Abstract
Background
Homeodomain transcription factors play critical roles in metazoan development. BELLRINGER (BLR), one such transcription factor, is involved in diverse developmental processes in Arabidopsis, acting in vascular differentiation, phyllotaxy, flower and fruit development. BLR also has a redundant role in meristem maintenance. Cell wall remodelling underpins many of these processes, and BLR has recently been shown to regulate expression of PECTIN METHYL-ESTERASE 5 (PME5), a cell wall modifying enzyme in control of phyllotaxy. We have further explored the role of BLR in plant development by analysing phenotypes and gene expression in a series of plants over-expressing BLR, and generating combinatorial mutants with blr, brevipedicellus (bp), a member of the KNOX1 family of transcription factors that has previously been shown to interact with blr, and the homeodomain transcription factor revoluta (rev), required for radial patterning of the stem.
Results
Plants over-expressing BLR exhibited a wide range of phenotypes. Some were defective in cell size and demonstrated misregulation of genes predominantly affecting cell wall development. Other lines with more extreme phenotypes failed to generate lateral organs, consistent with BLR repressing transcription in the shoot apex. Cell wall dynamics are also affected in blr mutant plants, and BLR has previously been shown to regulate vascular development in conjunction with BP. We found that when bp and blr were combined with rev, a set of defects was observed that were distinct from those of bp blr lines. In these triple mutants xylem development was most strikingly affected, resulting in an almost complete lack of vessels and xylem parenchyma with secondary thickening.
Conclusions
Our data support a role for BLR in ordering the shoot apex and, in conjunction with BP and REV, playing a part in determining the composition and organisation of the vascular system. Microarray analysis strongly indicates that the striking vascular phenotypes of blr bp rev triple mutants and plants over-expressing BLR result from the misregulation of a suite of genes, targets of BLR in wild type plants, that determine cell size and structure in the developing vasculature.
Background
BELLRINGER (BLR), also known as VAAMANA, PENNYWISE, LARSON and REPLUMLESS, is a member of the BELL family of homeodomain transcription factors and functions in diverse processes in the development of Arabidopsis[1-5]. BLR was identified independently as a suppresser of the floral homeotic gene AGAMOUS (AG) [5], as a factor required in fruit development for specification of the replum [1] and as a gene necessary for normal phyllotactic patterning [2,4]. BELL transcription factors were shown to enhance DNA binding of maize KNOTTED1 [6], the founding member of the KNOX1 gene family [7], and BLR was subsequently shown to act synergistically with the KNOX1 homeodomain transcription factor BREVIPEDICELLUS (BP) [3]. BLR and KNOX1 proteins heterodimerize [3] and, in Nicotiana epidermal cell transient assays, BLR translocates to the nucleus on binding to KNOX1 proteins [2]. This event is likely to have evolved early in eukaryotic development as it is required for the haploid-diploid transition in the green algae Chlamydomonas[8].
Although the expression patterns of BLR and KNOX1 overlap, they are not identical [3] and blr and knox1 mutants show distinctive phenotypes, suggesting that BLR function is unlikely to be confined to acting with KNOX1 proteins alone. Nevertheless, similarities are apparent, such as the short stature of both blr and bp plants [2-4] and the ability of blr and bp to enhance weak alleles of shoot meristemless (stm) [2,4], a KNOX1 gene required for shoot apical meristem maintenance [9,10], which points to a redundant role for BLR and BP in stem cell fate establishment [2-4]. Reports of genetic interactions between blr and bp mutants have, however, been conflicting, with the short stature phenotype of the bp blr double mutation being variously interpreted as additive [4], synergistic [3], or novel - because inflorescence elongation is observed in the single mutants but not in the double [2]. Phenotypic differences between blr and bp mutants are unlikely to result from redundancy between family members because multiple knockout lines within the KNOX1 and BELL families exhibit diverse phenotypes. For example, vegetative to reproductive transition defects are observed when blr mutants are combined with mutations in poundfoolish (pnf), the gene with which BLR shares most similarity in Arabidopsis[11], which are not observed in multiple knox1 mutants [12,13]. Similarly, multiple knox1 mutants do not resemble blr plants as one role of BP is repression of KNAT2 and KNAT6[12]. BLR is thus likely to have functions both dependent on and independent of KNOX1.
One KNOX1 independent function of BLR may include control of phyllotaxy as similar defects are not present in knox1 mutants. Normal phyllotaxy requires correct patterning of cell divisions [14,15] which depend in turn on strictly regulated cell-wall remodelling events [16] themselves driven by mechanical constraints [17]. Primordia outgrowth is accompanied by the de-methyl-esterification of pectic polysaccharides in the cell wall, and perturbation of the methyl-esterification status of pectins within the meristem by altering expression levels of PECTIN METHYL-ESTERASE 5 has been shown to alter phyllotactic pattern [18]. PME5 has been demonstrated to be negatively regulated by BLR in the meristem dome and this interaction is essential for maintenance of phyllotaxy, as incorrect positioning of organ initiation in blr mutants is suppressed in blr pme5 double mutants. The relationship between BLR and PME5 appears to be context specific as BLR promotes PME5 expression in internodes [19].
The functions of BP and BLR do overlap in vascular development. Typically in Arabidopsis stems, vascular tissue is radially arranged in collateral bundles with xylem present towards the centre of the stem and phloem positioned towards the outside, such that the xylem, vascular meristem (procambium) and phloem are positioned along the mediolateral axis [20]. Xylem is characterised by the presence of large vessels for water transport and smaller xylary fibres, both of which have secondary cell walls with the adjacent procambium forming an arc of dividing cells. Within the stem, the vascular tissue represents a developmental series with most recently derived tissue at the top and the oldest at the base [20]. A continuous vascular ring has been described in blr lines, a phenotype enhanced in bp blr double mutants [3] by the appearance of undeveloped vascular bundles [3] containing cells previously only observed occurring in small, isolated strips overlying some vascular bundles of bp plants [21,22].
Here we have exploited transgenic plants over-expressing BLR as a tool to identify putative downstream targets of this transcription factor. In addition to the previously described target of BLR, PME5[19], our microarray analysis has identified misregulation of many other genes encoding cell-wall-associated proteins following BLR over-expression. Defects in cell wall metabolism can affect cell size [23] and we therefore examined blr mutant vascular tissue, where cell size defects have previously been described [3]. We uncovered a genetic interaction between blr, bp and revoluta (rev), a homeodomain transcription factor of the HD-ZipIII family that has previously been shown to specify adaxial identity, radial patterning in the stem [24] and xylem differentiation [25]. Our results add further support to the notion [19] that in addition to regulating genes at the shoot apex, BLR plays a wider role in development through its control of the extracellular matrix.
Results and discussion
Over-expression of BELLRINGER reduces organ size
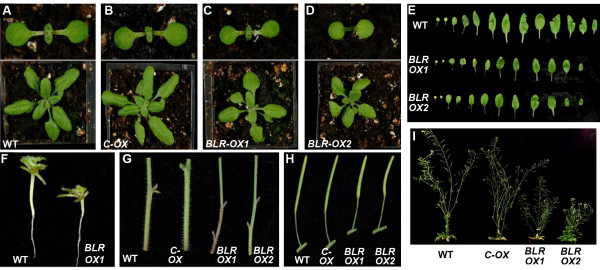
Over-expression studies have been informative in functional analysis of KNOX1[26-28]. Using a similar strategy, BLR cDNA was used to generate 35S::BLR constructs from which BLR over-expression (BLR-OX) transgenic lines were generated. Plants were observed with short hypocotyls, small rosette leaves, narrow cauline leaves, and short siliques (Figure 1A1F, 1H). BLR-OX plants also had shorter, thinner stems than wild type with more axiliary branches (Figure 1G, 1I). Northern blot analysis showed eight lines to exhibit increased BLR levels; BLR transcript levels in two of these lines, BLR-OX1 and BLR-OX2, are shown in Figure 2A.
Figure 1.
Phenotype of BLR-OX lines. 7 day-old (top panel) and 21 day-old (lower panel) wild type (A), transformed control (B), BLR-OX1 (C) and BLR-OX2 (D). (E) Dissected rosette leaves of 4 week-old WT, BLR-OX1 and BLR-OX2 plants. (F) WT and BLR-OX1 hypocotyls at 2 weeks-old. (G-I) Comparison of 5 week-old BLR-OX1, BLR-OX2 and control plant inflorescence stems (G), mature siliques (H) and stature (I).
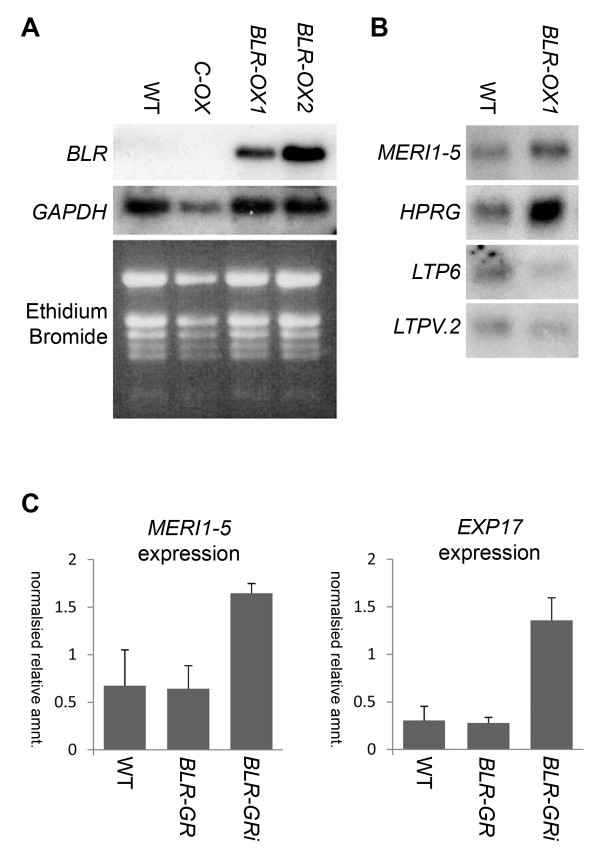
Figure 2.
Gene expression changes in plants over expressing BLR. (A) Comparison of BLR and GAPDH expression in wild type (WT), transformed control (C-OX), BLR-OX1 and BLR-OX2 lines by Northern blot. (B) Comparison of At4g30270/MERI1-5, At5g65660/HPRG, At3g08770/LTP6 and At3g53980/LTPV.2 expression by Northern blot. (C) Expression of MERI-5, and EXP17 in wild type, BLR-GR and BLR-GRi (72 hour induction) determined by qRT-PCR and shown as relative amount normalised to expression of ACT2.
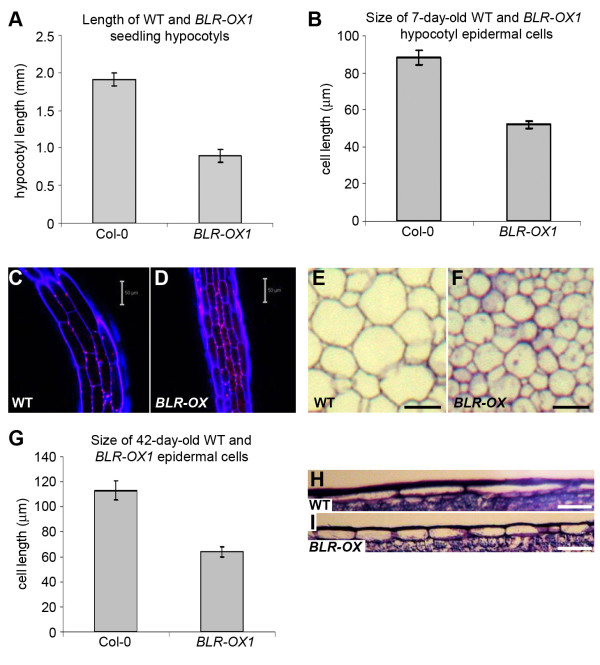
The organs of BLR-OX lines were smaller than wild type, so differences in cell size and number were investigated in 7 day epidermal hypocotyl cells from BLR-OX and wild type plants (Figure 3A-D). BLR-OX epidermal cells were significantly shorter than controls (Figure 3B); similar reductions were observed in longitudinal sections of inflorescence stem epidermis (Figure 3G-I) and transverse sections of pith parenchyma (Figure 3E-F). This reduction in cell size is thus likely to contribute to the smaller organ size of BLR-OX plants. Other histological features, including those in vascular tissue were indistinguishable from wild type.
Figure 3.
Cell size defects of BLR-OX seedlings. (A) Average hypocotyl length of 7 day-old wild type and BLR-OX1 lines (n = 14). (B) Mean epidermal cell size in hypocotyls of 7 day-old plants (n = 33). 7 day-old wild type (C) and BLR-OX (D) hypocotyl epidermis stained with propidium iodide. 200 μM2 cross-section from 42 day-old wild type (E) and BLR-OX1 (F) stems showing parenchyma cells. (G) Mean epidermal cell size of 42 day-old plants (n = 41). 100 μM row of epidermal cells from 42 day-old wild type (H) and BLR-OX (I) stems. Error bars show standard error. Scales are 50 μm (E-F), or 10 μm (H-I).
Expression changes in BLR-OX plants
To explore the changes in transcription underlying the BLR-OX phenotype, transcripts from hypocotyls of BLR-OX and control plants were subject to microarray analysis at stage 1.01 [29] of development. Using the ‘Significance Analysis of Microarrays’ strategy [30], 3109 genes demonstrated consistent expression changes and acceptable spot confidence of which 2957 had appropriate q-values (<0.1%). 927 genes demonstrated >1.5 fold expression change (Additional file 1: Table S1). Three lines of evidence point to the microarray data reflecting gene expression changes in planta; first, BLR was up-regulated 3.6 fold in BLR-OX compared to Col-0 (Table 1). Second, the expression of PME5, previously shown to be negatively regulated by BLR is reduced 2.3 fold (Table 1) and third, the promoters of 70 of the 100 genes showing the greatest fold change carried predicted BLR consensus DNA binding sites (Table 1). In contrast, the DNA binding sequence for ATHB2, which in common with BLR has a 9 base pair consensus sequence, occurs in only 9 of the 100 promoter regions. The array data was validated by testing expression of MERI-5 (At4g30270; 4.3 fold increase in BLR-OX), a xyloglucan endotransglucosylase which acts to hydrolyze glycosyl bonds, HPRG (At5g65660; 1.6 fold increase), a hydroxyproline-rich plasma membrane localised glycoprotein, and LPT6 (At3g08770; 1.9 fold decrease) and LTPV.2 (At3g53980; 6.3 fold decrease), both of which encode endomembrane localised lipid binding/transport proteins, by northern blot (Figure 2B). In all instances expression reflected the array data.
Table 1.
Genes prominently mis-regulated in BLR-OX
|
AGI number |
Common name |
Expression |
||
|---|---|---|---|---|
| BLR-OX | WT | Fold Change | ||
|
UPREGULATED IN BLR-OX | ||||
| At1g10070 1,4 |
Branched-chain amino acid aminotransferase 2 (BCAT-2) |
469 ± 320 |
47 ± 12 |
10.0 |
| At4g30270 1,2,4 |
Xyloglucan endotransglucosylase/hydrolase 24 (MERI-5) |
2269 ± 601 |
522 ± 38 |
4.3 |
| At3g47340 1,2,4 |
Glutamine dependent aparagine synthetase 1 |
1,231 ± 470 |
281 ± 41 |
4.4 |
| At1g75750 1,2,4 |
GA-responsive GAST1 protein homolog (GASA1) |
10890 ± 4769 |
2641 ± 390 |
4.1 |
| At5g02030 1,4 |
BELLRINGER |
2507 ± 452 |
706 ± 1,23 |
3.6 |
| At2g19800 1,4 |
Myo-inositol oxygenase 2 (MIOX2) |
2104 ± 759 |
700 ± 357 |
3.0 |
| At5g56870 1,4 |
Beta-galactosidase 4 (BGAL4) |
842 ± 140 |
265 ± 57 |
3.2 |
| At2g30600 2,4 |
BTB/POZ domain protein involved in cell adhesion |
2624 ± 839 |
1110 ± 353 |
2.4 |
| At5g49360 1,4 |
β-D-xylosidase/α-L-arabinofuranosidase (BXL1) |
2105 ± 591 |
983 ± 484 |
2.1 |
| At4g35770 2 |
Senescence-associated/Dark inducible protein (SEN1/DIN1) |
2393 ± 413 |
1,206 ± 374 |
2.0 |
| At2g45170 2,4 |
Autophagy 8e (APG8e) |
3685 ± 133 |
2007 ± 193 |
1.8 |
| At4g36850 2,4 |
PQ-loop repeat family/transmembrane family protein |
503 ± 88 |
270 ± 45 |
1.9 |
| At5g07440 2,4 |
Glutamate dehydrogenase 2 (GDH2) |
4403 ± 685 |
2364 ± 237 |
1.9 |
| At4g01630 1,4 |
Expansin, putative (EXP17) |
162 ± 24 |
88 ± 17 |
1.8 |
| At5g11720 1 |
Glycoside hydrolase family 31 |
1054 ± 113 |
578 ± 85 |
1.8 |
| At1g53910 1,4 |
AP2 domain-containing protein RAP2.12 |
9167 ± 47 |
5043 ± 441 |
1.8 |
| At1g12780 1,2 |
UDP-D-glucose/UDP-D-galactose 4-epimerase 1 (UGE1) |
239 ± 43 |
136 ± 35 |
1.8 |
| At4g14130 1 |
xyloglucan endotransglycosylase-related protein (XTR7) |
41,2 ± 156 |
272 ± 105 |
1.5 |
| At1g35250 1,4 |
Thioesterase superfamily protein |
1164 ± 132 |
769 ± 91 |
1.5 |
|
DOWNREGULATED IN BLR-OX | ||||
| At3g53980 1,4 |
Protease inhibitor/seed storage/lipid transfer protein family (LTPV.2) |
282 ± 35 |
1776 ± 616 |
6.3 |
| At1g73620 1 |
Thaumatin-like protein, putative |
268 ± 23 |
741 ± 23 |
2.8 |
| At1g21890 1,4 |
Nodulin MtN21/EamA-like transporter family protein |
165 ± 51 |
403 ± 81 |
2.4 |
| At5g06390 1,4 |
FASCICLIN-like arabinogalactan protein 17 (FLA17) |
65 ± 11 |
161 ± 6 |
2.5 |
| At5g67400 1,4 |
Root hair specific 19/Peroxidase 73 (RHS19/PER73) |
153 ± 8 |
377 ± 37 |
2.5 |
| At5g47500 1,4 |
Pectin lyase-like superfamily protein (PME5) |
302 ± 93 |
687 ± 136 |
2.3 |
| At3g05600 1,4 |
α/β-Hydrolase superfamily protein |
58 ± 1,2 |
116 ± 13 |
2.0 |
| At1g62560 1,3 |
Flavin-monooxygenase glucosinylate S-oxygenase 3 (FMO GS-OX3) |
406 ± 102 |
775 ± 194 |
1.9 |
| At3g08770 1,4 |
Lipid transfer protein 6 (LTP6) |
1475 ± 1,25 |
2758 ± 98 |
1.9 |
| At3g05470 1,4 |
Actin-binding FH2 (formin homology 2) family protein |
275 ± 160 |
516 ± 311 |
1.9 |
| At5g01870 1 |
Lipid transfer protein 10 (LTP10) |
360 ± 24 |
638 ± 58 |
1.8 |
| At5g61020 1,4 |
evolutionarily conserved C-terminal region 3 (ECT3) |
421 ± 98 |
736 ± 160 |
1.7 |
| At1g20930 1,4 | Cyclin-dependent kinase B2;2 (CDKB2;2) | 359 ± 61 | 598 ± 66 | 1.7 |
Genes with putative roles in extracellular matrix regulation (GO analysis), altered expression in bp mutants [36] or having a BLR consensus binding site in the promoter ( http://arabidopsis.med.ohio-state.edu/AtcisDB/) are shown.
1Role, or putative role in modification of the extracellular matrix.
2Reduced expression in bp compared to wild-type plants (Mele et al., 2003).
3Increased expression in bp compared to wild-type plants (Mele et al., 2003).
4BLR DNA binding consensus sequence present in promoter.
Expression values are presented as the mean ± standard error for three biological replicates.
To understand better the significance of transcriptional differences between BLR-OX and control plants, a gene ontogeny (GO) analysis was carried out. Strikingly, the only significant change proved to be in cell wall associated gene expression, which was enriched from 1.3% in controls to 6.8% in BLR-OX lines (Table 1) pointing to a role for BLR in regulating cell wall metabolism or organisation. Changes in cell size are regularly accompanied by expression changes in cell wall associated genes and the GO analysis thus draws together the gain-of-function phenotypes (Figures 1, 3) and the array data (Table 1). Lipid transfer proteins (LTPs) [31] and peroxidases [32] loosen cell walls and induce cell expansion; thus a 6 fold decrease in expression of LTP and a 2.5 fold reduction in peroxidise expression in BLR-OX plants (Table 1) is consistent with the reduction in cell size observed in BLR-OX lines. PME5, which is negatively regulated by BLR (2.3 fold down-regulated in BLR-OX), alters the gelling properties of cell walls [33], while other genes with altered expression are implicated by homology to have roles in cell expansion. For example members of the thaumatin family (2.8 fold down-regulated) have potential endo-β-1,3 glucanase activity [34].
Extreme BLR gain-of-function plants lack organ initiation
Organised phyllotaxy, lacking in blr mutants [2-4] requires accurate cell expansion and division during organogenesis [15], processes reliant on close regulation of cell-wall formation and expansion [16,18]. We therefore anticipated that phenotypes observed in BLR-OX lines could be strengthened to include disruption of organ initiation due to altered cell wall metabolism, particularly as BLR-OX lines analysed above exhibited only a relatively small expression increase (3.6 fold). Furthermore, in some contexts, BLR is localised in the cytoplasm, and its nuclear localisation is dependent on an interacting partner [2,35]. Plants were therefore generated expressing a steroid-inducible version of BLR in the form of a BLR-GR fusion regulated by the 35S CaMV promoter which would result in nuclear localisation of BLR in a steroid inducible manner, showing independence of an interacting partner. To confirm that expression changes of cell wall modifying enzymes following BLR-GR induction were consistent with those observed in BLR-OX lines, plants were subjected to dexamethasone (dex) induction for 72 h, as maximum activity of direct transcriptional targets in this system has been demonstrated as occurring between 64 and 128 hours [36]. Following dex induction, expression of both MERI-5 and EXP17 was elevated (Figure 2C), as in BLR-OX lines (Table 1) providing further evidence for BLR regulation of the extracellular matrix.
When BLR-GR seeds were germinated on 10 μm dex, only cotyledons were visible after 14 days (Figure 4A), suggesting that high levels of BLR in the nucleus lead to extremely limited growth. This observation may explain the relatively low increases in BLR expression in BLR-OX as plants with higher expression levels may not have been recovered in the T1 generation. Seedlings germinated for 3 days in the absence of steroid and subsequently transferred to 10 μm dex (BLR–GRi), produced leaves with severely reduced expansion (Figure 4B-E). Visible leaves no longer emerged after 21 days and the apex became enlarged (Figure 4F), suggesting that growth continued in the absence of organ initiation. Sections cut through these enlarging BLR–GRi apices revealed an absence of lateral organ primordia or any other clearly differentiated cell types (Figure 4G-H), suggesting that high levels of nuclear BLR results in anisotropic growth.
Figure 4.

Phenotypes of 35S::BLR-GR Plants. (A) Plants expressing BLR-GR did not generate post-embryonic structures when germinated on plates containing Dex. (D-H) Plants treated with dex, 3 days post-germination. BLR-GRi (D, F) did not initiate new lateral organs but developed a bulbous structure (arrow in F) at the apex, whereas dex treated wild type (E) and untreated BLR-GR (B) or wild type (C) developed normally. Longitudinal sections through apices (G-H) shows that in wild type plants at 3 weeks-old, the programme of inflorescence development had initiated (G), but induced 35S::BLR::GR plants lateral organ initiation had ceased (H), Scales are 100 μm.
Normal organ initiation and phyllotaxy in Arabidopsis requires interplay between BLR repression of PME5 in the shoot apical meristem [19], and loss of this repression during formation of organ primordia [4]. Loss of PME5 repression has been proposed to allow other cell wall modifying components to access the wall. Our results support this hypothesis for, in our BLR-GRi plants, BLR would continue to negatively regulate PME5 with the consequent inhibition of organ initiation we observed.
Organ initiation requires both the correct patterning of cell divisions [14,15] and strictly regulated cell wall remodelling [16,18]. The organisation of cell divisions in the apex has been shown to depend on mechanical pressures [17], which are likely to be influenced by the cell wall [37]. The current model for cell-wall remodelling during organ initiation at the apex holds that the breakage and annealing of cell-wall components is controlled by connections with microtubules, possibly through regulation of cellulose-microfibril length [38]. Treatment of Arabidopsis apices with oryzalin, which depolymerises microtubules and prevents ordered division, results in a bulbous apex and loss of lateral-organ initiation when combined with disrupted auxin signalling [39]. The cell wall plays a central part in the transmission of chemical and mechanical signals between cells, and the positioning of developmental cues at the apex of sunflowers is reported to be sensitive to mechanical interventions [17]. Our data suggest that BLR modulates the expression of genes encoding cell wall associated proteins, and our observation that plants strongly over-expressing BLR-GRi fusions generate bulbous apices incapable of forming lateral organs is consistent with the view that closely-regulated cell wall development is essential for ordered initiation of lateral organs at the shoot apex.
rev enhances the bp blr phenotype
The array experiments identified genes involved in cell expansion to be misregulated in BLR-OX plants. Expression changes in genes involved in cell expansion are also a feature of ectopic knox1 (STM) expression in poplar stem vascular tissue [40], and several putative BLR targets identified in BLR-OX arrays are reciprocally regulated in bp mutants [41] (Table 1). BP is required for patterning the inflorescence stem as sectors of differentiation-defective epidermal, cortical and vascular cells are present in bp mutant stems [22,42,43]. Furthermore, BP negatively regulates biosynthesis of lignin [41], a cell wall polymer, that is present in cells that have secondary walls including those of the vascular tissue. Despite exhaustive analysis, blr and wild type hypocotyls proved indistinguishable (not shown); however we saw clear differences in the vascular tissue of the inflorescence stem where BLR is expressed [4]. Loss of function blr phenotypes have been described in this region [3] in the relatively newly formed vascular tissue at the top of the stem (between the third and fourth siliques). Here blr mutants were found to form a continuous ring of vascular tissue, while bp blr double mutants demonstrated an enhanced blr phenotype as cells in the continuous vascular ring failed to differentiate and contained small, densely stained cells where xylem cells with secondary cell walls formed in wild type plants [3].
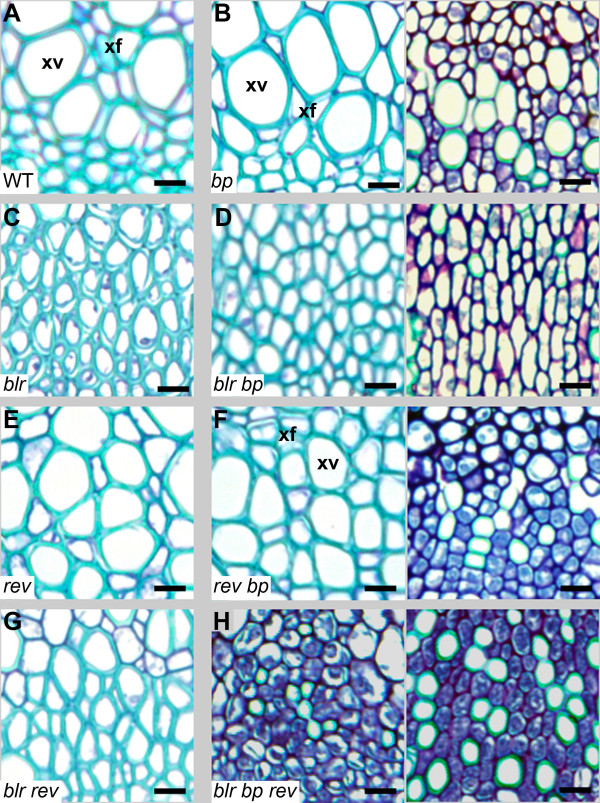
We analysed vascular tissue at a later stage in development, adjacent to the first cauline leaf on the primary inflorescence stem. blr vascular bundles were patterned differently from those of wild type, as the arc of procambial cells present in wild type appeared shallower in blr (Figure 5A, C). blr mutants with the strongest phenotypes also had vascular bundles lacking large xylem vessels and containing apparently only xylary fibres, suggesting that there was either a failure of vessel specification, or a defect in vessel cell size (Figure 6A, C). ‘Stripes’ of differentiation-defective cells overlying some vascular bundles in bp mutants gave rise to two distinct vascular morphologies (Figures 5B, 6B); Within the stripes xylem differentiation was altered because as with blr mutants, xylem lacked large vessels, however, in contrast to blr mutants, many of the smaller cells had not generated secondary cell walls [22,42] (Figure 6B). In areas outside the differentiation defective stripe, vascular patterning was indistinguishable from wild type (Figure 5B).
Figure 5.
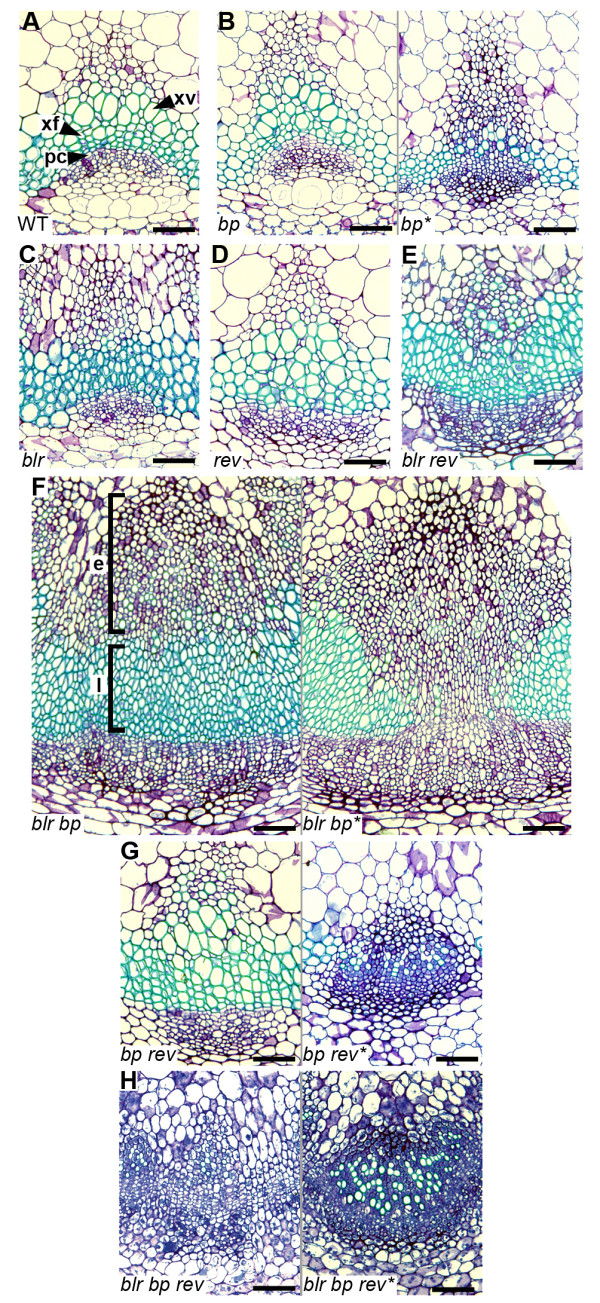
Vascular tissue in blr, bp and rev mutant combinations. Toluidine blue stained sections from inflorescence stem vascular tissue adjacent to the first cauline leaf. (A) Wild type vascular bundle with arrowheads pointing to a xylem vessel (xv), xylary fibres (xf) and vascular meristem (procambium; pc). (B) bp mutants are indistinguishable from wild type, except in differentiation defective stripes (bp*) where the arc of procambium cells is flattened, xylem vessels are absent or reduced in size and many cells are small and darkly stained suggesting they lack secondary walls (green in wild type xylem; A). blr (C) and blr rev (E) vascular tissue in which the arc of procambium is flattened and xylem vessels appear absent. (D) rev. (F) Large vascular expansion in blr bp. In vascular tissue derived early in development (square bracket; e) all cells are small and many lack secondary walls. In contrast, later derived tissue (square bracket; l) contains differentiated xyary fibres but no vessels, except in differentiation defective regions of the stem (blr bp*) where very few cells with secondary wall are present. (G) bp rev double mutants are indistinguishable from rev in most regions of the stem, however, in differentiation defective regions (bp rev*) cells were smaller than those observed in bp*. (H) Cells in blr bp rev inflorescence stems were extremely small and in the vast majority of cases secondary walls appeared absent. Scales are 50 μm.
Figure 6.
blr, bp and rev mutant xylem. Xylem from vascular bundle in inflorescence stems. (A) Wild type xylem with vessels (xv) and xylary fibres (xf) marked. (B) bp mutant xylem is indistinguishable from wild type (left hand side; LHS), except in differentiation defective tissue (right hand side; RHS) where vessels are absent or reduced in size and many cells lack secondary walls (green in wild type xylem; A). (C) Extreme blr phenotype where xylem vessels appear absent. (D) blr bp xylem in later derived tissue (see Figure 5F) with xyary fibres but no vessels (LHS). In differentiation defective regions of the stem (RHS) Few cells with secondary wall are present. (E) rev xylem, similar to wild type. (F) bp rev xylem is like that of rev (LHS) except in differentiation defective regions (RHS) where cells were smaller than those observed in bp. (G) rev blr xylem. (H) Most blr bp rev xylem lacked secondary walls. Scales are 10 μm.
bp blr vascular tissue has been described as reminiscent of that present in differentiation defective areas of bp mutants [3]. In bp blr bundles outside the differentiation-defective stripe, small darkly staining cells as described by Smith et al.[3], were observed towards the centre of the stem (Figure 5F) suggesting that these cells were derived early in vascular development. However, xylem cells generated later in development (closest to the procambium) underwent significant differentiation in bp blr double mutants because secondary cell walls were present, but as with extreme blr mutants, all xylem cells were reminiscent of xylary fibres (Figure 6D). In contrast, we observed very few xylem cells with secondary walls in bp blr vascular bundles within the differentiation-defective tissue associated with the bp mutation, even at this late stage of development (Figures 5F, 6D). Consequently, and consistent with other reports [3], blr enhances bp in the context of the vascular tissue and this interaction is particularly evident in mature bp blr vascular tissue within differentiation-defective regions of the stem (Figures 5A-C, 5F, 6A-D). We reasoned that analysis of bp and blr mutant combinations in a background of altered xylem identity may reveal redundant functions of bp and blr. REV has been associated with radial patterning in the stem [24] and has previously been shown to be required for development of interfascicular fibres [25,44], for xylem differentiation and maintenance of xylem cell number [25]. The role of REV in the xylem is likely to be partially redundant as phenotypes appear enhanced in rev phb/+ or rev phv mutants [45]. Furthermore in the shoot apex, as with bp[13] and blr[2], rev enhances the phenotype of weak alleles of stm[46] suggesting that REV may have overlapping functions with BLR. We therefore examined inflorescence stem vascular tissue in bp, blr and rev combinatorial mutants. Within rev vascular bundles xylem tissue resembled wild type in that both vessels and parenchyma were present, however the arc of procambium was shallower as in blr plants (Figure 5D), a phenotype also occurring in bp rev mutant vascular tissue, outside of differentiation defective tissue (Figure 5G). Within differentiation defective tissue, the xylem cells were generally similar to those of bp single mutants except that the relatively few cells that developed secondary walls were extremely small compared with those of bp (Figure 6F). blr rev and blr vascular bundles were similar but, strikingly, the xylem in vascular bundles of blr bp rev triple mutants comprised small and darkly stained xylem cells, whether or not the bundles lay within the stripe of differentiation-defective cells (Figures 5H, 6G). In extreme cases, no cells with secondary walls were seen in vascular bundles (Figure 5H).
Genetic interactions between BLR, REV and BP in the stem vary depending on tissue type. Vascular cell division between vascular bundles is one of the hallmarks of secondary growth (radial expansion). BP and BLR repress this process as bp blr mutants have a continuous ring of vascular tissue [3]. However, REV appears to be required for the large increase in vascular tissue present in bp blr because bp blr rev lines demonstrate a reduction in vascular tissue compared to bp blr (Figure 5F, H). This observation supports a role for REV (in contrast to BP and BLR) in promoting secondary growth, a function that has previously been suggested for its poplar orthologue [47]. BLR, REV and BP do not have contrasting roles in regulating xylem differentiation. The presence of xylem cells with secondary walls was reduced in double and triple mutant combinations (Figure 6) suggesting that BLR, REV and BP act together in this process. Consequently, BLR, REV and BP interactions are context dependent.
Although the primary defect observed in BLR-OX was one of reduction in cell size, similar to that observed in vascular tissue of blr mutant combinations, the relationship between cell size and expression of particular cell wall modifying genes is not always straightforward [48-53]. For example, over-expression of an E2Ff transcription factor causes a reduction in cell size, accompanied by a decrease in expression of expansins (EXP3, 7 and 9), a UDP-glucose-glucosyl transferase (UGT) and MERI-5[52] (up-regulated 4.6 fold in BLR-OX). However, over-expression of MERI-5 is reported elsewhere as promoting precocious elongation [54]. BLR control of the extracellular matrix is thus likely to be similarly complex. BLR-OX plants have smaller cells than those of wild-type, but blr mutants, when combined with bp and rev also resulted in vascular tissue with smaller cells.
Conclusions
Control of cell expansion is essential for normal plant development. Our results demonstrate that BLR controls cell expansion and differentiation in vascular development in a process also involving BP and REV transcription factors. Microarray analysis of plants over-expressing BLR indicates that BLR targets many genes involved in cell wall regulation. As dynamic control of the cell wall is essential for cell expansion, we suggest that the striking phenotypes of plants over- and under-expressing BLR may result from the disruption of this process.
Closely regulated cell wall dynamics are also required for a normal pattern of organ initiation at the shoot apex and this process has previously been shown to be regulated by BLR repression of PME5[19]. Our experiments with BLR-GR suggest that further transcriptional targets of BLR may also play important parts in this process.
Methods
BLR overexpression
Full-length BLR cDNA was cloned into pGEM-T Easy (Promega), subcloned as an EcoRI fragment into pART7 [55] and sequenced to identify plasmids with BLR in the sense orientation. The resultant CaMV35S::BLR::OCS-terminator cassette was subsequently excised from pART7 using NotI and subcloned into pMLBART. The resultant BLR-OX binary vector was introduced into Arabidopsis (Col-0) using standard methods [56]. For steroid-inducible BLR lines, two PCR products were amplified, one containing BLR cDNA and a second containing the glutocoticoid receptor (GR). The GR forward primer (AAAAAGCAGGCTTAATGACCACACTCAACATG) contained a partial attB1 site, while the reverse primer (GCTCCACCTCCACCTCCATACTCATGG) contained an Ala–Gly linker and part of the 5′ BLR sequence. Similarly, the forward BLR primer (GGAGGTGGAGGTGGAGCATGGCTGATGCAT) contained the Ala–Gly linker while the reverse primer (AGAAAGCTGGGTTCAACCTACAAAATCATG) contained an attB2 site. The two PCR products were mixed in equimolar concentrations and used as templates for overlap PCR with attB1 and attB2 primers resulting in a BLR-GR in-frame fusion, which was cloned into pDONR Zeo, sequenced, and subcloned into pEarleyGate100 using Gateway technology. BASTA-resistant Col-0 transgenic lines were generated as described above, and Arabidopsis transformants that had a wild-type phenotype when uninduced but responded to 10-μM dexamethasone were used for subsequent analysis.
Gene expression analysis
For Microarray experiments total RNA was extracted from Col-0 and BLR-OX plants using an RNeasy Plant Mini Kit (Qiagen). RNA integrity was checked on a 2100 BioAnalyzer (Agilent, CA, USA) prior to cDNA synthesis from 2 μg total RNA using a 3DNA Array 900TM kit (Genisphere Inc., Hatfield, PA, USA) with RT-primers that contained a dendrimer [57] capture sequence. Labelled cDNAs were hybridised for 16 hours to the Arabidopsis Atv3.3.2.z Array printed with the Operon Arabidopsis thaliana version 3 Array-Ready Oligo Set (Galbraith, USA) using a Slidebooster (Advalytix, Brunnthal, Germany) with a power setting of 27 and a pulse setting of 7:3. Array slides were washed in 2× SSC, 0.2% SDS (10 min, 55°C), 2× SSC (10 min, Room temp.) and 0.2× SSC (10 min, room temp.). Fluorescent-labelled Genisphere 3DNA dendrimers containing approximately 850 Cy3 or Cy5 dyes were hybridized for 4 hours to the arrays. Following a set of washes as above, arrays were scanned using a ScanArray Express HT Microarray Scanner (Perkin Elmer).
After calibration to obtain scans within the optimum dynamic range, image analysis and spot feature quantification were performed using BlueFuse version 3.1 (BlueGnome, Cambridge, UK). Data from each slide were confirmed to be essentially linear and globally median-normalized before further analysis. Spots were excluded when the combined confidence values of biological replicates was <0.005. Microarray data were median-normalized in Cy5 and Cy3 channels. Spots that showed expression changes in the same direction in all three replicates were analysed further.
Normalized Cy5/Cy3 log2 ratios (three values for each gene for BLR-OX lines and three for Col-0) were used to perform a significance analysis of microarray (SAM) [30] using the TIGR Multi Experiment Viewer (TMEV V3.0.3). The imputation engine was set as 10-nearest neighbour imputer and the number of permutations was 100. The delta value in the Significance Analysis was adjusted so that estimated false discovery rate (FDR) was 0% for significant genes. The expression-fold change was set as 1.5 as a threshold for changes of potential biological importance. The microarray data described here have been submitted to MIAMExpress ( http://www.ebi.ac.uk/arrayexpress/experiments/E-MEXP-852). GO annotation analysis was performed using http://www.arabidopsis.org/tools/bulk/go/index.jsp.
For northern blot analysis of primary transformants, two micrograms of total RNA extracted from leaf tissue was subjected to electrophoresis on a 1.2% formaldehyde agarose gel, which was subsequently blotted onto Hybond-NX membrane and hybridised with a full-length 32PdCTP-labelled cDNA probe. RNA was also visualised by ethidium bromide staining and the filter hybridised with GAPDH as loading controls. Validation of microarray targets was carried out on RNA from 6–7 day-old seedlings. Primers were used to amplify the 3′ UTR and adjoining exons for specific probes, which were 32PdCTP-labelled.
qRT-PCR analysis was carried out using SYBR Green JumpStart Taq ReadyMix (Sigma) and an ABI Prism 7000 machine (Applied Biosystems) with the standard sybr green detection programme. A melting curve was produced at the end of every experiment to ensure that only single products were formed. Gene expression was determined using a version of the comparative threshold cycle (Ct) method. The average amplification efficiency of each target was determined using LinReg [58] and samples were normalised to 18S rRNA (not shown) and ACT2 (shown). Results were similar independent of control used. Samples were measured in technical triplicates on biological triplicates.
Plant materials, growth conditions and imaging
Plants were grown in long-day conditions at 20°C. Mutant lines, blr (van-1) [2], rev-6[46], and bp-1[22,42], have been previously described. Digital images of the plants were analysed in Scion Image (Scion Corporation, Maryland, USA) for hypocotyl measurements. For histological analysis of BLR-OX and BLR-GR, plants were fixed, wax-embedded and sectioned using standard methods. Sections between 6 and 10 micrometers were stained with toluidine blue for light microscopy. For visualisation of hypocotyl epidermal cells, seedlings were stained with 10 μg ml−1 propidium iodide (Molecular Probes Europe BV, Netherlands) for 10 minutes in the dark before transfer to water. Seedlings were viewed by confocal microscopy on a Zeiss (Welwyn, UK) CLSM 510.
Toluidine blue stained resin sections for visualisation of transverse vascular bundle sections in inflorescence stems were taken from 5 week old plants. Sections were taken above the first cauline leaf and prepared as described in [59].
Authors’ contributions
AMB and HGD designed the experiments. JPE, LM, WZJ, HP, NC, NS and AMB carried out the experiments. JPE, AMB and HGD wrote the manuscript. All authors read and approved the final manuscript.
Authors’ information
Anuj M Bhatt and Hugh G Dickinson Joint last authors.
Supplementary Material
Genes changed in expression in BLR-OX in the same direction in all 3 replicates with an average of >1.5-fold change, arranged in order of spot confidence.
Contributor Information
J Peter Etchells, Email: Peter.Etchells@manchester.ac.uk.
Lucy Moore, Email: loofylu@hotmail.com.
Wen Zhi Jiang, Email: wjiang2@unl.edu.
Helen Prescott, Email: Helen.Prescott@plants.ox.ac.uk.
Richard Capper, Email: richard_capper@agilent.com.
Nigel J Saunders, Email: Nigel.Saunders@brunel.ac.uk.
Anuj M Bhatt, Email: Anuj.Bhatt@bbsrc.ac.uk.
Hugh G Dickinson, Email: Hugh.Dickinson@plants.ox.ac.uk.
Acknowledgements
We thank Andrey Lagodienko for preliminary BLR-OX analysis, Ian Moore for confocal microscopy assistance, John Baker for photography and Jo Dowding for technical help. Seeds were from NASC and materials for microarrays were provided by the University of Arizona. This study was supported by BBSRC (grant BB/G18339). Microarray facilities were established with financial support from the EPA Cephalosporin Trust (NS).
References
- Roeder AHK, Ferrandiz C, Yanofsky MF. The Role of the REPLUMLESS Homeodomain: Protein in Patterning the Arabidopsis Fruit. Curr Biol. 2003;13(18):1630–1635. doi: 10.1016/j.cub.2003.08.027. [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H. VAAMANA–a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene. 2004;328:103–111. doi: 10.1016/j.gene.2003.12.033. [DOI] [PubMed] [Google Scholar]
- Smith HMS, Hake S. The Interaction of Two Homeobox Genes, BREVIPEDICELLUS and PENNYWISE, Regulates Internode Patterning in the Arabidopsis Inflorescence. Plant Cell. 2003;15(8):1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Groover AT, Fontana JR, Martienssen RA. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development. 2003;130(17):3941–3950. doi: 10.1242/dev.00620. [DOI] [PubMed] [Google Scholar]
- Bao X, Franks RG, Levin JZ, Liu Z. Repression of AGAMOUS by BELLRINGER in Floral and Inflorescence Meristems. Plant Cell. 2004;16(6):1478–1489. doi: 10.1105/tpc.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HMS, Boschke I, Hake S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci USA. 2002;99(14):9579–9584. doi: 10.1073/pnas.092271599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350(6315):241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Lin H, Joo S, Goodenough U. Early Sexual Origins of Homeoprotein Heterodimerization and Evolution of the Plant KNOX/BELL Family. Cell. 2008;133(5):829–840. doi: 10.1016/j.cell.2008.04.028. [DOI] [PubMed] [Google Scholar]
- Barton MK, Poethig RS. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development. 1993;119(3):823–831. [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379(6560):66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Smith HMS, Campbell BC, Hake S. Competence to Respond to Floral Inductive Signals Requires the Homeobox Genes PENNYWISE and POUND-FOOLISH. Curr Biol. 2004;14(9):812–817. doi: 10.1016/j.cub.2004.04.032. [DOI] [PubMed] [Google Scholar]
- Ragni L, Belles-Boix E, Gunl M, Pautot V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis Inflorescences. Plant Cell. 2008;20(4):888–900. doi: 10.1105/tpc.108.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development. 2002;129(8):1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131(17):4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jonsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM. et al. Developmental Patterning by Mechanical Signals in Arabidopsis. Science. 2008;322(5908):1650–1655. doi: 10.1126/science.1165594. [DOI] [PubMed] [Google Scholar]
- Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C. Induction of Leaf Primordia by the Cell Wall Protein Expansin. Science. 1997;276(5317):1415–1418. doi: 10.1126/science.276.5317.1415. [DOI] [Google Scholar]
- Hernandez LF, Green PB. Transductions for the Expression of Structural Pattern: Analysis in Sunflower. Plant Cell. 1993;5(12):1725–1738. doi: 10.1105/tpc.5.12.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Höfte H, Laufs P, Pelloux J, Mouille G. Arabidopsis Phyllotaxis Is Controlled by the Methyl-Esterification Status of Cell-Wall Pectins. Curr Biol. 2008;18(24):1943–1948. doi: 10.1016/j.cub.2008.10.065. [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Salsac F, Morin H, Fournet F, Belcram K, Gillet F, Höfte H, Laufs P. et al. The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development. 2011;138(21):4733–4741. doi: 10.1242/dev.072496. [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Deyholos MK. Vascular development: the long and winding road. Curr Opin Plant Biol. 2006;9(1):48–54. doi: 10.1016/j.pbi.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Cartwright HN, Humphries JA, Smith LG. PAN1: A Receptor-Like Protein That Promotes Polarization of an Asymmetric Cell Division in Maize. Science. 2009;323(5914):649–651. doi: 10.1126/science.1161686. [DOI] [PubMed] [Google Scholar]
- Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD. KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell. 2002;14(3):547–558. doi: 10.1105/tpc.010391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L. Plant-cell expansion - Regulation of cell-wall mechanical-properties. Annu Rev Plant Physiol Plant Mol Biol. 1984;35:585–657. doi: 10.1146/annurev.arplant.35.1.585. [DOI] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial Patterning of Arabidopsis Shoots by Class III HD-ZIP and KANADI Genes. Curr Biol. 2003;13(20):1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye Z-H. IFL1, a Gene Regulating Interfascicular Fiber Differentiation in Arabidopsis, Encodes a Homeodomain-Leucine Zipper Protein. Plant Cell. 1999;11(11):2139–2152. doi: 10.1105/tpc.11.11.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugis G, Giannino D, Mele G, Nicolodi C, Chiappetta A, Bitonti MB, Innocenti AM, Dewitte W, Van Onckelen H, Mariotti D. Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 2001;126(4):1370–1380. doi: 10.1104/pp.126.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127(24):5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A Knotted1-Like Homeobox Gene in Arabidopsis Is Expressed in the Vegetative Meristem and Dramatically Alters Leaf Morphology When Overexpressed in Transgenic Plants. Plant Cell. 1994;6(12):1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. Growth Stage-Based Phenotypic Analysis of Arabidopsis: A Model for High Throughput Functional Genomics in Plants. Plant Cell. 2001;13(7):1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BAH, Fasolino A, Hilbers CW, Derksen J, Mariani C. Lipid Transfer Proteins Enhance Cell Wall Extension in Tobacco. Plant Cell. 2005;17(7):2009–2019. doi: 10.1105/tpc.105.032094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 2004;9(11):534–540. doi: 10.1016/j.tplants.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Imoto K, Yokoyama R, Nishitani K. Comprehensive approach to genes involved in cell wall modifications in Arabidopsis thaliana. Plant Mol Biol. 2005;58(2):177–192. doi: 10.1007/s11103-005-5344-7. [DOI] [PubMed] [Google Scholar]
- Grenier J, Potvin C, Trudel J, Asselin A. Some thaumatin-like proteins hydrolyse polymeric β-1,3-glucans. Plant J. 1999;19(4):473–480. doi: 10.1046/j.1365-313X.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- Cole M, Nolte C, Werr W. Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 2006;34(4):1281–1292. doi: 10.1093/nar/gkl016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft J, Samalova M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 2005;41(6):899–918. doi: 10.1111/j.1365-313X.2005.02342.x. [DOI] [PubMed] [Google Scholar]
- Castle ES. Membrane tension and orientation of structure in the plant cell wall. J Cell Comp Physiol. 1937;10(1):113–121. doi: 10.1002/jcp.1030100110. [DOI] [Google Scholar]
- Wasteneys GO. Progress in understanding the role of microtubules in plant cells. Curr Opin Plant Biol. 2004;7(6):651–660. doi: 10.1016/j.pbi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Corson F, Hamant O, Bohn S, Traas J, Boudaoud A, Couder Y. Turning a plant tissue into a living cell froth through isotropic growth. Proc Nat Acad Sci USA. 2009;106(21):8453–8458. doi: 10.1073/pnas.0812493106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groover A, Mansfield S, DiFazio S, Dupper G, Fontana J, Millar R, Wang Y. The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol Biol. 2006;61(6):917–932. doi: 10.1007/s11103-006-0059-y. [DOI] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S. The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 2003;17(17):2088–2093. doi: 10.1101/gad.1120003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(7):4730–4735. doi: 10.1073/pnas.072626099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SJ, Riggs CD. Pedicel development in Arabidopsis thaliana: Contribution of vascular positioning and the role of the BREVIPEDICELLUS and ERECTA genes. Dev Biol. 2005;284(2):451–463. doi: 10.1016/j.ydbio.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye ZH. Disruption of Interfascicular Fiber Differentiation in an Arabidopsis Mutant. Plant Cell. 1997;9(12):2159–2170. doi: 10.1105/tpc.9.12.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17(1):61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25:223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- Robischon M, Du J, Miura E, Groover A. The Populus Class III HD ZIP, popREVOLUTA, Influences Cambium Initiation and Patterning of Woody Stems. Plant Physiol. 2011;155(3):1214–1225. doi: 10.1104/pp.110.167007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9(5):701–713. doi: 10.1046/j.1365-313X.1996.9050701.x. [DOI] [Google Scholar]
- Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA. 2000;97(2):942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G-T, Shoda K, Tsuge T, Cho K-H, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J. 2002;21(6):1267–1279. doi: 10.1093/emboj/21.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Vuylsteke M, Florquin K, Rombauts S, Maes S, Ormenese S, Van Hummelen P, Van de Peer Y, Inze D, De Veylder L. Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J Cell Sci. 2003;116(20):4249–4259. doi: 10.1242/jcs.00715. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Lopez-Matas MA, Frundt C, Gutierrez C. Role of an Atypical E2F Transcription Factor in the Control of Arabidopsis Cell Growth and Differentiation. Plant Cell. 2004;16(9):2350–2363. doi: 10.1105/tpc.104.023978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Hiratsu K, Koyama T, Tanaka H, Ohme-Takagi M. A Chimeric AtMYB23 Repressor Induces Hairy Roots, Elongation of Leaves and Stems, and Inhibition of the Deposition of Mucilage on Seed Coats in Arabidopsis. Plant Cell Physiol. 2005;46(1):147–155. doi: 10.1093/pcp/pci005. [DOI] [PubMed] [Google Scholar]
- Verica JA, Medford JI. Modified MERI5 expression alters cell expansion in transgenic Arabidopsis plants. Plant Sci. 1997;125(2):201–210. doi: 10.1016/S0168-9452(97)00068-X. [DOI] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20(6):1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Stears RL, Getts RC, Gullans SR. A novel, sensitive detection system for high-density microarrays using dendrimer technology. Physiol Genomics. 2000;3(2):93–99. doi: 10.1152/physiolgenomics.2000.3.2.93. [DOI] [PubMed] [Google Scholar]
- Hardstedt M, Finnegan CP, Kirchhof N, Hyland KA, Wijkstrom M, Murtaugh MP, Hering BJ. Post-transplant upregulation of chemokine messenger RNA in non-human primate recipients of intraportal pig islet xenografts. Xenotransplantation. 2005;12(4):293–302. doi: 10.1111/j.1399-3089.2005.00228.x. [DOI] [PubMed] [Google Scholar]
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development. 2008;135(7):1315–1324. doi: 10.1242/dev.016469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes changed in expression in BLR-OX in the same direction in all 3 replicates with an average of >1.5-fold change, arranged in order of spot confidence.