.3), 110 mm KOAc, 2 mm MgOAc, 5 mm NaOAc, 0.5 mm EGTA, 2 mm DTT, and 1 μg/ml each of aprotinin, pepstatin A, and leupeptin) twice, permeabilized through incubation with 40 μg/ml digitonin in TB for 4 min, and then rinsed twice and incubated in TB for 5 min. The resulting permeabilized cells were pretreated with TB containing 4 μm RanGDP and an ATP regeneration system for 20 min at 30 °C to remove residual Imp-βs before the transport reaction. After rinsing with TB twice, the transport reaction was carried out through incubation of the cells in 250 μl of transport mixture (50% Imp-depleted cytosolic extract, 10% Imp- and RCC1-depleted nuclear extract, 1 μm p10/NTF2, and ATP regeneration system in TB) with (+Trn) or without (control) 0.3 μm Trn for 20 min at 30 °C. Reactions with and without Trn were always performed simultaneously. After the reaction, the cells were rinsed with TB twice and then incubated in 250 μl of extract mixture (50% Imp-depleted cytosolic extract and ATP regeneration system in TB) for 20 min at 30 °C to remove proteins binding non-specifically to the permeabilized cells. The cells were rinsed with NaCl-TB (110 mm NaCl instead of KOAc in TB) three times and suspended in 100 μl of nuclear buffer (20 mm Tris-HCl (pH 8.0, 4 °C), 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 2 mm DTT, and 1 μg/ml each of aprotinin, pepstatin A, and leupeptin). Proteins were extracted from the permeabilized cells via sonication and clarified via centrifugation for 15 min at 20,000 × g. Evaluation of the transport system is shown in Fig. 1B and supplemental Figs. S1–S3.
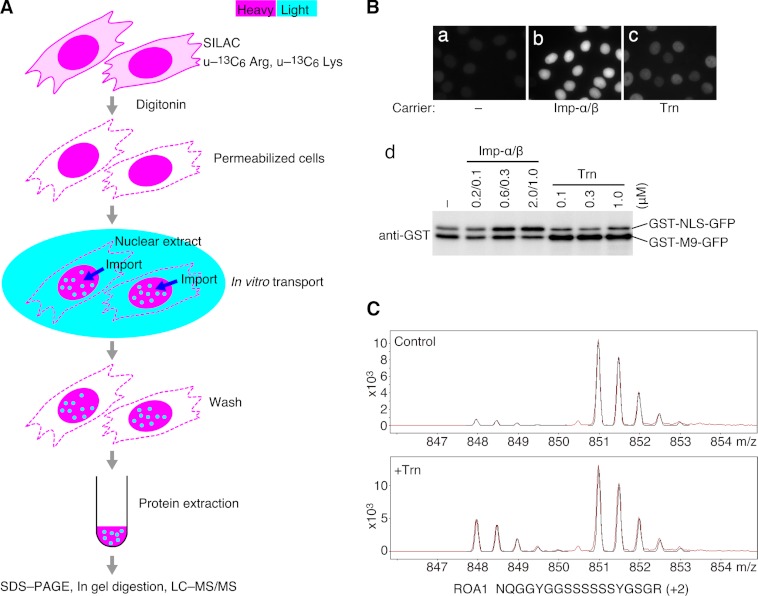
Fig. 1.
Transport-based cargo identification. A, method outline. Cells containing heavy proteins (magenta) were generated via SILAC and permeabilized with digitonin. Light proteins (cyan) in unlabeled nuclear extract were imported into the nuclei by Trn. Then, nuclear proteins were extracted and analyzed by means of LC-MS/MS. The light/heavy peptide ratios were measured concurrently with protein identification. Experiments with (+Trn) and without (control) Trn were performed simultaneously, and their results were compared. B, evaluation of the import assay. The reporter cargoes GST-NLS-GFP and GST-M9-GFP, which are specific for Imp-β plus Imp-α and Trn, respectively, were mixed and added to the transport reaction without carriers (a), with 0.6 μm Imp-α and 0.3 μm Imp-β (b), or with 0.3 μm Trn (c). The cells were observed via fluorescence microscopy. (d) After the transport reactions with the indicated concentrations of carriers, proteins were extracted and cargoes were examined using Western blotting with an antibody specific for GST. The transport of proteins in nuclear extract is evaluated in supplemental Fig. S2. C, typical mass spectra for a peptide derived from a Trn cargo (ROA1). The peaks corresponding to the light (m/z: 848–850) and heavy (m/z: 851–853) peptides were detected. The relative levels of the light peaks were compared between the control and +Trn spectra as follows: T/C = (L/HTrn)/(L/HCtl). Red, actual measurement; black, predicted spectrum.
Protein Identification and Quantification via LC-MS/MS
Two hundred micrograms of proteins collected from five to six transport reactions were separated via SDS-PAGE in one lane of a 7.5%–12.5% gradient polyacrylamide gel (height = 16 cm, thickness = 1 mm). The gel was sliced into 36 pieces per lane and incubated in 100 mm ammonium bicarbonate (pH 8.0) buffer containing 10 mm DTT for 1 h at 60 °C. After alkylation with 55 mm iodoacetamide in 100 mm ammonium bicarbonate buffer for 30 min at room temperature, the proteins were in-gel-digested with 1 pmol of trypsin (Promega, Madison, Wisconsin) in 50 μl of the same buffer for 16 h at 37 °C. Half the amount of peptides recovered from each gel piece was analyzed separately using a nanoflow LC linear ion-trap time-of-flight (TOF) mass spectrometer (NanoFrontier LD, Hitachi, Tokyo, Japan). The liquid chromatography was performed on a PicoFrit C18 column (75 μm × 100 mm, particle size = 3 μm; NewObjective, Wobum, Massachusetts) with elution (200 nl/min) using a 2%–49% acetonitrile gradient over 45 min in the presence of 0.3% formic acid. The mass spectrometer was operated in the data-dependent MS/MS mode. Precursor ions were measured by the TOF mass analyzer and subjected to fragmentation in the linear ion trap. For protein identification, peaks in the spectra were extracted by Mascot Distiller (version 2.3, Matrix Science, London, UK) with the correlation threshold set at 0.7, and the resultant peak lists were queried against the Swiss-Prot database (release 2010_04) using Mascot (version 2.2, Matrix Science). The taxonomy of interest was set as Homo sapiens (20,350 sequences). The search parameters used were as follows: enzyme cleavage specificity, trypsin; number of missed cleavages permitted, ≤2; fixed modification, Cys-carbamidomethylation; variable modification, Met-oxidation; quantitation, SILAC K+6 R+6 [MD]; mass tolerances for precursor and fragment ions, ±0.5 Da. Quantitative analysis of light/heavy (L/H) ratios of precursor ions was performed using Mascot Distiller, in which protein hits of the Mascot search were passed through the filter of significance threshold p < 0.05 (the default value of Mascot Distiller), and the L/H ratio of each precursor ion peptide was calculated from the peak areas in the MS spectra. To ensure reliable quantification, only high-quality peaks (intensity > 300, charge ≥ +2, standard error < 0.1, fraction > 0.5, and correlation > 0.8, as defined by Mascot Distiller) were considered for further analysis; proteins with no peptides meeting the above criteria in either the control or +Trn sample were excluded. For each parent protein, arithmetic mean L/H ratios were calculated separately for peptides from the control and +Trn samples (L/HCtl and L/HTrn, respectively). The T/C value (calculated as (L/HTrn)/(L/HCtl)) was defined to indicate the cargo potentiality of each protein. Proteins and peptides that satisfied the above criteria are listed in descending T/C value order in supplemental Table S1 and presented graphically in supplemental Fig. S4. In order to select proteins with highly reliable T/C values, we also applied two arbitrary thresholds to the L/H ratios. To discount spectra from proteins that bound non-specifically to the permeabilized cells (see supplemental Fig. S2), the L/HCtl value was required to be <1; to avoid inaccuracies near the lower limit of relative quantification, the values of L/HCtl and L/HTrn were required to be >0.03. The proteins that passed each test are listed in Table I. For each quantified protein, one representative MS/MS spectrum with annotation was depicted by Peptide View of the Mascot search program, saved in the mht file format of Internet Explorer (Microsoft, Redmond, Washington) using a homemade Visual Basic program, and shown in the “spectra” folder of the supplemental data. To further demonstrate the validity of our protein identification, the Mascot search results are presented in the peptide summary format with a cutoff score of 20 for accepting individual MS/MS spectra (supplemental Table S2A). False discovery rates determined by decoy searches were less than 10%. Because a few proteins, especially those who are members of large protein families, are missing in this table because of the different attributions of common peptides among family proteins, the result of querying with a cutoff score of 10 is summarized (supplemental Table S2B) to show that the protein identification with a cutoff score of 10 was consistent with that in supplemental Table S1.
Table I. Summary of the results; identified proteins for which the quantitation data passed the quality thresholds are listed in descending order of T/C value.
| Entry namea | Mass | L/H (Ctl)b | L/H (+Trn)c | T/Cd | Bead haloe | NLSh (reference) | |
|---|---|---|---|---|---|---|---|
| 1 | RA1L3_HUMAN | 34373 | 0.051 | 0.337 | 6.602 | (pseudogene)i | |
| 2 | ROA2_HUMAN | 37464 | 0.040 | 0.244 | 6.153 | ++ | PYj (30) |
| 3 | ROA1_HUMAN | 38837 | 0.086 | 0.423 | 4.910 | ++ | PY (30) |
| 4 | HNRPD_HUMAN | 38581 | 0.133 | 0.413 | 3.104 | + | PY (37) |
| 5 | ROAA_HUMAN | 36316 | 0.143 | 0.414 | 2.901 | ++ | PYj (this study) |
| 6 | RL3_HUMAN | 46365 | 0.098 | 0.229 | 2.339 | ++(++) | BIB (this study) |
| 7 | HNRH1_HUMAN | 49484 | 0.334 | 0.615 | 1.839 | ++ | PY (29) |
| 8 | CPSF6_HUMAN | 59344 | 0.243 | 0.328 | 1.349 | Degr.f | PY (14, 24) |
| 9 | SF3B2_HUMAN | 100279 | 0.492 | 0.578 | 1.176 | Degr.f(+) | BIB (this study) |
| 10 | RU2A_HUMAN | 28512 | 0.714 | 0.806 | 1.129 | ++(+) | BIB (this study) |
| 11 | RL27_HUMAN | 15788 | 0.120 | 0.134 | 1.121 | ++(++) | BIB (this study) |
| 12 | SNRPA_HUMAN | 31259 | 0.122 | 0.135 | 1.111 | + | PYj (24) |
| 13 | SF3B3_HUMAN | 136575 | 0.698 | 0.752 | 1.077 | Not exp.g | |
| 14 | SMC1A_HUMAN | 143771 | 0.132 | 0.137 | 1.039 | Not exp.g | – |
| 15 | PARP1_HUMAN | 113811 | 0.149 | 0.151 | 1.013 | Degr.f | – |
| 16 | IMB1_HUMAN | 98420 | 0.041 | 0.041 | 1.001 | ||
| 17 | RS12_HUMAN | 14905 | 0.296 | 0.289 | 0.976 | + | |
| 18 | HNRDL_HUMAN | 46580 | 0.177 | 0.168 | 0.946 | ++ | PY (38) |
| 19 | WBP11_HUMAN | 69954 | 0.197 | 0.183 | 0.928 | ||
| 20 | RL7_HUMAN | 29264 | 0.105 | 0.097 | 0.928 | ||
| 21 | PPIB_HUMAN | 23785 | 0.114 | 0.104 | 0.914 | ± | |
| 22 | RL26_HUMAN | 17248 | 0.118 | 0.107 | 0.907 | ||
| 23 | RL18A_HUMAN | 21034 | 0.129 | 0.117 | 0.905 | ||
| 24 | SK2L2_HUMAN | 118756 | 0.135 | 0.122 | 0.903 | ||
| 25 | ERP29_HUMAN | 29032 | 0.086 | 0.078 | 0.903 | ||
| 26 | NHP2_HUMAN | 17532 | 0.104 | 0.093 | 0.889 | ||
| 27 | RL12_HUMAN | 17979 | 0.136 | 0.118 | 0.868 | ||
| 28 | GRP78_HUMAN | 72402 | 0.243 | 0.211 | 0.868 | ||
| 29 | RAB7A_HUMAN | 23760 | 0.879 | 0.759 | 0.863 | ||
| 30 | PDIA3_HUMAN | 57146 | 0.109 | 0.094 | 0.861 | ||
| 31 | HNRPU_HUMAN | 91269 | 0.071 | 0.061 | 0.860 | + | |
| 32 | RL10A_HUMAN | 24987 | 0.138 | 0.118 | 0.855 | ||
| 33 | PRKDC_HUMAN | 473749 | 0.181 | 0.154 | 0.853 | ||
| 34 | RL14_HUMAN | 23531 | 0.142 | 0.121 | 0.849 | ||
| 35 | RL8_HUMAN | 28235 | 0.127 | 0.108 | 0.848 | ||
| 36 | RS4X_HUMAN | 29807 | 0.284 | 0.240 | 0.844 | ||
| 37 | RLA0L_HUMAN | 34514 | 0.195 | 0.163 | 0.834 | ||
| 38 | NPM_HUMAN | 32726 | 0.081 | 0.068 | 0.832 | − | |
| 39 | RL7A_HUMAN | 30148 | 0.130 | 0.107 | 0.822 | ||
| 40 | RL11_HUMAN | 20468 | 0.147 | 0.119 | 0.812 | ||
| 41 | RS11_HUMAN | 18590 | 0.311 | 0.251 | 0.808 | ||
| 42 | RL13A_HUMAN | 23619 | 0.129 | 0.104 | 0.808 | ||
| 43 | RL19_HUMAN | 23565 | 0.127 | 0.102 | 0.801 | ||
| 44 | RBBP7_HUMAN | 48132 | 0.959 | 0.768 | 0.800 | ||
| 45 | PDIA6_HUMAN | 48490 | 0.078 | 0.061 | 0.781 | ||
| 46 | RS10_HUMAN | 18886 | 0.306 | 0.238 | 0.777 | ||
| 47 | RS25_HUMAN | 13791 | 0.334 | 0.258 | 0.771 | ||
| 48 | RS29_HUMAN | 6900 | 0.295 | 0.226 | 0.767 | ||
| 49 | CPSM_HUMAN | 165975 | 0.047 | 0.036 | 0.764 | ||
| 50 | RS23_HUMAN | 15969 | 0.272 | 0.207 | 0.763 | ||
| 51 | RS13_HUMAN | 17212 | 0.304 | 0.232 | 0.763 | ||
| 52 | RS8_HUMAN | 24475 | 0.286 | 0.216 | 0.756 | ||
| 53 | DJB11_HUMAN | 40774 | 0.099 | 0.073 | 0.742 | − | |
| 54 | RL24_HUMAN | 17882 | 0.149 | 0.110 | 0.739 | ||
| 55 | RS7_HUMAN | 22113 | 0.347 | 0.249 | 0.719 | ||
| 56 | PAIRB_HUMAN | 44995 | 0.342 | 0.245 | 0.717 | ||
| 57 | RS18_HUMAN | 17708 | 0.328 | 0.235 | 0.717 | ||
| 58 | MCM3_HUMAN | 91551 | 0.350 | 0.250 | 0.714 | ||
| 59 | RS9_HUMAN | 22635 | 0.281 | 0.198 | 0.705 | ||
| 60 | RL5_HUMAN | 34569 | 0.253 | 0.177 | 0.698 | (39)k | |
| 61 | GBLP_HUMAN | 35511 | 0.351 | 0.245 | 0.697 | ||
| 62 | RS3_HUMAN | 26842 | 0.347 | 0.241 | 0.695 | ||
| 63 | PCNA_HUMAN | 29092 | 0.756 | 0.524 | 0.694 | − | |
| 64 | RS2_HUMAN | 31590 | 0.370 | 0.255 | 0.689 | (40) | |
| 65 | IF2A_HUMAN | 36374 | 0.912 | 0.628 | 0.689 | ||
| 66 | HNRPQ_HUMAN | 69788 | 0.729 | 0.498 | 0.683 | − | |
| 67 | RL4_HUMAN | 47953 | 0.143 | 0.098 | 0.682 | ||
| 68 | ENPL_HUMAN | 92696 | 0.104 | 0.070 | 0.675 | ||
| 69 | HNRPK_HUMAN | 51230 | 0.597 | 0.399 | 0.668 | − | |
| 70 | XRCC6_HUMAN | 70084 | 0.405 | 0.269 | 0.666 | ||
| 71 | APEX1_HUMAN | 35931 | 0.948 | 0.631 | 0.666 | ||
| 72 | NSUN2_HUMAN | 87214 | 0.403 | 0.266 | 0.661 | ||
| 73 | RS3A_HUMAN | 30154 | 0.294 | 0.194 | 0.660 | ||
| 74 | ALDR_HUMAN | 36230 | 0.323 | 0.213 | 0.659 | ||
| 75 | DHX15_HUMAN | 91673 | 0.555 | 0.364 | 0.656 | ||
| 76 | RL18_HUMAN | 21735 | 0.116 | 0.075 | 0.647 | ||
| 77 | BTF3_HUMAN | 22211 | 0.851 | 0.548 | 0.644 | ||
| 78 | HMCS1_HUMAN | 57828 | 0.266 | 0.169 | 0.637 | ||
| 79 | CBX3_HUMAN | 20969 | 0.189 | 0.120 | 0.632 | ||
| 80 | PSME3_HUMAN | 29602 | 0.904 | 0.571 | 0.631 | ||
| 81 | MCM2_HUMAN | 102516 | 0.577 | 0.363 | 0.628 | ||
| 82 | ACTN4_HUMAN | 105245 | 0.489 | 0.299 | 0.612 | ||
| 83 | PRP19_HUMAN | 55603 | 0.274 | 0.167 | 0.610 | ||
| 84 | PABP1_HUMAN | 70854 | 0.644 | 0.370 | 0.575 | ||
| 85 | DNMT1_HUMAN | 185388 | 0.556 | 0.315 | 0.566 | ||
| 86 | NUCL_HUMAN | 76625 | 0.139 | 0.076 | 0.549 | ||
| 87 | MCM6_HUMAN | 93801 | 0.590 | 0.320 | 0.542 | ||
| 88 | SERA_HUMAN | 57356 | 0.547 | 0.296 | 0.541 | − | |
| 89 | VINC_HUMAN | 124292 | 0.644 | 0.328 | 0.509 | ||
| 90 | DEK_HUMAN | 42933 | 0.125 | 0.052 | 0.415 | ||
a UniProtKB/Swiss-Prot entry name.
b Light/heavy ratio of the control reaction.
c Light/heavy ratio of the +Trn reaction.
d The light/heavy ratio of the +Trn reaction was divided by the light/heavy ratio of the control reaction.
e Results of the bead halo assay: ++ and +, positive; −, negative; ±, unclear; blank, not done; (++) and (+), positive for Trn binding when GFP-Lys/Arg-rich segment fusions were assayed (Fig. 3D).
f,g The assays were not done or the results were obscure, because the proteins were not expressed (g) or degraded extensively (f) in E. coli.
h Potential NLS for Trn-binding: PY, PY-NLS; BIB, BIB-domain-like NLS; –, undetermined; blank, not examined.
i Three peptides were matched in the Mascot search, but their sequences exist in ROA1 also.
j Predicted in this study or the indicated reference.
k Result of Xenopus L5.
Bead Halo Assay
Normalized extracts of E. coli expressing a green fluorescent protein (GFP) fusion protein (see “Experimental Procedures” in the supplemental material), glutathione (GSH)-Sepharose, and GST, GST-Trn, or GST-Imp-β were mixed in EHBN buffer (10 mm EDTA, 0.5% 1,6-hexanediol, 10 mg/ml bovine serum albumin, 125 mm NaCl) as described elsewhere (29). GTP-fixed mutant Q69L-Ran (30), which inhibits Trn-cargo interactions, was added when appropriate. After the mixtures had been incubated for 20 to 30 min, they were observed via fluorescence microscopy.
RESULTS
Transport-based Cargo Identification System
Screening for candidate cargo proteins relies on the relative quantification of stable isotope-labeled “heavy” and unlabeled “light” peptides by means of MS, together with an in vitro reconstituted transport system (Fig. 1). HeLa cells were labeled with u-13C6 Lys and u-13C6 Arg via SILAC, and their cell membranes were permeabilized by digitonin. Residual Imp-βs associated with nuclei of the permeabilized cells were removed through incubation with Ran and ATP. Nuclear and cytosolic extracts (the former a source of cargo proteins) were prepared from unlabeled HeLa cells and depleted of Imp-βs using phenyl-Sepharose (31) (supplemental Figs. S1A–S1C). Imp-βs are almost completely removed from the extracts by this method (32). The nuclear extract was further depleted of RCC1 (a guanine nucleotide exchange factor for Ran) using a Ran affinity method (supplemental Fig. S1D) because cytoplasmic RCC1 impairs the import reaction. The transport reaction was carried out using these materials in the absence (control) or presence (+Trn) of recombinant Trn. Unlabeled proteins from the extracts that bound non-specifically to structural components of the permeabilized cells were removed after the transport reaction through treatment with ATP and cytosolic extract depleted of Imp-βs and subsequent washing with a salt solution (supplemental Fig. S3). Proteins were extracted from the permeabilized cells, and their tryptic peptides were analyzed via LC-MS/MS to identify the proteins and quantify their light and heavy peptides (Fig. 1C). The performance of the transport system was evaluated with reporter cargoes: GST-NLS-GFP specific for Imp-β with Imp-α, and GST-M9-GFP for Trn (Fig. 1B). From a mixture of the two cargoes, both the Imp-β with Imp-α and Trn imported their specific cargo selectively. Likewise, natural cargo proteins labeled with biotin in the nuclear extract were imported selectively by these carriers (supplemental Fig. S2). Thus, our transport system provides import specificity sufficient for cargo identification.
Identification of Trn-specific Cargo Proteins
The MS/MS spectra were queried against the Swiss-Prot database, and the L/H ratio in the MS spectra for each precursor peptide ion was calculated using Mascot Distiller. Over 650 proteins each were identified in the control and Trn-reacted permeabilized cell extracts, with about 480 proteins detected in both extracts (supplemental Table S2A). Applying quality thresholds for quantification to the spectrum profiles reduced the number of proteins with valid L/H ratios to 298. The arithmetic mean L/H ratios for the peptides derived from each protein were calculated separately for the control (L/HCtl) and +Trn (L/HTrn) samples, and the T/C ratio ((L/HTrn)/(L/HCtl)) for each protein was used as an index of cargo potentiality. The 298 proteins are listed in descending T/C value order in supplemental Table S1 and presented graphically in supplemental Fig. S4. Highly ranked proteins showing outstanding T/C values are expected to be Trn cargoes, but the T/C values might be inaccurate as a result of experimental limitations. We thus further confined the proteins to those with more reliable T/C values by excluding proteins that might have bound non-specifically to the permeabilized cells or that had extremely low L/H ratios (see “Experimental Procedures”). Ultimately, 90 proteins were selected, as listed in descending T/C value order in Table I and presented in Fig. 2E. Hereinafter, the proteins are referred to by their numbers as presented in Table I. Proteins 1–8 that were assigned very high T/C values include four previously reported Trn cargo proteins. Proteins 9–18, with lower but still significantly high T/C values, include a predicted and an experimentally defined cargo protein. We postulate that these highly ranked proteins are potential Trn cargoes, but with two clear exceptions. RA1L3 (hnRNP A1-like) is a pseudogene, mistakenly identified because of the mis-assignment of ROA1 (hnRNP A1) derived peptides, and the high T/C value for IMB1 (Imp-β) resulted from export inhibition of residual Imp-β (supplemental Fig. S5). In some cases, proteins may be identified as different proteins of the same or related families possessing common amino acid sequences, depending on the parameters for database search (supplemental Tables S2A and S2B). Among the highly ranked proteins, one hnRNP H family protein was identified as HNRH1 (hnRNP H) or HNRPF (hnRNP F), depending on the attribution of a common peptide. Both of these hnRNPs have been reported as Trn cargoes (33). We adopted HNRPH1, as the representative of the family, for further analysis.
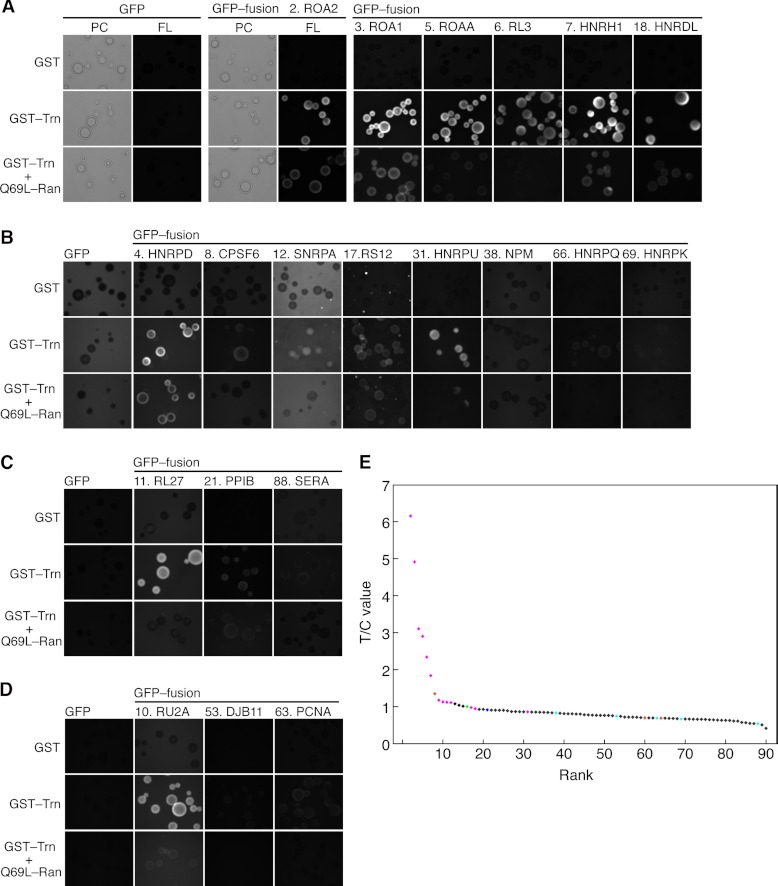
Fig. 2.
Interaction of candidate cargoes with Trn. A–D, interactions examined via bead halo assay. Bacterial extracts containing GFP-fusion proteins were mixed with GSH-Sepharose beads coated with GST or GST-Trn fusion proteins and then observed via fluorescence microscopy. Q69L-Ran, which inhibits the Trn-cargo specific interaction, was added as indicated. The protein numbering is identical to that in Table I. The images within each panel are comparable. PC, phase contrast; FL, fluorescence micrograph. A, B, the assay and microscopy conditions were appropriate for proteins expressed at high levels in E. coli. B, images taken with the same exposure time as in A were enhanced. C, D, proteins expressed at lower levels. The GFP moiety concentrations were half (C) and one-fifth (D) of those in A and B. Accordingly, the exposure time was longer. E, T/C values predict Trn cargoes. Proteins quantified via MS were ranked in descending T/C value order (Table I). Horizontal, protein rank; vertical, T/C value. Magenta, protein (or segment, Figs. 3C and 3D) that bound to Trn in the bead halo assays; blue, unclear; cyan, not bound; orange, not examined here but reported to be Trn cargo; green, IMB1; black, others. RA1L3 (sequence from pseudogene) is not shown.
Most of the 16 cargo candidates that we selected were not identified by means of a pull-down method reported previously (14). In that report, 15 proteins that bound to Trn in the absence of Ran but not in its presence were isolated from HeLa cell extract. The cargo candidates identified in this study differ from them, except for only a few (proteins 3, 7, and 8).
To further clarify the relative strengths of our transport assay vis-à-vis conventional pull-down assays, we carried out a pull-down-based assay designed to detect Ran-regulated Trn-cargo interactions using the same depleted nuclear extract as used in the transport-based methods. From the extract, Trn-binding proteins were precipitated by GST-Trn with GSH-Sepharose beads, and after washing, proteins were eluted by the GTP-fixed mutant Q69L-Ran or a control buffer (see supplemental “Experimental Procedures ” and supplemental Fig. S6), mimicking the protein–protein interactions in Trn mediated protein import. Proteins enriched in the Q69L-Ran eluate were identified via LC-MS/MS. Many proteins were identified in the LC-MS/MS analysis for each band in the SDS-PAGE, and the ten proteins with the highest scores are listed in supplemental Table S4A. Of the 16 cargo candidates obtained via the transport-based method, eight (proteins 6, 8, 10–14, and 17) are absent in supplemental Table S4A. These results indicate that the cargo proteins identified in the transport-based method partly overlap with those from pull-down methods but differ considerably, indicating that our transport-based method can complement conventional pull-down methods.
Interaction between Identified Cargoes and Trn
Next, we confirmed the physical interactions between the identified candidate cargoes and Trn through a bead halo assay (29). The highly ranked proteins and randomly selected lower ranked proteins in Table I were expressed as GFP fusion proteins in Escherichia coli (supplemental Table S3). The bacterial extracts were mixed with GST-Trn fusion protein–coated GSH-Sepharose and then observed via fluorescence microscopy (Fig. 2). The GFP fusion proteins were grouped according to expression level, and the bacterial extracts containing them were normalized within each group (see supplemental “Experimental Procedures ” and supplemental Fig. S7A). GFP fusions of proteins 2, 3, 5–7, 10, 11, and 18 displayed brighter halos, which disappeared after the addition of Q69L-Ran. Neither the combination of GFP alone mixed with GST-Trn nor GFP fusion proteins mixed with GST alone produced halos. Therefore, the observed binding was specific. Halos for GFP fusions of proteins 4, 12, 17, and 31 were evident in the enhanced images and disappeared after the addition of Q69L-Ran. In contrast, GFP fusions of proteins 21, 38, 53, 63, 66, 69, and 88 produced faint or undetectable halos. GFP fusion of protein 8, which was heavily degraded in E. coli (supplemental Fig. S7A), gave ambiguous results. GFP fusions of proteins 9 and 13–15 were either not expressed or severely degraded in E. coli extracts. In summary, proteins with higher T/C values bound to Trn, whereas those with values less than 0.85 did not. Consequently, we believe that 16 proteins (proteins 2–15, 17, and 18) are potential Trn cargoes. Protein 21 and lower-ranked proteins did not bind to Trn, with the exception of protein 31, HNRPU (hnRNP U). Given that many hnRNPs are reported to be Trn cargoes, HNRPU is most likely a specific cargo. As this example illustrates, in the intermediate T/C value range, some disagreement between the T/C value and cargo potentiality might be unavoidable.
BIB Domain-like Lys/Arg-rich NLS
Eight highly ranked proteins (proteins 2–5, 7, 8, 12, and 18) contained sequences identical or similar to the well-established Trn-binding sequence PY-NLS (24) (Fig. 3A), which was originally identified in ROA1 as the M9 sequence (25). Other highly ranked proteins (proteins 6, 9–11, 13–15, and 17) do not contain similar sequences, though they bound to Trn in the bead halo assays. Another type of Trn-binding sequence, the BIB domain in RL23A, which is enriched in basic residues and binds to other carriers as well, was reported previously (10). We examined the possibility that the highly ranked proteins contain a BIB-domain-like NLS instead of the PY-NLS that is responsible for Trn binding. The defining features of the BIB domain are not clear, except that it overlaps cNLS motifs, in which Lys and Arg are the key elements. We therefore hypothesized that the presence of a cNLS motif might be a prerequisite for this type of NLS and searched for the six classes of cNLS motifs (8) shown in Fig. 3B. Among the eight highly ranked proteins lacking the PY-NLS, six contained one or more matches to the cNLS motifs (19 in total) generally found in Lys/Arg-rich regions. Because the BIB domain of RL23A is longer than the cNLS motifs, we selected nineteen 50-residue segments with the cNLS motifs placed at the center (Fig. 3C). Each segment was fused to GFP (supplemental Fig. S7B) and examined for Trn binding using a bead halo assay (Fig. 3D). The GFP fusion to one segment from RL27 (RL27-I) was heavily degraded and could not be measured. Of the remaining 18 segments, 5 bound to Trn, including the first Lys/Arg-rich segment of SF3B2 (SF3B2-I), for which Trn binding could not be measured for the full-length protein (Table I). Four out of the five segments were found to bind directly to Imp-β (Fig. 3D), a feature shared with the reported BIB domain. Interestingly, the segments that bound to both Trn and Imp-β differed in amino acid composition from the other 13 segments, showing a high ratio of Arg to Asp+Glu content (Fig. 3E).
Fig. 3.
Two types of Trn-binding sequences. A, PY-NLS motif matches in the identified cargoes. Five proteins (proteins 2–5 and 18) contain typical PY-NLS sequences, whereas the hydrophobic segments of three proteins (proteins 7, 8, and 12) diverge. Φ, hydrophobic; X, any amino acid; magenta, conserved Pro-Tyr dipeptide; cyan, conserved Arg, His, or Lys residue; green, hydrophobic motif with a conserved Gly, Ala, or Ser residue. The sequences shown are from the UniProt database. Identifier: ROA2, P22626–1; ROA1, P09651–2; HNRPD, Q14103–1; ROAA, Q99729–2; HNRDL, Q14979; HNRH1, P31943; CPSF6, Q16630; SNRPA, P09012. B, cNLS motifs. Six classes of cNLS motifs defined by Kosugi et al. (8) were searched in the identified cargoes not carrying PY-NLS sequences. We expanded the definition of bipartite cNLS to derive more candidate sequences. K+R≥3/5, three or more Lys or Arg within five residues; [ ], any one of the amino acids within the brackets is allowed; [∧DE], any residue other than Asp or Glu is allowed; green, Lys; cyan, Arg. C, segments fused to GFP. Nineteen cNLS motif matches (underscored) were found in the cargoes, and segments consisting of 50 residues centered on them were expressed as GFP fusion proteins. Some segments were shorter than 50 residues, because the cNLS motif match was near the N- or C-terminus of the protein. The segments are shown in five groups ordered, from the top, as follows: (1) the BIB domain of RL23A reported to bind to both Trn and Imp-β (not assayed here), (2) the four fragments that bound to both GST-Trn and GST-Imp-β, (3) RU2A-I, which bound only to GST-Trn (weakly), (4) the fragments that bound to neither GST-Trn nor GST-Imp-β, and (5) the heavily degraded RL27-I. Green, Lys; cyan, Arg; magenta, Asp or Glu. The numbers displayed on the left and right of each segment indicate amino acid positions in the protein sequences listed in supplemental Table S3 and denote the co-ordinates of that segment in its UniProt protein sequence. The one exception is PARP1-II (marked with an asterisk). The sequence of PARP1 isoform (supplemental Table S3) that we cloned diverges from the one in UniProt in the last half of this segment, and DNA for the segment was cloned separately from a HeLa cDNA library. D, interactions between the Lys/Arg-rich segments and Trn or Imp-β. The segments fused to GFP were analyzed with GST-Trn or GST-Imp-β in the bead halo assay. Q69L-Ran was added as indicated. The images in the right-hand panel were recorded with a longer exposure. E, the Asp+Glu content of the segments in groups (1)–(4) in C plotted against their Arg content. (1) cyan diamond, (2) magenta diamond, (3) magenta square, and (4) black diamond.
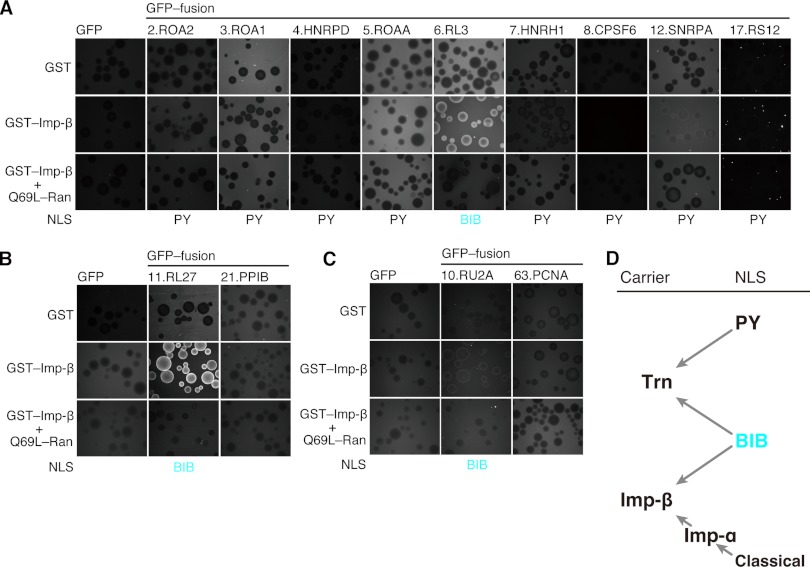
Next, we analyzed Imp-β binding of the full-length proteins containing the BIB-domain-like NLSs and PY-NLSs (Fig. 4). All of the proteins with BIB-domain-like NLSs bound to Imp-β, including RU2A, whose Lys/Arg-rich segment (RU2A-I) failed to bind to Imp-β (Fig. 3D). The extensive degradation of GFP-RU2A-I (supplemental Fig. S7B) might explain this discrepancy. In contrast, the PY-NLS-containing proteins did not bind to Imp-β, with the exception of SNRPA, which bound only weakly. These results confirm that PY-NLSs generally bind to Trn exclusively, whereas BIB-domain-like NLSs bind to both Trn and Imp-β (Fig. 4D).
Fig. 4.
Interaction between Imp-β and full-length Trn cargoes. A–C, Imp-β binding of the full-length proteins. The Trn cargoes carrying BIB-domain-like NLSs or PY-NLSs were analyzed for binding to Imp-β in the bead halo assay. Proteins are grouped as in Fig. 2, and the images are comparable within each panel. The results of PPIB and PCNA are presented as negative controls. The class of NLS (PY-NLS or BIB-domain-like NLS) is indicated. D, Trn NLSs and carriers. The BIB-domain-like NLS binds to both Trn and Imp-β directly, whereas PY-NLS binds to Trn exclusively. Undiscovered NLSs differing from these two might also exist.
To be certain that the BIB-domain-like segments are truly distinct from PY-NLSs, we conducted an additional analysis based on a very broadly defined PY-NLS motif (supplemental Fig. S8A) that covers all PY-NLSs summarized by Süel et al. (34), as well as many spurious matches (supplemental Fig. S8B). Even with this broad definition, SF3B2-I did not overlap with a PY-NLS motif match, and RL3-I and RL3-II overlapped only by one or two residues to the N-terminal tail of PY-NLS motif matches. So we conclude that the Trn-binding shown by these segments is not attributable to a PY-NLS. RL27-II partially overlapped a basic PY-NLS motif match with an atypical proline-leucine instead of Pro-Tyr, but the N-terminal basic epitope part of this match is outside of the segment tested in the fusion protein (whose sequence does not fully match either type of PY-NLS). So these four Trn-binding segments cannot be explained in terms of binding to PY-NLSs. In contrast, the weakly Trn-binding segment RU2A-I overlaps entirely with a PY-NLS motif match, and so without further experimental analysis, we cannot rule out the possibility that the Trn binding of that segment is due to a PY-NLS. Nevertheless, our results suggest that the BIB-domain-like NLSs, which had been previously demonstrated only in RL23A, can be classified as more general NLSs found in several proteins.
DISCUSSION
We developed a transport-based system for identifying cargo proteins specific for individual Imp-βs and used it for large-scale screening of Trn-specific cargoes. As a result, 298 proteins produced adequate MS spectra for calculating T/C values, the index for cargo potentiality (supplemental Table S1 and supplemental Fig. S4). Because the L/H ratios of some proteins binding non-specifically to the permeabilized cells might not reflect transport efficiency, we removed proteins with high L/HCtl ratios (>1) and also ruled out proteins with low L/HTrn and L/HCtl ratios (<0.03) to ensure the accuracy of the quantification. Notably, only a few of the proteins with high T/C values failed to pass the L/H ratio criteria (compare Fig. 2E and supplemental Fig. S4), and the yield of cargo candidates was not greatly affected by this selection. Ultimately we obtained 90 proteins with highly reliable T/C values as listed in Table I, and among those, proteins 1–18, except for IMB1 and RA1L3, are possible Trn cargoes. Of those 16 proteins, 12 bound to Trn in the bead halo assay (Figs. 2 and 3), 8 are reported or predicted Trn cargoes (Table I), and 8 contain the PY-NLSs or analogous sequences (Fig. 3A). These facts support the likelihood that other proteins with similar T/C values are also bona fide Trn cargoes, including proteins 13–15, which lack additional corroborating evidence.
Although we successfully identified a considerable number of known and novel Trn cargoes, we also missed many known cargoes. To elucidate the reason for this, we traced the progression through our transport assay of 44 previously reported (either demonstrated or predicted) Trn cargoes (supplemental Table S5A). Among the 44 proteins, 21 are included in the 650 proteins identified in one or both of the control and +Trn cell extracts (supplemental Table S2), but only 13 proteins are included in the 298 proteins quantified in both the extracts adequately enough to calculate T/C values (supplemental Table S1). Thus, many reported cargoes, and those predicted by Lee et al. (24) (supplemental Table S5B), were not identified or quantified in the present LC-MS/MS analysis. We speculate that the absence of these defined or predicted cargoes in our LC-MS/MS result is attributable to the current performance limitations of the instrument, rather than to the sample preparation. Western blotting (supplemental Fig. S9A) revealed that both the control and +Trn extracts applied to LC-MS/MS analysis contained the well-characterized Trn cargoes EWS, TAP/NXF1, FUS, SAM68, and HuR, but they were not detected by the LC-MS/MS (see supplemental Table S5). The mass spectrometer was operated in the data-dependent acquisition mode. Although this currently is a standard operation mode for attaining high-throughput quantification with accuracy, it often fails to identify proteins or peptides that are not separated from abundant ones in SDS-PAGE or LC. Because LC-MS/MS analysis is a generic technique that can be substituted by an improved method, the numbers of proteins that we identified (650) and quantified (298), and consequently the number of cargoes that can be assigned T/C values, are expected to increase significantly with the use of advanced MS techniques that are rapidly developing. Thus, we believe our method is promising for higher-throughput identification of cargoes, although the identification of those with very low copy numbers or extensive post-translational modification might remain difficult without an alternative approach.
In Western blotting (supplemental Fig. S9A), the cargoes were detected at almost the same level in the control and +Trn extracts. This does not necessarily indicate low transport activities, because the signal intensity represents the sum of the imported and endogenous proteins. For example, the expected signal intensity ratio of protein 10, RU2A, with L/HCtl: 0.714 and L/HTrn: 0.806 is +Trn/control = (1 + 0.806)/(1 + 0.714) ≈ 1.05, and the 5% difference might be hardly apparent in Western blotting. Nonetheless, Trn efficiently imported recombinant GFP-RU2A in our transport system (supplemental Fig. S9B). Likewise, Trn imports GFP fusion proteins of the reported Trn cargoes, TAP/NXF1, SAM68, HuR, and PABP2, in our system (supplemental Fig. S9B), though they were not detected in the LC-MS/MS analysis. Given that many of the known cargo proteins, albeit not all, were present in the depleted nuclear extract, and because Trn imports the ones we tested, it is likely that many more imported cargoes could be identified with the use of a more advanced instrument.
From the 298 proteins successfully quantified, 198 with L/HCtl > 1 and 10 with L/HCtl or L/HTrn < 0.03 were removed from consideration, leaving the 90 proteins in Table I. In this step, the number of previously reported cargoes decreased from 13 to 8 (supplemental Table S5A). However, the ratio of previously reported cargoes increased from 90/298 (30%) to 8/13 (62%). Thus, despite the extensive elimination of proteins from consideration, our method has the potential to identify cargoes at a practical level.
Anecdotal analysis justifies our elimination criteria in many cases. For example, we eliminated the heat shock proteins Hsp70 and Hsp90, which interact with a variety of proteins, might well adhere to the permeabilized cells, and often exhibit non-specific binding in conventional protein–protein interaction assays. So the inability to provide clear results for heat shock proteins is a common problem for many assay systems, not just ours. One known Trn cargo, Histone H2B, was discarded on the basis of its low L/H ratio, but it would be an exceptional case. L/H ratios of histones will be low even if they are imported in the transport system, because endogenous histones in the permeabilized cells are exceptionally abundant. The effect of the threshold L/H: 0.03 on the final number of cargoes is negligible, because only ten proteins (e.g. VIME and MDHM, supplemental Table S1) were eliminated from the total protein list based on the threshold. Thus, only three known cargoes were dismissed because of the threshold L/HCtl: 1.0, and the non-specific binding of proteins influences the cargo identification less severely than might be expected.
Significantly, our transport-based method identified a set of cargoes different from that identified by the conventional pull-down method carried out in this and other studies (14). The major merits of our transport-based method are that it fully involves the cellular transport system and accumulates cargoes in the nuclei. The pull-down experiment carried out in this study also involves the biochemical function of Trn–Ran interaction, mimicking the cargo release from Trn within a nucleus. In our hands, however, the separation of specific cargoes from the background of non-specific proteins was not effective. Many proteins were readily released into both the control and Q69L-Ran eluted fractions (supplemental Fig. S6), probably because the cargo dissociation from Trn is rapid. A washing condition that removes nonspecific proteins but preserves specific cargoes on Trn is difficult to establish, and specific cargoes dissociate from Trn even in the control buffer during the elution. Although our pull-down-based method leaves room for improvement, it is clear that the transport-based method has the advantage of accumulating cargoes into the nuclei even though their interaction with their carriers is short-lived. Thus the transport-based method can complement pull-down methods by identifying a different subset of cargoes.
The cargoes that we identified are mainly RNA metabolism factors, including ribosomal proteins. This observation agrees with the features of Trn cargoes that have been reported to date and demonstrates that RNA interaction is a general attribute of many Trn cargoes. We expect that cargoes transported by the same carrier might share a specific function, and our method should be useful in elucidating common properties of cargoes transported by other Imp-β family carriers.
Our results are consistent with the view of PY-NLS as a Trn specific signal. None of the PY-NLS carrying cargoes tested here bound to Imp-β (Fig. 4). Some PY-NLS carrying Trn cargoes, such as TAP/NXF1 and SNRPA, are known to bind to other import carriers, but the binding to the other carriers is not necessarily attributable to their PY-NLSs. The N-terminal 100 residue region of TAP/NXF1 binds to Trn and some other Imp-βs. However, both a divergent cNLS and a hydrophobic PY-NLS can be assigned separately within this region, with amino acid substitutions in the former primarily hampering binding to Imp-β, whereas in the latter binding to Trn is affected (35). SNRPA carries a PY-NLS (Fig. 3A) and binds to Trn (Fig. 2B), but it also carries a cNLS and binds to Imp-α (36).
Eight of the Trn cargo proteins identified here contain PY-NLSs, supporting the view that this well-established signal is the major Trn NLS. However, our results also suggest that many Trn cargoes may be recognized by a distinct NLS. We identified five segments in four Trn cargoes that match cNLS motifs and bind to both Trn and Imp-β. As these characteristics are reminiscent of the BIB domain (10), we refer to these segments as BIB-domain-like. The number of instances (our five segments plus the original BIB domain) of this potential new NLS is still too limited for rigorous statistical inference, but we note that the ratio of Arg to Asp+Glu is higher for those segments than for other cNLS-like segments that did not bind Trn (Fig. 3E).
As described above, our study has shed new light on Trn cargo recognition, potentially breathing new life into the decade-old notion of the BIB domain as a general NLS. Unfortunately, for many Imp-βs, few if any cargoes are known, and no specific NLSs have even been proposed. We expect our method will be useful for identifying novel NLSs through the identification of carrier specific cargoes.
Supplementary Material
Acknowledgments
We thank Yuriko Morinaka, Ai Watanabe, Yuumi Oda, and Seishi Iguchi for technical assistance. We thank the Support Unit for Bio-material Analysis, RIKEN BSI Research Resources Center, especially Kaori Otsuki, Masaya Usui, and Aya Abe for MS analysis.
Footnotes
* This work was supported by RIKEN Special Project Funding for Basic Science in Cellular System Project Research and funding awarded to N.I. by the Japan Society for the Promotion of Science (JSPS) through the “Funding Program for Next Generation World-Leading Researchers (NEXT Program),” initiated by the Council for Science and Technology Policy (CSTP).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- BIB
- β-like import receptor binding
- cNLS
- classical nuclear localization signal
- GFP
- green fluorescent proteins
- GSH
- glutathione
- Imp
- importin
- Imp-βs
- importin-β family protein
- L/H
- light/heavy
- NLS
- nuclear localization signal
- SILAC
- stable isotope labeling by amino acids in cell culture
- TB
- transport buffer
- Trn
- transportin.
REFERENCES
- 1. Terry L. J., Shows E. B., Wente S. R. (2007) Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416 [DOI] [PubMed] [Google Scholar]
- 2. Poon I. K., Jans D. A. (2005) Regulation of nuclear transport: central role in development and transformation? Traffic 6, 173–186 [DOI] [PubMed] [Google Scholar]
- 3. Pemberton L. F., Paschal B. M. (2005) Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6, 187–198 [DOI] [PubMed] [Google Scholar]
- 4. Mingot J. M., Kostka S., Kraft R., Hartmann E., Görlich D. (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J. 20, 3685–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gontan C., Güttler T., Engelen E., Demmers J., Fornerod M., Grosveld F. G., Tibboel D., Görlich D., Poot R. A., Rottier R. J. (2009) Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J. Cell Biol. 185, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 7. Blazek E., Meisterernst M. (2006) A functional proteomics approach for the detection of nuclear proteins based on derepressed importin alpha. Proteomics 6, 2070–2078 [DOI] [PubMed] [Google Scholar]
- 8. Kosugi S., Hasebe M., Matsumura N., Takashima H., Miyamoto-Sato E., Tomita M., Yanagawa H. (2009) Six classes of nuclear localization signals specific to different binding grooves of importin α. J. Biol. Chem. 284, 478–485 [DOI] [PubMed] [Google Scholar]
- 9. Yang S. N., Takeda A. A., Fontes M. R., Harris J. M., Jans D. A., Kobe B. (2010) Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-α using oriented peptide library screening. J. Biol. Chem. 285, 19935–19946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jäkel S., Görlich D. (1998) Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bohnsack M. T., Regener K., Schwappach B., Saffrich R., Paraskeva E., Hartmann E., Görlich D. (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21, 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stüven T., Hartmann E., Görlich D. (2003) Exportin 6: a novel nuclear export receptor that is specific for profilin·actin complexes. EMBO J. 22, 5928–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mingot J. M., Bohnsack M. T., Jäkle U., Görlich D. (2004) Exportin 7 defines a novel general nuclear export pathway. EMBO J. 23, 3227–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Güttinger S., Mühlhäusser P., Koller-Eichhorn R., Brennecke J., Kutay U. (2004) Transportin2 functions as importin and mediates nuclear import of HuR. Proc. Natl. Acad. Sci. U.S.A. 101, 2918–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brownawell A. M., Macara I. G. (2002) Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol. 156, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mosammaparast N., Pemberton L. F. (2004) Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556 [DOI] [PubMed] [Google Scholar]
- 17. Harel A., Forbes D. J. (2004) Importin beta: conducting a much larger cellular symphony. Mol. Cell 16, 319–330 [DOI] [PubMed] [Google Scholar]
- 18. Tahara K., Takagi M., Ohsugi M., Sone T., Nishiumi F., Maeshima K., Horiuchi Y., Tokai-Nishizumi N., Imamoto F., Yamamoto T., Kose S., Imamoto N. (2008) Importin-β and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J. Cell Biol. 180, 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 20. Adam S. A., Marr R. S., Gerace L. (1990) Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111, 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Melchior F., Sweet D. J., Gerace L. (1995) Analysis of Ran/TC4 function in nuclear protein import. Methods Enzymol. 257, 279–291 [DOI] [PubMed] [Google Scholar]
- 22. Pollard V. W., Michael W. M., Nakielny S., Siomi M. C., Wang F., Dreyfuss G. (1996) A novel receptor-mediated nuclear protein import pathway. Cell 86, 985–994 [DOI] [PubMed] [Google Scholar]
- 23. Imasaki T., Shimizu T., Hashimoto H., Hidaka Y., Kose S., Imamoto N., Yamada M., Sato M. (2007) Structural basis for substrate recognition and dissociation by human transportin 1. Mol. Cell 28, 57–67 [DOI] [PubMed] [Google Scholar]
- 24. Lee B. J., Cansizoglu A. E., Süel K. E., Louis T. H., Zhang Z., Chook Y. M. (2006) Rules for nuclear localization sequence recognition by karyopherin β2. Cell 126, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Siomi H., Dreyfuss G. (1995) A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129, 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmidt C., Lipsius E., Kruppa J. (1995) Nuclear and nucleolar targeting of human ribosomal protein S6. Mol. Biol. Cell 6, 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Russo G., Ricciardelli G., Pietropaolo C. (1997) Different domains cooperate to target the human ribosomal L7a protein to the nucleus and to the nucleoli. J. Biol. Chem. 272, 5229–5235 [DOI] [PubMed] [Google Scholar]
- 28. Lau C. K., Delmar V. A., Chan R. C., Phung Q., Bernis C., Fichtman B., Rasala B. A., Forbes D. J. (2009) Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol. Biol. Cell 20, 4043–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel S. S., Rexach M. F. (2008) Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol. Cell. Proteomics 7, 121–131 [DOI] [PubMed] [Google Scholar]
- 30. Bischoff F. R., Ponstingl H. (1995) Catalysis of guanine nucleotide exchange of Ran by RCC1 and stimulation of hydrolysis of Ran-bound GTP by Ran-GAP1. Methods Enzymol. 257, 135–144 [DOI] [PubMed] [Google Scholar]
- 31. Ribbeck K., Görlich D. (2002) The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21, 2664–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kose S., Furuta M., Imamoto N. (2012) Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell 149, 578–589 [DOI] [PubMed] [Google Scholar]
- 33. Van Dusen C. M., Yee L., McNally L. M., McNally M. T. (2010) A glycine-rich domain of hnRNP H/F promotes nucleocytoplasmic shuttling and nuclear import through an interaction with transportin 1. Mol. Cell. Biol. 30, 2552–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Süel K. E., Gu H., Chook Y. M. (2008) Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 6, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Z. C., Satterly N., Fontoura B. M., Chook Y. M. (2011) Evolutionary development of redundant nuclear localization signals in the mRNA export factor NXF1. Mol. Biol. Cell 22, 4657–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hieda M., Tachibana T., Fukumoto M., Yoneda Y. (2001) Nuclear import of the U1A splicesome protein is mediated by importin α/β and Ran in living mammalian cells. J. Biol. Chem. 276, 16824–16832 [DOI] [PubMed] [Google Scholar]
- 37. Suzuki M., Iijima M., Nishimura A., Tomozoe Y., Kamei D., Yamada M. (2005) Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. FEBS J. 272, 3975–3987 [DOI] [PubMed] [Google Scholar]
- 38. Kawamura H., Tomozoe Y., Akagi T., Kamei D., Ochiai M., Yamada M. (2002) Identification of the nucleocytoplasmic shuttling sequence of heterogeneous nuclear ribonucleoprotein D-like protein JKTBP and its interaction with mRNA. J. Biol. Chem. 277, 2732–2739 [DOI] [PubMed] [Google Scholar]
- 39. Clauβen M., Rudt F., Pieler T. (1999) Functional modules in ribosomal protein L5 for ribonucleoprotein complex formation and nucleocytoplasmic transport. J. Biol. Chem. 274, 33951–33958 [DOI] [PubMed] [Google Scholar]
- 40. Antoine M., Reimers K., Wirz W., Gressner A. M., Müller R., Kiefer P. (2005) Identification of an unconventional nuclear localization signal in human ribosomal protein S2. Biochem. Biophys. Res. Commun. 335, 146–153 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.