Abstract
Vesicular tethers and SNAREs are two key protein components that govern docking and fusion of intracellular membrane carriers in eukaryotic cells. The COG (conserved oligomeric Golgi) complex has been specifically implicated in the tethering of retrograde intra-Golgi vesicles. Using yeast two hybrid and co-immunoprecipitation approaches, we show that the COG6 subunit of the COG complex is capable of interacting with a subset of Golgi SNAREs, namely STX5, STX6, GS27, and SNAP29. Interaction with SNAREs is accomplished via the universal SNARE-binding motif of COG6. Overexpression of COG6, or its depletion from cells, disrupts the integrity of the Golgi complex. Importantly, COG6 protein lacking the SNARE binding domain is deficient in Golgi binding, and is not capable of inducing Golgi complex fragmentation when overexpressed. These results indicate that COG6-SNARE interactions are important for both COG6 localization and Golgi integrity.
Keywords: COG complex, Golgi apparatus, SNARE, GS27, intra-Golgi retrograde traffic, vesicle tethering
Introduction
Membrane trafficking in eukaryotic cells is governed by modular protein machinery that regulates the formation and precise delivery of intracellular vesicular transport carriers(1). Vesicular tethers, as a part of the docking/fusion module, regulate the initial docking of a transport carrier to its acceptor compartment, and thereby determine the targeting specificity of vesicles(2).
The Conserved Oligomeric Golgi (COG) complex belongs to the CATCHR (complexes associated with tethering containing helical rods) family of protein complexes (3) and is found in all eukaryotes from yeasts to humans (4) as a peripheral membrane complex that is localized at the Golgi apparatus and regulates vesicular trafficking within this organelle(4, 5). The COG complex consists of eight subunits named COG1–8 (6, 7), which are grouped into two subcomplexes: COG1–4 (Lobe A) and COG5–8 (Lobe B) (8, 9). Lobe A subunits are essential for growth in yeast, and cause severe glycosylation defects in mammalian cells when mutated or deleted (10-12). Conversely, deletions of Lobe B subunits cause rather mild growth defects in yeast (13), indicating either a redundant, or regulatory role of this protein subcomplex.
The COG complex functions in the tethering of vesicles that recycle resident Golgi proteins such as glycosidases and glycosylation enzymes (14). Consequently, defects in the COG complex subunits result in congenital disorders of glycosylation (CDG) type II (15). In humans the deficiency of Lobe B subunits COG6 and COG7 show more severe phenotypes as compared to the deficiency of Lobe A subunits COG2 and COG4 (16). In particular, a COG6 mutation (G549V) (designated as CDG-IIL (COG6-CDG)), is characterized by combined N- and O-glycosylation deficiency and results in a lethal phenotype.(17). The reason for this specific importance of COG complex Lobe B subunits in higher eukaryotes is not clear. We and others have shown previously that depletion of COG6 in HeLa cells results in mislocalization of Golgi enzymes (14), and significant delays in retrograde transport of both Shiga and Subtilase toxins (18, 19).
Similarly to the other CATCHRs, the COG complex has been shown to interact both genetically and physically with other members of the Golgi docking/fusion module including SNAREs. We and others have shown previously that the yeast and mammalian COG complex functionally interacts with the Sed5/STX5 containing SNARE complex (20, 21) through a direct interaction between COG4 and Sed5/STX5, while COG6 directly interacts with STX6 (18) but the complete network and the exact nature of COG-SNARE interactions has not been explored.
In this study, we have investigated Golgi phenotypes in cells that either under- or over-express the COG6 subunit of the COG complex. In both cases the Golgi ribbon becomes fragmented, indicating that a proper cellular concentration of COG6 is important for the integrity of the Golgi complex morphology. We extended our initial investigation of COG-SNARE interactions to examine the interaction of COG6 with 14 known Golgi-localized SNARE proteins. Our results demonstrate specific interactions of COG6 with the four Golgi SNAREs STX5, STX6, GS27, and SNAP29 by yeast two hybrid and native co-immunoprecipitation approaches. Furthermore, we also uncovered the specific SNARE-interacting motif in COG6, and demonstrated that COG6 lacking the SNARE interacting motif failed to localize to the Golgi, or induce Golgi fragmentation when overexpressed.
Results
Cog6 retains its Golgi localization in cells depleted of COG8
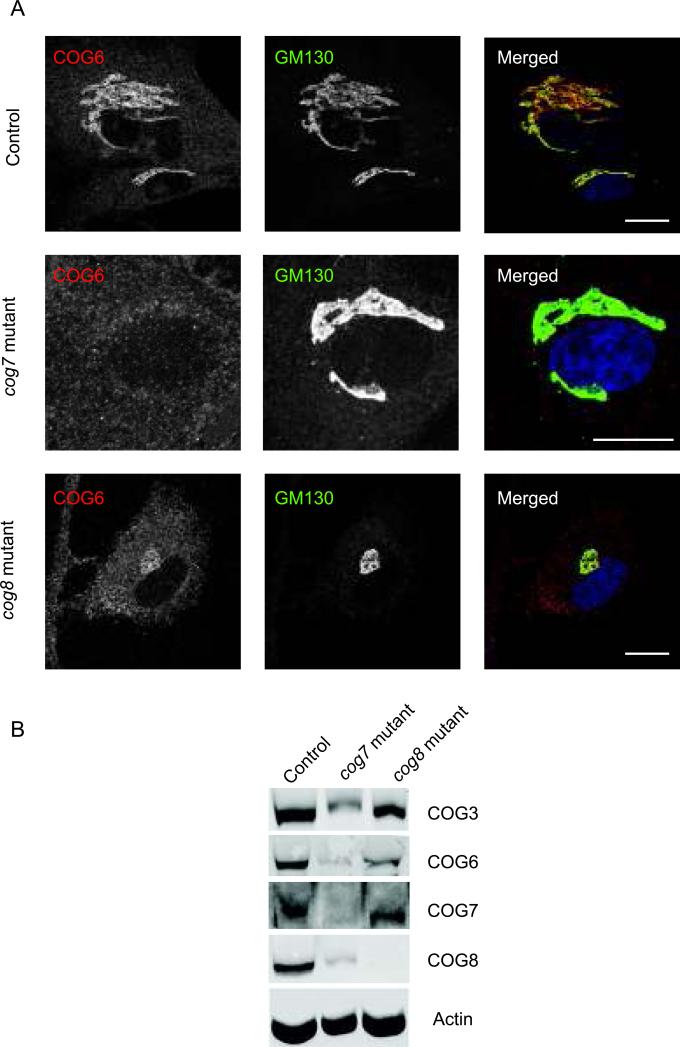
In mammalian cells the COG6 protein exists in at least two complexes: as a part of the large octameric COG complex, and in a smaller Lobe B sub-complex (7). We have shown previously that in HeLa cells COG6 localizes not only to the Golgi ribbon, but is also present on trafficking intermediates that carry Golgi glycosyltransferases (14). To test if the subcellular localization of COG6 is dependent on expression of other Lobe B subunits we examined human fibroblasts isolated from control, cog7 (22) and cog8 (23) mutant patients (Figure 1). In agreement with data obtained with HeLa cells, in control fibroblasts the COG6 signal was concentrated in a perinuclear area partially co-localizing with cis-Golgi marker GM130 (Figure 1A, WT). Additional peripheral staining represents either soluble COG6 protein, or COG6 that is present on trafficking intermediates which cannot be resolved by standard confocal microscopy. In cog7 mutant cells the level of COG6 protein was significantly decreased (Figure 1B). The remaining COG6 signal was completely dislocated from the Golgi membranes (Figure 1A, cog7 mutant), indicating that the integrity of the Lobe B subcomplex is important for the stability and proper localization of COG6 protein. Surprisingly, in cog8 mutant fibroblasts a sizable fraction of the COG6 signal was found associated with Golgi membranes (Figure 1A, cog8 mutant). COG8 is known to form a bridge between Lobe A and Lobe B sub-complexes (9). These results indicate that COG6 may associate with Golgi membranes independently of the Lobe A sub-complex. Since the COG6 sequence lacks any trans-membrane motifs or known signals for lipid modification, its association with the membrane is likely facilitated through protein-protein interactions with other trans-membrane or peripheral membrane proteins, like Rabs and SNAREs. Indeed COG6 was shown to interact with several Golgi Rabs, including Rab6a, Rab6b and Rab1 (24), but COG6 Golgi localization was not severely affected in cells deficient for Rab6 and Rab1 (Suppl. Figure 1 and data not shown), suggesting that COG-SNARE interactions could hold a key for the COG6 membrane attachment.
Figure 1. COG6 localization in Human fibroblasts.
(A) Human (WT) fibroblasts, cog7 deficient mutant fibroblasts, and cog8 mutant fibroblasts were grown on coverslips and stained with antibodies against COG6 and GM130. Scale bar, 10 μm. (B) Lysates from WT, cog7 deficient mutant, and cog8 mutant fibroblasts were analyzed by SDS-PAGE followed by western blot with antibodies against COG3, COG6, COG7, COG8, and actin. COG6 localization and stability is severely altered in COG7 deficient mutant fibroblasts.
COG complex interacts with multiple Golgi SNAREs
To find all Golgi SNAREs that are capable of interacting with the COG6 subunit, we performed a directed yeast two hybrid screen testing the potential interaction of COG6 with 14 Golgi localized SNARE proteins. Full-length COG6 was fused to the C-terminus of the Gal4 transcription factor activation domain (AD) (9). The cytoplasmic portion of the mammalian Golgi SNAREs Bet1, GS15, GS27/membrin, GS28, Sec22, SNAP29, STX5a, STX6, STX10, STX16, VAMP4, Vti1a, Vti1b, YKT6 and the ER SNARE STX18 were fused to the Gal4 binding domain (BD). COG6, in addition to previously reported interactions with Qa SNARE STX5 (21) and Qc SNARE STX6 (18), also demonstrated positive interactions with Qb SNARE GS27 and Qab SNARE SNAP29 (Figure 2). In a parallel study we also confirmed that STX5 is the only SNARE partner for COG4 and discovered that COG8 displayed a novel set of interactions with the SNARE's STX5, STX6, STX16, and GS27 (Willett and Lupashin, submitted). These results demonstrate that only a subset of the COG subunits interacts with a subset of Golgi SNAREs.
Figure 2. Yeast two hybrid assay.
Yeast diploids co-expressing Gal4 AD-hCOG6 fusions and GAL4 BD-SNARE fusions were grown in liquid selective media to OD600 = 1.0. Diploids were titrated (0.1, 0.01, 0.001, 0.0001; total 5 μl) and plated on agar plates lacking uracil and leucine (–URA/–LEU) for diploid growth, adenine (–ADE) for strong interaction, or histidine (–HIS) for weak interaction. Yeast diploids were scored for growth after 72 h at 30°C.
To validate the COG6-SNARE interactions in vivo we performed co-immunoprecipitation (co-IP) experiments in HeLa cells co-expressing myc tagged COG6 and either GFP-STX5, CFP-GS27, CFP-STX6, GFP-STX16, CFP-SNAP29, or (as negative controls) GFP or CFP-BET1. IP of the SNARE with GFP antibodies generally confirmed the interactions detected by the yeast two hybrid approach. COG6-myc strongly interacted with SNAP29 and GS27 (~8% and 6% co-immunoprecipitation, respectively) (Figure 3 A, B). Weaker, yet reproducible, interactions were also observed for the COG6-STX6, COG6-STX16 and COG6-STX5 pairs. To demonstrate that these interactions are also occurring with endogenous proteins, we performed a native IP using COG3, COG4, and COG6 antibodies, followed by detection of the endogenous GS27 co-immunoprecipitated (Figure 3C). Under these conditions only the COG6-GS27 interaction was detected, confirming the specificity of the COG6-GS27 interaction.
Figure 3. COG6 interacts with a subset of intra-Golgi SNARE proteins.
(A) HeLa cells were transiently cotransfected with plasmids encoding fluorescent tagged SNAREs and COG6-myc. 24 h post-transfection, cells were collected, lysed, and GFP-SNARE interacting proteins were precipitated with anti-GFP antibodies. Immuno-precipitates, along with 10% of total input, were separated on a 12% SDS-PAGE gel, transferred to a nitrocellulose membrane, and probed with antibodies against myc (upper panel) and GFP (lower panel). Band densitometry was evaluated using Odyssey software. Co-immunoprecipitation efficiency values were calculated by dividing Co-IP by input. (B) COG6-myc demonstrated its strongest interaction with CFP-SNAP29 (8%) and CFP-GS27 (6%). (C) NRK cells were lysed and COG-interacting proteins were immunoprecipitated with affinity-purified anti-COG3, COG4 and COG6 IgGs. Immuno-precipitates, along with 10% of total input and flow through, were separated on a 12% SDS-PAGE gel, transferred to a nitrocellulose membrane, and probed with antibodies against GS27, COG3, COG4, and COG6.
The SNARE-interacting domain of COG6 is required for COG6 Golgi targeting
To identify the specific region of COG6 involved in the interaction with its cognate SNARE proteins, we created a set of yeast two hybrid plasmids encoding COG6 fragments fused with the AD domain. Yeast two hybrid experiments showed that a peptide, consisting of residues 76-150 (denoted herein after as SNARE-binding motif (SBM)), was capable of interacting with the SNAREs GS27, SNAP29, STX5, and STX6, while no interaction was detected for the other domains of the protein (Figure 4A, Supplemental Figure 2). During preparation of this manuscript, the Lev lab identified a region of COG6 which is capable of interaction with STX6 (aa81-165) (18), consistent with our results. To verify that the SBM region of COG6 is capable of binding to COG6 interacting SNARE proteins, recombinant GS27-His was incubated with GST-COG6-76-150 or with GSTSec22 as a negative control and GST was pulled by incubation of glutathione-sepharose beads. GS27-His was pulled by GST-COG6-76-150 but not with GSTSec22 (Figure 4B), indicating that the SBM of COG6 is sufficient and specific for interaction with COG6 interacting SNARE proteins. Interaction with GSTCOG6-76-150 was also detected with STX6-his (Supplemental Figure 3). Furthermore, Co-IP of COG6-myc mutants with CFP-GS27 confirmed the yeast two hybrid and GST pull-down results. Interaction of CFP-GS27 with COG6 carrying a deletion of the SBM peptide was significantly (greater than 3-fold) reduced as compared to the full-length protein, indicating that this region of COG6 is essential for the COG6-SNARE interaction (Figure 4C). Two partial deletions in the SBM domain of the COG6 protein also showed a significantly decreased interaction with CFP-GS27 when compared to full length, indicating that the entire SBM region is necessary for the COG6-GS27 interaction. Importantly, COG6 carrying the internal deletion was stable upon expression in HeLa cells.
Figure 4. Characterization of COG6 SBM domain.
(A) Summary of yeast two hybrid interaction data for diploids expressing BD-SNAREs and AD-hCOG6 truncated mutants. (B) Recombinant GST-COG6 76-150 or GST-Sec22 were incubated with GS27-His. After incubation a pulldown assay using glutathione-sepharose beads was performed. Eluates were separated on a 12% SDS-PAGE gel and then stained with coomassie. * indicates non specific band from His-GS27 purification. (C) HeLa cells transiently co-transfected with plasmids encoding CFP-GS27 and COG6-myc deletion mutants. 24 h post-transfection, cells were collected, lysed, and precipitated with anti-GFP antibodies. COG6-myc missing amino acid residues 76-150 resulted in ~3 fold reduction of interaction with CFP-GS27 as compared to full length COG6-myc, ~7 fold reduction of interaction when amino acids 76-112 or 112-150 were removed. (D) HeLa cells grown on cover slips were transiently transfected with DNA constructs encoding either full length COG6-myc, or truncated COG6-myc Δ76-150 for 24 h, fixed, and stained with myc-specific antibodies to visualize COG6-myc and with anti-Giantin to image Golgi. In mildly expressing cells, full length COG6-myc localizes to Golgi membranes (arrows) whereas COG6 Δ76-150-myc is not concentrated in the perinuclear region. Scale bar, 10 μm.
To test whether the COG6 SNARE-binding domain is important for COG6 localization, we expressed either full-length COG6-myc, or COG6-Δ76-150myc in HeLa cells. 48 h post transfection, the full-length COG6 colocalized with the Golgi marker Giantin, while COG6-Δ76-150 was diffusely distributed through the cell (Figure 4D), indicating that the SNARE-binding domain of COG6 is essential for its proper Golgi localization.
Placement of COG6 to outer mitochondrial membranes causes mitochondrial aggregation and the relocalization of COG6 partner proteins to the vicinity of the mitochondria
To gain a better understanding of the nature and hierarchy of COG-SNARE interactions, a mitochondrial mislocalization strategy (25) was employed. COG6 was fused to the mCherry fluorescent protein and the mitochondrial-targeting ActA sequence from Listeria (Figure 5A). Transient expression of mCherry-ActA (mChActA) demonstrated intense labeling of the mitochondrial network without any visible effect on Golgi structure (stained with GM130 in Figure 5B, C). Remarkably, expression of COG6-mChActA caused dramatic changes to mitochondrial morphology (Figure 5E). In COG6-mChActA expressing cells the majority of the mitochondria were aggregated, while in some cells Golgi membranes appeared to be fragmented. Importantly, aggregation of the mitochondria was not the result of possible crosslinking via COG6 dimerization, since COG6 did not dimerize in either yeast two hybrid or co-expression experiments (9).
Figure 5. Mitochondria-targeted COG6 specifically relocates COG8 and CFP-GS27.
(A) A schematic diagram of the mitochondrial targeting protein constructs. HeLa cells were co-transfected with mChActA and CFP-GS27(B), mChActA and CFP-STX6 (C), COG6-mChActA and YFP-COG3 (D), COG6-mChActA and COG8-GFP (E), COG6-mChActA and CFP-STX6 (F), COG6-mChActA and CFP-GS27 (G), COG6Δ76-150-mChActA and CFP-GS27 (H), COG6Δ76-150-mChActA and GFP-Rab6Q72L (I), and then fixed and analyzed 24 h after transfection using confocal microscopy. Cells were stained with antibodies to GM130 (B-D, F- I) to identify Golgi, or mitochondria resident protein Ox Phos ComplexV (E). Mitochondria are not clustered in cells expressing mChActA (A, B), and are clustered in cells expressing COG6-mChActA. Line plots for overlap between red, green and blue channels are shown. Scale bar, 10 μm.
We next determined the fate of the other COG subunits upon re-localization of COG6 to the mitochondria. A small fraction of YFP-COG3 (Figure 5D) and the entire population of GFP-COG8 (Figure 5E) were colocalized with COG6-mChActA-labelled mitochondria, indicating that mitochondria-located COG6 can redirect its binding partners to a new cellular location. This relocalization was specific since both the cis-Golgi resident proteins GM130 (Figure 5D, F, G, I) and p115 (Figure 5H) remained associated with fragmented Golgi membranes.
To test if the displacement of the COG complex onto mitochondria would influence the location of COG6 SNARE partner proteins, we co-expressed CFP-tagged SNAREs with COG6-mChActA and analyzed the intracellular localization of the SNAREs by confocal microscopy (Figure 5). In both control and mChActA expressing cells CFP-tagged SNAREs are preferentially localized to the Golgi area (Figure 5B, C). In agreement with COG6-STX6 protein-protein interactions, upon expression of COG6-mChActa some of the CFP-STX6 signal relocalizes to the vicinity of COG6 labeled mitochondria (Figure 5F). A significantly larger fraction of the CFP-GS27 signal was found to be co-localized with COG6-mChActA-decorated mitochondria (Figure 5G), indicating that either the CFP-GS27 protein or the GS27-bearing trafficking intermediates become re-routed from the Golgi to another cellular location (presumably to the organelle possessing the highest concentration of COG6 protein). Importantly, relocalization of CFP-GS27 to the mitochondria vicinity was dramatically reduced in cells expressing COG6- Δ76-150-mChActA (Figure 5H), indicating that the SBM region is essential for rerouting COG6 SNARE partners to mitochondria. This SBM region is specifically regulating COG6-SNARE interactions, evidenced by another COG6 partner protein, Rab6Q72L (24), which was still recruited to mitochondrial membranes decorated with COG6-Δ76-150-mChActA (Figure 5I).
Golgi complex fragments upon over-expression or depletion of COG6 protein
To gain additional insights about the role of the COG6 subunit we have used an over-expression strategy. Using multiple C and N terminally fluorescently tagged versions of COG6, we observed that the Golgi integrity (intact Golgi ribbon) was compromised in cells that over-express tagged COG6 protein, resulting in Golgi mini-stacks labeled with Golgi markers GalNAcT2-GFP and GM130 (Figure 6B, C). This Golgi phenotype was especially evident in cells that express even a modest amount of RFP-or CFP-tagged COG6 (Figure 6 B, C and data not shown). In contrast, Golgi integrity was preserved in cells that over-express simply RFP (Figure 6 A), or tagged versions of other COG subunits (not shown) suggesting that excess COG6 is specifically titrating out particular Golgi factors that are important for the maintenance of the Golgi structure. Interestingly, RFP-COG6 itself was concentrated in multiple structures colocalizing with CFP-GS27 (Figure 6B), suggesting that COG6-GS27 dis-regulated protein-protein interactions could be the cause for Golgi fragmentation. To test this hypothesis we expressed RFP-tagged COG6 lacking the SBM (RFP-COG6-Δ76-150) and observed that this hybrid protein indeed failed to induce Golgi fragmentation (Figure 6D), confirming the importance of COG-SNARE interactions for the Golgi integrity.
Figure 6. COG6 overexpression or depletion disrupts Golgi structure.
HeLa cells stably expressing GalNacT2-GFP were transiently transfected with RFP (A), RFP-COG6 and CFP-GS27 (B), RFP-COG6 (C) or RFP-COG6 Δ76-150 (D), and then analyzed 24 h after transfection using confocal microscopy. Cells were stained with antibodies to GM130 (C, D). Note Golgi disruption in cells transfected with full length RFP-COG6 but not when transfected with RFP-COG6 Δ76-150 or RFP. Scale bar, 10 μm. Electron micrographs of HeLa cells transfected with control (E) and COG6 (F) siRNA. 72 h after transfection cells were fixed and processed for TEM. Arrows indicate Golgi membranes. Scale bar, 500nm.
Furthermore, we found that it is not only overexpression of COG6 which is harmful for the Golgi integrity, but its depletion results in a similar phenotype. Electron microscopy examination of HeLa cells showed a long Golgi ribbon in control cells (Figure 6E). In contrast, in cells depleted for the COG6 subunit, the Golgi ribbon was fragmented to multiple Golgi ministacks located in a perinuclear area (Figure 6F).
Discussion
The COG complex belongs to a CATCHR sub-family of multi-subunit protein complexes that has long been implicated in the tethering of membrane trafficking intermediates at different steps of the secretory pathway (3) but the precise mechanism by which these complexes orchestrate vesicular approach, recognition, tethering, and fusion is still unclear. It is becoming increasingly evident that different subunits of CATCHR complexes are playing distinct roles in tethering processes, and the COG complex appears to be following the same trend. In addition to COG-SNARE interactions (18, 20, 21, 26), COG subunits are known to interact with coiled-coil tethers, such as COG7 interactions with golgin-84 (27) or COG2 interactions with p115 (28). Furthermore, the COG subunits COG4 and COG7 have been shown to have a major impact on the stability of Golgi glycosyltransferases (14, 29). The bulk of the COG complex is associated with the Golgi membrane, however the exact nature of the COG membrane receptor (or receptors) is also not known. Most obvious candidates for the COG membrane receptors are SNAREs and small GTP binding proteins associated with the Golgi membrane.
It was shown that SNAREs are the major partner protein family for all CATCHR complexes (18, 21, 30-32), and subsequently it was proposed that one of the evolutionary conserved functions of vesicle tethering complexes is to assist and/or stabilize SNARE complex formation. Other possible roles for direct CATCHR-SNARE interaction may include the recycling of SNARE-containing vesicles and the arrangement of multiple t-SNARE complexes on the acceptor membrane that is necessary for the efficient membrane fusion reaction. In this work we have discovered that in addition to STX5 and STX6 that were previously shown to interact with COG6 (18, 21), the STX5 partner SNARE GS27 and another Golgi SNARE SNAP29 binds to COG6. SNAP29 will also bind bind to STX6 (33) forming a complex that is distinct from the well-studied STX16/STX6/Vti1/VAMP4 SNARE assembly, suggesting that the COG complex may regulate an additional SNAP29/STX6-containing SNARE complex. In support of this hypothesis it was recently found that both SNAP29 and COG physically interact with the BLOC1 complex that is responsible for specialized cargo sorting in the endosome-to-Golgi retrograde trafficking pathway (34).
Our results indicate that COG6 interacts with all SNARE partners via the universal N-terminally located SNARE Binding Motif (SBM). Crystal structure of the COG6 protein has not been solved for any organism yet, therefore we used the Phyre2 protein structure prediction server www.sbg.bio.ic.ac.uk/phyre2 (35) to model the COG6 protein based on the known structure of other CATCHR family members. This prediction utilized known CATCHR proteins as a template, and predicted 75% of the COG6 residues with more than 90% confidence. The resulted 3D model (Suppl. Figure 2) indicates that the SBM region (red) indeed could form a separate domain at the base of the elongated tower-like structure. The COG6 tower-like structure is similar to rod-like elements found in other CATCHR proteins (3), and is likely assisting in capturing and tethering transport vesicles. Interestingly, Phyre2 prediction program found a significant homology between SBM domain and the independently folded Habc domain (36) of SNARE proteins STX12, STX10 and STX1. The Habc domain is known to interact with the SNARE domain (37), indicating that COG6 SBM domain could use the same mechanism for its interaction with SNARE partners. The SBM domain also shares structural similarity with the GAT domain of GGA1 protein that is responsible for the attachment of GGA1 to TGN (38). Furthermore, based on our results, we hypothesize that the SBM is the domain which is responsible for attachment of COG6 to membranes. By attaching to membranes at its base, the stalk domain of COG6 would then be available for interaction with incoming vesicles. An alternative model is that the SBM containing domain will interact with SNARE proteins attached to vesicles, tethering the vesicles to Golgi membranes for subsequent fusion.
While we clearly show that the SBM of COG6 is responsible for interaction with SNAREs, this domain is not responsible for all COG6 interactions. COG6 lacking this domain will recruit another known binding partner, Rab6. We can hypothesize then that COG6 functions by interacting with multiple binding partners, either simultaneously or sequentially. Further in vitro experiments with protein partners identified in this study will shed new light on the exact sequence of protein-protein interactions that lead to COG-regulated tethering and subsequent fusion of retrograde intra-Golgi vesicles.
In this study, we have begun the molecular dissection of the COG6 subunit of the COG complex by uncovering a complete set of COG6-interacting Golgi SNARE proteins, defining a universal COG6 SBM sequence, and providing evidence that the SBM is playing an important role in COG6 Golgi localization and function.
Materials and Methods
Reagents and antibodies
Protein G agarose beads were purchased from Roche. Antibodies used for immunofluorescence (IF) microscopy or western blotting (WB) were purchased through commercial sources, gifts from generous individual investigators, or generated by us via affinity purification. Antibodies (and their dilutions) were as follows:
Rabbit polyclonal antibodies
Myc (Bethyl Laboratories) WB 1:10000, IF 1:4000; COG3 (this lab) IP 1ug/rxn; COG4 (this lab) IP 1ug/rxn; COG6 (this lab) IP 1ug/rxn, IF 1:1000, WB 1:1000; COG7 (this lab) WB: 1:1000; COG8 (this lab) WB 1:1000; GS27 (Synaptic Systems) WB 1:400; Giantin (Covance) IF 1:2000.
Mouse monoclonal antibodies
GFP (Molecular Probes) IP 1ug/rxn; GFP (Covance) WB 1:2000; GM130 (BD Biosciences) IF 1:400; COG3 (this lab) WB 1:1000; p115 (tissue culture supernatant) IF 1:100; Ox Phos ComplexV (Invitrogen) IF 1:400; Myc (Cell Signaling) IF 1:2000; β-Actin (Sigma) WB 1:10000.
Secondary IRDye 680 goat anti-rabbit, IRDye 700 goat anti-mouse and IRDye 800 donkey anti-goat for WB were obtained from LI-COR Biosciences. Anti-rabbit HiLyte 488, HiLyte 555, and DyLight647 for IF were obtained from AnaSpec and Jackson ImmunoResearch, Inc.
Cell culture
Control, cog7 and cog8 mutant fibroblasts were obtained from Dr. Hudson Freeze (Burnham Medical Research Institute). HeLa cells were purchased from ATCC. Cells cultured in DMEM/F-12 50/50 medium supplemented with 15 mM HEPES, 2.5 mM L-glutamine, 10% FBS, with or without 1% antibiotic/antimycotic (100 U/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B). Cells were grown at 37°C and 5% CO2 in a 90% humidified incubator. YFP-COG3 stable cell lines were generated as previously described (21). HeLa cells stably expressing GalNAcT2-GFP were obtained from Dr. Brian Storrie (UAMS) and were cultured in DMEM/F-12 50/50 medium supplemented with 15 mM HEPES, 2.5 mM L-glutamine, 10% FBS, and 0.4 mg/ml G418 sulfate. Cell culture media was obtained from Thermo Scientific and serum from Atlas Biologicals.
siRNA-induced knockdowns
siRNA duplexes for COG6 (14, 19) or Rab6 ((39) was obtained from Dharmacon (Chicago, IL). Transfection was performed using either oligofectamine or lipofectamine RNAiMAX siRNA Transfection Reagent (Invitrogen), following a protocol recommended by Invitrogen and cells were analyzed 72h after transfection.
Plasmid preparation and transfection
Yeast and mammalian expression constructs were generated using standard molecular biology techniques or obtained as generous gifts.
For yeast two hybrid
cDNA constructs expressing SNARE proteins were purchased from Open Biosystems. DNA fragments encoding cytoplasmic domains of SNAREs were subcloned into pGBDU vector as EcoR1/Sal1 or BglII/Xho1 inserts and verified by sequencing.
For IF/IP
Human COG6 with deletions of residues 76-150, 76-112, or 112-150 were generated by PCR using either hCOG6-3myc, RFP-hCOG6, or hCOG6-mCherry-C1-ActA as templates. To generate fluorescently-tagged chimeras full-length proteins were inserted into pCFP-C1, pEGFP-C1 (BspE1/BamHI) or pmCherry-C1-ActA (BspE1/NdeI) vectors. pCFP-GS27 and pCFP-STX6 were gifts from Dr. R. Scheller, Plasmids were isolated from bacterial cells using the QIAprep Spin Miniprep Kit (Qiagen). Plasmid DNA transfections into tissue culture cells were performed with Lipofectamine 2000 (Invitrogen) or with FuGene (Roche), according to the manufacturer's protocol. Transfection efficiency averaged 75%. Expression of tagged COG6 ranged from 2.5X higher to equal concentration compared to endogenous COG6. The expression of tagged COG6 did not correlate to its co-transfection partners, indicating that fluorescent tagged SNAREs had no affect on COG6 expression levels.
Immunofluorescence microscopy (IF)
Cells were grown on 12 mm glass coverslips (#1, 0.17 mm thickness) one day before transfection. After transfection cells were fixed and stained as described previously(14). In short, cells were fixed in 4% paraformaldehyde (16% stock solution; Electron Microscopy Sciences). Cells were then treated with 0.1% Triton X-100 for one minute. After that the cells were treated with 50 mM ammonium chloride for 5 min. Cells were washed with PBS and blocked twice for 10 min with 1% BSA, 0.1% saponin in PBS. Cells then were incubated for 1 hour with primary antibody diluted in antibody buffer (1% cold fish gelatin, 0.1% saponin in PBS) at room temperature. Cells were washed four times with PBS and incubated for 30 min with fluorescently tagged secondary antibody (1:400 HiLyte; AnaSpec) in antibody buffer at room temperature. After that, coverslips were washed four times with PBS, rinsed with ddH2O, and mounted on glass microscope slides using Prolong® Gold antifade reagent (Invitrogen). Cells were imaged with the 63X oil 1.4 numerical aperture (NA) objective of a LSM510 Zeiss Laser inverted microscope outfitted with confocal optics. Image acquisition was controlled with LSM510 software (Release Version 4.0 SP1). The “RGB profiler” plug-in of Image J (http://rsbweb.nih.gov/ij) was used to generate line plots for individual channels.
SDS–PAGE and Western blotting (WB)
SDS–PAGE and WB were performed as described previously (20). The blots were incubated first with primary antibodies, and then with a secondary immunoglobulin G (IgG) antibody conjugated with IRDye 680 or IRDye 800 dyes. The blots were scanned and analyzed with an Odyssey Infrared Imaging System (LI-COR). At least three independent experiments were performed to calculate both mean and standard deviation values.
Yeast two-hybrid assay
The following constructs were used in yeast two-hybrid assay: COG6 subunit of mammalian COG complex cloned as C-terminal fusion construct with Gal4 AD (prey) and SNARE constructs lacking transmembrane domain were cloned as C-terminal fusion constructs with Gal4 BD (bait) by recombination cloning in yeast. The Gal4 two-hybrid system was used as previously described (40). Reporter strains of opposite mating types LY612 (Mat a) and LY613 (Mat α; trp1–901 leu2–3,112 ura3–52 his3–200 gal4 Δgal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) were transformed with bait and prey fusion constructs. The growth of independent transformants was assessed on media lacking leucine (prey fusions) and uracil (bait fusions). Diploid yeasts containing combinations of the human COG6 and SNAREs, as well as empty vectors (negative controls) and pair-wise combinations of human COG2/4 (positive control; (9)) were created by mass mating. After mass mating, yeast diploids were selected on –LEU/–URA plates. Diploids were grown in liquid –Leu/–Ura medium to the same optical density (D600 = 0.8) and titrated 1:10, 1:100, 1:1,000, and 1:10,000 in ddH2O. 5 μl of each dilution was applied on a selective medium lacking adenine, histidine, and leucine/uracil, incubated at 30°C for 72 h, and scored for growth. Expression of all hybrid molecules was verified by immunoblotting. The relative stringency of direct protein–protein interaction was estimated by growth assay on agar plates lacking either histidine (–HIS; leaky regulation, suitable for weak protein–protein interactions) or adenine (–ADE; high stringency regulation, suitable for strong protein– protein interactions).
GST pulldown assay
Recombinant GST-COG6 76-150 or GST-Sec22 were incubated with either GS27-His or STX6-His overnight in a 50mM Tris-HCl 150mM NaCl pH 7.4 buffer. After incubation the reactions were spun at 14,000k RPM to remove any large precipitates. The supernatant was then mixed with 10uL of Glutathione sepharose beads and rotated at room temperature for 30min. After rotation, beads were washed 3 times in TBS, and then proteins were eluted in 30uL of 10mM glutathione pH8. Aliquot of protein load, and eluates were separated on a 12% SDS-PAGE gel and then stained with coomassie stain.
Immunoprecipitations (IP)
Cells were collected by trypsinization, and lysed in a 1% Triton X100 PBS buffer with 10μL/mL of 100X Halt protease inhibitor cocktail and 2 μL/mL PMSF for 1hr on ice. Cell lysate (500 μL) was spun at 20,000×g and 90% of the supernatant was added to GFP antibodies and incubated on ice overnight. The next day, 20 μL of protein G agarose beads (Roche) (or 60 μL of 30% beads) were added to each reaction and the samples incubated at 4°C on a rotator for 2h. Unbound material was removed and the beads were washed 4X in 0.05% TritonX100 in PBS and eluted in 2X sample buffer to a total volume of 50 μL. GFP IP efficiencies average 50-75%. Each IP experiment was repeated a minimum of three times.
Transmission Electron Microscopy (TEM)
Samples were treated according to Valdivia's lab protocol (41) with some modifications. In short, cells were fixed for 20 min on ice with 2.5% glutaraldehyde and 0.05% malachite green (EMS) in 0.1M sodium cacodylate buffer, pH 6.8. Samples were post-fixed for 30 min at room temperature with 0.5% osmium tetroxide and 0.8% potassium ferricyanide in 0.1 M sodium cacodylate, for 20 min on ice in 1% tannic acid, and for 1 h in 1% uranyl acetate at room temperature. Specimens were dehydrated with a graded ethanol series, and embedded in Araldite 502/Embed 812 resin (EMS). Ultrathin sections were imaged at 80 kV on a FEI Technai G2 TF20 transmission electron microscope. Digital images were acquired with a FEI Eagle 4kX USB Digital Camera.
Supplementary Material
Acknowledgements
We are thankful to R.Duden, H. Freeze, F. Hughson, A.Linstedt, S. Munro, R. Scheller, A. Shestakova, B. Storrie, D.Ungar and others who provided reagents and critical reading of the manuscript. We also would like to thank Bridgette Rooney and Jacob Szwedo for technical support. This work was supported by the National Science Foundation (MCB-0645163) (V.L) and the National Institute of Health (1R01GM083144-01A1) (V.L.).
Abbreviations
- COG
conserved oligomeric Golgi
- mCh
mCherry
- SBM
SNARE binding motif
- SNARE
soluble N-ethylmalemide-sensitive fusion attachment protein receptor
Footnotes
Author contributions
Experiments were designed by R.W., T.K., and V.L.; T.K. performed yeast two-hybrid screen; R.W. and T.K. performed co-IP and IF analysis; I.P. performed electron microscopy; R.W. and V.L. prepared the manuscript.
Conflict of interest statement
None declared.
References
- 1.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116(2):153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 2.Cottam NP, Ungar D. Retrograde vesicle transport in the Golgi. Protoplasma. 2012;249(4):943–955. doi: 10.1007/s00709-011-0361-7. [DOI] [PubMed] [Google Scholar]
- 3.Yu IM, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annual Reviews Cell Develompental Biology. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- 4.Miller VJ, Ungar D. Re'COG'nition at the Golgi. Traffic. 2012;13(7):891–897. doi: 10.1111/j.1600-0854.2012.01338.x. [DOI] [PubMed] [Google Scholar]
- 5.Lupashin V, Ungar D. COG complex. In: Mironov A, Pavelka M, editors. The Golgi Appartus. SpringerWien; New York: 2008. pp. 120–129. [Google Scholar]
- 6.Whyte JR, Munro S. The Sec34/35 Golgi Transport Complex Is Related to the Exocyst, Defining a Family of Complexes Involved in Multiple Steps of Membrane Traffic. Developmental Cell. 2001;1(4):527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- 7.Ungar D, Oka T, Brittle EE, Vasile E, Lupashin VV, Chatterton JE, Heuser JE, Krieger M, Waters MG. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. Journal of Cell Biology. 2002;157(3):405–415. doi: 10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fotso P, Koryakina Y, Pavliv O, Tsiomenko AB, Lupashin VV. Cog1p plays a central role in the organization of the yeast conserved oligomeric golgi complex. Journal of Biological Chemistry. 2005;280(30):27613–27623. doi: 10.1074/jbc.M504597200. [DOI] [PubMed] [Google Scholar]
- 9.Ungar D, Oka T, Vasile E, Krieger M, Hughson FM. Subunit architecture of the conserved oligomeric golgi complex. Journal of Biological Chemistry. 2005;280(38):32729–32735. doi: 10.1074/jbc.M504590200. [DOI] [PubMed] [Google Scholar]
- 10.Shite S, Seguchi T, Yoshida T, Kohno K, Ono M, Kuwano M. A new class mutation of low density lipoprotein receptor with altered carbohydrate chains. Journal of Biological Chemistry. 1988;263(36):19286–19289. [PubMed] [Google Scholar]
- 11.Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. Journal of Cell Biology. 2005;168(5):747–759. doi: 10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson BC, Smith RD, Ungar D, Nakamura A, Jeffrey PD, Lupashin VV, Hughson FM. Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proceedings of the National Academy of Science U S A. 2009;106(32):13329–13334. doi: 10.1073/pnas.0901966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40(Database issue):D700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pokrovskaya ID, Willett R, Smith RD, Morelle W, Kudlyk T, Lupashin VV. Conserved oligomeric Golgi complex specifically regulates the maintenance of Golgi glycosylation machinery. Glycobiology. 2011;21(12):1554–1569. doi: 10.1093/glycob/cwr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foulquier F. COG defects, birth and rise! Biochim Biophys Acta. 2009;1792(9):896–902. doi: 10.1016/j.bbadis.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Reynders E, Foulquier F, Annaert W, Matthijs G. How Golgi glycosylation meets and needs trafficking: the case of the COG complex. Glycobiology. 2011;21(7):853–863. doi: 10.1093/glycob/cwq179. [DOI] [PubMed] [Google Scholar]
- 17.Lubbehusen J, Thiel C, Rind N, Ungar D, Prinsen BH, de Koning TJ, van Hasselt PM, Korner C. Fatal outcome due to deficiency of subunit 6 of the conserved oligomeric Golgi complex leading to a new type of congenital disorders of glycosylation. Hum Mol Genet. 2011;19(18):3623–3633. doi: 10.1093/hmg/ddq278. [DOI] [PubMed] [Google Scholar]
- 18.Laufman O, Hong W, Lev S. The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. Journal of Cell Biol. 2011;194(3):459–472. doi: 10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith RD, Willett R, Kudlyk T, Pokrovskaya I, Paton AW, Paton JC, Lupashin VV. The COG complex, Rab6 and COPI define a novel Golgi retrograde trafficking pathway that is exploited by SubAB toxin. Traffic. 2009;10(10):1502–1517. doi: 10.1111/j.1600-0854.2009.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. Journal of Cell Biology. 2002;157(4):631–643. doi: 10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V. Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. Journal of Cell Biology. 2007;179(6):1179–1192. doi: 10.1083/jcb.200705145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng BG, Kranz C, Hagebeuk EE, Duran M, Abeling NG, Wuyts B, Ungar D, Lupashin V, Hartdorff CM, Poll-The BT, Freeze HH. Molecular and clinical characterization of a Moroccan Cog7 deficient patient. Mol Genet Metab. 2007;91(2):201–204. doi: 10.1016/j.ymgme.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kranz C, Ng BG, Sun L, Sharma V, Eklund EA, Miura Y, Ungar D, Lupashin V, Winkel RD, Cipollo JF, Costello CE, Loh E, Hong W, Freeze HH. COG8 deficiency causes new congenital disorder of glycosylation type IIh. Hum Mol Genet. 2007;16(7):731–741. doi: 10.1093/hmg/ddm028. [DOI] [PubMed] [Google Scholar]
- 24.Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics. 2008;7(6):1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Sengupta D, Truschel S, Bachert C, Linstedt AD. Organelle tethering by a homotypic PDZ interaction underlies formation of the Golgi membrane network. Journal of Cell Biology. 2009;186(1):41–55. doi: 10.1083/jcb.200902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laufman O, Kedan A, Hong W, Lev S. Direct interaction between the COG complex and the SM protein, Sly1, is required for Golgi SNARE pairing. Embo J. 2009;28(14):2006–2017. doi: 10.1038/emboj.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sohda M, Misumi Y, Yamamoto A, Nakamura N, Ogata S, Sakisaka S, Hirose S, Ikehara Y, Oda K. Interaction of Golgin-84 with the COG complex mediates the intra-Golgi retrograde transport. Traffic. 2010;11(12):1552–1566. doi: 10.1111/j.1600-0854.2010.01123.x. [DOI] [PubMed] [Google Scholar]
- 28.Sohda M, Misumi Y, Yoshimura S, Nakamura N, Fusano T, Ogata S, Sakisaka S, Ikehara Y. The interaction of two tethering factors, p115 and COG complex, is required for Golgi integrity. Traffic. 2007;8(3):270–284. doi: 10.1111/j.1600-0854.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 29.Peanne R, Legrand D, Duvet S, Mir AM, Matthijs G, Rohrer J, Foulquier F. Differential effects of lobe A and lobe B of the Conserved Oligomeric Golgi complex on the stability of {beta}1,4-galactosyltransferase 1 and {alpha}2,6-sialyltransferase 1. Glycobiology. 2011;21(7):864–876. doi: 10.1093/glycob/cwq176. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Victoria FJ, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-golgi network. Mol Cell Biol. 2009;29(19):5251–5263. doi: 10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivaram MV, Saporita JA, Furgason ML, Boettcher AJ, Munson M. Dimerization of the exocyst protein Sec6p and its interaction with the t-SNARE Sec9p. Biochemistry. 2005;44(16):6302–6311. doi: 10.1021/bi048008z. [DOI] [PubMed] [Google Scholar]
- 32.Kraynack BA, Chan A, Rosenthal E, Essid M, Umansky B, Waters MG, Schmitt HD. Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol Biol Cell. 2005;16(9):3963–3977. doi: 10.1091/mbc.E05-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong SH, Xu Y, Zhang T, Griffiths G, Lowe SL, Subramaniam VN, Seow KT, Hong W. GS32, a novel Golgi SNARE of 32 kDa, interacts preferentially with syntaxin 6. Mol Biol Cell. 1999;10(1):119–134. doi: 10.1091/mbc.10.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gokhale A, Larimore J, Werner E, So L, Moreno-De-Luca A, Lese-Martin C, Lupashin VV, Smith Y, Faundez V. Quantitative Proteomic and Genetic Analyses of the Schizophrenia Susceptibility Factor Dysbindin Identify Novel Roles of the Biogenesis of Lysosome-Related Organelles Complex 1. J Neurosci. 2012;32(11):3697–3711. doi: 10.1523/JNEUROSCI.5640-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94(6):841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 37.Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. Embo J. 1999;18(16):4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins BM, Watson PJ, Owen DJ. The structure of the GGA1-GAT domain reveals the molecular basis for ARF binding and membrane association of GGAs. Dev Cell. 2003;4(3):321–332. doi: 10.1016/s1534-5807(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y, Shestakova A, Hunt L, Sehgal S, Lupashin V, Storrie B. Rab6 regulates both ZW10/RINT-1 and conserved oligomeric Golgi complex-dependent Golgi trafficking and homeostasis. Mol Biol Cell. 2007;18(10):4129–4142. doi: 10.1091/mbc.E07-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144(4):1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proceedings of the National Academy of Science U S A. 2008;105(27):9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.