Abstract
Objective
To assess the usefulness of 2 rapid molecular diagnostic techniques, polymerase chain reaction (PCR) and loop-mediated isothermal amplification (LAMP), in Clostridium difficile infection (CDI).
Methods
We conducted a systematic review and meta-analysis to evaluate the accuracy of PCR and LAMP in diagnosis of CDI, including studies that used toxigenic culture or cytotoxicity assay as reference standard.
Results
A search of PubMed and CinAHL medical databases yielded 25 PCR studies, including 11,801 samples that met inclusion criteria and 6 heterogeneous studies that evaluated LAMP. With toxigenic culture as a standard, pooled sensitivity was 0.92 (95% confidence interval [CI], 0.91-0.94); specificity, 0.94 (95% CI, 0.94-0.95); and diagnostic odds ratio, 378 (95% CI, 260-547). With cytotoxicity as a standard, pooled sensitivity was 0.87 (95% CI, 0.84-0.90); specificity, 0.97 (95% CI, 0.97-0.98); and diagnostic odds ratio, 370 (95% CI, 226-606).
Conclusion
Polymerase chain reaction is a highly accurate test for identifying CDI. Heterogeneity in LAMP studies did not allow meta-analysis; however, further research into this promising method is warranted.
Abbreviations and Acronyms: CDI, Clostridium difficile infection; CI, confidence interval; DOR, diagnostic odds ratio; FN, false-negative; FP, false-positive; LAMP, loop-mediated isothermal amplification; LR+, positive likelihood ratio; LR−, negative likelihood ratio; NPV, negative predictive value; PCR, polymerase chain reaction; PPV, positive predictive value; PRISMA, preferred reporting items for systematic meta-analysis; SROC, summary receiver operating curve; TN, true-negative; TP, true-positive
Clostridium difficile is the most common bacterial cause of health care–associated diarrhea, accounting for 15% to 25% of antibiotic-associated diarrhea.1 In the past several years, a rapid increase in the incidence of C difficile infection (CDI) has occurred with recognition of new, highly virulent strains causing global outbreaks.2-6 Each year, CDI affects an estimated 500,000 persons, accounting for more than $1 billion in costs and 20,000 deaths.7
Rapid and accurate diagnosis of CDI is essential both for improving outcomes of patients with CDI and for reducing horizontal transmission in health care facilities. However, diagnosis of CDI remains challenging. Most tests currently in use are either insensitive, such as the rapid enzyme immunoassay,8-11 or labor intensive and not readily available, such as the cell cytotoxicity assay and toxigenic culture.11,12 More recently, rapid molecular assays such as the polymerase chain reaction (PCR)11,13-34 and the technically simpler loop-mediated isothermal amplification (LAMP)35-40 have become readily available for the diagnosis of CDI. Although more expensive than traditional assays,17 these tests have potential for rapid and accurate diagnosis and have been supported by recent guidelines by the American Society of Microbiology.41,42 The most recent Infectious Diseases Society of America guideline states that, although promising, current data are insufficient to define the role of PCR in diagnosing CDI and makes no mention of LAMP.43
We performed a meta-analysis to investigate the performance of PCR and LAMP assays for diagnosis of CDI when compared with reference standards of cytoxicity assays or toxigenic culture.
Methods
Search Strategy
We conducted a search of 11 medical databases (Supplemental Appendix, available online at http://www.mayoclinicproceedings.com), including CinAHL and PubMed, from May 1, 2011, to January 27, 2012, using the keywords Clostridium difficile and polymerase chain reaction, Clostridium difficile and PCR, Clostridium difficile and LAMP, and Clostridium difficile, along with each of the following words: diagnosis, infection, and microbiology. The search was limited to clinical studies involving human patients, either children or adults, with a diagnosis or suspected diagnosis of CDI. No language or publication date restrictions were applied to the search. Studies were included if toxigenic culture or cytotoxicity assay was used as reference standard. Studies were excluded if the PCR was part of a multistep testing algorithm (eg, screening only of samples positive for glutamate dehydrogenase).
A standard form was used to extract relevant data on the basis of the preferred reporting items for systematic meta-analysis (PRISMA).44 Study validity was assessed on the basis of the Standards for Reporting of Diagnostic Accuracy Initiative and the Review of Methodological Standards.45
Data Abstraction
From each study, data were abstracted on the type of PCR and reference standard used in the test. The number of true-positive (TP), false-positive (FP), true-negative (TN), and false-negative (FN) results were abstracted as well. These were summarized as sensitivity, TP/(TP + FN); specificity, TN/(TN + FP); positive predictive value (PPV), TP/(TP + FP); negative predictive value (NPV), TN/(TN + FN); and prevalence, (TP + FN)/(TP + FN + TN + FN).
Positive likelihood ratio (LR+) (Sensitivity/1 − Specificity) and negative likelihood ratio (LR−) (1 − Sensitivity/Specificity) are used to evaluate how a study measure influences posttest probability using the Bayes theorem. For a positive test result, Pretest probability × LR+ = Posttest probability, and for a negative test result, Pretest probability × LR− = Posttest probability. The effect a test has on posttest probability can be summarized by using the diagnostic odds ratio (DOR), defined as LR+/LR−, where higher values denote a better discriminatory diagnostic test.46
Sensitivity and specificity are true performance statistics for a test independent of disease prevalence in a population. The major determinant for these values is that the cutoff differentiates positive from negative test results, that is, the defining optical density for the PCR at which CDI is diagnosed. A high cutoff will have a low FP rate (high specificity), but more cases will be missed (low sensitivity), whereas a low cutoff will have the opposite effect. This cutoff value is termed the diagnostic threshold.
Statistical Analyses
Sensitivity and specificity for PCR and the reference standard were calculated from the data in each study. Pooled PPV, NPV, sensitivity, specificity, LR+, LR−, and DOR were calculated for PCR with the use of the DerSimonian-Laird random effects model.47 For each statistic, the 95% confidence interval (CI) was calculated on the basis of the F distribution method for the binomial proportion.
Heterogeneity was assessed with the use of I2 analysis, where 0% indicates low heterogeneity and 100% indicates high discordance between studies.48 Subgroup analyses were conducted using metaregression to determine the contribution of individual factors such as prevalence and PCR brand on heterogeneity, where P<.05 indicates a contribution to heterogeneity. One source of heterogeneity unique to diagnostic meta-analysis is the threshold effect, which occurs when studies implicitly or explicitly use different thresholds to define a positive test result. The presence of threshold effect is tested by calculating the Spearman coefficient between sensitivity and specificity, where values ≤0.5 or >0.5 indicate possible threshold effect.49
A summary measure of accuracy (Q*) was calculated, which corresponds to the upper left–most point on the summary receiver operating characteristic (SROC) curve, where sensitivity equals specificity. This value can be between 0 and 1, with 1 indicating the highest sensitivity/specificity. This value has been recommended over the area under the receiver operating characteristic curve region of greatest interest.50,51 Statistics were calculated manually and with use of Meta-DiSc software.52
Results
The PCR search strategy identified 802 potential studies, of which 733 were excluded as duplicates, basic science studies, or addressing a different research question. The full text of the remaining 69 articles was reviewed by all authors, and after excluding animal studies, review articles, case reports, and studies that used PCR for ribotyping rather than diagnosis, 19 articles were identified as meeting criteria. Repeated search and manual inspection of references yielded 6 additional studies meeting criteria, for a total of 25 included studies. This study strategy is summarized in the PRISMA flow diagram (Supplemental Figure, available online at http://www.mayoclinicproceedings.com).
The LAMP search strategy yielded 6 articles, all of which were relevant to diagnosis of CDI. This is not included in the PCR searches summarized in the PRISMA flow diagram.
The studies meeting testing criteria included a combined total of 11,801 clinical samples. Two studies were restricted to adults,28,31 1 was restricted to children,53 1 included adults and children older than 2 years,29 and the remaining studies specified neither age nor target population from which samples were obtained. Testing criteria were routine laboratory test samples in 13 studies,13,15,16,18,19,23-25,29,30,33,54,55 patients with symptoms in 5,20,26,28,31,32 part of routine screening on a long-term care ward in 1,14 and not specified in the remainder. Thirteen studies were performed outside of the United States.13-16,18-20,23,30-32,34,53 One study was a multicenter trial with 6 US locations and 1 Canadian hospital.54 Most of the studies were recent, with 17 having been performed after 2005.15,16,18,20,21,23-30,32,34,54,55 Study characteristics are given in Table 1.
TABLE 1.
Characteristics of Included Studies
| Reference, year | Site | Patient population | Testing criteria | Multiple PCRs |
|---|---|---|---|---|
| Arzese et al,14 1995 | Udine, Italy | Long-term care ward | Admission to long-term care ward | No |
| Alonso et al,13 1999 | Madrid, Spain | NR | Routine testing | No |
| Guilbault et al,19 2002 | Montreal, Canada | NR | Routine testing for suspected CDI | No |
| Bélanger et al,17 2003 | Manassas, VA, and Brussels, Belgium | NR | NR | No |
| Zheng et al,33 2004 | Blacksburg, VA, Portland, OR, and Albuquerque, NM | NR | Routine testing, with antibiotic-associated diarrhea | No |
| van den Berg et al,31 2005 | The Netherlands | Adults | Patients with diarrhea, in hospital >72 h | No |
| Peterson et al,26 2007 | Evanston, IL | NR | Presence of symptoms | No |
| van den Berg et al,32 2007 | The Netherlands | NR | Patients with diarrhea, in hospital >72 h | No |
| Sloan et al,27 2008 | Rochester, MN | NR | NR | No |
| Barbut et al,15 2009 | Paris, France | NR | Suspected CDI | No |
| Eastwood et al,18 2009 | Leeds, UK | NR | Samples routinely tested | No |
| Huang et al,20 2009 | Cytotoxin assay | NR | Presence of symptoms | No |
| Stamper et al,29 2009 | Baltimore, MD | Adults | Presence of symptoms | No |
| Stamper et al,28 2009 | Baltimore, MD | Age ≥2 y | Routine testing | No |
| Terhes et al,30 2009 | Hungary | NR | Suspected CDI | No |
| de Boer et al,55 2010 | The Netherlands | NR | Suspected CDI | No |
| Kvach et al,24 2010 | New Haven, CT | NR | Routinely tested samples | No |
| Novak-Weekly et al,25 2010 | Los Angeles, CA, and Houston, TX | Adults and children | Suspected CDI | No |
| Tenover et al,54 2010 | 7 Centers in North America | NR | Routine testing | No |
| Barbut et al,16 2011 | Paris, France | NR | Suspected CDI | No |
| Karre et al,21 2011 | Rochester, MN | NR | NR | Yes |
| Knetsch et al,23 2011 | Leeds, UK | NR | Routine testing | Yes |
| Selvaraju et al,56 2011 | Kansas City, MO | NR | Convenience sampling of previously frozen stool samples | Yes |
| Zidarič et al,34 2011 | Slovenia | NR | NR | Yes |
| Kim et al,53 2012 | Korea | NR | Routine testing | No |
CDI = Clostridium difficile infection; NR = not reported; PCR = polymerase chain reaction.
The most commonly used PCR was BD GeneOhm (BD Diagnostics–Infectious Disease, La Jolla, CA) in 9 studies,15,18,21,23,24,28,30,34,56 followed by Cepheid Xpert (Cepheid, Sunnyvale, CA) in 4 studies,20,25,34,54 LightCycler (Roche Applied Science, Indianapolis, IN) in 2 studies,21,27 and Progastro (Gen-Probe, Inc, San Diego, CA) in 3 studies.21,29,56 The remaining studies used in-house sequences or less commonly known PCR tests or did not specify the sequence used. For a reference standard, 9 studies used cytotoxicity assays alone,13,17,19,20,25,30-32,55 11 used toxigenic culture alone,16,21,23,24,27,29,33,34,53,54,56 and the remaining studies used both reference standards. Prevalence of CDI in each study was highly variable, ranging from 6% to 37% (Supplemental Table 1, available online at http://www.mayoclinicproceedings.com).
When toxigenic culture was used as a reference standard, pooled sensitivity was 0.92 (95% CI, 0.91-0.94), with I2 of 77.9%, and pooled specificity was 0.94 (95% CI, 0.94-0.95), with I2 of 93.4%. With cytotoxicity as reference standard, pooled sensitivity was 0.87 (95% CI, 0.84-0.90), with I2 of 76.1%, and pooled specificity was 0.97 (95% CI, 0.97-0.98), with I2 of 79.9%. There was no evidence of a threshold effect in either the studies using toxigenic culture or those using cytotoxicity assay.
Summary receiver operator curves are displayed using toxigenic culture as the reference standard and cytotoxicity assay as the reference standard (Figure 1). With a toxigenic culture standard, the area under the SROC curve is 0.99, and Q* is 0.95. Using cytotoxicity assay as the criterion standard, area under curve is 0.99, and Q* is 0.95. Overall, these statistics are consistent with an accurate diagnostic test.
FIGURE 1.
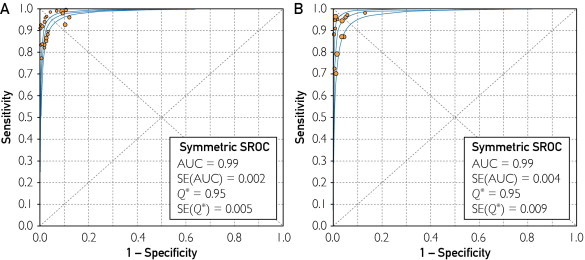
Summary receiver operating curves (SROC) of studies using toxigenic culture (A) and cytotoxicity assay (B) as reference standard. With toxigenic culture reference standard, area under curve (AUC) is 0.99, with Q* of 0.95. With cytotoxicity assay reference standard, AUC is 0.99, with Q* 0.95. Individual studies and their relative weights are designated by the size of circles. Pooled results are represented by the diamond. Figure generated using Meta-DiSc software.
With median sensitivity and specificity of the various commercial assays of the test, a plot of PPV and NPV according to prevalence is depicted in Figure 2. Under typical conditions (prevalence, 0%-20%), the NPV of the assay is quite high, whereas the PPV performance is variable.
FIGURE 2.
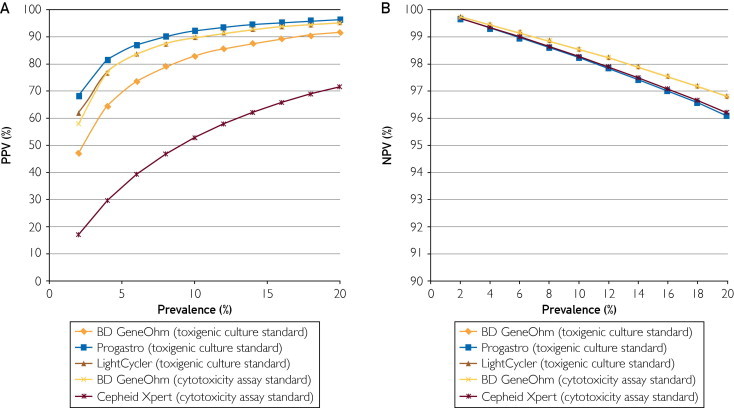
Estimated positive predictive value (PPV) (A) and negative predictive value (NPV) (B) of assays according to disease prevalence of stool tested. Median values for specificity and sensitivity were used to calculate these values. Reference standards are given in parentheses.
With toxigenic culture as a reference standard, the LR+ ratio was 33 (95% CI, 20-53), with moderate heterogeneity (I2 = 67.4%), and the LR− ratio was 0.09 (0.07-0.13), with moderate heterogeneity (I2 = 54.4%). At cytotoxicity assay, the LR+ ratio was 32.6 (95% CI, 20.2-52.5), with high heterogeneity (I2 = 76.6%), and the LR− ratio was 0.11 (95% CI, 0.07-0.18), with moderate heterogeneity (I2 = 71.0%).
The DOR using toxigenic culture and cytotoxicity assay as the criterion standard are illustrated in Figure 3. With toxigenic culture as a standard, DOR was 378 (95% CI, 260-547), with moderate heterogeneity (I2 = 37.3%). With cytotoxicity as a standard, the pooled DOR was 370 (95% CI, 226-606), with low heterogeneity (I2 = 18%).
FIGURE 3.
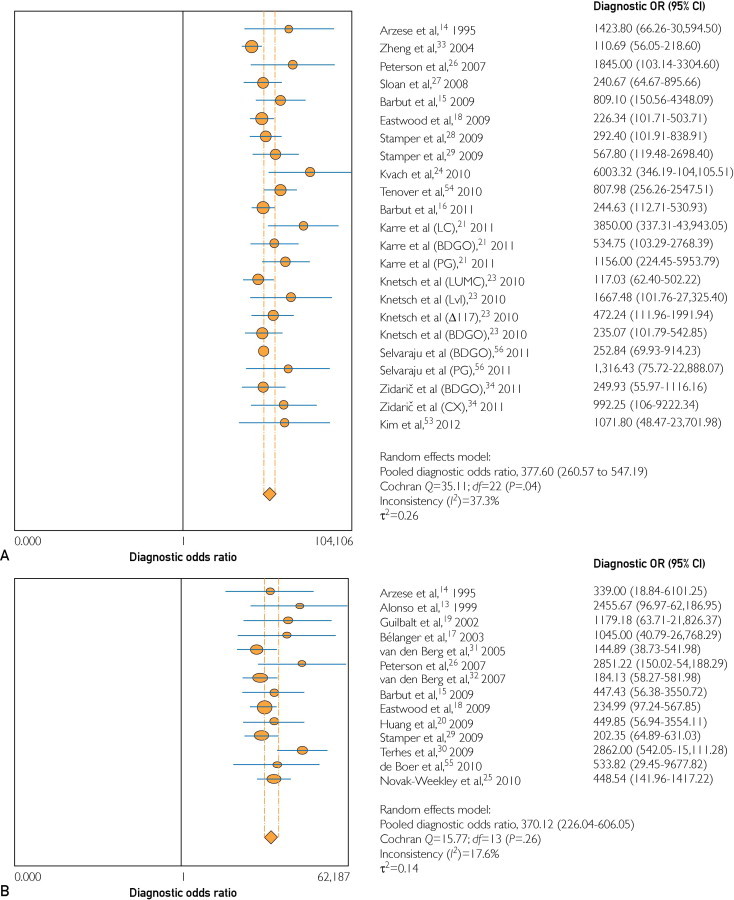
Diagnostic odds ratio (OR) of polymerase chain reaction (PCR) tests as compared with toxigenic culture (A) and cytotoxin assay (B). Studies that evaluated more than one PCR are noted in parentheses. BDGO = BD GeneOhm; CI = confidence interval; CX = Cepheid Xpert; LC = LightCycler; PG = Progastro.
Subgroup analyses were performed using the Progastro, LightCycler, and BD GeneOhm PCRs, with toxigenic culture as reference standard, and the BD GeneOhm and Cepheid Xpert (Supplemental Table 2, available online at http://www.mayoclinicproceedings.com). Metaregression did not find that brand significantly contributed to heterogeneity (Progastro, P=.41; LightCycler, P=.65; and BD GeneOhm, P=.97). At cytotoxicity assay, metaregression again did not demonstrate that brand contributed to heterogeneity (BD GeneOhm, P=.95; and Cepheid Xpert, P=.89).
The effect of prevalence on the heterogeneity of the results was modeled using metaregression. This was not significant with either toxigenic culture (P=.58) or cytotoxicity assay (P=.24) as reference standard.
The 6 studies identified at LAMP used heterogeneous reference methods. Two studies used agreement between other molecular methods (eg, PCR as the criterion standard),35,37 2 studies used a toxigenic culture standard,39,40 and 2 studies used a cytotoxicity assay.36,38 Of the latter 2 studies, 1 used LAMP only in a 2-step algorithm,36 thus making it difficult to examine independently. The other noted a high FP rate with LAMP, which the authors attributed to superiority of LAMP over cytotoxicity assay.38 Combined sensitivity was 0.92 (95% CI, 0.88-0.95), and specificity was 0.97 (95% CI, 0.96-0.98), with I2 of 44.3% and 87.4%, respectively (Table 2). In all likelihood, the differing reference standards contributed to this inconsistency. The LAMP studies included 1685 participants, with individual studies ranging in size from 74 to 472 samples. Five studies used consecutive samples submitted to laboratory testing for suspected C difficile, and 1 study36 was restricted to children. All studies ranged in sample size, from 139 to 472 participants, with a total of 1685 individuals.
TABLE 2.
Results of LAMP
| Reference, year | Reference Standard | No. of studies | Sensitivity | Specificity | Notes |
|---|---|---|---|---|---|
| Kato et al,38 2005 | Cytotoxicity assay | 74 | 0.97 | 0.71 | Authors attribute high false-positive rate to superiority of LAMP, eg, more sensitive than cytotoxicity assay |
| Lalande et al,39 2011 | Toxigenic culture | 472 | 0.92 | 0.99 | Study indicates LAMP to be more sensitive than cytotoxicity assay, to which it was compared |
| Doing and Hintz,37 2012 | Agreement with ProGastro PCR test | 446 | 0.98 | 0.99 | Cases without agreement tested with cytotoxicity assay |
| Boyanton et al,35 2012 | Agreement between 4 different molecular tests for CDI | 139 | 0.95 | 0.97 | Unresolved cases tested with toxigenic culture |
| Norén et al,40 2011 | Toxigenic culture | 272 | 0.98 | 0.98 | … |
| Ota and McGowan,36 2012 | Cytotoxicity assay | 141 | 0.89 | 0.98 | LAMP tests were performed as part of a 2-step glutamate dehydrogenase assay |
LAMP = loop-mediated isothermal amplification; PCR = polymerase chain reaction.
Discussion
The increasing incidence of CDI, and in particular hospital-acquired CDI, is a major challenge faced by health care institutions. The spectrum of infections caused by C difficile ranges from asymptomatic colonization to toxic megacolon with septic shock.57 Clinical findings such as stool odor, consistency, and frequency are unreliable for diagnosis because they have poor sensitivity and specificity.58,59 Endoscopy, while specific if pseudomembranes are detected, is insensitive and costly to perform routinely. Thus, diagnosis of CDI hinges on microbiologic and molecular testing. A number of tests are available for diagnosis of CDI, such as enzyme immunoassay for toxins A, B, or both; cytoxicity assays for toxin B; and toxigenic culture. These tests all have limitations, either in test performance, availability, or ease of performance.10 This uncertainty is reflected in the debatable practices of multiple-test algorithms60 or repeated testing of stool61 to optimize performance. Given these challenges and the variability across institutions in choice of diagnostic test,62 the availability of PCR is a promising approach to diagnosis of CDI.
Our findings indicate that PCR is a useful diagnostic test with a high degree of accuracy on the basis of DOR and Q* statistic. Likelihood ratios, in particular when compared with a toxigenic culture reference standard, indicate that the test is useful in determining posttest probability of CDI. The SROC curves, DOR, and LR data all support the use of PCR for diagnosis of CDI as a highly discriminatory test. A negative test result is adequate to rule out the presence of the disease for both clinical and epidemiologic purposes. The predictive values depend on prevalence, which was variable in the included studies. The PPV increases as prevalence increases and reaches 95%, for a prevalence of ≥20%.
The present study has limitations that stem from heterogeneity in the design of the studies analyzed. To minimize this, we included only studies that used toxigenic culture or cytoxicity assay as the reference standard. A possible contribution to this is variability in the technique and performance of the toxigenic cultures and cytotoxicity assays, because cytotoxicity assays, in particular, may be inferior to PCR as a diagnostic method.39 The most significant probable contributor to heterogeneity was the baseline criteria for accepting stool samples for testing. Most studies did not specify the clinical criteria used to submit stool samples to the laboratory for testing. Inasmuch as PCR may detect colonization without infection, this effect would not be reflected in prevalence calculated using the criterion standard. If some studies restricted use of laboratory testing to cases with high probability, for example, accepting only unformed stools and using ≥3 loose stools in 24 hours to define diarrhea,63 and others included cases with low probability, this could account for the heterogeneity observed.
We did not include studies that used PCR as part of a multistep algorithm and cannot address the role of PCR in that setting. Given the need to rapidly diagnose CDI, multistep testing algorithms may not offer an advantage but may be used for cost savings.
Our analysis extends the results of a recently published meta-analysis that compared PCR for diagnosis of CDI.64 Deshpande et al64 conducted a search of 4 databases and identified 19 articles published between January 1995 and September 2010. Inclusion and exclusion criteria were similar to those in the present study, although our search identified 2 additional studies that were published after the search dates used by Deshpande and colleagues. The reference standards used were also the same, although toxigenic culture and cytotoxicity assay were combined for the main analysis by Deshpande and colleagues, whereas we opted to separate these 2 groups because of their heterogeneity and to prevent double counting of populations in studies that used 2 reference standards.
LAMP seems to be a promising test according to current data; however, the high degree of heterogeneity in the study designs tempers any conclusions drawn from aggregate data. Nonetheless, the high degree of performance in each of the 6 studies makes a strong case for continued research into this diagnostic tool.
Conclusion
Polymerase chain reaction is a promising test for diagnosis of CDI, with high sensitivity, specificity, PPV, and NPV. We recommend PCR as a preferred diagnostic test for CDI and further investigation of LAMP as an alternative molecular diagnostic tool. Additional studies are needed to clarify the role of PCR for detection of asymptomatic colonization and to determine whether infection control interventions directed at isolation of the asymptomatic carrier would reduce nosocomial CDI.
Supplemental Online Material
References
- 1.Bartlett J.G. Antibiotic-associated diarrhea. Clin Infect Dis. 1992;15(4):573–581. doi: 10.1093/clind/15.4.573. [DOI] [PubMed] [Google Scholar]
- 2.Warny M., Pepin J., Fang A. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366(9491):1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 3.Pépin J., Valiquette L., Alary M.E. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ. 2004;171(5):466–472. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald L.C., Killgore G.E., Thompson A. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353(23):2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett J.G. Narrative review: the new epidemic of Clostridium difficile-associated enteric disease. Ann Intern Med. 2006;145(10):758–764. doi: 10.7326/0003-4819-145-10-200611210-00008. [DOI] [PubMed] [Google Scholar]
- 6.Cherifi S., Delmee M., Van Broeck J., Beyer I., Byl B., Mascart G. Management of an outbreak of Clostridium difficile-associated disease among geriatric patients. Infect Control Hosp Epidemiol. 2006;27(11):1200–1205. doi: 10.1086/507822. [DOI] [PubMed] [Google Scholar]
- 7.Kyne L., Hamel M.B., Polavaram R., Kelly C.P. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin Infect Dis. 2002;34(3):346–353. doi: 10.1086/338260. [DOI] [PubMed] [Google Scholar]
- 8.Guerrero D.M., Chou C., Jury L.A., Nerandzic M.M., Cadnum J.C., Donskey C.J. Clinical and infection control implications of Clostridium difficile infection with negative enzyme immunoassay for toxin. Clin Infect Dis. 2011;53(3):287–290. doi: 10.1093/cid/cir361. [DOI] [PubMed] [Google Scholar]
- 9.Chapin K.C., Dickenson R.A., Wu F., Andrea S.B. Comparison of five assays for detection of Clostridium difficile toxin. J Mol Diagn. 2011;13(4):395–400. doi: 10.1016/j.jmoldx.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Planche T., Aghaizu A., Holliman R. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect Dis. 2008;8(12):777–784. doi: 10.1016/S1473-3099(08)70233-0. [DOI] [PubMed] [Google Scholar]
- 11.Kufelnicka A.M., Kirn T.J. Effective utilization of evolving methods for the laboratory diagnosis of Clostridium difficile infection. Clin Infect Dis. 2011;52(12):1451–1457. doi: 10.1093/cid/cir201. [DOI] [PubMed] [Google Scholar]
- 12.Delmée M., Van Broeck J., Simon A., Janssens M., Avesani V. Laboratory diagnosis of Clostridium difficile-associated diarrhoea: a plea for culture. J Med Microbiol. 2005;54(pt 2):187–191. doi: 10.1099/jmm.0.45844-0. [DOI] [PubMed] [Google Scholar]
- 13.Alonso R., Muñoz C., Gros S., García de Viedma D., Peláez T., Bouza E. Rapid detection of toxigenic Clostridium difficile from stool samples by a nested PCR of toxin B gene. J Hosp Infect. 1999;41(2):145–149. doi: 10.1016/s0195-6701(99)90052-x. [DOI] [PubMed] [Google Scholar]
- 14.Arzese A., Trani G., Riul L., Botta G.A. Rapid polymerase chain reaction method for specific detection of toxigenic Clostridium difficile. Eur J Clin Microbiol Infect Dis. 1995;14(8):716–719. doi: 10.1007/BF01690883. [DOI] [PubMed] [Google Scholar]
- 15.Barbut F., Braun M., Burghoffer B., Lalande V., Eckert C. Rapid detection of toxigenic strains of Clostridium difficile in diarrheal stools by real-time PCR. J Clin Microbiol. 2009;47(4):1276–1277. doi: 10.1128/JCM.00309-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbut F., Monot M., Rousseau A. Rapid diagnosis of Clostridium difficile infection by multiplex real-time PCR. Eur J Clin Microbiol Infect Dis. 2011;30(10):1279–1285. doi: 10.1007/s10096-011-1224-z. [DOI] [PubMed] [Google Scholar]
- 17.Bélanger S.D., Boissinot M., Clairoux N., Picard F.J., Bergeron M.G. Rapid detection of Clostridium difficile in feces by real-time PCR. J Clin Microbiol. 2003;41(2):730–734. doi: 10.1128/JCM.41.2.730-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eastwood K., Else P., Charlett A., Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47(10):3211–3217. doi: 10.1128/JCM.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilbault C., Labbé A.C., Poirier L., Busque L., Béliveau C., Laverdière M. Development and evaluation of a PCR method for detection of the Clostridium difficile toxin B gene in stool specimens. J Clin Microbiol. 2002;40(6):2288–2290. doi: 10.1128/JCM.40.6.2288-2290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H., Weintraub A., Fang H., Nord C.E. Comparison of a commercial multiplex real-time PCR to the cell cytotoxicity neutralization assay for diagnosis of Clostridium difficile infections. J Clin Microbiol. 2009;47(11):3729–3731. doi: 10.1128/JCM.01280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karre T., Sloan L., Patel R., Mandrekar J., Rosenblatt J. Comparison of two commercial molecular assays to a laboratory-developed molecular assay for diagnosis of Clostridium difficile infection. J Clin Microbiol. 2011;49(2):725–727. doi: 10.1128/JCM.01028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato N., Ou C.Y., Kato H. Detection of toxigenic Clostridium difficile in stool specimens by the polymerase chain reaction. J Infect Dis. 1993;167(2):455–458. doi: 10.1093/infdis/167.2.455. [DOI] [PubMed] [Google Scholar]
- 23.Knetsch C.W., Bakker D., de Boer R.F. Comparison of real-time PCR techniques to cytotoxigenic culture methods for diagnosing Clostridium difficile infection. J Clin Microbiol. 2011;49(1):227–231. doi: 10.1128/JCM.01743-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kvach E.J., Ferguson D., Riska P.F., Landry M.L. Comparison of BD GeneOhm Cdiff real-time PCR assay with a two-step algorithm and a toxin A/B enzyme-linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection. J Clin Microbiol. 2010;48(1):109–114. doi: 10.1128/JCM.01630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novak-Weekley S.M., Marlowe E.M., Miller J.M. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol. 2010;48(3):889–893. doi: 10.1128/JCM.01801-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson L.R., Manson R.U., Paule S.M. Detection of toxigenic Clostridium difficile in stool samples by real-time polymerase chain reaction for the diagnosis of C. difficile-associated diarrhea. Clin Infect Dis. 2007;45(9):1152–1160. doi: 10.1086/522185. [DOI] [PubMed] [Google Scholar]
- 27.Sloan L.M., Duresko B.J., Gustafson D.R., Rosenblatt J.E. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J Clin Microbiol. 2008;46(6):1996–2001. doi: 10.1128/JCM.00032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamper P.D., Alcabasa R., Aird D. Comparison of a commercial real-time PCR assay for tcdB detection to a cell culture cytotoxicity assay and toxigenic culture for direct detection of toxin-producing Clostridium difficile in clinical samples. J Clin Microbiol. 2009;47(2):373–378. doi: 10.1128/JCM.01613-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamper P.D., Babiker W., Alcabasa R. Evaluation of a new commercial TaqMan PCR assay for direct detection of the Clostridium difficile toxin B gene in clinical stool specimens. J Clin Microbiol. 2009;47(12):3846–3850. doi: 10.1128/JCM.01490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terhes G., Urbán E., Sóki J., Nacsa E., Nagy E. Comparison of a rapid molecular method, the BD GeneOhm Cdiff assay, to the most frequently used laboratory tests for detection of toxin-producing Clostridium difficile in diarrheal feces. J Clin Microbiol. 2009;47(11):3478–3481. doi: 10.1128/JCM.01133-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Berg R.J., van Coppenraet L.S.Bruijnesteijn, Gerritsen H.J., Endtz H.P., van der Vorm E.R., Kuijper E.J. Prospective multicenter evaluation of a new immunoassay and real-time PCR for rapid diagnosis of Clostridium difficile-associated diarrhea in hospitalized patients. J Clin Microbiol. 2005;43(10):5338–5340. doi: 10.1128/JCM.43.10.5338-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Berg R.J., Vaessen N., Endtz H.P., Schülin T., van der Vorm E.R., Kuijper E.J. Evaluation of real-time PCR and conventional diagnostic methods for the detection of Clostridium difficile-associated diarrhoea in a prospective multicentre study. J Med Microbiol. 2007;56(pt 1):36–42. doi: 10.1099/jmm.0.46680-0. [DOI] [PubMed] [Google Scholar]
- 33.Zheng L., Keller S.F., Lyerly D.M. Multicenter evaluation of a new screening test that detects Clostridium difficile in fecal specimens. J Clin Microbiol. 2004;42(8):3837–3840. doi: 10.1128/JCM.42.8.3837-3840.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zidarič V., Kevorkijan B.K., Oresic N., Janezic S., Rupnik M. Comparison of two commercial molecular tests for the detection of Clostridium difficile in the routine diagnostic laboratory. J Med Microbiol. 2011;60(pt 8):1131–1136. doi: 10.1099/jmm.0.030163-0. [DOI] [PubMed] [Google Scholar]
- 35.Boyanton B.L., Jr, Sural P., Loomis C.R. Loop-mediated isothermal amplification compared to real-time pcr and enzyme immunoassay for toxigenic C. difficile detection. J Clin Microbiol. 2012;50(3):640–645. doi: 10.1128/JCM.01014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ota K.V., McGowan K.L. Clostridium difficile testing algorithms using glutamate dehydrogenase antigen and C. difficile toxin enzyme immunoassays with C. difficile nucleic acid amplification testing increase diagnostic yield in a tertiary pediatric population. J Clin Microbiol. 2012;50(4):1185–1188. doi: 10.1128/JCM.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doing K.M., Hintz M.S. Prospective evaluation of the Meridian Illumigene loop-mediated amplification assay and the Gen Probe ProGastro Cd polymerase chain reaction assay for the direct detection of toxigenic Clostridium difficile from fecal samples. Diagn Microbiol Infect Dis. Jan 2012;72(1):8–13. doi: 10.1016/j.diagmicrobio.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Kato H., Yokoyama T., Kato H., Arakawa Y. Rapid and simple method for detecting the toxin B gene of Clostridium difficile in stool specimens by loop-mediated isothermal amplification. J Clin Microbiol. 2005;43(12):6108–6112. doi: 10.1128/JCM.43.12.6108-6112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lalande V., Barrault L., Wadel S., Eckert C., Petit J.C., Barbut F. Evaluation of a loop-mediated isothermal amplification assay for diagnosis of Clostridium difficile infections. J Clin Microbiol. 2011;49(7):2714–2716. doi: 10.1128/JCM.01835-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norén T., Alriksson I., Andersson J., Akerlund T., Unemo M. Rapid and sensitive loop-mediated isothermal amplification test for Clostridium difficile detection challenges cytotoxin B cell test and culture as gold standard. J Clin Microbiol. 2011;49(2):710–711. doi: 10.1128/JCM.01824-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carroll K.C., Loeffelholz M. Conventional versus molecular methods for the detection of Clostridium difficile. J Clin Microbiol. 2011;49(9):S49–S52. [Google Scholar]
- 42.American Society for Microbiology A practical guidance document for the laboratory detection of toxigenic Clostridium difficile [update of September 9, 2010, version] September 21, 2010. http://www.asm.org/images/pdf/Clinical/clostridiumdifficile9-21.pdf May 31, 2012.
- 43.Cohen S.H., Gerding D.N., Johnson S. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31(5):431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 44.Moher D., Liberati A., Tetzlaff J., Altman D.G., PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62(10):1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Bossuyt P.M., Reitsma J.B., Bruns D.E. ; Standards for Reporting of Diagnostic Accuracy: The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. Ann Intern Med. 2003;138(1):W1–W12. doi: 10.7326/0003-4819-138-1-200301070-00012-w1. [DOI] [PubMed] [Google Scholar]
- 46.Glas A.S., Lijmer J.G., Prins M.H., Bonsel G.J., Bossuyt P.M. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56(11):1129–1135. doi: 10.1016/s0895-4356(03)00177-x. [DOI] [PubMed] [Google Scholar]
- 47.Fleiss J.L. The statistical basis of meta-analysis. Stat Methods Med Res. 1993;2(2):121–145. doi: 10.1177/096228029300200202. [DOI] [PubMed] [Google Scholar]
- 48.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Devillé W.L., Buntinx F., Bouter L.M. Conducting systematic reviews of diagnostic studies: didactic guidelines. BMC Med Res Methodol. 2002;2:9. doi: 10.1186/1471-2288-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moses L.E., Shapiro D., Littenberg B. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Stat Med. 1993;12(14):1293–1316. doi: 10.1002/sim.4780121403. [DOI] [PubMed] [Google Scholar]
- 51.Walter S.D. Properties of the summary receiver operating characteristic (SROC) curve for diagnostic test data. Stat Med. 2002;21(9):1237–1256. doi: 10.1002/sim.1099. [DOI] [PubMed] [Google Scholar]
- 52.Zamora J., Abraira V., Muriel A., Khan K., Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim H., Jeong S.H., Kim M., Lee Y., Lee K. Detection of Clostridium difficile toxin A/B genes by multiplex real-time PCR for the diagnosis of C. difficile infection. J Med Microbiol. 2012;61(pt 2):274–277. doi: 10.1099/jmm.0.035618-0. [DOI] [PubMed] [Google Scholar]
- 54.Tenover F.C., Novak-Weekley S., Woods C.W. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol. 2010;48(10):3719–3724. doi: 10.1128/JCM.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Boer R.F., Wijma J.J., Schuurman T. Evaluation of a rapid molecular screening approach for the detection of toxigenic Clostridium difficile in general and subsequent identification of the tcdC Δ117 mutation in human stools. J Microbiol Methods. 2010;83(1):59–65. doi: 10.1016/j.mimet.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 56.Selvaraju S.B., Gripka M., Estes K., Nguyen A., Jackson M.A., Selvarangan R. Detection of toxigenic Clostridium difficile in pediatric stool samples: an evaluation of Quik Check Complete Antigen assay, BD GeneOhm Cdiff PCR, and ProGastro Cd PCR assays. Diagn Microbiol Infect Dis. 2011;71(3):224–229. doi: 10.1016/j.diagmicrobio.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 57.Trudel J.L. Clostridium difficile colitis. Clin Colon Rectal Surg. 2007;20(1):13–17. doi: 10.1055/s-2007-970195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burdette S.D., Bernstein J.M. Does the nose know?: The odiferous diagnosis of Clostridium difficile-associated diarrhea [letter] Clin Infect Dis. 2007;44(8):1142. doi: 10.1086/513033. [DOI] [PubMed] [Google Scholar]
- 59.Johansen A., Vasishta S., Edison P., Hosein I. Clostridium difficile associated diarrhoea: how good are nurses at identifying the disease? Age Ageing. 2002;31(6):487–488. doi: 10.1093/ageing/31.6.487-a. [DOI] [PubMed] [Google Scholar]
- 60.Wilcox M.H., Planche T., Fang F.C., Gilligan P. What is the current role of algorithmic approaches for diagnosis of Clostridium difficile infection? J Clin Microbiol. 2010;48(12):4347–4353. doi: 10.1128/JCM.02028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo R.F., Banaei N. Is repeat PCR needed for diagnosis of Clostridium difficile infection? J Clin Microbiol. 2010;48(10):3738–3741. doi: 10.1128/JCM.00722-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barbut F., Delmée M., Brazier J.S., ESCMID Study Group on Clostridium difficile (ESGCD) A European survey of diagnostic methods and testing protocols for Clostridium difficile. Clin Microbiol Infect. 2003;9(10):989–996. doi: 10.1046/j.1469-0691.2003.00683.x. [DOI] [PubMed] [Google Scholar]
- 63.Peterson L.R., Robicsek A. Does my patient have Clostridium difficile infection? Ann Intern Med. 2009;151(3):176–179. doi: 10.7326/0003-4819-151-3-200908040-00005. [DOI] [PubMed] [Google Scholar]
- 64.Deshpande A., Pasupuleti V., Rolston D.D. Diagnostic accuracy of real-time polymerase chain reaction in detection of Clostridium difficile in the stool samples of patients with suspected Clostridium difficile infection: a meta-analysis. Clin Infect Dis. 2011;53(7):e81–e90. doi: 10.1093/cid/cir505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


