Abstract
One of the most formidable impediments to clinical translation of RNA interference (RNAi) is safe and effective delivery of the siRNAs to the desired target tissue at therapeutic doses. We previously described in vivo cell type-specific delivery of anti-HIV small-interfering RNAs (siRNAs) through covalent conjugation to an anti-gp120 aptamer. In order to improve the utility of aptamers as siRNA delivery vehicles, we chemically synthesized the gp120 aptamer with a 3′ 7-carbon linker (7C3), which in turn is attached to a 16-nucleotide 2′ OMe/2′ Fl GC-rich bridge sequence. This bridge facilitates the noncovalent binding and interchange of various siRNAs with the same aptamer. We show here that this aptamer-bridge-construct complexed with three different Dicer substrate siRNAs (DsiRNAs) results in effective delivery of the cocktail of DsiRNAs in vivo, resulting in knockdown of target mRNAs and potent inhibition of HIV-1 replication. Following cessation of the aptamer-siRNA cocktail treatment, HIV levels rebounded facilitating a follow-up treatment with the aptamer cocktail of DsiRNAs. This follow-up injection resulted in complete suppression of HIV-1 viral loads that extended several weeks beyond the final injection. Collectively, these data demonstrate a facile, targeted approach for combinatorial delivery of antiviral and host DsiRNAs for HIV-1 therapy in vivo.
Introduction
Combinatorial antiretroviral therapy which combines two or more antiretroviral drugs that act on different stages of the HIV life cycle is currently used to suppress viral replication. Although highly effective, there are often undesirable side effects and compliance problems as well as the emergence of drug resistance that complicate the use of this therapy.1 Because of these issues, a number of investigators have been exploring other approaches for inhibiting HIV-1 infection. Nucleic acid-based therapeutics have been considered as a promising alternative to the small molecule antiviral to overcome these limitations. One of the most potent of these is the use of RNA interference (RNAi).2,3 In the case of HIV-1, either the entire genome of the virus or a large number of HIV-1 host-dependency factors (HDFs) are potential targets for RNAi.4,5,6 Numerous RNAi applications targeting HIV-1 or cellular factors have demonstrated efficient suppression of viral replication both in cell culture and in animal models.7,8 The entire repertoire of HIV-1–encoded genes (tat, rev, gag, pol, nef, vif, env, vpr, and the long terminal repeat) are susceptible to RNAi-induced gene silencing in cell lines.9,10,11 Moreover, host genes such as the chemokine receptor CCR5 can be targeted alone or in combination with viral sequences in a tailored fashion to reduce the emergence of viral escape mutants.12,13,14
Although siRNA-based therapeutic strategies for combating HIV/AIDS hold considerable promise, the actual clinical applications of RNAi for treatment of HIV/AIDS has been limited due to concerns about inefficient systemic delivery of siRNAs to the appropriate target cells or tissues; and viral escape from the RNAi mechanism.15,16 Efforts to improve the therapeutic performance of siRNAs for the treatment of HIV/AIDS should include delivery formulations with targeting moieties as well as combinatorial approaches with other therapeutics. Recent targeted delivery approaches have provided encouragement that this obstacle may be surmountable. For example, single chain antibody-siRNA chimeras targeting either the T-cell CD7 (Cluster of Differentiation 7) receptor17 or HIV-1 glycoprotein gp12018 have been demonstrated to functionally deliver anti-HIV siRNAs in vivo. By using the lymphocyte function-associated antigen-1 (LFA-1) integrin-targeted immunoliposome system, selective targeting and delivery of an anti-CCR5 siRNA to lymphocytes in vivo has been demonstrated.19 As mentioned above, the rapid emergence of viral escape mutants often abrogates RNAi efficacy. A single nucleotide mutation in a critical position within the HIV target sequence relative to the siRNA site of interaction could diminish and even eliminate RNAi activity.20,21,22,23 Therefore, a dual functioning aptamer-mediated cell type-specific delivery of combinatorial RNAi therapeutics also has been developed. For example, HIV-neutralizing aptamers specific to either the CD4 (cluster of differentiation 4) receptor24 or HIV-1 glycoprotein gp12025,26,27 have been harnessed for targeting delivery of multiple anti-HIV siRNAs in vitro as well as in humanized mice.
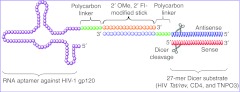
We previously described a noncovalent strategy to assemble a single HIV-1 gp120 aptamer with three different Dicer substrate siRNAs (DsiRNAs) targeting HIV-1 tat/rev or the HDFs (CD4 and Transportin-3 (TNPO3)).26 The resulting aptamer-siRNA conjugates were internalized into cells expressing the HIV-1 envelope and the combination of aptamer-DsiRNAs effectively inhibited viral replication in HIV-1–infected cells. In this design format, both the aptamer and siRNA portions are chemically synthesized and subsequently conjugated via a “sticky bridge”, which is composed of a 16-base 2′ OMe, 2′ Fl-modified GC-rich sequence attached to the aptamer via a polycarbon linker (Figure 1). The physical combination of the HIV-neutralizing aptamer and combination of anti-HIV siRNAs has the potential for large-scale chemical production along with the facile delivery of combinations of siRNAs to mitigate the emergence of viral escape mutants.
Figure 1.
Schematic of anti-HIV-1 gp120 aptamer-DsiRNA conjugates (N-1 design). The aptamer portion of the conjugate binds to gp120. A GC-rich “sticky bridge” facilitates the interchange of three different DsiRNAs (one against the HIV-1 tat/rev transcripts, and two siRNAs targeting the HIV-1 host-dependency factors (CD4 and TNPO3)) with the same aptamer. The 3′-end of the aptamer and one of the two siRNA strands were chemically conjugated with complementary sticky sequences, thereby, allowing the aptamer and siRNA portions to be noncovalently conjugated via Watson-Crick base-pairing by simple mixing. A 7 unit three-carbon linker (C3) between the aptamer/siRNA and stick is indicated in green. For clarity, the N-1 design is shown as an example, in which the 3′-end of the antisense strand of the DsiRNA was appended to a bridge complementary sequence. DsiRNA, Dicer substrate small-interfering RNA.
In the present study, we utilized a humanized mouse model25,28,29,30,31 to evaluate the in vivo antiviral efficacy of the chemically synthesized anti-gp120 aptamer-sticky bridge-DsiRNA conjugates. Humanized Rag2−/−γc−/− mice (RAG-hu) were infected with HIV-1 and subsequently given weekly intravenous injections of the anti-gp120 aptamer-sticky bridge-siRNA conjugates targeting both the virus and cellular transcripts. The results obtained demonstrate functional delivery of all three siRNAs and potent suppression of HIV-1 infection as well as protection against viral-induced CD4+ T-cell depletion. A follow-up treatment of the aptamer-DsiRNAs conjugates following viral rebound resulted in complete, long-term suppression of HIV-1 viral loads. Collectively, these data demonstrate a facile, cell type-specific aptamer-mediated delivery of DsiRNAs in a combinatorial fashion for targeting both viral and cellular transcripts in vivo.
Results
Dicer processing of anti-gp120 aptamer-delivered DsiRNA conjugates
As we previously described, a GC-rich “sticky bridge” facilitates the interchange of different DsiRNAs with a given aptamer.26 We have used this approach to target three different transcripts (HIV-1 tat/rev and two HIV-1 HDFs12 CD4 and TNPO3) with an aptamer that targets HIV gp120. The 3′-end of the aptamer and one of the two siRNA strands were chemically conjugated with 16 bases of complementary GC-rich sequences, thereby, allowing the aptamer and siRNA portions to be noncovalently joined via Watson-Crick base-pairing by simple mixing of the two reactants (Figure 1). Two different design formats (named N-1 and N-2) were constructed. In the N-1 design, the 3′-end of the antisense strand of the DsiRNA was appended to a bridge complementary sequence (Figure 1), whereas the 3′-end of sense stand was joined with this sequence in the N-2 design.
Although both designs showed inhibition of HIV-1 infection in human T cell lymphoblast-like cell line (Supplementary Figure S1a), the N-1 design mediated more efficient knockdown of the target mRNA and exhibited better strand selectivity (Supplementary Figure S1b,c). The Tat/rev N-1 and TNPO3 N-1 conjugates were observed to have somewhat better efficacy in gene silencing than the corresponding Tat/rev N-2 and TNPO3 N-2 conjugates, consistent with the comparison of their HIV-1 p24 data. In vitro Dicer assays indicated that Dicer preferentially enters these substrates from the two base 3′-overhang.26 Since the orientation of the sense and antisense strands differs between N-1 and N-2 with respect to the two base 3′-overhang, Dicer cleavage results in predictably different 21-23 siRNA products from these two configurations which can have different potencies.26
In order to validate that the siRNAs released from the conjugates were actually triggering RNAi, we next investigated siRNA-directed mRNA cleavage using a modified 5′-RACE (Rapid amplification of cDNA ends) PCR assay (as described in Supplementary Materials and Methods). It has been established that Ago2 mediates cleavage between bases 10 and 11 relative to the 5′ end of each siRNA.32,33 Thus the RACE PCR product sequence analyses of the target should reveal a 3′ linker addition at the base positioned 11 nucleotides from the 5′ end of the siRNA. The experimental RNAs (Tat/rev 27-mer siRNA, Tat/rev N-1 or N-2) and a HIV pNL4-3 luc were first cotransfected into HEK293 cells with Lipofectamine 2000. PCR bands of the predicted lengths based upon Dicer cleavage of the DsiRNAs were detected in the total RNA samples from HEK293 cells transfected with the conjugates following three nested PCR reactions (Supplementary Figure S2a). The gel-purified bands of the predicted lengths were TA cloned and individual clones were characterized by DNA sequencing to verify the expected PCR products Tat/rev N-1 and N-2 treatment produced two distinct cleavage sites (Supplementary Figure S1a,b). For Tat/rev N-1, the cleavage takes place at the predicted position for the siRNA duplex between positions 10 and 11 from the 5′ end of the siRNA antisense strand. However, Tat/rev N-2 produced some non-predicted cleavage positions, suggesting a secondary Dicer cleavage of the siRNA portion might exist. Similarly, TNPO3 N-1 produced the predicted cleavage RACE product for the siRNA duplex between positions 10 and 11 from the 5′ end of the siRNA antisense strand. Two distinct non-expected cleavages took place in the treatment with the TNPO3 N-2 conjugate (Supplementary Figure S2c,d). Together, these results suggest that more predictable and potent Dicer cleavage of the DsiRNA species is generated from the N-1 design.
Anti-gp120 aptamer-DsiRNA conjugate is partially resistant to nuclease degradation in mouse serum
In the conjugates, all the RNA components were chemically synthesized with 2′-fluoropyrimidine modifications since the original aptamer selection was carried out using this backbone modification. To test the stability in serum, one aptamer-tat/rev siRNA conjugate (Tat/rev N-1) was incubated in 50 or 10% normal mouse serum for different lengths of times at 37 °C followed by denaturing polyacrylamide gel electrophoretic analyses (Supplementary Figure S3, experimental details were detailed in Supplementary Materials and Methods). After 8 hours of incubation, ~30% of the full-length 2′-fluoro–modified conjugate was detectable in 50% serum and more than 50% was detectable in 10% serum, validating the stabilizing effects of the 2′-Fl modification.
Systemic administration of aptamer-DsiRNA conjugates suppresses HIV-1 viral load in viremic RAG-hu mice
Given the positive results of anti-gp120 aptamer-mediated DsiRNA delivery in HIV-1–infected human T cell lymphoblast-like cell line and primary human peripheral blood mononuclear cells (PBMCs), we next sought to determine whether the aptamer-cocktailed DsiRNA conjugates (N-1 design format) with a combination of three siRNAs (targeting HIV-1 tat/rev and HIV-dependency factors CD4 and TNPO3) are functional in vivo. The ability of the conjugates to suppress HIV-1 viremia was evaluated in Rag2−/−γc−/− mice (RAG-hu) infected with HIV-1 NL4.3 via intraperitoneal injection. Three weeks post-infection, the mice became viremic and were subsequently injected via the tail vein with preparations of the aptamer-cocktailed siRNA conjugates as described in the Materials and Methods section. Control groups were administered with buffer or naked cocktailed DsiRNAs.
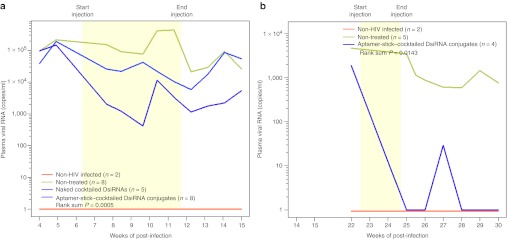
Five to eight animals in each treatment group were given five weekly injections of 0.25 nmol of aptamer-cocktailed DsiRNAs or 0.25 nmol total of a combination of the anti-tat/rev, anti-CD4, and anti-TNPO3 DsiRNAs. To determine whether the established viral loads could be impacted by the treatments, the mice were monitored by quantitative real-time PCR for plasma viral loads on a weekly basis (Figure 2a and Supplementary Table S1a). We observed a general pattern of decreased viral loads in the majority of aptamer-cocktailed DsiRNA conjugate-treated mice. The suppression of viral loads was on average three logs relative to the control groups and this difference reached statistical significance with a rank sum P = 0.0005 (Figure 2a and Supplementary Table S1c). The viral loads were suppressed to below detectable levels within 3 weeks in five of eight conjugate-treated mice (at week 9–11 of post-treatment, as shown in Supplementary Table S1a). This suppression persisted for as long as 3 weeks following the termination of the injections, indicating sustained efficacy of the conjugates. There was no suppression of viral loads in the animals treated with the DsiRNAs alone (Figure 2a and Supplementary Table S1a).
Figure 2.
Aptamer-cocktailed DsiRNA conjugates suppress viral loads in HIV-1–infected RAG-hu mice. HIV-1 viral loads at different weeks post-infection and treatment are indicated. The treatment period is indicated by the yellow framed in region. Weeks post-injection and the time point of treatment start and end are indicated. (a) The first treatment includes five weekly injections: The viral loads of uninfected mice (n = 2), non-treated mice (n = 8), naked cocktail DsiRNA-treated mice (n = 5), and aptamer-cocktailed DsiRNA conjugates treated mice (n = 8) are indicated. (b) The re-treatment including twice weekly injections with aptamer-cocktailed DsiRNA conjugates: The viral loads of uninfected mice (n = 2), non-treated mice (n = 5), and aptamer-cocktailed DsiRNA conjugates treated mice (n = 4) are indicated. P values for both experiments were determined as described in Materials and Methods and Supplementary Table S1c,d. The viral RNA was detected through qRT-PCR as described in Materials and Methods. If there was no detectable viral RNA, we established this as a value of 1 (100) to allow for the use of logarithmic values on the Y-axis. DsiRNA, Dicer substrate small-interfering RNA; qRT-PCR, quantitative reverse transcription-PCR.
Once the aptamer-DsiRNA treatments were terminated, viral loads increased in the majority of the treated animals. To determine whether or not the treatment had enriched for viral mutants resistant to the aptamer-DsiRNA conjugates, we asked whether or not re-treatment with the conjugates could restore inhibition of viral levels to those seen with the first set of treatments. Experimental animals in which the viral loads had become elevated were re-treated with two separate injections of the aptamer-cocktailed DsiRNA conjugates at 12.5 and 13.5 weeks following the last administration from the previous treatment period (Figure 2b and Supplementary Table S1b). The re-treatment resulted in a statistically significant (P = 0.0143) suppression of HIV-1 levels (Supplementary Table S1d). Complete suppression persisted for 2 weeks beyond the re-treatment period in the aptamer-DsiRNA conjugate-treated mice. Three of the four treated mice had undetectable viral loads even up to 6 weeks following the re-treatment as shown in Supplementary Table S1b.
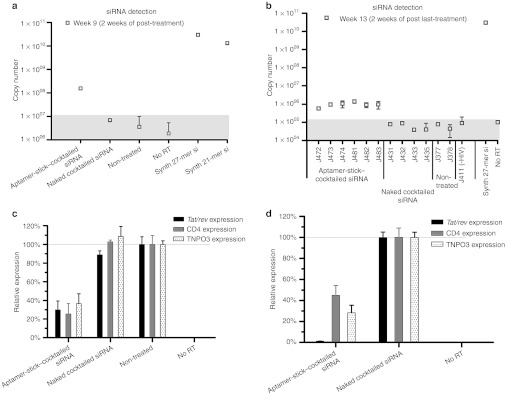
We validated the aptamer-mediated DsiRNA delivery to infected T lymphocytes using a TaqMan quantitative PCR (qPCR) assay on RNA isolated from peripheral blood cells collected following both the second treatment and 2 weeks subsequent to the final treatment in the first experimental set of animals (Figure 2). The results presented in Figure 3a reveal that the tat/rev siRNA was detectable in all of the aptamer-DsiRNA conjugate-treated mice at 2 weeks of treatment. Even at 2 weeks following the last injection, the siRNAs were still detectable in all the conjugate-treated mice (Figure 3b), indicating the stability and retention of the siRNAs delivered by the aptamer in vivo. In contrast, no siRNA was detected in the cellular fractions from mice treated with the naked DsiRNA.
Figure 3.
The detection and function of tat/rev siRNA in PBMCs from treated RAG-hu mice. (a,b) Detection of the tat/rev siRNA sequences at weeks (a) 9 and (b) 13 post-infection using naked cocktailed DsiRNAs versus aptamer-cocktailed DsiRNA conjugates treated animals from Figure 2a. The background copy number of siRNA is <107 or 105 (gray). Error bars indicate SD (n = 4 measurement per sampe). (c,d) Expression levels of targeted tat/rev, CD4, and TNPO3 gene transcripts at weeks 8 and 9 post-infection are shown relative to (c) HIV-1–infected, non-treated animal samples and (d) naked siRNA-treated animal samples, respectively. DsiRNA, Dicer substrate small-interfering RNA; PBMC, peripheral blood mononuclear cell; RT, reverse transcriptase.
We next asked whether the siRNAs triggered sequence-specific mRNA degradation in the HIV-1–infected, aptamer-DsiRNA–treated Rag-hu mice. Reduction in the mRNA levels of the three targeted transcripts (HIV tat/rev, CD4, and TNPO3) in blood cells of infected and treated mice were assessed by quantitative real-time PCR. The aptamer-DsiRNA conjugates substantially decreased the corresponding targeted mRNA levels in the treated mice following the secondary and last injection with the aptamer-DsiRNA conjugates (weeks 9 and 13) (Figure 3c,d).
HIV-1–induced CD4 T-cell depletion is prevented in aptamer-DsiRNA conjugate-treated animals
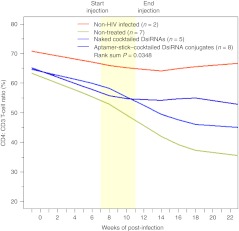
HIV-1 infection is associated with the progressive loss of CD4+ T cells through apoptosis or decreased production.1,34 The CD4 cell count is a standard tool for monitoring the progression of HIV-1/AIDS. Significant loss of CD4+ helper T cells could accelerate immunodeficiency. Considering the significant suppression of HIV-1 viral replication, we next determined whether treatment of HIV-1–infected RAG-hu mice with the aptamer-DsiRNA conjugates could protect against HIV-1–mediated CD4+ T-cell depletion. We evaluated CD4+ T-cell levels in peripheral blood collected biweekly during and after treatment (Figure 4 and Supplementary Table S2a). For the HIV-negative mice, the level of CD4+ T cells remained stable within a 5% variation throughout the course of the experiments. In the animals that were infected with HIV-1 but not receiving treatment, viral infection lead to a steady decline in CD4+ T cells beginning at week 4 post-infection falling from initial levels of about 65% to below 35% at 18 weeks. When infected mice were treated with the aptamer-DsiRNA conjugates, the levels of CD4+ T cells remained near the levels of the uninfected mice and the CD4+ T cells remained stable well beyond the last treatment (~55%), indicating that the treatment also significantly impacted on CD4+ cell survival (P = 0.0348) (Supplementary Table S2b).
Figure 4.
Aptamer-cocktailed DsiRNA complexes protect against CD4+ T-cell loss in RAG-hu mice. CD4+ T-cell levels were assessed by FACS at each indicated week pre- and post-siRNA treatment. Start and end of treatments are indicated by the yellow framed in region. Mice from the first treatment are depicted in Figure 2a. Uninfected mice (n = 2), non-treated mice (n = 7), naked cocktailed DsiRNA-treated mice (n = 5), and aptamer-cocktailed DsiRNA conjugates treated mice (n = 8) are indicated. P values for the experiment are indicated and were determined as described in Materials and Methods and Supplementary Table S2b. DsiRNA, Dicer substrate small-interfering RNA; FACS, fluorescence-activated cell sorting.
Absence of type I IFN induction by aptamer-DsiRNA conjugates
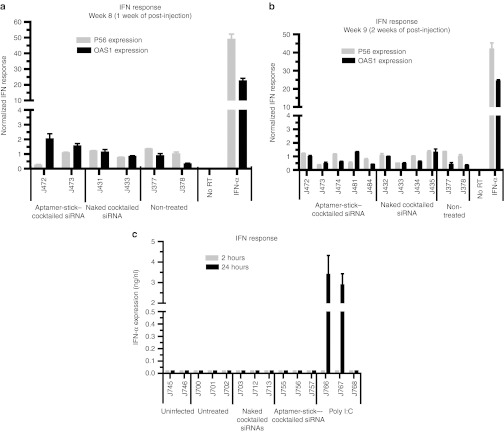
It had been reported earlier that synthetic siRNAs delivered via systemic administration can nonspecifically activate innate inflammatory cytokine production (tumor necrosis factor-α, interleukin-6, and interleukin-12) as well as interferon (IFN)-responsive genes, and this, in turn, can trigger undesirable cellular toxicity.35,36 Hence, we assessed the potential induction of type I IFN-mediated gene expression using a quantitative reverse transcription-PCR (qRT-PCR) expression assay on total RNAs derived from PBMCs isolated from the aptamer-siRNA conjugate treated and control mice. IFN-α treatment of PBMCs was used as a positive control to confirm upregulation of p56 and OAS1 gene expression. No significant differences in IFN-responsive gene expression were observed in RNAs isolated from treated or control animals at different times post-treatment (Figure 5a,b).
Figure 5.
In vivo administration of aptamer-cocktailed DsiRNA conjugates does not induce type I interferon (IFN). (a,b) The expression of type I IFN response genes (P56 and OAS1) at week (a) 8 and (b) 9 post-infection after treatment with naked cocktailed siRNAs and aptamer-cocktailed DsiRNA conjugates. IFN-α–treated, HIV-1–infected human PBMCs were used as a positive control. Gene expression was normalized to the gapdh mRNA. Error bars indicate SD (n = 4). (c) The expression levels of IFN-α at 2 or 24 hours after aptamer-cocktailed DsiRNA conjugates injections as measured by an ELISA are shown. Poly (I:C)-treated infected Rag-hu mice were used as a positive control. Error bars indicate SD (n = 3). DsiRNA, Dicer substrate small-interfering RNA; PBMC, peripheral blood mononuclear cell; RT, reverse transcription.
We also evaluated the innate IFN response within 2 and 24 hours after the injection of the conjugates by using an ELISA. IFN-α levels in HIV-1 uninfected and HIV-1–infected animals with or without treatment were monitored directly and no significant elevation of IFN-α was observed (Figure 5c). In contrast, IFN-α (>3 ng/ml) was induced in the control animals treated with poly I:C (polyinosine-polycytosine), a double-stranded RNA activator of innate IFN response.
Deep sequence analyses of viral envelope and tat/rev mRNAs
A major challenge in the treatment of HIV-1 infection is the rapid generation of drug-resistant mutations. It has been demonstrated that prolonged culturing of HIV-infected cells harboring expressed short hairpin RNAs often results in the evolution of escape variants that are resistant to the expressed short hairpin RNA.20,21,23,37 To determine whether or not treatment of HIV-1–infected animals with the aptamer-DsiRNA conjugates resulted in the evolution of escape mutants, we carried out deep sequencing analyses of the entire envelope gene as well as the HIV-1 tat/rev siRNA target sequence. Pooled viruses obtained from serum were collected from aptamer-DsiRNAs conjugate-treated animals at weeks 7 through 9 post-treatment. Illumina deep sequence analyses of viral RNAs (Supplementary Table S3) revealed multiple point mutations in the envelope sequences of the conjugate-treated mouse compared with the parental virus. The average frequency of unique point mutations compared with the parental virus was <3% (frequency is defined as the number of reads with mutations as a percentage of total reads). Supplementary Table S3 lists all the point mutations with a frequency above 1% in both the HIV-1 env-targeted domain and tat/rev-targeted domain, in which six and five mutations were detected in the gp120 and gp41 domain, respectively. Only four point mutations were found in the tat and rev shared exon, and no mutations were observed in the tat/rev siRNA target sequence (5982–6008 nt). These data suggest that the aptamer-multiplexed DsiRNA conjugate treatment did not select for viral escape mutants.
Discussion
The efficacy and sequence specificity of RNAi make this an attractive approach for the therapeutic treatment of a wide variety of human maladies ranging from cancer to infectious diseases.38,39,40,41 RNAi-based therapeutics can be multiplexed to prevent viral escape and both viral and cellular sequences can be targeted. Another class of therapeutic nucleic acids, aptamers, show promise as potential for targeted siRNA delivery.42,43 Aptamers specific to either CD424 or HIV-1 gp12025,26,27 have been successfully combined with anti-HIV-1 siRNAs to obtain a dual-inhibitory drug that is able to functionally deliver anti-HIV-1 siRNAs for target mRNA knockdown and either neutralize the virus (anti-gp120 aptamer) or block viral entry (CD4 aptamer).
We previously demonstrated that systemic administration of a dual functioning anti-gp120 aptamer-tat/rev siRNA chimera efficiently suppressed HIV-1 infection prevented helper CD4+ T cell decline in humanized mice.25 This aptamer-siRNA chimera was produced by T7 transcription, the conventional method for RNA aptamer production. For each siRNA fused to the aptamer, a different DNA transcription template has to be synthesized. In the present study, we took advantage of a chemically synthesized gp120 aptamer with a 3′ polycarbon linker fused to a backbond modified “sticky” sequence that allows the annealing of any siRNA duplex with a complementary sequence on one of the siRNA strands. Although several preclinical studies of aptamer-mediated siRNA targeting have been successful in the animal models of cancer and other diseases, siRNA activity is often compromised when it is covalently fused to an aptamer. It is necessary to develop guiding rules for designing active conjugates. To find out a better design configuration, we simply constructed two different design formats (named N-1 and N-2) and assessed their antiviral and gene-silencing activities using qRT-PCR and 5′-RACE PCR to monitor target knockdown. We observe that the orientation of the siRNA conjugated with aptamer impacts Dicer processing and mRNA cleavage, thus resulting in different RNAi activity and antiviral efficiency. The N-1 design in which 3′-end of the antisense strand of siRNA is appended to the complementary strand of the sticky bridge sequence was more effectively processed into a functional siRNA product with better RNAi potency than the N-2 design. Thus, we used the N-1 format with better RNAi potency to evaluate the in vivo efficacy of the aptamer-mediated delivery of three different DsiRNAs in vivo in a humanized mouse model of HIV-1 infection.
In order to avoid the ineffectiveness in RNAi-based HIV-1 therapy due to the generation of viral escape mutants, one of the strategies is to mitigate viral escape from RNAi by using multiple RNAi effectors. Our recent study44 has demonstrated that a dendrimer-mediated cocktail of anti-host and -viral DsiRNAs nanoparticle delivery system provided more viral suppression and T-cell protection in vivo than the dendrimer-individual siRNA nanoparticle. Herein, in addition to targeting conserved viral target (HIV Tat/rev transcript), we therefore chose to transiently inhibit the expression of two HDFs (CD4 and TNPO3) in combinatorial mode since CD4 is absolutely required for HIV-1 entry, and TNPO3 is required for viral integration. We have demonstrated that intravenous injections with the anti-gp120 aptamer-DsiRNA conjugates consisting of a cocktail of these three DsiRNAs downregulated all three targeted transcripts in vivo, suggesting that a combination of DsiRNAs can be functionally delivered in vivo by the same chemically synthetic RNA aptamer. Furthermore, the inhibition of these targets resulted in a marked suppression of HIV-1 viral loads and protection of CD4+ T-cells from HIV-mediated depletion. Five of the eight mice tested had undetectable viral loads even up to 3 weeks after the final treatment (Supplementary Table S1a), indicating sustained efficacy of the conjugates. Moreover, we show that viral rebounds which took place 3 months after cessation of the first treatment was fully suppressed by re-injection of the aptamer-DsiRNA conjugates. Importantly, this suppression lasted for an additional 3 weeks beyond the re-treatment period. Taken together, these data demonstrate a functional in vivo delivery of multiplexes anti-HIV-1 DsiRNAs via a chemically synthesized aptamer with a sticky bridge. Compared to a time-consuming, covalent aptamer-single siRNA chimera, the chemically synthesized aptamer and this “sticky brigde” strategy, the advantage of the synthetic RNA aptamer offer many favorable advantages, such as (i) large-scale manufacture with a relative lower cost; (ii) facile, noncovalent interchange of various siRNAs with the same/different aptamer portions; (iii) combinatorial RNAi effectors providing a safe and effective antiviral efficiency in the absence of viral resistance.
Materials and Methods
Materials. Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St Louis, MO), all restriction enzymes were obtained from New England BioLabs (Ipswich, MA) and all cell culture products were purchased from GIBCO (Gibco BRL/Life Technologies, a division of Invitrogen, Grand Island, NY). Random primers (Invitrogen, Carlsbad, CA); Lipofectamine 2000 (Invitrogen); Random primers (Invitrogen); HEK 293 and CCRF-CEM (ATCC); CHO-Env Transfectants (CHO-WT and CHO-EE); the HIV-1 NL4.3 and HIV-1 IIIB viruses were obtained from the AIDS Research and Reference Reagent Program.
siRNAs. DsiRNAs were purchased from Integrated DNA Technologies (Coralville, IA).
Anti-tat/rev Site I 27-mer: Sense: 5′-GCG GAG ACA GCG ACG AAG AGC UCA UCA-3′ antisense: 5′-UGA UGA GCU CUU CGU CGC UGU CUC CGC dTdT-3′.
Anti-CD4 27-mer: Sense: 5′-GAU CAA GAG ACU CCU CAG UGA GAA G-3′ antisense: 5′-CUU CUC ACU GAG GAG UCU CUU GAU CUG-3′ (2′-OMe modified U was underlined.)
Anti-TNPO3 27-mer: Sense: 5′-CGA CAU UGC AGC UCG UGU ACC AG dGdC-3′ antisense: 5′-GCC UGG UAC ACG AGC UGC AAU GUC GUU-3′.
Aptamer-siRNA conjugates. A-1-stick, stick-sense, and stick-antisense were chemically synthesized by the Synthetic and Biopolymer Chemistry Core in the City of Hope. And the corresponding stick-siRNAs and aptamer-stick-siRNAs were formed as previously described.45
A-1-stick: 5′-GGGAGGACGAUGCGGAAUUGAGGGACCACGCGCUGCUUGUUGUGAUAAGCAGUUUGUCGUGAUGGCAGACGACUCGCCCGA XXXXXXXGUACAUUCUAGAUAGCC-3′
Tat/rev N-1:
Antisense-stick: 5′-UGAUGAGCUCUUCGUCGCUGUCUCCGCXXXXXGGCUAUCUAGAAUGUAC-3′
Sense strand: 5′-GCGGAGACAGCGACGAAGAGCUCAUCAUU-3′
Tat/rev N-2:
Sense-stick: 5′-GCGGAGACAGCGACGAAGAGCUCAUCAXXXXXAGGAAGUGGCGCACCU-3′
Antisense strand: 5′-UGAUGAGCUCUUCGUCGCUGUCUCCGC UdT-3′
TNPO3 N-1:
Antisense-stick: 5′-UACACGAGCUGCAAUGUCGGCUUUGCU XXXXXGGCUAUCUAGAAUGUAC-3′
Sense strand: 5′-CAAAGCCGACAUUGCAGCUCGUGUAUU-3′
TNPO3 N-2:
Sense-stick: 5′-CGACAUUGCAGCUCGUGUACCAGGCXXXXXGGCUAUCUAGAAUGUAC-3′
Antisense strand: 5′-GCCUGGUACACGAGCUGCAAUGUCG UU-3′
CD4 N-2:
Stick-sense: 5′-UCAAGAGACUCCUCAGUGAGAAGAA XXXXXGGCUAUCUAGAAUGUAC-3′
Antisense strand: 5′-UUCUUCUCACUGAGGAGUCUCUUGAUU-3′
The underlined nucleotides corresponds to the stick sequence. The stick sequence contains 2′-OMe–modified A and G and 2′-F–modified U and C. The 2′-F–modified U and C are highlighted as bolded text. The italic X indicates the 3 carbon linker (C3) between the aptamer/siRNA and stick sequences.
Generation and HIV-1 infection of humanized Rag2−/−γc−/− mice (RAG-hu mice). Humanized BALB/c-Rag2−/−γc−/− mice were prepared as previously described using human fetal liver-derived CD34+ cells.28,46 Briefly, neonatal mice were conditioned by irradiating at 350 rads and then injected intrahepatically with 0.5–1 × 106 human CD34+ cells. Approximately 12 weeks post-reconstitution, mice were screened for human cell engraftment. Blood was collected by tail bleeds, and RBCs were lysed using the Whole Blood Erythrocyte Lysing Kit (R&D Systems, Minneapolis, MN). The white blood cell fraction was stained with antibodies against the human pan-leukocyte marker CD45 (Caltag, a division of Invitrogen, Grand Island, NY) and FACS analyzed as described. To infect human cell reconstituted RAG-hu mice, HIV-1 NL4-3 (1.2 × 105 viral load IU/ml) in a 100 µl volume was injected intraperitoneally at least 12 weeks after cell engraftment. Viral loads were examined weekly and viremia was established in all the mice by 4 weeks. Treatment was done by intravenous injection on the last day of week 4 with 0.25 nmol experimental RNAs (4.6 µg cocktailed DsiRNAs including equal amount of three DsiRNAs: tat/rev DsiRNA, TNPO3 DsiRNA, and CD4 DsiRNA, or 14.1 µg A-1-stick–cocktailed DsiRNAs conjugates with equal amount of three DsiRNA portions) in a 40 µl volume, followed by another the next day. Later, the injections were continued on a weekly basis for 4 weeks. In the second in vivo treatment experiment, 0.25 nmol conjugates in a 40 µl volume were administered at 23 and 24 weeks of post-infection like above.
Measurement of viral load in plasma. To quantify cell-free HIV-1 by qRT-PCR, RNA was extracted from 25 to 50 µl of EDTA-treated plasma using the QIAamp Viral RNA kit (QIAGEN, Valencia, CA). cDNAs were produced with Superscript III reverse transcriptase (Invitrogen) using a primer set specific for the HIV-1 long terminal repeat sequence, and qPCR was performed with the same primer set and a long terminal repeat-specific probe using Supermix UDG (Invitrogen) as described.
Flow cytometry. Whole blood was collected and RBCs were lysed as reported previously. Peripheral blood cells were stained for hCD3-PE and hCD4-PECy5 (Caltag) markers and analyzed using a Coulter EPICS XL-MCL FACS analyzer (Beckman Coulter, Brea, CA). CD4+ T-cell levels were calculated as a ratio of the entire CD3 population (CD4 + CD3+:CD4 − CD3+). To establish baseline CD4+ T-cell ratios, all mice were analyzed before infection.
Detection of tat/rev siRNA. At 1, 3, and 9 weeks post-injection, blood samples were collected and small RNAs were isolated with MirVana miRNA isolation Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instruction. The siRNA quantification was performed using TaqMan MicroRNA Assay according to manufacturer's recommended protocol (Applied Biosystems). Ten nanogram of small RNA, 0.2 µmol/l stem-loop RT primer, RT buffer, 0.25 mmol/l dNTPs, 3.33 units/ml MultiScribe RT, and 0.25 units/ml RNase inhibitor were used in 15 µl RT reactions for 30 minutes at 16 °C, 30 minutes at 42 °C, and 5 minutes at 85 °C, using the TaqMan MicroRNA reverse transcription Kit (Applied Biosystems). For real-time PCR, 1.33 µl of cDNA, 0.2 mmol/l TaqMan Probe, 1.5 mmol/l forward primer, 0.7 mmol/l reverse primer, and TaqMan Universal PCR Master Mix were added in 20 µl reactions for 10 minutes at 95 °C and 40 cycles of 15 seconds at 95 °C and 1 minute at 60 °C. All real-time PCR experiments were done using an iCycler iQ system (Bio-Rad, Hercules, CA). Primers were as follows: Site I Looped RT primer: 5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA CAG CG-3′ Site I forward primer: 5′-GCT GAT GAG CTC TTC GTC G-3′ Site I reverse primer: 5′-GTG CAG GGT CCG AGG T-3′ Site I probe primer: 5′-6- FAM- TCG CAC TGG ATA CGA CAC AGC GAC GA –BHQ1-3′. In this case, a synthetic 27-mer duplex RNA was used as positive control.
Assays of targeted gene expression. Human PBMCs were obtained from transplanted mice at 1 and 3 weeks post-injection and total RNAs were isolated with STAT-60 (TEL-TEST “B”) according to the manufacturer's instructions. Residual DNA was digested using the DNA-free kit per the manufacturer's instructions (Ambion, a division of Invitrogen, Grand Island, NY). cDNA was made using 2 µg of total RNA. RT was carried out using Moloney murine leukemia virus reverse transcriptase and random primers in a 15 µl reaction according to the manufacturer's instructions (Invitrogen). Expression of the tat/rev coding RNAs was analyzed by qRT-PCR using 2× iQ SyberGreen Mastermix (BIO-RAD) and specific primer sets at a final concentration of 400 nmol/l. Gapdh expression was used for normalization of the qPCR data. Primers were as follows: IIIB or NL4-3 tat/rev forward primer: 5′-GGC GTT ACT CGA CAG AGG AG -3′ IIIB or NL4-3 tat/rev reverse primer: 5′-TGC TTT GAT AGA GAA GCT TGA TG-3′ CD4 forward primer: 5′-GCT GGA ATC CAA CAT CAA GG-3′ CD4 reverse primer: 5′-CTT CTG AAA CCG GTG AGG AC-3′ TNPO3 forward primer: 5′-CCT GGA AGG GAT GTG TGC-3′ TNPO3 reverse primer: 5′-AAA AAG GCA AAG AAG TCA CAT CA-3′ gapdh forward primer 1: 5′-CAT TGA CCT CAA CTA CAT G-3′ gapdh reverse primer 2: 5′-TCT CCA TGG TGG TGA AGA C-3′.
Interferon assays. Total RNA was isolated from PBMCs of treated mice using STAT-60. Expression of mRNAs encoding p56 (CDKL2) and OAS1 were analyzed by qRT-PCR using 2× iQ SyberGreen Mastermix (BIO-RAD) as described above and specific primer sets for these genes at final concentrations of 400 nmol/l. Primers were as follows: P56 (CDKL2) forward, 5′-TCA AGT ATG GCA AGG CTG TG-3′ P56 (CDKL2) reverse, 5′-GAG GCT CTG CTT CTG CAT CT-3′ OAS1 forward, 5′-ACC GTC TTG GAA CTG GTC AC-3′ OAS1 reverse, 5′-ATG TTC CTT GTT GGG TCA GC-3′ gapdh expression was used for normalization of the qPCR data.
To measure any induced IFN-α directly, human IFN-α1 ELISA Ready-SET-Go! (eBioscience, San Diego, CA) was used. Mice were injected with the naked cocktailed DsiRNA or ptamer-cocktailed DsiRNA conjugates. At 2 and 24 hours post-treatment, 25–50 µl of EDTA-treated plasma was collected from three mice per treatment group and from three positive control mice which had been intravenously injected with 5 µg of poly (I:C) (Sigma-Aldrich) in a 50 µl volume. Plasma levels were evaluated as per instructions supplied in the kit.
Statistical methods. The mouse viral loads and CD4:CD3 T-cell ratios were plotted by using a Lowess smoother across values. Viral loads were first log-transformed before smoothing and then anti-transformed for plotting. Missing values were imputed with a last observation carried forward scheme. The calculations were conducted as previously described.25,47
SUPPLEMENTARY MATERIAL Figure S1. HIV-1 suppression by chemically synthesized aptamer (A-1-stick) and aptamer-stick-siRNA conjugates. Figure S2. Chemically synthesized aptamer-stick-siRNA conjugates are processed by Dicer and induce mRNA cleavage. Figure S3. 2′-Fluoro–modified aptamer-stick-siRNA conjugate in mouse serum. Table S1. HIV-1 viral loads in individual RAG-hu mice. Table S2. CD4 T-cell levels in individual treated and control RAG-hu mice. Table S3. Sequence analysis of mutant virus from animals treated with aptamer-stick–cocktailed DsiRNA conjugates. Materials and Methods.
Acknowledgments
We thank National Institutes of Health AIDS Research and Reference Reagents Program for HIV-1 related reagents used in this work. The authors would like to thank City of Hope DNA sequencing core (Harry Gao and Jinhui Wang) for Solexa Deep Sequencing, City of Hope Bioinformatics Core facility (Xiwei Wu and Haiqing Li) for prompt data analyses, and John Burnett for critical reading of the manuscript. This work was supported by National Institutes of Health RO1 grants AI057066 and AI073255 to R.A., AI29329, AI42552, and HL07470 to J.J.R. This work has also been facilitated by the infrastructure and resources provided by the Colorado Center for AIDS Research grant P30 AI054907. J.J.R. and J.Z. have an issued patent on “Cell-type specific aptamer-siRNA delivery system for HIV-1 therapy”. Patent no.: US8,222,116 B2, issued date: 17 July 2012. The other authors declared no conflict of interest.
Supplementary Material
HIV-1 suppression by chemically synthesized aptamer (A-1-stick) and aptamer-stick-siRNA conjugates.
Chemically synthesized aptamer-stick-siRNA conjugates are processed by Dicer and induce mRNA cleavage.
2′-Fluoro–modified aptamer-stick-siRNA conjugate in mouse serum.
HIV-1 viral loads in individual RAG-hu mice.
CD4 T-cell levels in individual treated and control RAG-hu mice.
Sequence analysis of mutant virus from animals treated with aptamer-stick–cocktailed DsiRNA conjugates.
REFERENCES
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D., and, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE., and, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA., and, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Rossi JJ. RNAi as a treatment for HIV-1 infection. BioTechniques. 2006. pp. 25–29. [DOI] [PubMed]
- Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK.et al. (2002siRNA-directed inhibition of HIV-1 infection Nat Med 8681–686. [DOI] [PubMed] [Google Scholar]
- Boden D, Pusch O., and, Ramratnam B. HIV-1-specific RNA interference. Curr Opin Mol Ther. 2004;6:373–380. [PubMed] [Google Scholar]
- Martínez MA. Progress in the therapeutic applications of siRNAs against HIV-1. Methods Mol Biol. 2009;487:343–368. doi: 10.1007/978-1-60327-547-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK., and, Gaur RK. Progress towards therapeutic application of RNA interference for HIV infection. BioDrugs. 2009;23:269–276. doi: 10.2165/11317120-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Tsygankov AY. Current developments in anti-HIV/AIDS gene therapy. Curr Opin Investig Drugs. 2009;10:137–149. [PubMed] [Google Scholar]
- Podlekareva D, Mocroft A, Dragsted UB, Ledergerber B, Beniowski M, Lazzarin A.et al. (2006Factors associated with the development of opportunistic infections in HIV-1-infected adults with high CD4+ cell counts: a EuroSIDA study J Infect Dis 194633–641. [DOI] [PubMed] [Google Scholar]
- Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J.et al. (2005Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy Mol Ther 12900–909. [DOI] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ.et al. (2008Identification of host proteins required for HIV infection through a functional genomic screen Science 319921–926. [DOI] [PubMed] [Google Scholar]
- König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT.et al. (2008Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication Cell 13549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC.et al. (2008Genome-scale RNAi screen for host factors required for HIV replication Cell Host Microbe 4495–504. [DOI] [PubMed] [Google Scholar]
- Haasnoot J, Westerhout EM., and, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL.et al. (2008T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice Cell 134577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM.et al. (2005Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors Nat Biotechnol 23709–717. [DOI] [PubMed] [Google Scholar]
- Kim SS, Peer D, Kumar P, Subramanya S, Wu H, Asthana D.et al. (2010RNAi-mediated CCR5 silencing by LFA-1-targeted nanoparticles prevents HIV infection in BLT mice Mol Ther 18370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopman NC, ter Brake O., and, Berkhout B. Anticipating and blocking HIV-1 escape by second generation antiviral shRNAs. Retrovirology. 2010;7:52. doi: 10.1186/1742-4690-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eije KJ, ter Brake O., and, Berkhout B. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J Virol. 2008;82:2895–2903. doi: 10.1128/JVI.02035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R.et al. (2004Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition J Virol 782601–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden D, Pusch O, Lee F, Tucker L., and, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler LA, Trifonova R, Vrbanac V, Basar E, McKernan S, Xu Z.et al. (2011Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras J Clin Invest 1212401–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H.et al. (2011An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice Sci Transl Med 366ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R.et al. (2009Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells Nucleic Acids Res 373094–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J., and, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges BK, Akkina SR, Folkvord JM, Connick E., and, Akkina R. Mucosal transmission of R5 and X4 tropic HIV-1 via vaginal and rectal routes in humanized Rag2-/- gammac -/- (RAG-hu) mice. Virology. 2008;373:342–351. doi: 10.1016/j.virol.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton PW., and, Garcia JV. Novel humanized murine models for HIV research. Curr HIV/AIDS Rep. 2009;6:13–19. doi: 10.1007/s11904-009-0003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand N, Ploss A, Balling R, Becker PD, Borsotti C, Brezillon N.et al. (2009Humanized mice for modeling human infectious disease: challenges, progress, and outlook Cell Host Microbe 65–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne R, Pedati C, Guendel I, Carpio L, Kehn-Hall K, Saifuddin M.et al. (2009The utilization of humanized mouse models for the study of human retroviral infections Retrovirology 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP., and, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G., and, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Moir S., and, Fauci AS. B cells in HIV infection and disease. Nat Rev Immunol. 2009;9:235–245. doi: 10.1038/nri2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S.et al. (2005Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7 Nat Med 11263–270. [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K., and, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Shah PS, Pham NP., and, Schaffer DV. HIV develops indirect cross-resistance to combinatorial RNAi targeting two distinct and spatially distant sites. Mol Ther. 2012;20:840–848. doi: 10.1038/mt.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH., and, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- de Fougerolles A, Vornlocher HP, Maraganore J., and, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D., and, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer L, Rossi JJ., and, Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther. 2007;14:1057–1064. doi: 10.1038/sj.gt.3302977. [DOI] [PubMed] [Google Scholar]
- Zhou J., and, Rossi JJ. Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides. 2011;21:1–10. doi: 10.1089/oli.2010.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., and, Rossi JJ. Aptamer-targeted RNAi for HIV-1 therapy. Methods Mol Biol. 2011;721:355–371. doi: 10.1007/978-1-61779-037-9_22. [DOI] [PubMed] [Google Scholar]
- Zhou J, Neff CP, Liu X, Zhang J, Li H, Smith DD.et al. (2011Systemic administration of combinatorial dsiRNAs via nanoparticles efficiently suppresses HIV-1 infection in humanized mice Mol Ther 192228–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R.et al. (2009Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells Nucleic Acids Res 373094–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges BK, Wheat WH, Palmer BE, Connick E., and, Akkina R. HIV-1 infection and CD4 T cell depletion in the humanized Rag2-/-gamma c-/- (RAG-hu) mouse model. Retrovirology. 2006;3:76. doi: 10.1186/1742-4690-3-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitberg B., and, Röhmel J. Exact distributions for permutation and rank tests: an introduction to some recently published algorithms. Statistical Software Newsletter. 1986;12:10–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HIV-1 suppression by chemically synthesized aptamer (A-1-stick) and aptamer-stick-siRNA conjugates.
Chemically synthesized aptamer-stick-siRNA conjugates are processed by Dicer and induce mRNA cleavage.
2′-Fluoro–modified aptamer-stick-siRNA conjugate in mouse serum.
HIV-1 viral loads in individual RAG-hu mice.
CD4 T-cell levels in individual treated and control RAG-hu mice.
Sequence analysis of mutant virus from animals treated with aptamer-stick–cocktailed DsiRNA conjugates.