Abstract
Major depressive disorder (MDD) is a widespread and debilitating mental disorder. However, there are no biomarkers available to aid in the diagnosis of this disorder. In this study, a nuclear magnetic resonance spectroscopy–based metabonomic approach was employed to profile urine samples from 82 first-episode drug-naïve depressed subjects and 82 healthy controls (the training set) in order to identify urinary metabolite biomarkers for MDD. Then, 44 unselected depressed subjects and 52 healthy controls (the test set) were used to independently validate the diagnostic generalizability of these biomarkers. A panel of five urinary metabolite biomarkers—malonate, formate, N-methylnicotinamide, m-hydroxyphenylacetate, and alanine—was identified. This panel was capable of distinguishing depressed subjects from healthy controls with an area under the receiver operating characteristic curve (AUC) of 0.81 in the training set. Moreover, this panel could classify blinded samples from the test set with an AUC of 0.89. These findings demonstrate that this urinary metabolite biomarker panel can aid in the future development of a urine-based diagnostic test for MDD.
Major depressive disorder (MDD)1 is a debilitating mental disorder affecting up to 15% of the general population and accounting for 12.3% of the global burden of disease (1, 2). Currently, the diagnosis of MDD still relies on the subjective identification of symptom clusters rather than empirical laboratory tests. The current diagnostic modality results in a considerable error rate (3), as the clinical presentation of MDD is highly heterogeneous and the current symptom-based method is not capable of adequately characterizing this heterogeneity (4). An approach that can be used to circumvent these limitations is to identify disease biomarkers to support objective diagnostic laboratory tests for MDD.
Metabonomics, which can measure the small molecules in given biosamples such as plasma and urine without bias (5), has been extensively used to characterize the metabolic changes of diseases and thus facilitate the identification of novel disease-specific signatures as putative biomarkers (6–10). Nuclear magnetic resonance (NMR) spectroscopy–based metabonomic approaches characterized by sensitive, high-throughput molecular screening have been employed previously in identifying novel biomarkers for a variety of neuropsychiatric disorders, including stroke, bipolar disorder, and schizophrenia (11–13).
Specifically with regard to MDD, several animal studies have already characterized the metabolic changes in the blood and urine (14–19). These studies provide valuable clues as to the pathophysiological mechanism of MDD. However, no study has been designed with the aim of diagnosing this disease. Recently, using an NMR-based metabonomic approach, this research group identified a unique plasma metabolic signature that enables the discrimination of MDD from healthy controls with both high sensitivity and specificity (20). These findings motivated further study on urinary diagnostic metabolite biomarkers for MDD, which would be more valuable from a clinical applicability standpoint, as urine can be more non-invasively collected. Moreover, previous studies have also demonstrated the feasibility of identifying diagnostic metabolite biomarkers of psychiatric disorders in the urine. For example, using an NMR-based metabonomics approach, Yap et al. (21) identified a unique urinary metabolite signature that clearly discriminated autism patients from healthy controls. As systemic metabolic disturbances have been observed in the urine of a depressed animal model, it is likely that diagnostic metabolite markers for MDD can be detected in human urine.
Therefore, in this study, NMR spectroscopy combined with multivariate pattern recognition techniques were used to profile 82 first-episode drug-naïve MDD subjects and 82 healthy controls (the training set) in order to identify potential metabolite biomarkers for MDD. Furthermore, 44 unselected MDD subjects and 52 healthy controls (the test set) were employed to independently validate the diagnostic performance of these urinary metabolite biomarkers.
EXPERIMENTAL PROCEDURES
Participants
Prior to the collection of urine samples, written informed consents were obtained from all subjects. The protocol of this study was reviewed and approved by the Ethical Committee of Chongqing Medical University. A total of 126 depressed subjects were recruited from the psychiatric center of the First Affiliated Hospital at Chongqing Medical University. All diagnoses were carried out according to the Structured Psychiatric Interview using DSM-IV-TR criteria (22). The 17-item version of the observer-rated Hamilton Depression Rating Scale (HDRS) was used to assess depression severity (23). The depressed subjects with HDRS scores of greater than 17 were recruited. The majority of these MDD subjects (n = 95) were first episode and drug naïve, and the remaining MDD subjects (n = 31) were being treated with various anti-depressants. The detailed individual demographic and clinical data of the recruited subjects are presented in supplemental Table S1. Exclusion criteria for the MDD subjects included any pre-existing physical or other mental disorders and/or illicit drug use. During the same time period, 134 healthy control subjects were recruited from the medical examination center of First Affiliated Hospital at Chongqing Medical University. Healthy controls were required to have no previous lifetime history of neurological, DSM-IV Axis I/II, or systemic medical illness.
The recruited MDD subjects and healthy controls were divided into a training set and a test set. The training set, including 82 first-episode drug-naïve MDD subjects and 82 healthy controls, was used to identify potential urinary metabolite markers for MDD; the remaining subjects were used to construct the test set to independently validate the diagnostic generalizability of these urinary metabolite markers. The use of wholly independent samples in the validation is an essential step prior to moving ahead with the use of identified biomarkers in clinical studies (24).
The clinical characteristics of the recruited MDD subjects and healthy controls (HCs) are shown in Table I. All MDD subjects scored higher on the HDRS than healthy controls in both training and test sets. The MDD group and HC group did not significantly differ in terms of gender and body mass index in either set. As for age, the MDD subjects and HCs were matched in the training set but not in the test set.
Table I. Demographic and clinical details of recruited subjects.
| Training set |
Test set |
|||||
|---|---|---|---|---|---|---|
| HC | MDD | Pa | HC | MDD | Pa | |
| Sample size | 82 | 82 | – | 52 | 44 | – |
| Medication (Y/N) | N | N | – | N | 13/31 | – |
| Sex (M/F) | 53/29 | 46/36 | 0.26 | 27/25 | 17/27 | 0.19 |
| Age (years)b | 34.2 ± 10.1 | 32.2 ± 10.3 | 0.20 | 28.8 ± 9.8 | 34.1 ± 9.1 | 0.01 |
| BMIb | 20.9 ± 2.6 | 21.6 ± 2.7 | 0.15 | 21.3 ± 2.4 | 22.1 ± 3.1 | 0.14 |
| HDRS scoresb | 0.2 ± 0.6 | 22.4 ± 4.6 | 0.00 | 0.3 ± 0.7 | 25.7 ± 3.9 | 0.00 |
HC, healthy control; MDD, major depressive disorder; Y/N, yes/no; M/F, male/female; BMI, body mass index; HDRS, Hamilton Depression Rating Scale.
Chi-square analyses for categorical variables (sex).
a Two-tailed Student's t test for continuous variables (age, BMI, and HDRS scores).
b Values expressed as the mean ± S.D.
Sample Preparation and NMR Acquisition
After overnight fasting of the subjects, morning urine samples were collected in sterile cups and transferred into sterile tubes. All urine samples were then centrifuged at 1500 g for 10 min. The resulting supernatant was divided into equal aliquots and stored at −80 °C until later analysis.
For NMR analysis, urine samples were thawed and centrifuged at 1500 g for 10 min to remove precipitation. Then, 500 μl of urine was mixed with 100 μl of phosphate buffer (90% D2O, 1 mm 3-trimethylsilyl-1-[2,2,3,3-2H4] propionate (TSP), and 3 mm sodium azide; pH 7.4). After centrifugation at 12,000 rpm for 10 min, 500 μl samples of supernatant were transferred into 5 mm NMR tubes.
The proton spectra were collected on a Bruker AVANCE II 600 spectrometer (Bruker Biospin, Rheinstetten, Germany) operating at 600.13 MHz 1H frequency. A standard one-dimensional pulse sequence was used (recycle delay-90°-t1–90°-tm-90°-acquire free induction decay (FID)). Typically, 64 transients and 16K data points were collected with a spectral width of 8000 Hz, an acquisition time of 0.945 s, and a relaxation delay of 2 s. The FID was zero-filled, and an exponential line-broadening function of 0.3 Hz was applied to the FID prior to Fourier transformation. Urine resonance assignments were performed according to references from existing literature and public and in-house NMR databases (20, 25, 26).
Metabonomics Data Analysis
All spectra were manually phased and baseline referenced to TSP resonance at δ0.0. The NMR spectra (0.5–9.5 ppm) were segmented into equal widths (0.005 ppm) using the AMIX package (Bruker Biospin, Germany). Spectral regions of the water and urea resonances (δ4.13–6.8) were removed in order to eliminate baseline effects of imperfect water saturation. The remaining spectral segments in each NMR spectrum were normalized to the total sum of the spectral intensity to partially compensate for differences in concentration among the numerous metabolites. The normalized integral values were imported into SIMCA-p + 12.0 software (Umetrics, Umeå, Sweden) as variables.
A supervised multivariate approach, termed orthogonal partial least-squares discriminant analysis (OPLS-DA), was performed on the unit-variance-scaled spectral data to visualize discrimination between HCs and MDD subjects (27, 28). The quality of the OPLS-DA models was described in terms of three parameters (R2X, R2Y, and Q2Y), which were calculated by the default leave-one-out procedure. R2X and R2Y were used to quantify the goodness-of-fit; Q2Y was employed to assess the predictability of the model (29). To rule out the non-randomness of separation between groups, a 300-iteration permutation test was performed (30). If the values of Q2 and R2 resulting from the original model were higher than the corresponding values from the permutation test, the model was considered valid (29).
The coefficient loading plots of the OPLS-DA model were used to identify the spectral variables responsible for sample differentiation on the scores plot (31). Based on the number of samples used to construct the OPLS-DA models, a correlation coefficient of r > 0.276 was adopted as a cut-off value for statistical significance based on the discrimination significance at the level of p = 0.01. The colors of spectral signals projected onto the coefficient plot are proportional to the magnitude of the metabolites attributed to the discrimination between groups based on r values. Red coloring denotes a high correlation, and blue coloring denotes no correlation. Moreover, because of the intrinsic quantitative property of NMR technology, the intensity of each peak in the NMR spectra was proportional to the concentration in the samples (the concentrations of the metabolites were calculated relative to that of TSP, which was quantitatively added into the samples as a reference.). Thus, the metabolites identified as contributing to the discrimination between MDD subjects and HCs using multivariate analysis were manually calculated by means of peak integration. The nonparametric Mann–Whitney U test was used to detect statistically significant differences between the two groups.
Procedure for Identification of Urinary Biomarkers for MDD
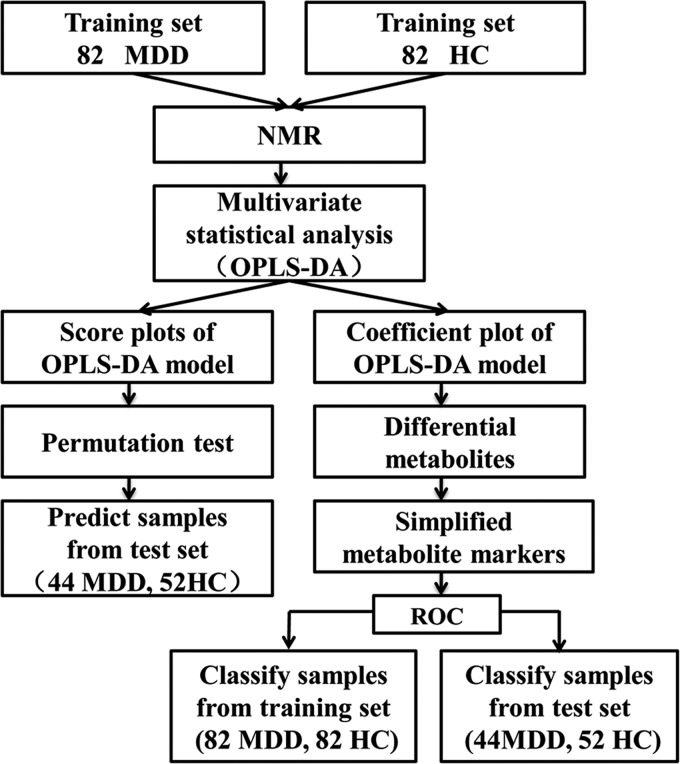
The overall workflow involved in identifying a simplified set of urinary metabolite biomarkers for MDD is summarized in Fig. 1. Because diagnosis based on the quantification of a small number of metabolites would be more feasible and convenient in clinical practice, a stepwise optimization algorithm based on Akaike's information criterion was employed to optimize the metabolite biomarker combination (32). To further evaluate the diagnostic performance of this simplified set of MDD biomarkers, a receiver-operating characteristic (ROC) curve analysis was carried out to quantify the ability of this metabolite biomarker panel to discriminate between MDD subjects and HCs in both training and test sets (33).
Fig. 1.
An overview of the NMR-based metabonomic workflow identifying urine metabolite biomarkers for MDD.
Statistical Analysis
As appropriate, comparisons of demographic characteristics between groups were performed using the parametric Student's t test, the nonparametric Mann–Whitney U test, or the chi-square test (SPSS 13.0). A p value of less than 0.05 was considered statistically significant.
RESULTS
Metabonomic Analysis of Urine Obtained from MDD Subjects and HCs
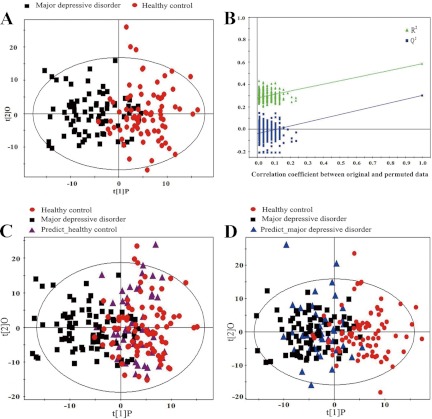
In the training set, OPLS-DA analysis was carried out to explore the metabolic differences between MDD subjects and HCs. Representative 600 1H NMR spectra of urine obtained from an MDD subject and an HC are shown in supplemental Fig. S1. The spectrum resonances assigned to the key metabolites are noted. The score plots of the OPLS-DA model showed that the MDD subjects were distinguishable from HCs with mild overlap (R2(X) cum = 12.7%, R2(Y) cum = 58.6%, Q2 = 47.2%; Fig. 2A). The values of the parameters (R2X, R2Y, and Q2Y) describing the OPLS-DA model were positive, demonstrating a robust metabolic difference between MDD subjects and HCs. Furthermore, a permutation test was employed to validate the OPLS-DA model. The validation plot demonstrated that the OPLS-DA model was valid, as the original Q2 and R2 values to the right were significantly higher than all corresponding permutated Q2 and R2 values to the left (Fig. 2B). To independently validate the diagnostic performance of the OPLS-DA model, the model was then used to predict class membership in the test set. The T-predicted scatter plot from the OPLS-DA model demonstrated that 34 of the 44 MDD subjects and 42 of the 52 HCs were correctly predicted by the OPLS-DA model, yielding a predictive accuracy of 79.2% (Fig. 2C). These results show that this OPLS-DA model generated by urinary metabolite profiling holds promise as an empirical diagnostic tool for MDD.
Fig. 2.
Metabonomic analysis of urine samples from MDD subjects and HCs. A, the OPLS model was used to discriminate between 82 first-episode drug-naïve depressed subjects (black boxes) and 82 HCs (red dots) in the treating set. The OPLS-DA score plots show a clear discrimination between MDD subjects and demographically matched HCs. B, permutation test showing the original R2 and Q2 values (top right) as significantly higher than corresponding permuted values (bottom left), demonstrating the OPLS-DA model's robustness. C, the OPLS-DA model constructed with 82 MDD patients (black squares) and 82 HCs (red dots) was used to predict the class membership of 52 HCs (purple triangle) from the test set; 42 of 52 HCs were correctly predicted. D, the OPLS-DA model generated with 82 MDD patients (black squares) and 82 HCs (red dots) was used to classify 44 depressed patients (blue triangles) from the test set; 34 of 44 depressed patients were correctly classified.
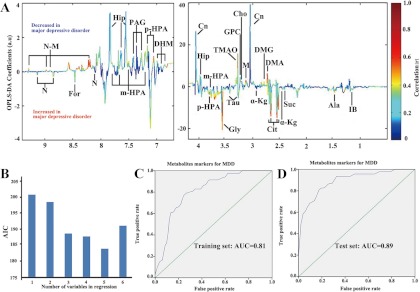
Analysis of the OPLS-DA loading coefficient plots resulted in the identification of 23 differential metabolites with a correlation coefficient of r > 0.276. The relative concentration of these 23 metabolites responsible for discriminating between MDD subjects and HCs is presented in supplementary Table S2. Relative to HCs, MDD subjects were characterized by higher levels of alanine, citrate, formate, glycine, isobutyrate, methylmalonate, nicotinate, succinate, taurine, and α-ketoglutarate, and lower levels of 3,4-dihydroxymandelate, choline, creatinine, dimethylamine, dimethylglycine, glyceroylphosphocholine, hippurate, malonate, m-hydroxyphenylacetate, N-methylnicotinamide, phenylacetyglycine, p-hydroxyphenylacetate, and trimethylamine-N-oxide (Fig. 3A). Univariate statistical analysis was then applied to validate the metabolic changes identified through multivariate statistical analysis; the majority of differential metabolites remained significantly changed (Table II). In addition, the other metabolites that were not considered as differential metabolites between the MDD and HC groups (r < 0.276) are listed in supplemental Table S3. The relative concentrations of these non-differential metabolites are presented in supplementary Table S4.
Fig. 3.
Identification and validation of urinary metabolite markers of MDD. A, to identify the spectral variables responsible for discrimination between MDD patients and HCs, the corresponding loading coefficient plots of the OPLS-DA model were analyzed. In all, 23 metabolites were identified with a correlation coefficient of r > 0.276. Peaks in the positive direction indicate that metabolite levels are increased in HCs; peaks in the negative direction indicate that metabolite levels are increased in MDD subjects. B, different combinations of urine metabolites were used to construct various logistical regression models. Akaike's information criterion (AIC) of each model is presented. The model constructed with five select urine metabolites—N-methylnicotinamide, m-hydroxyphenylacetate, malonate, alanine, and formate—showed the highest predictive ability. C, D, receiver operating characteristic curve analysis was performed to evaluate the diagnostic performance of these five metabolite biomarkers, obtaining area under the curve (AUC) values of 0.81 (95% confidence interval (0.74, 0.87)) in the training set and 0.89 (95% confidence interval (0.83, 0.95)) in the test set.
Table II. Key urinary metabolites responsible for the discrimination between MDD subjects and HCs.
| Chemical shift/ppm multiplicitya | Metabolites | rb | p valuec |
|---|---|---|---|
| 1.14(d) | Isobutyrate | −0.306 | 0.594 |
| 1.48(d) | Alanine | −0.510 | 0.106 |
| 2.45(t), 3.01(t) | α-ketoglutarate | −0.328 | 0.015 |
| 2.54(d), 2.69(d) | Citrate | −0.500 | 0.325 |
| 2.72(s) | Dimethylamine | 0.539 | 0.000 |
| 2.78(s) | Dimethylglycine | 0.407 | 0.000 |
| 3.05(s), 4.06(s) | Creatinine | 0.336 | 0.000 |
| 3.12(s) | Malonate | 0.480 | 0.000 |
| 3.20(s) | Choline | 0.428 | 0.000 |
| 3.23(s) | Glyceroylphosphocholine | 0.487 | 0.000 |
| 3.27(t), 3.43(t) | Taurine | −0.430 | 0.001 |
| 3.30(s) | Trimethylamine-N-oxide | 0.373 | 0.000 |
| 3.57(s) | Glycine | −0.430 | 0.084 |
| 3.78(s), 7.14(d), 7.21(d) | p-hydroxyphenylacetate | 0.359 | 0.000 |
| 3.80(s), 7.32(d), 7.49(m), 7.69(s), 7.76(d) | m-hydroxyphenylacetate | 0.491 | 0.000 |
| 3.97(d), 7.55(t), 7.64(t), 7.84(d) | Hippurate | 0.311 | 0.000 |
| 6.87(d), 6.92(s), 6.98(d) | 3,4-dihydroxymandelate | 0.328 | 0.000 |
| 7.28(d), 7.36(t), 7.42(dd) | Phenylacetyglycine | 0.359 | 0.000 |
| 8.46(s) | Formate | −0.460 | 0.054 |
| 8.21(d), 8.90(d), 8.97(d), 9.29(s) | N-methylnicotinamide | 0.529 | 0.000 |
| 1.24(d) | Methylmalonate | −0.345 | 0.003 |
| 8.03(m), 8.58(d), 8.85(d), 9.13(s) | Nicotinate | −0.355 | 0.482 |
| 2.41(s) | Succinate | −0.343 | 0.048 |
a Multiplicity: s, singlet; d, doublet; t, triplet; q, quartet; dd, doublet of doublets; m, multiplet.
b Positive correlation coefficients indicate significantly lower levels in MDD subjects relative to HCs; negative values indicate significantly higher levels in MDD subjects relative to HCs.
c p values were derived from the non-parametric Mann–Whitney U test.
Identification and Validation of Urinary Metabolite Biomarker Panel for Diagnosis of MDD
In order to identify a simplified metabolite biomarker panel for MDD diagnosis, a stepwise optimization algorithm based on Akaike's information criterion (AIC) was performed. A stepwise regression analysis demonstrated that the most significant deviations between MDD subjects and HCs could be described by five metabolites: malonate, formate, N-methylnicotinamide, m-hydroxyphenylacetate, and alanine. Therefore, these five biomarkers yielded the highest predictive power for future diagnostic applications (Fig. 3B). An ROC analysis was further performed to quantify the diagnostic performance of this panel in both training and test sets. The area under the curve (AUC) of this panel was 0.81 (95% confidence interval: 0.74–0.87) in the training samples (82 MDD subjects and 82 HCs) and 0.89 (95% confidence interval: 0.83–0.95) in the test samples (44 MDD subjects and 52 HCs) (Figs. 3C and 3D). The diagnostic performance of this panel is similar to that of the OPLS-DA model constructed with all the differential metabolites, demonstrating the efficacy of this simplified urinary metabolite panel in MDD detection.
DISCUSSION
MDD is a widespread and debilitating mental disorder. Currently, no biomarkers are available to aid clinicians in diagnosing this disorder. In this study, an NMR-based metabonomic approach was employed to identify potential urinary metabolite biomarkers for MDD. A panel consisting of five urinary metabolite biomarkers—malonate, formate, N-methylnicotinamide, m-hydroxyphenylacetate, and alanine—was identified. This panel enabled the discrimination of MDD subjects from HCs with AUCs of 0.81 and 0.89 in the training set and test set, respectively. These findings demonstrate that urinary metabolite biomarkers can facilitate MDD detection and could aid the development of objective laboratory-based diagnostic tools for MDD.
In order to capture the urinary metabolite biomarkers that truly reflect the pathophysiologic changes inherent in the MDD disease state, only first-episode drug-naïve MDD subjects were recruited into the training set. However, given that medication use is common in most MDD patient populations, unselected subjects—both medicated and unmedicated—from the test set were used to independently validate the diagnostic generalizability of the biomarkers. Furthermore, the MDD subjects and HCs were not age-matched in this test set. Under these circumstances, the panel still distinguished blinded MDD subjects from HCs with an AUC of 0.89 in the test set, highlighting the diagnostic robustness of the biomarker panel.
In this study, 23 differentially expressed metabolites were initially identified that distinguished MDD subjects from HCs. The combination of these 23 differential metabolites enabled discrimination between MDD subjects and HCs in the test set with an accuracy of 79.2%. This finding suggests a robust diagnostic performance of these 23 metabolites. However, in clinical practice, it is not feasible, convenient, or economical to simultaneously measure a large number of metabolites in order to diagnose a disease state. Therefore, using a stepwise optimization algorithm based on AIC, a simplified biomarker panel of only five metabolites was constructed to discriminate MDD subjects from HCs while retaining high accuracy. Because of this analytically selective strategy, the smaller biomarker panel is likely to be of more clinical value than those from previous metabolomic studies on MDD (20, 34).
Among the five select biomarkers in the panel, levels of alanine and formate were not significantly perturbed in the univariate statistical analysis. However, these metabolites were included in the simplified diagnostic signature, as they were identified by multivariate analysis. This was done because the stepwise regression analysis showed that the addition of these two amino acid metabolites resulted in the highest predictive power. This result shows the advantage of a multivariate statistical approach in detecting the potential significance of subtle metabolic differences between experimental groups related to an associated univariate analysis (35). To better understand the underlying pathogenesis of MDD, the differential metabolites were comprehensively analyzed in terms of in vivo metabolic activity. These metabolites were found to be primarily involved in (i) energy metabolism, (ii) gut microbial metabolism, and (iii) tryptophan-nicotinic acid metabolism, which are discussed in detail below.
Energy Metabolites
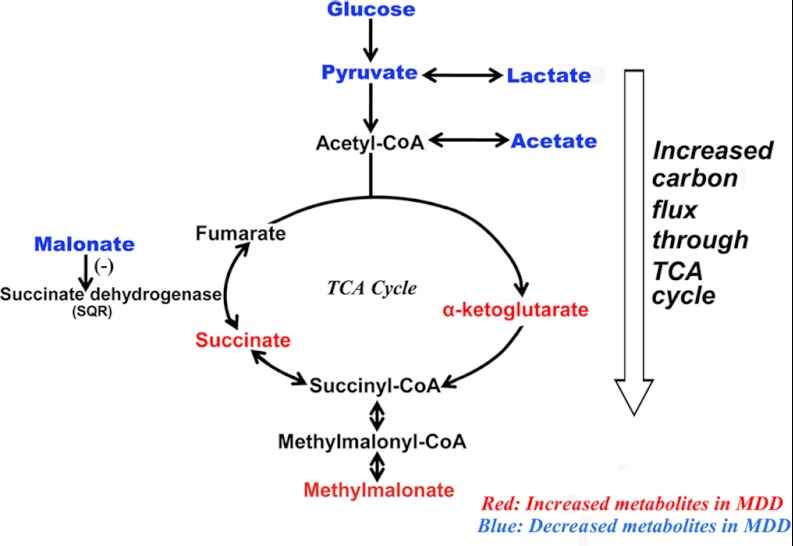
Urinary levels of three tricarboxylic acid (TCA) cycle–associated metabolites—α-ketoglutarate, succinate, and malonate—were significantly perturbed in MDD subjects relative to HCs. α-ketoglutarate and succinate are the two metabolic precursors immediately preceding succinate dehydrogenase (or succinate-coenzyme Q reductase (SQR)), the TCA cycle enzyme complex that converts succinate into fumarate. The significantly increased α-ketoglutarate and succinate levels in the urine of MDD subjects found here, in conjunction with the significantly decreased levels of plasma acetate, glucose, lactate, and pyruvate found in MDD subjects from this group's previous study (20), likely indicate greater carbon flux through the TCA cycle accompanied by reduced SQR activity (Fig. 4). Levels of malonate, which competitively inhibits SQR, were significantly decreased in MDD subjects relative to HCs. This decrease in malonate levels might be a corrective feedback mechanism to compensate for an underlying decrease in SQR activity. Moreover, the increased excretion of the related metabolite methylmalonate also observed in MDD subjects here is produced from methylmalonyl-CoA, a metabolite in equilibrium with succinyl-CoA. This increased methylmalonate excretion likely indicates an increased succinyl-CoA level in MDD subjects, which is consistent with the aforementioned increased TCA cycle flux in MDD subjects (Fig. 4).
Fig. 4.
Summary of the urinary metabolites involved in disturbance of energy metabolism. These differential metabolites suggest increased three tricarboxylic acid (TCA) cycle flux in MDD subjects.
More holistically, given that glucose is the primary carbon source for glycolysis, this increased TCA cycle flux may be associated with the upstream deficiencies in circulating glucose levels previously observed in MDD subjects. Glycolysis coupled with the TCA cycle fully oxidizes glucose and supplies energy for the brain. Although the adult human brain constitutes a mere 2% of total body weight, this energy-intensive organ consumes 25% of total body glucose (36). Therefore, the reduction in circulating glucose levels might lead to chronic glucose deficiencies in the brains of MDD subjects. Accordingly, previous studies have shown a reduction in glucose metabolism in several brain regions of MDD patients (37, 38). When the decreased glucose metabolism was reversed in these patients, the depressive symptoms were ameliorated (39). Moreover, the increased urinary excretion of formate, an electron acceptor, from MDD subjects in the current study implies a reduced capacity for energy production. These findings suggest that decreased central energy production in MDD subjects may be mirrored by peripheral metabolic perturbations.
Gut Microbial Metabolites
Urinary levels of five microbiotic metabolites—m-hydroxyphenylacetate, hippurate, dimethylamine, dimethylglycine, and trimethylamine-N-oxide—were significantly decreased in MDD subjects relative to HCs. These metabolites are uniquely produced by bacterial metabolism in the intestinal tract, indicating that MDD may be associated with variations in intestinal microflora. Accordingly, previous urinary metabonomic analysis in a depressed animal model has shown that depressed behavior is associated with changes in gut microflora (17). Interestingly, several clinical studies have demonstrated that MDD patients display a high comorbidity of irritable bowel syndrome (IBS) (40, 41), a disorder involving gut microflora. The fecal microflora in IBS patients show abnormally higher numbers of facultative organisms and lower numbers of lactobacilli and bifidobacteria (42). These combined findings highlight the potential involvement of gut microbiotic variation in the development of MDD.
Tryptophan–Nicotinic Acid Metabolism
N-methylnicotinamide (NMNA), an end-product of nicotinamide metabolism, was significantly increased in MDD subjects relative to HCs. Given that the NMNA precursor nicotinamide is involved in the tryptophan–nicotinic acid pathway (43), the increased excretion of urinary NMNA observed here suggests an up-regulation of tryptophan–nicotinic acid pathway activity in MDD subjects. Tryptophan is the biochemical precursor of both serotonin and nicotinic acid (21). Therefore, increases in the downstream metabolites of nicotinic acid metabolism might indicate decreased serotonin biosynthesis. This speculation is in concord with the well-established theory that serotonergic neurotransmission deficiencies contribute to the pathoetiology of MDD (44).
The results and conclusions of this study should be cautiously interpreted on account of several limitations. The diagnostic performance of the urinary metabolite biomarker panel was confirmed solely by discriminating MDD subjects from HCs. Future work should focus on whether or not these biomarkers can be applied to differentiate MDD from other psychiatric disorders. Moreover, all subjects were of the same ethnicity and were recruited from the same site; thus, ethno- and site-specific biases cannot be ruled out. Further studies involving heterogeneous populations from multiple clinical sites are required.
In conclusion, with the use of a 1H NMR-based metabonomic method, a panel of urinary metabolite biomarkers for MDD was identified using a homogeneous sample set. This panel was then independently validated in a diverse sample set. Five metabolite biomarkers—malonate, formate, N-methylnicotinamide, m-hydroxyphenylacetate, and alanine—could be used to accurately distinguish MDD subjects from HCs in both treating and test sets. These findings lay the groundwork for the future development of a urine-based diagnostic test for MDD.
Supplementary Material
Acknowledgments
Our sincere gratitude is extended to Professors Delan Yang and Hua Hu for their efforts in sample collection.
Footnotes
* This work was supported by the National Basic Research Program of China (973 Program) (Grant No. 2009CB918300), the National Natural Science Foundation of China (Grant No. 30900456), and the Natural Science Foundation Project of Chongqing (CSTC, 2008BB5238 and 2010BB5393).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HC
- healthy control
- MDD
- major depressive disorder
- NMR
- nuclear magnetic resonance
- OPLS-DA
- orthogonal partial least-squares discriminant analysis
- TCA
- tricarboxylic acid.
REFERENCES
- 1. Kessler R. C., Berglund P., Demler O., Jin R., Koretz D., Merikangas K. R., Rush A. J., Walters E. E., Wang P. S. (2003) The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). JAMA 289, 3095–3105 [DOI] [PubMed] [Google Scholar]
- 2. Reynolds E. (2003) Brain and mind: a challenge for WHO. Lancet 361, 1924. [DOI] [PubMed] [Google Scholar]
- 3. Mitchell A. J., Vaze A., Rao S. (2009) Clinical diagnosis of depression in primary care: a meta-analysis. Lancet 374, 609–619 [DOI] [PubMed] [Google Scholar]
- 4. Chen L. S., Eaton W. W., Gallo J. J., Nestadt G. (2000) Understanding the heterogeneity of depression through the triad of symptoms, course and risk factors: a longitudinal, population-based study. J. Affect. Disord. 59, 1–11 [DOI] [PubMed] [Google Scholar]
- 5. Nicholson J. K., Lindon J. C., Holmes E. (1999) Metabonomics: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 6. Nicholson J. K., Lindon J. C. (2008) Systems biology: metabonomics. Nature 455, 1054–1056 [DOI] [PubMed] [Google Scholar]
- 7. Huang Z., Lin L., Gao Y., Chen Y., Yan X., Xing J., Hang W. (2011) Bladder cancer determination via two urinary metabolites: a biomarker pattern approach. Mol. Cell. Proteomics 10, M111.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qi Y., Li P., Zhang Y., Cui L., Guo Z., Xie G., Su M., Li X., Zheng X., Qiu Y. (2011) Urinary metabolite markers of precocious puberty. Mol. Cell. Proteomics. 11, M111.011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fischer R., Trudgian D. C., Wright C., Thomas G., Bradbury L. A., Brown M. A., Bowness P., Kessler B. M. (2012) Discovery of candidate serum proteomic and metabolomic biomarkers in ankylosing spondylitis. Mol. Cell. Proteomics 11, M111.013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng T. M. K., Lu Y. E., Guest P. C., Rahmoune H., Harris L. W., Wang L., Ma D., Stelzhammer V., Umrania Y., Wayland M. T. (2010) Identification of targeted analyte clusters for studies of schizophrenia. Mol. Cell. Proteomics 9, 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jung J. Y., Lee H. S., Kang D. G., Kim N. S., Cha M. H., Bang O. S., Ryu do H., Hwang G. S. (2011) 1H-NMR-based metabolomics study of cerebral infarction. Stroke 42, 1282–1288 [DOI] [PubMed] [Google Scholar]
- 12. Sussulini A., Prando A., Maretto D. A., Poppi R. J., Tasic L., Banzato C. E., Arruda M. A. (2009) Metabolic profiling of human blood serum from treated patients with bipolar disorder employing 1H NMR spectroscopy and chemometrics. Anal. Chem. 81, 9755–9763 [DOI] [PubMed] [Google Scholar]
- 13. Yang J., Chen T., Sun L., Zhao Z., Qi X., Zhou K., Cao Y., Wang X., Qiu Y., Su M. (2011) Potential metabolite markers of schizophrenia. Mol. Psychiatry 10.1038/mp.2011.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang F., Jia Z., Gao P., Kong H., Li X., Lu X., Wu Y., Xu G. (2010) Metabonomics study of urine and plasma in depression and excess fatigue rats by ultra fast liquid chromatography coupled with ion trap-time of flight mass spectrometry. Mol. Biosyst 6, 852–861 [DOI] [PubMed] [Google Scholar]
- 15. Li Z. Y., Zheng X. Y., Gao X. X., Zhou Y. Z., Sun H. F., Zhang L. Z., Guo X. Q., Du G. H., Qin X. M. (2010) Study of plasma metabolic profiling and biomarkers of chronic unpredictable mild stress rats based on gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 24, 3539–3546 [DOI] [PubMed] [Google Scholar]
- 16. Zheng S., Zhang S., Yu M., Tang J., Lu X., Wang F., Yang J., Li F. (2011) An 1 H NMR and UPLC–MS-based plasma metabonomic study to investigate the biochemical changes in chronic unpredictable mild stress model of depression. Metabolomics. 7, 413–423 [Google Scholar]
- 17. Zheng S., Yu M., Lu X., Huo T., Ge L., Yang J., Wu C., Li F. (2010) Urinary metabonomic study on biochemical changes in chronic unpredictable mild stress model of depression. Clin. Chim. Acta 411, 204–209 [DOI] [PubMed] [Google Scholar]
- 18. Liu X. J., Li Z. Y., Li Z. F., Gao X. X., Zhou Y. Z., Sun H. F., Zhang L. Z., Guo X. Q., Du G. H., Qin X. M. (2012) Urinary metabonomic study using a CUMS rat model of depression. Magn. Reson. Chem. 50, 187–192 [DOI] [PubMed] [Google Scholar]
- 19. Wang X., Zhao T., Qiu Y., Su M., Jiang T., Zhou M., Zhao A., Jia W. (2009) Metabonomics approach to understanding acute and chronic stress in rat models. J. Proteome Res. 8, 2511–2518 [DOI] [PubMed] [Google Scholar]
- 20. Zheng P., Gao H. C., Li Q., Shao W. H., Zhang M. L., Cheng K., Yang D. Y., Fan S. H., Chen L., Fang L. (2012) Plasma metabonomics as a novel diagnostic approach for major depressive disorder. J. Proteome Res. 11, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 21. Yap I. K., Angley M., Veselkov K. A., Holmes E., Lindon J. C., Nicholson J. K. (2010) Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J. Proteome Res. 9, 2996–3004 [DOI] [PubMed] [Google Scholar]
- 22. American Psychiatric Association (2001) Diagnostic and Statistical Manual of Mental Disorders, 4th Ed., American Psychiatric Association, Washington, D.C [Google Scholar]
- 23. Williams J. B. W. (1988) A structured interview guide for the Hamilton Depression Rating Scale. Arch. Gen. Psychiatry 45, 742. [DOI] [PubMed] [Google Scholar]
- 24. Micheel C., Nass S., Omenn G. (2012) Evolution of Translational Omics: Lessons Learned and the Path Forward (Institute of Medicine Consensus Report), National Academies Press, Washington, D.C: [PubMed] [Google Scholar]
- 25. Yap I. K. S., Angley M., Veselkov K. A., Holmes E., Lindon J. C., Nicholson J. K. (2010) Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J. Proteome Res. 9, 2996–3004 [DOI] [PubMed] [Google Scholar]
- 26. Beckwith-Hall B., Nicholson J., Nicholls A., Foxall P., Lindon J., Connor S., Abdi M., Connelly J., Holmes E. (1998) Nuclear magnetic resonance spectroscopic and principal components analysis investigations into biochemical effects of three model hepatotoxins. Chem. Res. Toxicol. 11, 260–272 [DOI] [PubMed] [Google Scholar]
- 27. Trygg J., Wold S. (2002) Orthogonal projections to latent structures (O-PLS). J. Chemom. 16, 119–128 [Google Scholar]
- 28. Bylesj M., Rantalainen M., Cloarec O., Nicholson J. K., Holmes E., Trygg J. (2006) OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 20, 341–351 [Google Scholar]
- 29. Mahadevan S., Shah S. L., Marrie T. J., Slupsky C. M. (2008) Analysis of metabolomic data using support vector machines. Anal. Chem. 80, 7562–7570 [DOI] [PubMed] [Google Scholar]
- 30. Jung Y., Lee J., Kwon J., Lee K. S., Ryu D. H., Hwang G. S. (2010) Discrimination of the geographical origin of beef by 1H NMR-based metabolomics. J. Agric. Food Chem. 58, 10458–10466 [DOI] [PubMed] [Google Scholar]
- 31. Cloarec O., Dumas M. E., Trygg J., Craig A., Barton R. H., Lindon J. C., Nicholson J. K., Holmes E. (2005) Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal. Chem. 77, 517–526 [DOI] [PubMed] [Google Scholar]
- 32. Oikonomopoulou K., Li L., Zheng Y., Simon I., Wolfert R., Valik D., Nekulova M., Simickova M., Frgala T., Diamandis E. (2008) Prediction of ovarian cancer prognosis and response to chemotherapy by a serum-based multiparametric biomarker panel. Br. J. Cancer 99, 1103–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bradley A. P. (1997) The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 30, 1145–1159 [Google Scholar]
- 34. Paige L. A., Mitchell M. W., Krishnan K. R., Kaddurah-Daouk R., Steffens D. C. (2007) A preliminary metabolomic analysis of older adults with and without depression. Int. J. Geriatr. Psych. 22, 418–423 [DOI] [PubMed] [Google Scholar]
- 35. MacIntyre D. A., Jimenez B., Lewintre E. J., Martin C. R., Schafer H., Ballesteros C. G., Mayans J. R., Spraul M., Garcia-Conde J., Pineda-Lucena A. (2010) Serum metabolome analysis by 1H-NMR reveals differences between chronic lymphocytic leukaemia molecular subgroups. Leukemia 24, 788–797 [DOI] [PubMed] [Google Scholar]
- 36. Zauner A., Daugherty W. P., Bullock M. R., Warner D. S. (2002) Brain oxygenation and energy metabolism: part I-biological function and pathophysiology. Neurosurgery 51, 289. [PubMed] [Google Scholar]
- 37. Baxter L. R., Jr., Schwartz J. M., Phelps M. E., Mazziotta J. C., Guze B. H., Selin C. E., Gerner R. H., Sumida R. M. (1989) Reduction of prefrontal cortex glucose metabolism common to three types of depression. Arch. Gen. Psychiatry 46, 243. [DOI] [PubMed] [Google Scholar]
- 38. Videbech P. (2000) PET measurements of brain glucose metabolism and blood flow in major depressive disorder: a critical review. Acta Psychiatr. Scand. 101, 11–20 [DOI] [PubMed] [Google Scholar]
- 39. Kennedy S. H., Evans K. R., Kruger S., Mayberg H. S., Meyer J. H., McCann S., Arifuzzman A. I., Houle S., Vaccarino F. J. (2001) Changes in regional brain glucose metabolism measured with positron emission tomography after paroxetine treatment of major depression. Am. J. Psychiatry 158, 899–905 [DOI] [PubMed] [Google Scholar]
- 40. Ladep N. G., Obindo T. J., Audu M. D., Okeke E. N., Malu A. O. (2006) Depression in patients with irritable bowel syndrome in Jos, Nigeria. World J. Gastroenterol. 12, 7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gros D. F., Antony M. M., McCabe R. E., Swinson R. P. (2009) Frequency and severity of the symptoms of irritable bowel syndrome across the anxiety disorders and depression. J. Anxiety Disord. 23, 290–296 [DOI] [PubMed] [Google Scholar]
- 42. Madden J., Hunter J. (2002) A review of the role of the gut microflora in irritable bowel syndrome and the effects of probiotics. Br. J. Nutr. 88, 67–72 [DOI] [PubMed] [Google Scholar]
- 43. Lester G. (1971) End-product regulation of the tryptophan-nicotinic acid pathway in Neurospora crassa. J. Bacteriol. 107, 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Belmaker R., Agam G. (2008) Major depressive disorder. New Engl. J. Med. 358, 55–68 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.