Abstract
Bacteria, such as Escherichia coli and Caulobacter crescentus, are the most studied and perhaps best-understood organisms in biology. The advances in understanding of living systems gained from these organisms are immense. Application of single-molecule techniques in bacteria have presented unique difficulties owing to their small size and highly curved form. The aim of this review is to show advances made in single-molecule imaging in bacteria over the past 10 years, and to look to the future where the combination of implementing such high-precision techniques in well-characterized and controllable model systems such as E. coli could lead to a greater understanding of fundamental biological questions inaccessible through classic ensemble methods.
Keywords: bacteria, single molecule, imaging, biophysics, review
1. Introduction
While much of the early work pertaining to imaging single molecules in cells has been performed in mammalian cells, a growing amount of the single-molecule studies have begun to be performed in bacterial cells. Part of the allure of developing single-molecule techniques in bacteria, and in particular the Gram-negative bacteria Escherichia coli, is to be able to draw on the vast knowledge of these model organisms in designing experiments and interpreting results. Imaging single molecules in small bacterial cells does present technical challenges. Their small size requires higher observation rates as fast diffusing molecules tend to reach the boundaries of the cell in a short time [1]. Furthermore, single-molecule imaging of membrane-bound molecules requires an analysis that is mindful of the curvature of the outer envelope. As technology advances, such difficulties lessen and the advantages of the use of model organisms become more attractive.
This review will look at single-molecule imaging in live cells with an emphasis on the use of this technique in model bacterial cells, such as E. coli. This review will not try to be comprehensive, but instead give an overview of the breadth of single-molecule research in bacteria. The review will look at techniques used to image single molecules, and then look at some of the results that have been published regarding mobility and interactions measured in live cells with single-molecule sensitivities. Note that this review will mostly limit its scope to single-molecule imaging experiments performed in bacterial cells, and as such not mention much of the wide range of beautiful single-molecule experiments performed in eukaryotic cells and the seminal works performed in vitro on molecular motors, DNA mechanics, single-molecule bonds, protein folding/unfolding, etc.
2. A brief history of single-molecule imaging
The era of single-molecule experiments began in 1970 when Ehrenstein, Lecar and Nossal displayed the first, to our knowledge, trace of the discrete opening and closing of individual ion channels when they doped lipid bilayers with ‘sufficiently small amounts of excitability-inducing material’. As is stated in this publication, ‘ … we report on properties of membranes with two, three, and five levels. If membranes have individual channels that can be open or closed, the corresponding number of channels are one, two, and four’ [2, p. 123]. With this, the deterministic world of analysing the trajectory of the mean of an ensemble was supplemented with the probabilistic world of observing individual molecular events.
Direct single-molecule imaging experiments began with the seminal work of Moerner & Kador in 1989 [3], where single molecules of pentacene were observed in a solid p-terphenyl crystal at liquid-helium temperatures. This work showed that the actual observation of single molecules was possible. Over the next few years much progress was made. Single, immobilized, fluorescent dye molecules were first observed at room temperature under ambient conditions using near-field optical techniques in 1993 [4]. In 1996, the first wide-field imaging of single dye molecules labelling lipids in a fluid bilayer was achieved [5].
Around the same time, another approach was being employed to view molecular motions. Building on earlier techniques, gold nanoparticles were used to label single (or few) molecules and were observed in contrast enhanced brightfield microscopy [6,7]. While criticized for the size of the label (compared with the size of a protein) and for its possible multivalancy, these probes were much easier to image and allowed early work to extend the frequency of observation to several orders-of-magnitude higher than has yet been achieved by fluorescence labelling [8].
In general, early work in single-molecule imaging in live cells began with imaging in mammalian cells. Several reasons for this include not only the obvious direct relation between such cells and studies of human health but also technical reasons. Many standard, immortalized mammalian cells can be plated onto a coverglass and will grow into large (sizes approx. 20 µm), relatively flat, ‘pancake-like’ forms. This facilitated the use of advanced microscopy techniques such as total internal reflection fluorescence (TIRF) microscopy to reduce background, and thus enhance detection of dim single fluorescently labelled molecules in or near the flat membrane. As detection sensitivity increased through better camera sensors and imaging techniques, the need for ideal substrates was relaxed. Furthermore, the observation rate increased allowing high-speed imaging of single fluorescent molecules in live cells.
3. Single-molecule techniques
(a). Microscopy
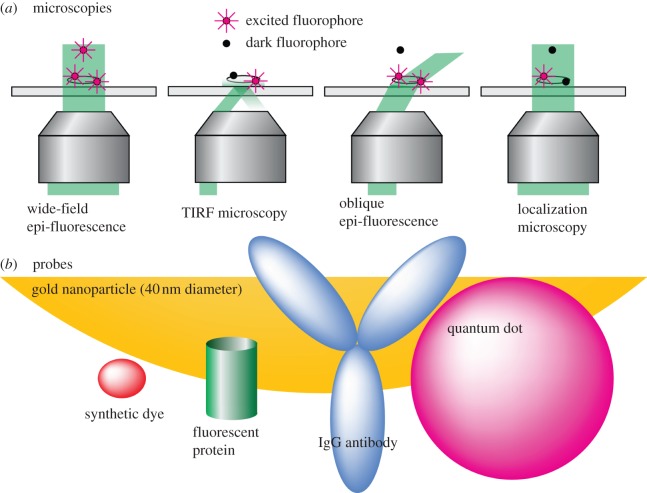
By far, most techniques developed for performing single-molecule imaging experiments in live cells employ some variant of light microscopy (figure 1). Many single-molecule studies, both in vitro and in vivo, use TIRF microscopy, where an illumination laser is brought to the glass–media interface at an angle greater than that required for total internal reflection. This produces an evanescent field in the media that exponentially decays over approximately 100–200 nm. However, the small, highly curved form of most bacteria precludes the use of TIRF microscopy owing to its limited depth of illumination.
Figure 1.
Schematic of microscopies and probes used in single-molecule experiments. (a) Microscopies: different types of microscopy have different depths of excitation and different methodologies for reducing excess fluorescence. Shown are standard epi-fluorescence, TIRF, oblique angle epi-fluorescence and localization microscopies. (b) Probes: shown to approximate scale are the sizes of common fluorescent probes, including synthetic dyes (such as cyanine, Alexa or Atto dyes), fluorescent proteins, quantum dots (approx. 5–10 nm diameter) and gold nanoparticles (greater than 40 nm diameter). For comparison, an IgG antibody is also shown to approximate scale.
For bacteria, usually a laser-based epi-illumination scheme is employed. Some alter the beam entering a TIRF microscope set-up such that the illumination beam is slightly outside the condition required for total internal reflection, thus illuminating the cell at a high angle [9,10]. This allows for full illumination of small cells such as bacteria and also allows for illumination of the apical membrane of larger eukaryotic cells without exciting excess fluorescent matter in the sample volume above the cell.
A technique that is being recently employed to many different systems is that of single-molecule localization or ‘super-resolution’ microscopy, where the high localization accuracy of single-molecule imaging [11] is exploited to build, over time, an image that seems to break the resolution limit. Among these techniques are stochastic reconstruction microscopy (STORM) [12], photoactivated localization microscopy (PALM) [13] and fluorescence PALM [14]. These techniques rely on the ability of certain fluorescent probes to be activated or re-activated by a pulse of light (see §3b). Overly labelled structures can be outlined by selective (though random) (re-)activation of a sparse number of the labels. These probes are imaged until bleached and a subsequent pulse turns on another random small set of probes. During each imaging phase, the centres of the well-spaced probes can be determined to a precision much greater than the resolution of the microscope. The image is built from a construction of the positions of the centres of the probes after many cycle of activation/image/bleach.
(b). Probes
The most common probes in single-molecule imaging are fluorescent dyes/fluorescent proteins, though nanoparticles such as quantum dots and even small colloidal gold and latex particles have been used (figure 1).
For labelling of externally expressed molecules such as on the outer leaflet of the outer membrane of Gram-negative bacteria, bath application of antibodies or ligands with attached synthetic dyes or nanoparticles can be used. This allows for a great range of dye characteristics to be specifically chosen for a specific experiment (i.e. excitation/emission wavelengths, brightness, photostability, etc.). For labelling of cytoplasmic molecules (or cytoplasmic domains of membrane molecules), one usually defaults to employing fluorescent proteins [15] as the label of choice. Fluorescent proteins have the advantage that they may be directly linked to the protein under consideration, and its expression can be finely controlled genetically. Currently, the range of characteristics available for fluorescent proteins, though more limited than that for synthetic dyes, is still large enough for their widespread use in both single-molecule and bulk multi-colour imaging.
Photoactivatable (or re-activatable) and photoswitchable fluorescent dyes and proteins are key to many localization microscopy techniques [16] and have recently found use in classic single-molecule imaging experiments. Photoactivatable dyes allow for activation or re-activation after photobleaching when excited by a specific wavelength usually different from their fluorescence excitation maxima, usually in the UV region of the spectrum. Photoswitchable dyes switch between two or more states with different excitation/emission profiles. Such dyes allow for over-labelling or expression since the experimenter has control of turning a small portion on when desired. While being used extensively in localization microscopy techniques, they are being used in traditional single-molecule imaging studies in bacteria where overlapping of the point-spread function of probes can become a difficulty at even small probe numbers in the confined space of the bacteria.
(c). Analyses
Central to most single-molecule imaging techniques is the ability to localize the position of a single molecule in an image obtained from the microscope. It has been shown that, given enough photons above background noise, a single molecule can be localized to nanometre precision [11]. After localization, either an image is developed as in localization microscopies or the dynamics of the molecule is tracked between subsequent frames of a video of the dye's motion.
Analysis of single-molecule motions is more complicated in the small, highly curved bacteria cell. Perhaps the best method at present to analyse the motion of molecules in either the membranes or cytoplasm of a small bacteria cell is through comparison with computer simulations of such motion, to remove effects of confinement to the cell surface/interior [9,17]. As experiments continue to test more complicated interactions between molecules within bacterial membranes and in the cytoplasm, it is expected that the use of more sophisticated simulations will be required to understand the measured results.
4. Mobility in bacteria
It is interesting to note that one simplification that arises with the use of bacteria as a model cell type is that owing to the small size of bacteria cells, molecules find each other through simple random walks in either the two-dimensional membranes or the three-dimensional cytoplasm, without the need to have developed active transport systems such as have been developed by cells from higher organisms. Thus before one expects to understand interactions in bacteria, one must first look at molecular mobility in the membranes and cytoplasm.
(a). Outer bacterial membrane
The study of the mobility of membrane proteins in E. coli began with imaging the diffusion of outer membrane proteins, most probably owing to the ease of access to their extracellular domains for labelling. The outer membrane of Gram-negative bacteria presents an interesting and unique bilayer consisting primarily of phospholipid and lipopolysaccharide in its inner and outer leaflet, respectively [18]. The outer membrane acts as the first barrier between the cell and the environment and as such plays critical roles in protecting the cell, in detecting its environment and in scavenging scarce nutrients. Many outer membrane proteins contain a β-barrel membrane spanning region in order to allow passage of small molecules into the periplasm where they can access the inner membrane [19].
The earliest study of outer membrane protein mobility was performed by Oddershede et al. [20], observing the diffusion of λ-receptor (LamB), the maltodextrin transport channel in E. coli, using single-particle tracking in conjunction with a weak optical trap. The local short-time diffusion coefficient for the λ-receptor was measured by attaching a 530 nm microsphere to the protein, and imaging the probe's motion at 25 Hz in the presence of weak laser optical tweezers. They found a short-time diffusion coefficient of 0.15 ± 0.10 μm2 s−1. In the absence of the laser trap, no long-range motion was observed. Instead, the λ-receptor displayed confinement in a region of diameter 29 nm.
In 2004, Gibbs et al. [21] also looked at the mobility of the λ-receptor employing 20-nm diameter gold labels imaged at 1 Hz. In this study, Gibbs et al. classified the motion of the λ-receptor into two populations, a slow (approx. 60%) and a fast (approx. 40%)-moving population. The slow population displayed confinement in domains of size 20–50 nm for the entire 5 min of observation, consistent with the findings of Oddershede et al. [20]. The faster population made excursion of 100–300 nm during the 5 min of observation, implying a long-time diffusion coefficient of 10–4–10–5 μm2 s–1.
Recently, our group published a study of the diffusion of the outer membrane cobalamin receptor, BtuB, and the outer membrane pore, OmpF, which are implicated in the dynamic formation of a translocon for the bacteriotoxin colicin E3 [10]. In this, BtuB and OmpF were labelled by fluorescently tagged polyclonal antibodies to the outer membrane proteins and imaged at 40 Hz. The diffusion coefficients of antibody labelled BtuB and OmpF were measured to be 0.05 ± 0.01 and 0.006 ± 0.002 μm2 s–1, respectively (figure 2). Furthermore, a mutation to BtuB to hinder its interaction with its cytoplasmic membrane protein partner, TonB, increased the diffusion coefficient of BtuB to 0.27 ± 0.06 μm2 s–1 implying interactions between TonB and BtuB influence the mobility of BtuB. BtuB was found to diffuse an average distance of 190 nm in 0.25 s showing a high long-range mobility, while OmpF was found to be confined in a domain of diameter approximately 100 nm consistent with the earlier results on λ-receptor confinement.
Figure 2.
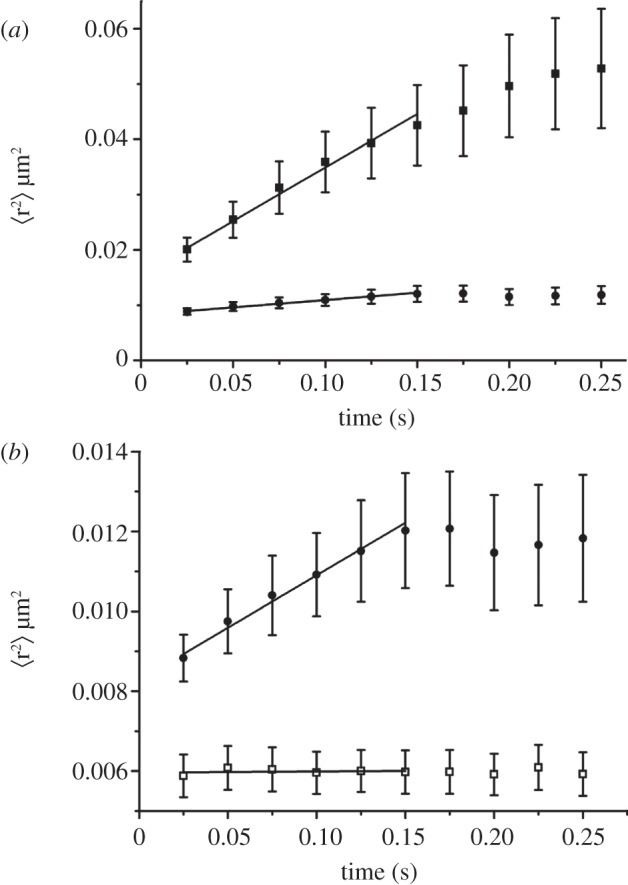
Mean square displacement of proteins diffusing in the outer membrane of E. coli. Each curve is an average curve over many single-molecule experiments tracking individual molecules. The short-time linear region is used to estimate the molecule's diffusion coefficient. (a) Time dependence of the mean square displacement of the cobalamin receptor BtuB (closed squares) and the outer membrane pore OmpF (closed circles). Both molecules were imaged using a low concentration of antibodies labelled with Alexa Fluor 555. (b) Expanded view of the OmpF curve from (a) compared with the mean square displacement measured for anti-BtuB immobilized on the coverglass as a standard for the precision of the measurement. Reprinted with permission from Spector et al. [10].
(b). Inner bacterial membrane
The inner or cytoplasmic bacterial membrane is a regular phospholipid-based membrane, unlike the outer membrane. It is in close contact with both the periplasm and the cytoplasm and as such has a role in material transport into/out of the cell interior. The inner membrane contains proteins involved in small molecule transport, protein translocation and other membrane-bound processes [19].
The earliest single-molecule imaging of inner membrane proteins in Gram-negative bacteria was performed by the Moerner group in 2004 on the histidine kinase PleC labelled with enhanced yellow fluorescent protein (EYFP) in Caulobacter crescentus [17]. Therefore, freely diffusing PleC-EYFP molecules were measured to diffuse with a diffusion coefficient of 0.012 ± 0.002 μm2 s–1.
Leake et al. [22], as part of a study on the turnover of the flagella motor proteins (see also §6), determined the diffusion coefficient of the flagella motor protein MotB labelled by green fluorescent protein (GFP). In that study, the diffusion coefficient of free MotB-GFP (i.e. outside the motor complex) was found to be 0.0088 ± 0.0026 μm2 s–1. Furthermore, in another study, Leake et al. [23] monitored the mobility of the inner membrane protein TatA in the twin-arginine translocation (Tat) system while complexed to the other Tat proteins. By labelling TatA with YFP they found that the diffusion coefficient, when corrected for the curvature of the membrane, decreased with increasing size of the Tat complex, ranging from about 0.1 μm2 s–1 for complexes with about 10 TatA-YFP molecules to less than 0.01 μm2 s–1 for complexes of near 100 TatA-YFP molecules.
(c). The cell cytoplasm
The cell cytoplasm is the inside of the cell and it contains a highly crowded [24] and heterogeneously distributed set of proteins, ribosomes, RNA and DNA. In this environment, molecules find each other to perform tasks simply through diffusion in bacteria, unlike in some larger cell types where active transport is used. As such, it is important to understand how proteins diffuse in the cytosol in order to understand reaction kinetics inside cells.
While earlier studies have looked at the mobility of proteins in the cytoplasm of E. coli and other bacteria using bulk techniques, such as fluorescence recovery after photobleaching (FRAP), recent advances in high-speed low-light-level video microscopy now allow direct imaging of the motion of individual molecules in the three-dimensional cytoplasm of the cell. For comparison, FRAP studies found that the diffusion coefficient of a fluorescent protein expressed in the cytoplasm of E. coli ranged from 6 to 14 μm2 s–1 which is an order-of-magnitude below the diffusion coefficient of GFP in pure water, 88 μm2 s–1 [25].
The earliest single-molecule study of diffusion in a bacterial cytoplasm looked at the mobility of the cytoplasmic structural protein MreB labelled with yellow fluorescent protein (YFP) in C. crescentus [26]. Using an observation rate of 65 Hz and correcting the results for the shape of the C. crescentus cells, Kim et al. [26] found that the MreB-YFP construct had a diffusion coefficient of 1.75 ± 0.17 μm2 s–1. MreB-YFP was also found to undergo simple Brownian motion over the observation time.
Niu & Yu [27] looked at the mobility of another bacterial structural protein, the tubulin homologue FtsZ tagged with the photoactivatable fluorescent protein dendra2. FtsZ-dendra2 in E. coli could be separated into a significant sized immobile fraction, most probably involved in Z-ring formation near the cell centre, and a mobile fraction. The mobile fraction was found to travel throughout the cell but was restricted to a helical path. The movement was analysed in terms of anomalous sub-diffusion with a time dependence of the mean square displacement that scales as time to the power 0.74. Niu & Yu remark that FtsZ explores the entire cell similarly to a free molecule with diffusion coefficient 0.1 μm2 s–1.
Recent single-molecule studies have measured diffusion of inert fluorescent proteins in cytoplasm at a higher sampling rate of 250 Hz. English et al. [28] employed a photoconvertible GFP variant, mEos2, as a reference to study RelA, a ribosome-dependent (p)ppGpp synthetase, and ribosomes. They obtained an apparent diffusion coefficient for mEos2 of 8–16 μm2 s–1 depending on the location within the cell. Using simulations taking into consideration the effect of cell geometry, they calculated the microscopic diffusion coefficient of mEos2 to be 13 μm2 s–1. They also found that the enzymatically active RelA had a similar diffusion behaviour to free mEos2 with confinement corresponding to the size of the cells, whereas inactive RelA diffuses like a ribosomal protein, L25, at a much slower rate, about 0.5 μm2 s–1, showing a confinement much smaller than the cytoplasm.
Bakshi et al. [29] reported a mean diffusion coefficient of 7.3 ± 1.1 μm2 s–1 for the photoactivatable fluorescent protein Kaede in the E. coli cytoplasm at 250 Hz. They used the localization microscopy technique of PALM to determine the shape of the cells and investigated the spatial dependency of the diffusion rate in the cytoplasm. They showed that while the distribution of Kaede is relatively homogeneous, the mobility of free diffusion is lower in the ribosome-rich region between nucleoid lobes especially in longer cells, which are probably in a later stage of their cell cycles.
We recently published the full mean square displacement versus time plot for the diffusion of free GFP in the E. coli cytoplasm from 1 ms to 0.2 s [9]. This was done by performing tracking at observation rates of 60, 120, 260, 400 and 1000 Hz and ‘stitching’ together the full plot (figure 3). Analysis was done through comparison with simulations of diffusion in model E. coli shells whose size was distributed as those used in the actual experiments. We found that the entire curve could be simulated assuming that the GFP molecules underwent simple Brownian motion at all time scales above 1 ms with an actual diffusion coefficient of 5.6 μm2 s–1, where the curvature seen in the mean square displacement was entirely accounted for by the geometry of the cells. Thus, at time scales over 1 ms or, equivalently, length scales over approximately 100 nm, a free GFP molecule ‘sees’ the cytoplasm of E. coli as a simple fluid with no internal structure in our study. Below this time scale, one must expect the granularity of the cytoplasm coupled with possibly short-time non-specific interactions is the reason for the 10-fold reduction in the diffusion coefficient of GFP in the cytoplasm compared with that in pure water.
Figure 3.
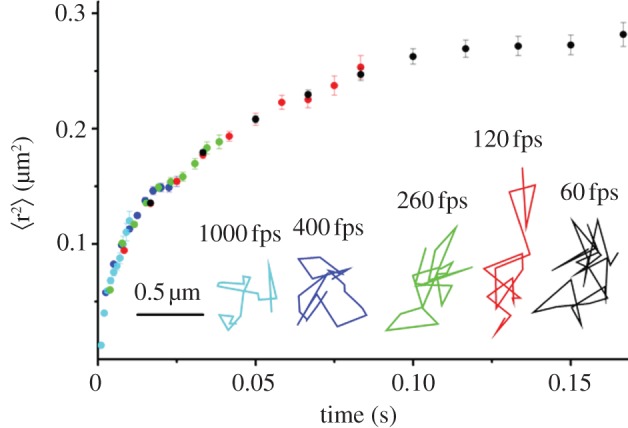
Averaged mean square displacement of free GFP in the cytoplasm of E. coli. The full plot was obtained by aligning the first 10 time delays obtained at five different time resolutions (60, 120, 260, 400, 1000 Hz in black, red, green, blue, cyan, respectively). Representative trajectories at each time resolution are also shown in black, red, green, blue, cyan, respectively. These trajectories show the locations of the GFP molecule in each frame at the frame rates shown. The effect of the confinement inside the cell boundaries can be seen in the fact that the step size does not scale with imaging rate as expected for unrestricted Brownian motion [1]. The entire curve could be simulated by only assuming that the GFP molecules underwent simple Brownian motion at all time scales above 1 ms confined in model E. coli shells whose size was distributed as those observed in the actual experiments.
Also in the cytoplasm, several groups have looked at the mobility of regions of the DNA in live E. coli. Notably, Elmore et al. [30] used a GFP fusion to the Lac repressor to bind the DNA near the oriC site and monitored its mobility using time-lapse imaging. They found, after correction for expansion due to cell growth, that the diffusion coefficient for oriC was only 4.3 ± 2.6 × 10−5 μm2 s–1 on minute time scales though one might expect much higher mobility on shorter time scales. Espeli et al. [31] viewed the mobility of several chromosomal loci throughout the cell cycle using time-lapse microscopy as well. The loci studied were distributed throughout the chromosome such that each of the four macrodomains (Ori, Ter, Right and Left) and the two non-structured regions (NSR and NSL) of the E. coli chromosome were sampled. They found loci in the macrodomains had an average diffusion coefficient of 1–2 × 10−4 μm2 s–1 while those in the non-structured region had a diffusion coefficient of 4 × 10−4 μm2 s–1 on a tens of seconds time scale [31].
5. Localization microscopy imaging in bacteria
Localization microscopy imaging has been performed to image the distribution of parts of the E. coli chemotaxis network, specifically, the aspartate chemoreceptor Tar labelled with monomer Eos (mEos), the chemotaxis adaptor protein CheW labelled by tandem-dimer Eos (tdEos)and the response regulator CheY labelled by tdEos [32]. Using the technique of PALM, the authors mapped out a total of 1.1 million individual proteins on 326 cells! From this immense amount of information, they analysed clustering of the protein and concluded that the cluster arose from stochastic self-assembly, implying that the complex, approximately periodic structures can be created and maintained in biological membranes without direct attachment to a cytoskeleton or using active processes.
More recent work in localization microscopy has looked at the distribution of nucleoid-associated proteins (NAPs) in C. crescentus [33] and E. coli [34] cells. Using the natural blinking of the fluorescent protein EYFP under high-intensity illumination bound to the protein HU, the most abundant NAP, the Moerner group showed that HU was mostly uniformly distributed throughout swarmer and stalk cell stages with clustering found at the midplane of nascent daughter pre-divisional cells in C. crescentus [33]. The Xie and Zhuang groups employed the photoactivation of mEos2 tagged to several major NAPs to investigate the organization of the chromosome in E. coli using the three-dimensional STORM technique. The distribution of most of the NAPs observed was essentially uniform throughout the nucleoid region, including HU as seen in C. crescentus, with the exception of the global transcription silencer, H-NS, which was found to be highly clustered in two clusters per chromosome [34].
Another recent application of localization microscopy in live C. crescentus has provided clear three-dimensional images of the Z-ring of the tubulin orthologue, FtsZ [35]. These images clearly show the doughnut-shaped ring of FtsZ molecules in stalked cells with an internal opening of diameter approximately 150 nm and roughly symmetrically constricted Z-ring in pre-divisional cells with no internal opening and an overall diameter of only 150 nm. Furthermore, owing to the rapid turnover of FtsZ molecules in the ring, the depolymerization rate could be directly estimated by measuring the average time of viewing individual fluorophores in the ring (which was much slower than the measured bleaching rate in fixed cells).
6. Stoichiometry and turnover
The Leake group has led the way in determining the structure and dynamics of clusters of molecules in live bacteria. As an example, in Leake et al. [22] the dynamics of the flagella motor protein MotB labelled with GFP in the motor complex was monitored through localized FRAP and fluorescence loss in photobleaching (FLIP) measurements with single-molecule sensitivity. By directly watching the photobleaching of a single motor complex, they found that 22 ± 6 copies of MotB-GFP were incorporated in the motor as had been expected. Furthermore, through monitoring the loss/gain of fluorescence between two motor complexes after one was photobleached, and measuring the diffusion coefficient of free MotB-GFP outside the complexes (0.0088 ± 0.0026 μm2 s−1), they calculated the turnover rate of MotB-GFP to be 0.04 ± 0.02 s−1. More recently, Reyes-Lamothe et al. [36] used these techniques to investigate the architecture of the DNA replisome, where they analysed the stoichiometry of 10 different protein components in the active replisome in live E. coli.
Similarly, Lia et al. investigated the active DNA replisome in live E. coli. In this study, a stroboscopic illumination was used to limit photobleaching and hence enable the direct imaging of longer-time polymerase exchange dynamics at the replication fork. Specifically, they showed exchange of the core polymerase Pol III in the replisome in vivo and determined the rates of exchange. Furthermore, by dual labelling a component of Pol III and the single-strand binding protein (SSB), they could show a coincidence between Pol III binding and SSB fluctuations implying that the Pol III exchanged is the one synthesizing the lagging strand.
7. Imaging gene expression
In a pair of landmark papers, the group of X. Sunney Xie took single-molecule imaging in bacteria in the direction of looking directly at cellular processes, in particular at gene expression and enzyme activity.
In Yu et al. [37], gene expression was directly monitored by placing a construct of the transmembrane serine chemoreceptor Tsr coupled to the fast-folding variant of YFP, Venus, behind the lac promoter (replacing the native lacZ gene) on the E. coli genome under low expression rate conditions. Here, the membrane-bound Tsr–Venus construct was used simply as an easy to localize probe for measuring protein production at the single-molecule level. The study shows conclusively that proteins are produced in bursts from the lac operon. Analysis further showed that the each protein burst originated from a single mRNA that was randomly transcribed.
In Cai et al. [38], the product of an enzymatic reaction was monitored in order to quantitate gene expression and protein production. Combining microfluidics with single-molecule imaging, the production of fluorescein from fluorescein-di-β-d-galactopyranoside by the enzyme β-galactosidase (β-gal) was employed as a monitor for translation of β-gal (which was also placed after the lac operon).
These two papers demonstrated that single-molecule, single-cell biology and enzymology in the model–system of E. coli (and in yeast as well) was possible.
8. The future
This review has tried to give an overview of the breadth of work that has been performed in bacteria at the single-molecule level. We have shown that single-molecule experiments in bacteria have certainly overcome the technical difficulties associated with the size and structure of bacteria. Above are examples of determining mobility in membranes and the cytoplasm, determining localization precisely, determining numbers of proteins in complexes and measuring cluster dynamics and the ability to quantify gene expression all in model bacterial systems. We are now in an era beyond the novelty of the technique (i.e. the era showing what experiments are possible) and the technique is a tool in the researcher's bag to be applied to understanding fundamental biological questions from the point of view of the individual molecules.
As an example of such an application where single-molecule techniques have been applied to understanding a fundamental question in biology that is hard to access through bulk techniques, the Elf group has very recently published a study directly determining the method through which a transcription factor searches for its binding site on DNA in live E. coli [39]. In brief, by labelling the lac repressor LacI with YFP, they could image the binding of individual LacI-YFP to the lac operator site, allowing them to test whether the lac repressor used a combination of three- (cytoplasmic) and one-dimensional (along the DNA) diffusion, known as facilitated diffusion, to find its binding site. After confirming that LacI-YFP did use facilitated diffusion, they went on to determine the number of base pairs on average LacI-YFP slid along the DNA before binding. This demonstrates the power of combining the single-molecule technique with one of the most-studied model organisms. True tests of basic, fundamental questions in biology can be performed.
Along the same idea, our group is currently using single-molecule techniques in E. coli to unravel the dynamics and interactions of molecules involved in signal transduction in two-component regulatory systems and to look at the molecular interactions required in the transduction of energy from the inner to the outer membrane in some transport systems.
For the future, we expect that applying single-molecule techniques to understand long-standing questions in biology (and new questions also) using well-characterized model cells such as E. coli will make great advances and, hopefully, pose new questions that have not yet been thought of.
Acknowledgements
The authors thank Prof. Brian Todd for many great discussions and for making them aware of the Ehrenstein, Lecar and Nossal paper showing single ion channel signatures. The Ritchie laboratory acknowledge funding from the National Institutes of Health GM83296 and the National Science Foundation MCB09522999.
References
- 1.Ritchie K, Shan XY, Kondo J, Iwasawa K, Fujiwara T, Kusumi A. 2005. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys. J. 88, 2266–2277 10.1529/biophysj.104.054106 (doi:10.1529/biophysj.104.054106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrenstein G, Lecar H, Nossal R. 1970. The nature of the negative resistance in bimolecular lipid membranes containing excitability-inducing material. J. Gen. Physiol. 55, 119–133 10.1085/jgp.55.1.119 (doi:10.1085/jgp.55.1.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moerner WE, Kador L. 1989. Optical detection and spectroscopy of single molecules in a solid. Phys. Rev. Lett. 62, 2535–2538 10.1103/PhysRevLett.62.2535 (doi:10.1103/PhysRevLett.62.2535) [DOI] [PubMed] [Google Scholar]
- 4.Betzig E, Chichester RJ. 1993. Single molecules observed by near-field scanning optical microscopy. Science 262, 1422–1425 10.1126/science.262.5138.1422 (doi:10.1126/science.262.5138.1422) [DOI] [PubMed] [Google Scholar]
- 5.Schmidt T, Schutz GJ, Baumgartner W, Gruber HJ, Schindler H. 1996. Imaging of single molecule diffusion. Proc. Natl Acad. Sci. USA 93, 2926–2929 10.1073/pnas.93.7.2926 (doi:10.1073/pnas.93.7.2926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Brabander M, Nuydens R, Geerts H, Hopkins CR. 1988. Dynamic behavior of the transferrin receptor followed in living epidermoid carcinoma (A431) cells with nanovid microscopy. Cell Motil. Cytoskeleton 9, 30–47 10.1002/cm.970090105 (doi:10.1002/cm.970090105) [DOI] [PubMed] [Google Scholar]
- 7.Kusumi A, Sako Y, Yamamoto M. 1993. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65, 2021–2040 10.1016/S0006-3495(93)81253-0 (doi:10.1016/S0006-3495(93)81253-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell. Biol. 157, 1071–1081 10.1083/jcb.200202050 (doi:10.1083/jcb.200202050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lill Y, Kaserer WA, Newton SM, Lill M, Klebba PE, Ritchie K. 2012. Single-molecule study of molecular mobility in the cytoplasm of Escherichia coli. Phys. Rev. E 86, 021907. 10.1103/PhysRevE.86.021907 (doi:10.1103/PhysRevE.86.021907) [DOI] [PubMed] [Google Scholar]
- 10.Spector J, Zakharov S, Lill Y, Sharma O, Cramer WA, Ritchie K. 2010. Mobility of BtuB and OmpF in the Escherichia coli outer membrane: implications for dynamic formation of a translocon complex. Biophys. J. 99, 3880–3886 10.1016/j.bpj.2010.10.029 (doi:10.1016/j.bpj.2010.10.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yildiz A, Selvin PR. 2005. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc. Chem. Res. 38, 574–582 10.1021/ar040136s (doi:10.1021/ar040136s) [DOI] [PubMed] [Google Scholar]
- 12.Rust MJ, Bates M, Zhuang X. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 10.1038/nmeth929 (doi:10.1038/nmeth929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 10.1126/science.1127344 (doi:10.1126/science.1127344) [DOI] [PubMed] [Google Scholar]
- 14.Hess ST, Girirajan TP, Mason MD. 2006. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 10.1529/biophysj.106.091116 (doi:10.1529/biophysj.106.091116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsien RY. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 10.1146/annurev.biochem.67.1.509 (doi:10.1146/annurev.biochem.67.1.509) [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Suarez M, Ting AY. 2008. Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol. 9, 929–943 10.1038/nrm2531 (doi:10.1038/nrm2531) [DOI] [PubMed] [Google Scholar]
- 17.Deich J, Judd EM, McAdams HH, Moerner WE. 2004. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc. Natl Acad. Sci. USA 101, 15 921–15 926 10.1073/pnas.0404200101 (doi:10.1073/pnas.0404200101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikaido H, Vaara M. 1987. Outer membrane. In Escherichia coli and Salmonella typhimurium: cellular and molecular biology (ed. Neidhardt FC.), pp. 7–22, Washington, DC: American Society for Microbiology [Google Scholar]
- 19.Ruiz N, Kahne D, Silhavy TJ. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4, 57–66 10.1038/nrmicro1322 (doi:10.1038/nrmicro1322) [DOI] [PubMed] [Google Scholar]
- 20.Oddershede L, Dreyer JK, Grego S, Brown S, Berg-Sorensen K. 2002. The motion of a single molecule, the lambda-receptor, in the bacterial outer membrane. Biophys. J. 83, 3152–3161 10.1016/S0006-3495(02)75318-6 (doi:10.1016/S0006-3495(02)75318-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbs KA, Isaac DD, Xu J, Hendrix RW, Silhavy TJ, Theriot JA. 2004. Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol. Microbiol. 53, 1771–1783 10.1111/j.1365-2958.2004.04242.x (doi:10.1111/j.1365-2958.2004.04242.x) [DOI] [PubMed] [Google Scholar]
- 22.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443, 355–358 10.1038/nature05135 (doi:10.1038/nature05135) [DOI] [PubMed] [Google Scholar]
- 23.Leake MC, Greene NP, Godun RM, Granjon T, Buchanan G, Chen S, Berry RM, Palmer T, Berks BC. 2008. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl Acad. Sci. USA 105, 15 376–15 381 10.1073/pnas.0806338105 (doi:10.1073/pnas.0806338105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmerman SB, Trach SO. 1991. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 222, 599–620 10.1016/0022-2836(91)90499-V (doi:10.1016/0022-2836(91)90499-V) [DOI] [PubMed] [Google Scholar]
- 25.Terry BR, Matthews EK, Haseloff J. 1995. Molecular characterisation of recombinant green fluorescent protein by fluorescence correlation microscopy. Biochem. Biophys. Res. Commun. 217, 21–27 10.1006/bbrc.1995.2740 (doi:10.1006/bbrc.1995.2740) [DOI] [PubMed] [Google Scholar]
- 26.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. 2006. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc. Natl Acad. Sci. USA 103, 10 929–10 934 10.1073/pnas.0604503103 (doi:10.1073/pnas.0604503103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu L, Yu J. 2008. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys. J. 95, 2009–2016 10.1529/biophysj.108.128751 (doi:10.1529/biophysj.108.128751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J. 2011. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc. Natl Acad. Sci. USA 108, E365–E373 10.1073/pnas.1102255108 (doi:10.1073/pnas.1102255108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakshi S, Bratton BP, Weisshaar JC. 2011. Subdiffraction-limit study of Kaede diffusion and spatial distribution in live Escherichia coli. Biophys. J. 101, 2535–2544 10.1016/j.bpj.2011.10.013 (doi:10.1016/j.bpj.2011.10.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elmore S, Muller M, Vischer N, Odijk T, Woldringh CL. 2005. Single-particle tracking of oriC-GFP fluorescent spots during chromosome segregation in Escherichia coli. J. Struct. Biol. 151, 275–287 10.1016/j.jsb.2005.06.004 (doi:10.1016/j.jsb.2005.06.004) [DOI] [PubMed] [Google Scholar]
- 31.Espeli O, Mercier R, Boccard F. 2008. DNA dynamics vary according to macrodomain topography in the E-coli chromosome. Mol. Microbiol. 68, 1418–1427 10.1111/j.1365-2958.2008.06239.x (doi:10.1111/j.1365-2958.2008.06239.x) [DOI] [PubMed] [Google Scholar]
- 32.Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. 2009. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 7, e1000137. 10.1371/journal.pbio.1000137 (doi:10.1371/journal.pbio.1000137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SF, Thompson MA, Schwartz MA, Shapiro L, Moerner WE. 2011. Super-resolution imaging of the nucleoid-associated protein HU in Caulobacter crescentus. Biophys. J. 100, L31–L33 10.1016/j.bpj.2011.02.022 (doi:10.1016/j.bpj.2011.02.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang WQ, Li GW, Chen CY, Xie XS, Zhuang XW. 2011. Chromosome organization by a nucleoid-associated protein in live bacteria. Science 333, 1445–1449 10.1126/science.1204697 (doi:10.1126/science.1204697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biteen JS, Goley ED, Shapiro L, Moerner WE. 2012. Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astigmatism. Chemphyschem 13, 1007–1012 10.1002/cphc.201100686 (doi:10.1002/cphc.201100686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reyes-Lamothe R, Sherratt DJ, Leake MC. 2010. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328, 498–501 10.1126/science.1185757 (doi:10.1126/science.1185757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J, Xiao J, Ren X, Lao K, Xie XS. 2006. Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 10.1126/science.1119623 (doi:10.1126/science.1119623) [DOI] [PubMed] [Google Scholar]
- 38.Cai L, Friedman N, Xie XS. 2006. Stochastic protein expression in individual cells at the single molecule level. Nature 440, 358–362 10.1038/nature04599 (doi:10.1038/nature04599) [DOI] [PubMed] [Google Scholar]
- 39.Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. 2012. The lac repressor displays facilitated diffusion in living cells. Science 336, 1595–1598 10.1126/science.1221648 (doi:10.1126/science.1221648) [DOI] [PubMed] [Google Scholar]