Abstract
Trs130 is a specific component of the TRAPP II (Transport protein particle II) complex, which functions as a guanine exchange factor (GEF) for Rab GTPases Ypt31/32. Ypt31/32 is known to be involved in autophagy, although the precise mechanism has not been thoroughly studied. In this study, we investigated the potential involvement of Trs130 in autophagy and found that both the cytoplasm-to-vacuole targeting (Cvt) pathway and starvation-induced autophagy were defective in a trs130ts (trs130 temperature-sensitive) mutant. Mutant cells could not transport Atg8 and Atg9 to the preautophagosomal structure/ phagophore assembly site (PAS) properly, resulting in multiple Atg8 dots and Atg9 dots dispersed in the cytoplasm. Some dots were trapped in the trans-Golgi. Genetic studies showed that the effect of the Trs130 mutation was downstream of Atg5 and upstream of Atg1, Atg13, Atg9 and Atg14 on the autophagic pathway. Furthermore, overexpression of Ypt31 or Ypt32, but not of Ypt1, rescued autophagy defects in trs130ts and trs65ts (Trs130-HA Trs120-myc trs65Δ) mutants. Our data provide mechanistic insight into how Trs130 participates in autophagy and suggest that vesicular trafficking regulated by GTPases/GEFs is important in the transport of autophagy proteins from the trans-Golgi to the PAS.
Keywords: TRAPP II, GEF complex, Ypt31/32, Rab GTPases, autophagy
Introduction
Rab (Ypt in yeast) GTPases are critical for vesicle trafficking in eukaryotic cells (1, 2). Rab/Ypt is activated from the inactive GDP state to the active GTP state by guanine nucleotide exchange factor (GEF) (1, 3). Three TRAPP complexes (I, II and III) are reported for their GEF activities for Ypt1 and Ypt31/32 (4–11). TRAPP I, with subunits Bet3, Bet5, Trs20, Trs23 and Trs31, is required for endoplasmic reticulum-to-Golgi transport (12, 13). TRAPP III, which has the components of TRAPP I plus Trs85, functions in autophagy (6, 14). However, the composition and function of TRAPP II is controversial. TRAPP II was initially thought to contain components of TRAPP I plus Trs130, Trs120 and Trs65, but the precise subunit composition remains controversial. Trs33, originally reported to be a TRAPP I subunit (12, 13), is suggested to be TRAPP II-specific based on genetic data (7). Recently, Tca17 was also reported to be a TRAPP II-specific subunit (15, 16). Our genetic and biochemical data support the idea that TRAPP II functions with Ypt31/32 in Golgi-to-PM and endosome-to-Golgi transport (7, 9, 10). Mutation or deletion of a TRAPP II-specific subunit impairs the GEF activity of the TRAPP complex for Ypt31/32, but not Ypt1. Conversely, Ypt31 or Ypt32, but not Ypt1, suppresses the growth phenotype of a mutant in a TRAPP II-specific subunit (7, 9, 10). Trs130 has a key role in the specificity of GEF activity of TRAPP II for Ypt31/32 (10); and Trs65 affects the protein level of Trs130 (9). There are also reports that TRAPP II functions with Ypt1 (not Ypt31) in ER-to-Golgi and endosome-to-Golgi transport (5, 8, 17, 18).
Recently, several Ypt proteins have been reported to be involved in autophagy, including the functionally redundant pair Ypt31/32 (6, 14, 19–21). However, the precise mechanism by which Ypt31/32 functions in autophagy has not been thoroughly studied. In addition, the role of TRAPP II in autophagy has not been established. Here we study its role in autophagy regardless of with which Ypt it functions.
The hallmark of autophagy is the formation of double-membrane autophagosomes. After two decades of research, the various membrane and protein trafficking processes involved in the formation of autophagosomes are just beginning to be elucidated. Atg8 and Atg9 are two key proteins that function in this process. Through a reversible posttranslational modification, Atg8 becomes associated with the precursor membrane sac at the preautophagosomal structure/ phagophore assembly site (PAS), possibly promoting membrane sac expansion. Some Atg8 molecules later dissociate, while the rest become trapped in completed autophagosomes and are eventually degraded in the vacuole (the yeast analogue of the lysosome) (22–25). Atg9 acts upstream of Atg8 (26). Atg9 is unique among known autophagy-related proteins as the only conserved integral membrane protein. Atg9 normally travels between the PAS and other peripheral sites (27, 28). This shuttling process is proposed to link membrane sources and the PAS. The molecular machinery of autophagosome formation can adapt to different cellular needs. Under starvation/stress conditions, yeasts use the molecular machinery for non-selective autophagy. Under nutrient rich/normal conditions, the same core machinery carries out a selective autophagy process, the cytoplasm-to-vacuole targeting (Cvt) pathway (29). This pathway is responsible for the transport of several zymogens (including prApe1) from the cytoplasm to the vacuole. The effect of a particular gene in autophagy can be measured by monitoring the changes of the Cvt pathway and starvation-induced autophagy in its mutant.
Here, we investigated the connection between TRAPP II and autophagy by analyzing autophagy in TRAPP II-specific subunit mutants. By monitoring the transport of Atg8, Atg9, and Ape1, we uncovered a new function of Trs130 in autophagy. Mutation of Trs130 impaired both the Cvt pathway and starvation-induced autophagy. Furthermore, autophagy defects and growth defect phenotypes in the Trs130 mutant were suppressed by Ypt31 or Ypt32, but not by Ypt1. Our data suggested that Trs130 participates in autophagy as a subunit of TRAPP II, which mediates the post-Golgi vesicular transport of Atg8 and Atg9 from trans-Golgi to the PAS. This effect was mediated by Ypt31/32, the GTPase substrate of TRAPP II, but not by Ypt1.
Results
Trs130 and other TRAPP II-specific subunits are required for both the Cvt pathway and starvation-induced autophagy
Mutation of TRAPP II-specific subunit affects the GEF activity of the TRAPP complex on Ypt31/32 (7, 9, 10) and/or the assembly of the TRAPP II complex (7, 15). Recently, Ypt31/32 were reported to be involved in autophagy (20). To examine the potential participation of Trs130 and other TRAPP II-specific subunits in the regulation of autophagy, we constructed mutants of TRAPP II-specific subunits and monitored the transport of Ape1 and Atg8 to investigate Cvt pathway status and starvation-induced autophagy.
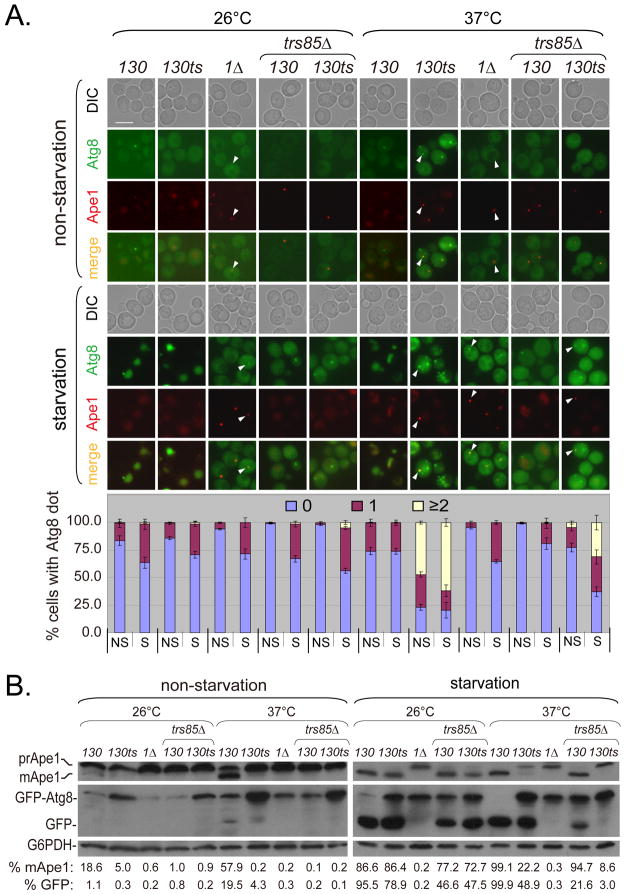
Under non-starvation conditions (in nutrient-rich medium) at a permissive temperature (26°C), Atg8 existed in the cytosol, while Ape1 entered the vacuole in both wild-type (WT) and trs130ts mutant cells. Occasionally, a single Atg8 dot and/or single Ape1 dot per cell can be seen. In contrast, percentages of cells with multiple (≥2) Atg8 dots per cell or with Ape1 dots increased in trs130ts mutant cells at the restrictive temperature (37°C) (Figure 1A upper). Similar results were found in WT and trs65ts mutant cells (Figure S1A upper). Cells with multiple Atg8 dots were observed in trs120Δ (trs120Δ+Ypt31) and trs33Δ (trs33Δ Trs130-HA) mutants under non-starvation conditions at 37°C, although few of these types of cells were seen at 26°C, especially in the trs33Δ mutant. In the tca17ts (tca17Δ Trs130-HA Trs120-myc) mutant, cells with multiple Atg8 dots were rarely observed under non-starvation conditions at 37°C (Figures S1A, C and E). In some trs130ts and trs65ts mutant cells at 37°C, one of the Atg8 dots was seen to colocalize with Ape1, indicating that trafficking of Atg8 to the PAS was not completely stopped (Figures 1A and S1A). As a control, we used the atg1Δ mutant. Atg1 is essential for autophagosome formation, but not for the targeting of Atg8 and Ape1 to the PAS. As expected, colocalization of Atg8 dots with Ape1 dots was observed in the atg1Δ mutant. Immunoblot assays showed that maturation of Ape1 in trs120Δ or trs33Δ mutants, similar to in trs130ts, was not as efficient as in WT at either temperatures under non-starvation conditions (in YPD) (Figures 1B left, S1D and S1F left). In trs65ts mutant cells, Ape1 maturation was decreased at 37°C but not at 26°C, while Ape1 maturation in tca17ts cells was different from the other TRAPP II-specific subunit mutants (Figure S1 Bleft). We also noticed that under non-starvation conditions, the level of GFP-Atg8 protein in most mutant cells was much higher than in WT cells (Figures 1B, S1B and S1F).
Figure 1. Cvt pathway and starvation-induced autophagy are impaired in trs130ts mutant cells at the restrictive temperature.
(A) Atg8 and Ape1 were mislocalized in trs130ts mutant cells at 37°C. WT and mutant cells were tagged with GFP-Atg8 and RFP-Ape1 integration plasmids. An atg1Δ strain from the same background was used as a control. TRS85 (TRAPP III-specific subunit Trs85) was deleted from TRS130 and trs130ts cells to compare the sites of action of Trs130 (TRAPP II) and Trs85 (TRAPP III). Cells were grown and treated as stated in Materials and Methods. Experiments were repeated twice and representative results from a single experiment are presented. Arrowheads indicate colocalization of Atg8 and Ape1. Bar, 5 μm. The percentage of cells with Atg8 dots in three categories: 0, 1 and multiple dots (≥2 dots per cell) was quantitated. NS, non-starvation; S, starvation. At least 300 cells were counted in at least three fields for each strain. Error bars represent standard deviation. (B) Ape1 maturation was blocked and GFP-Atg8 degradation was reduced in trs130ts mutant cells at 37°C. Cells grown as in (A) but with YPD instead of SD-Ura were subjected to immunoblot assay. Immunoblot assay was done and blots were quantitated as stated in Materials and Methods. Only mean values are presented because of space limitations. The standard deviations were less than 10% of mean values.
We further examined cells under nitrogen starvation conditions (SD-N) (Figures 1 and S1), which leads to enhanced autophagic flux (30). In WT cells, both Atg8 and Ape1 were efficiently delivered to the vacuole. In contrast, while Atg8 and Ape1 in trs130ts and trs65ts mutants were delivered to the vacuole at 26°C, both accumulated as dots outside the vacuole at 37°C. Similar to the results in nutrient-rich medium, Atg8 existed as multiple dots (Figures 1A and S1A). In the atg1Δ control cells, Ape1 appeared as a single dot and Atg8 dots accumulated outside the vacuole (Figure 1A). Immunoblot assays showed that under starvation conditions, the extent of Ape1 maturation in trs130ts and trs65ts was similar to in WT at 26°C, but this was partially blocked at 37°C. Consistent with the defect in vacuolar transport, GFP-Atg8 was not degraded efficiently at 37°C in trs130ts and trs65ts mutants (Figures 1B and S1B). In the control atg1Δ cells, Ape1 maturation and GFP-Atg8 degradation were blocked in all conditions tested (Figure 1B). Atg8 transport was not impaired in tca17ts, but was impaired in trs120Δ and trs33Δ mutant in cells treated under starvation conditions for a longer time (≥2 hours) (Figures S1C and S1E). When tca17ts cells were treated under starvation conditions for a shorter time (0.5 hour), the percentage of cells with multiple Atg8 dots significantly increased compared to WT (Figures S1A). Immunoblot data for GFP-Atg8 degradation in the mutants were consistent with fluorescent microscopy data (Figures S1B, S1D and S1F).
The TRAPP III-specific subunit Trs85 is involved in autophagy (6, 14). To compare the sites of action of TRAPP II to TRAPP III in the autophagic pathway, we deleted TRS85 in trs130ts and WT. In trs130ts trs85Δ double mutants, fewer cells had multiple Atg8 dots than in trs130ts mutants; however, in trs130ts trs85Δ double mutants, more cells had multiple Atg8 dots than in trs85Δ mutants at the restrictive temperature (Figures 1A). Inhibition of GFP-Atg8 degradation and maturation of prApe1 in trs130ts trs85Δ double mutants was greater than in either trs130ts or trs85Δ mutants (Figures 1B). This suggested that the relationship between TRAPP III and TRAPP II complexes was not a strict upstream-downstream relationship, but most likely a parallel relationship in the autphagic pathway.
We further examined the level of autophagy using a quantitative biochemical method, the Pho8Δ60 assay (31). The results showed that autophagy activity significantly decreased in trs130ts and trs65ts mutants after incubation at the restrictive temperature 36.5°C in SD-N. Partial and full recovery autophagy activity was achieved for trs130ts and trs65ts, respectively, after shifting to the permissive temperature for 2 hours, All samples consistently had low autophagy activity when cells were incubated in YPD (Figure S4A). Additionally, we verified that trs130ts and trs65ts mutants were viable after incubation in SD-N at 36.5°C for 2 hours (Figure S4B).
Together, these results indicated that the autophagic pathways (the Cvt pathway and starvation-induced autophagy) were severely defective in trs130ts mutant cells at a restrictive temperature. Other TRAPP II-specific subunit mutants were partially defective under the same conditions.
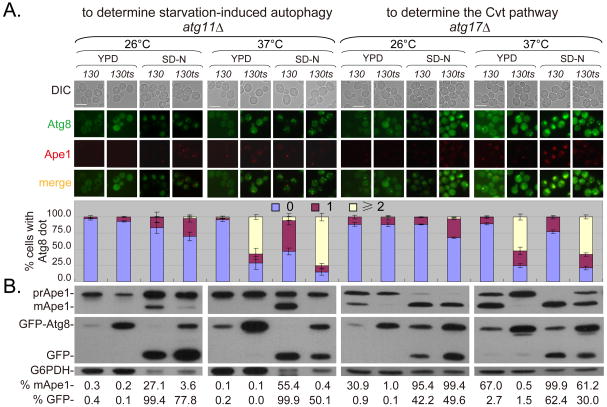
To further confirm the involvement of Trs130 in both the Cvt pathway and starvation-induced autophagy, we deleted the pathway-specific genes (ATG11 and ATG17) individually. In the absence of ATG11, only starvation-induced non-selective autophagy operated normally (32, 33), whereas in the absence of ATG17, the Cvt pathway operated normally (34). In WT-background cells with ATG11 knocked out, starvation led to significant vacuolar transport and degradation of GFP-Atg8 (Figure 2). In contrast, these processes were partially reduced at 26°C and more severely defective at 37°C in SD-N in trs130ts atg11Δ cells (Figure 2), indicating that Trs130 was required for starvation-induced autophagy. In WT-background cells with ATG17 knocked out, vacuolar transport and maturation of Ape1 occurred normally in YPD (Figure 2). However, in trs130ts cells with ATG17 knocked out, vacuolar transport and Ape1 maturation were defective at both 26°C and 37°C (Figure 2), indicating that Trs130 was involved in the Cvt pathway.
Figure 2. Involvement of Trs130 in the Cvt pathway and starvation-induced autophagy.
ATG11 and ATG17 were deleted from WT and trs130ts mutant cells and the resulting strains were incubated as in Figure 1A. Bar, 7 μm. The percentage of cell swith Atg8 dots was quantitated and presented as in Figure 1A. Immunoblot assay (bottom) was done as in Figure 1B and used to indicate change in GFP-Atg8 degradation and Ape1 maturation. Blots were quantified and presented as in Figure 1B.
Site of action for Trs130 in the autophagic pathway
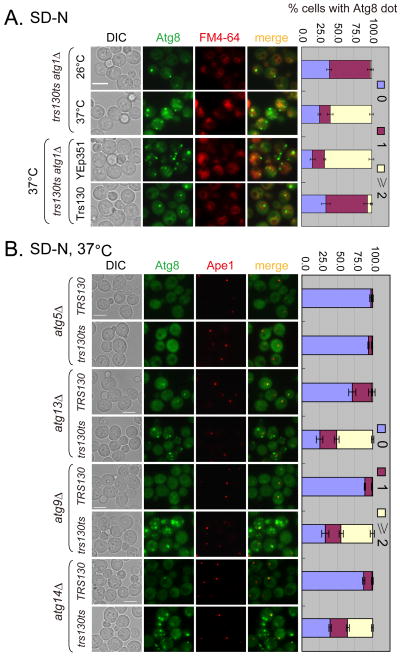
Next, we determined the step at which Trs130 functions in autophagy by genetic epistasis analysis. A hallmark of the trs130ts mutant is that Atg8 accumulates as multiple dots instead of entering the vacuole. If these dots represent trafficking intermediates before Atg8 reaches the PAS, deleting additional genes (such as ATG1) that act after Atg8 recruitment to the PAS would not change the phenotype. Alternatively, if the dots are completed autophagosomes unable to fuse with the vacuole, knocking out ATG1 would prevent their appearance (35). Consistent with the former possibility, we found that multiple Atg8 dots outside the vacuole persisted in trs130ts mutant cells at 37°C in all nutrient conditions, regardless of the presence of ATG1 (Figures 3A and S2A). Overexpression of Trs130 rescued autophagy defects in trs130ts atg1Δ mutant cells at 37°C to be like at 26°C (Figures 3A and S2A).
Figure 3. Trs130 acts in the autophagic pathway under starvation.
ATG1, ATG5, ATG13, ATG9 and ATG14 were deleted from WT and trs130ts mutants as indicated. (A) Multiple (≥2 dots per cell) GFP-Atg8 dots were formed in trs130ts atg1Δ under starvation at the restrictive temperature; this was rescued by Trs130. Cells were grown in rich medium (YPD or SD-Leu) to mid-log phase at 26°C and starved in SD-N as in Figure 1A and stained with FM4-64 during the last hour to detect the vacuole. Bar, 5 μm. (B) Multiple GFP-Atg8 dots were seen in the trs130ts mutant with deleted ATG genes as indicated except for deletion of ATG5 under starvation at the restrictive temperature. Cells were cultured and examined as in Figure 1A for starvation. Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A.
To further verify that the multiple dots in trs130ts were trafficking intermediates, we extended the analysis to delete ATG5, ATG13, ATG9 and ATG14. Atg5 is a component of the E3-like ligase complex essential for the association of Atg8 with the precursor membrane sac (36). Atg13 is a phosphoprotein in complex with Atg1; together they regulate the initiation of autophagy in yeast (28). Atg9 is a transmembrane protein shuttling between the PAS and other peripheral sites that is suggested to provide membrane resources for autophagosomes (27, 28). Atg14 is an autophagy-specific component of class III PIK3 complex I that targets the complex to the PAS (37). Deletion of ATG5 completely eliminated the formation of Atg8 dots (Figures 3B and S2B), indicating that the association of Atg8 with these dots happens after conjugation of Atg8 to phosphatidylethanolamine (PE). In contrast, the multiple-dot phenotype was not significantly affected by deleting ATG13, ATG9 or ATG14 (Figures 3B and S2B), indicating that Trs130 functioned after Atg5 and before Atg1, Atg13, Atg9 and Atg14.
Atg9 anterograde transport is defective in trs130ts and trs65ts mutants
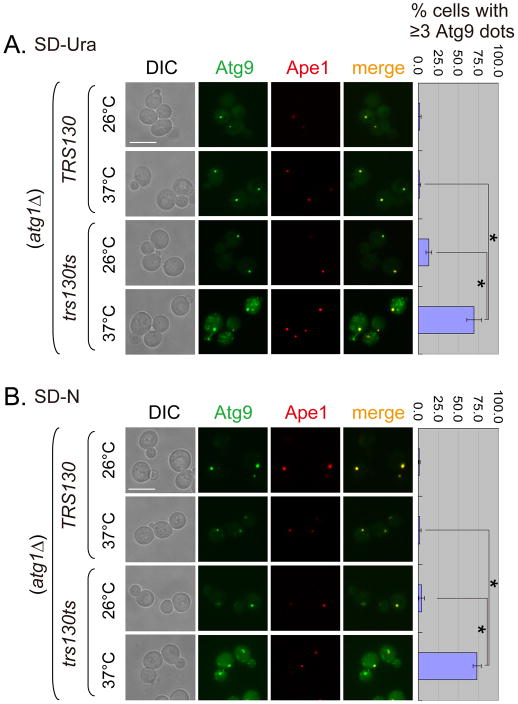
The trafficking defect of Atg8 in the trs130ts mutant prompted us to further examine Atg9 trafficking. Atg9 acts upstream of Atg8; it is required for the proper recruitment of Atg8 to the PAS (26). As mentioned above, Atg9 normally shuttles between the PAS and several non-PAS organelles (27, 28). The release of Atg9 from the PAS depends on several Atg proteins, including Atg1. Accordingly, transport of Atg9 to the PAS can be examined by knocking out ATG1 (28). When ATG1 was deleted from the WT background, Atg9 became concentrated as a bright perivacuolar dot that colocalized with the PAS marker Ape1 and the phenotype was independent of culture medium (Figure 4). In contrast, when ATG1 was deleted from the trs130ts mutant, a typical mutant cell contained multiple Atg9 puncta at 37°C, only one of which colocalized with Ape1 (Figure 4). The effect on Atg9 trafficking in the trs130ts mutant was also observed in the trs65ts mutant (Figure S3). The results suggested that the anterograde trafficking of Atg9 to the PAS was severely impaired in the Trs130 mutant.
Figure 4. Anterograde transport of Atg9 to the PAS is altered in trs130ts cells at the restrictive temperature.
WT and trs130ts mutant cells tagged with Atg9-3XGFP and RFP-Ape1 in an atg1Δ background were incubated under conditions similar to those described in Figure 1A and examined for fluorescence. (A) Multiple Atg9 dots were formed in trs130ts atg1Δ in rich medium at a restrictive temperature. WT and trs130ts mutant cells grown to mid-log phase in SD-Ura at 26°C were grown at 26°C for 1.5 hours or at 37°C for 2 hours. (B) Multiple Atg9 dots were formed in trs130ts atg1Δ under starvation at a restrictive temperature. Cells of (A) grown to mid-log phase in SD-Ura at 26°C were further starved in SD-N for 2 hours at the indicated temperatures. Bar, 7 μm. The percentage of cells with ≥3 Atg9 dots per cell was quantitated and presented as in Figure 1A. Asterisks indicate P < 0.001 as highly significant.
Subcellular localization of multiple Atg8 and Atg9 dots in the trs130ts mutant
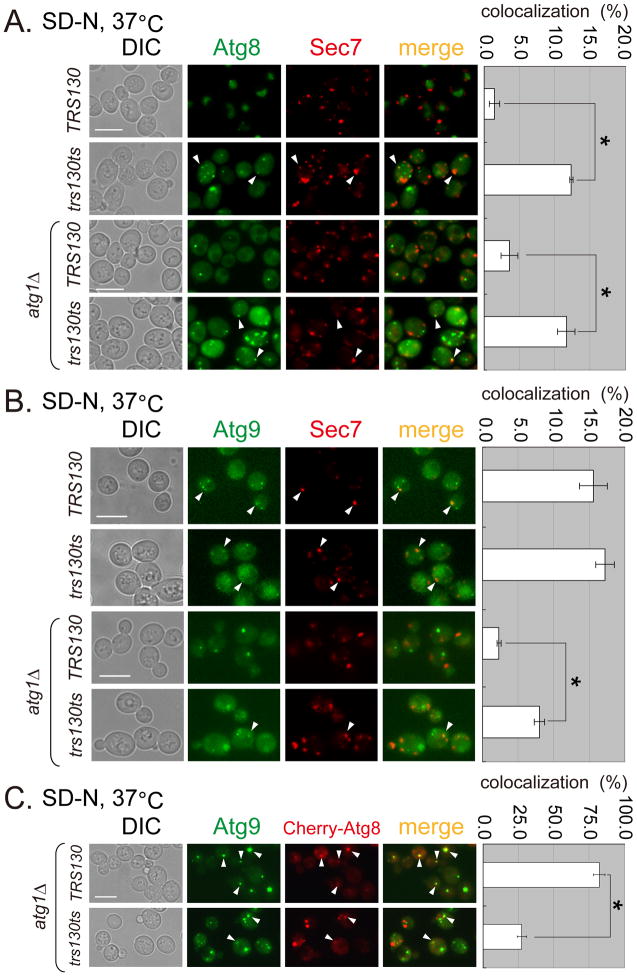
Trs130 is reported to regulate the function of Ypt31/32 in vesicles exiting the trans-Golgi (10). If Trs130-mediated vesicular trafficking connects the autophagic pathway with the secretory pathway, the loss of Trs130 function might cause mislocalization of autophagy components to the trans-Golgi. We therefore checked whether Atg8 or Atg9 colocalized with the trans-Golgi marker Sec7 in the trs130ts mutant. In WT cells cultured in SD-N at 37°C, most GFP-Atg8 was transported into the vacuole, with little if any colocalization with Sec7-DsRed (Figure 5A upper). In contrast, in the trs130ts mutant, more than 10% of the Atg8 puncta colocalized with Sec7 puncta. Colocalization of Atg8 with Sec7 in the trs130ts mutant was not significantly altered when ATG1 was deleted (Figure 5A), consistent with Trs130 acting upstream of Atg1.
Figure 5. Atg8 and Atg9 are mislocalized in trs130ts mutant cells under starvation at the permissive temperature.
WT and trs130ts mutant cells (with ATG1 or atg1Δ) carrying the indicated fluorescent fusion proteins were grown in starvation conditions as in Figure 1A and examined for fluorescence. (A) Atg8 was partly colocalized with Sec7-DsRed (trans Golgi marker) in trs130ts mutant cells at 37°C after starvation. (B) Atg9 was partly colocalized with Sec7-DsRed in WT and trs130ts mutant cells at 37°C after starvation. (C) Drastically reduced colocalization between Atg8 and Atg9 was observed in the trs130ts mutant in atg1Δ background at 37°C after starvation. Atg9-3XGFP tagged WT and trs130ts mutant cells in atg1Δ background were transformed with Cherry-Atg8 plasmid (CUP1 promoter, LEU2, 2′). Cells were grown as in Figure 1A for starvation with 25 mM CuSO4 to induce Cherry-Atg8. The percentage of colocalization was calculated as green puncta to red puncta in (A) and (B), and red puncta to green puncta in (C) for two repeated experiments for a total of about 500 cells in ten fields. Results are presented as average percentage with standard deviation (right). Bar, 7 μm. Asterisks indicate P < 0.001 as highly significant.
We then examined whether Atg9 was trapped in the trans-Golgi in the trs130ts mutant at 37°C. In WT cells, about 15% of the Atg9 dots colocalized with Sec7 puncta. Deletion of ATG1 in WT caused the redistribution of Atg9 to a single dot at the PAS that no longer colocalized with Sec7. In contrast, trs130ts mutant cells with ATG1 deleted had multiple Atg9 dots. In these cells, although the ratio of Atg9 puncta colocalized with Sec7 puncta decreased, it was still significantly higher than that in WT cells with ATG1 deleted (Figure 5B). These results indicated that for both Atg8 and Atg9, a small portion of the protein was trapped in the trans-Golgi in the trs130ts mutant at a restrictive temperature.
Since only a small portion of Atg8 and Atg9 protein was trapped in the trans-Golgi, we next examined whether the mislocalized Atg proteins assembled. We deleted ATG1 to immobilize Atg9 on the PAS and overexpressed Cherry-Atg8 from a plasmid to determine the degree of Atg8 and Atg9 colocalization. Based on Atg9 puncta, about 75% of Atg8 and Atg9 colocalized on the PAS in WT cells (Figure 5C). In contrast, the rate of colocalization decreased significantly to about 25% in trs130ts mutant cells (Figure 5C), which presumably also include those Atg8 and Atg9 proteins simultaneously trapped in the trans-Golgi.
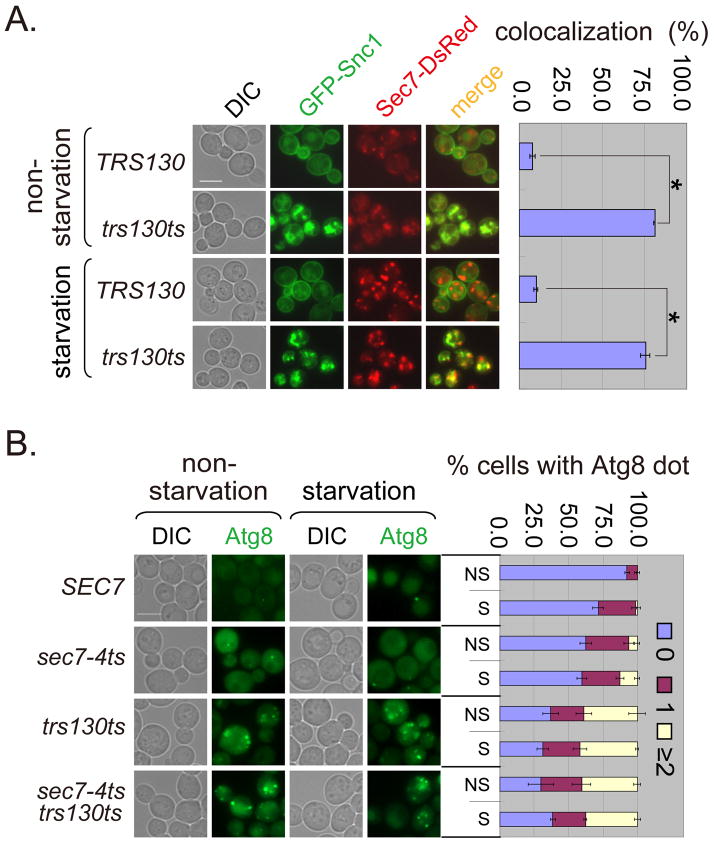
To determine whether proteins not involved in autophagy were also trapped in the trans-Golgi of trs130ts mutant at the restrictive temperature under different nutrient conditions, we examined a non-Atg protein, Snc1. Snc1 was trapped in the trans-Golgi independent of nutrient conditions in trs130ts mutant (Figure 6A). In general both Atg proteins and non-Atg proteins might need to pass through the trans-Golgi. As the Golgi is a source of yeast autophagosomal membranes and exiting the Golgi is important for autophagy (38–40), we compared the roles of Trs130 and the trans-Golgi protein Sec7 in autophagy. In the sec7-4ts mutant, Atg8 formed multiple dots per cell but at a low percentage, while the percentage of cells with multiple Atg8 dots in the trs130ts sec7-4ts double mutant was not significantly higher than in the trs130ts mutant (Figure 6B). We concluded that Trs130 functioned at or before Sec7 in the autophagic pathway.
Figure 6. Trs130 functions at the trans-Golgi for both Atg and non-Atg proteins.
(A) Non-Atg protein Snc1 accumulated at the trans-Golgi marked with Sec7-DsRed in trs130ts under both non-starvation and starvation conditions. Cells were grown as in Figure 1A. The percentage of cells with colocalization of Snc1 and Sec7 was calculated as green puncta to red puncta as in Figure 5A. (B) Trs130 did not function after Sec7 on the starvation-induced autophagy pathway. WT (SEC7 background) and mutant cells with chromosomally integrated GFP-Atg8 were grown as in Figure 1A for starvation. Cells were examined at 34°C because the SEC7 background makes mutant cells more temperature sensitive. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A. Bar, 5 μm.
Taken together, these results suggested that the loss of Trs130 function disrupted the proper assembly of Atg9 and Atg8 at the PAS, presumably by interfering with the transit of Atg8 and Atg9 through the trans-Golgi. A minor portion of Atg8 and Atg9 proteins were on the trans-Golgi, while a majority of Atg8 and Atg9 proteins were localized on structures in trs130ts mutant cells that are yet to be identified.
Ypt31, but not Ypt1, suppresses autophagy defects in trs130ts and trs65ts mutants
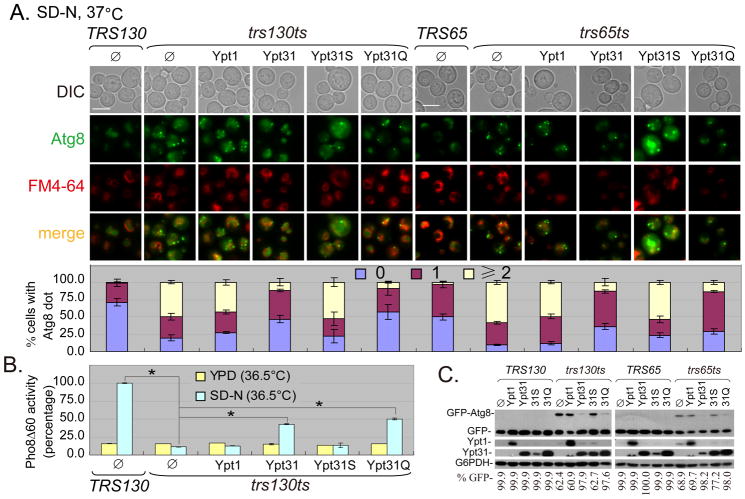
As mentioned previously, the TRAPP II complex functions as a GEF for Ypt31/32 (7, 9, 10). The temperature-sensitive growth phenotype of a trs130ts mutant can be rescued by both WT and GTP-bound forms of Ypt31/32, but not by Ypt1 (10, 41–43). A similar rescue of a temperature-sensitive growth phenotype is reported for the trs65ts mutant (9). As autophagy is impaired in both trs130ts and trs65ts mutants at the restrictive temperature, we examined whether the effect of Trs130 and Trs65 on autophagy was mediated by their role in serving as a GEF for Ypt31/32. We overexpressed WT and the GTP/GDP-bound forms of Ypt31 and Ypt1 in trs130ts and trs65ts mutants expressing GFP-Atg8, and examined the extent of suppression by small GTPases on growth and autophagy defects. Consistently, only the WT and the GTP-bound Ypt31 suppressed the growth defects of trs130ts and trs65ts mutants (Figures S4C and D). More importantly, the overexpression of either WT or GTP-bound Ypt31 suppressed autophagy defects in trs130ts and trs65ts mutants at the restrictive temperature restoring the phenotype to be similar to that of WT cells, as shown by the proper transport and degradation of GFP-Atg8. Neither Ypt1 nor the GDP-bound Ypt31 rescued the growth and autophagy defects of trs130ts and trs65ts mutants at the restrictive temperature (Figures 7A, S4C and D). A similar suppression of growth defects by Ypt1 and Ypt31 was observed in tca17ts and trs33Δ (trs33Δ Trs130-HA) mutants (Figures S4D and E). The suppression of the autophagy defect of trs130ts by WT and GTP-bound Ypt31 was confirmed with the quantitative Pho8Δ60 assay (Figure 7B). The suppression of the autophagy defects of trs130ts and trs65ts mutants was further confirmed with an immunoblot assay (Figure 7C). We verified the functionality of the control Ypt1 plasmid by its ability to suppress both the growth (Figure S4F) and autophagy defects of ypt1ts (data not shown). Like Ypt31, Ypt32 rescued the growth and autophagy defects in the trs130ts and trs65ts mutants (Figures S5A and B; data not shown for the trs65ts mutant). The overexpression of TRAPP II-specific subunits slightly suppressed the growth defect of ypt31Δ/32ts, although suppression of Atg8 transport was limited (Figures 5C and D). These data suggested that Trs130 participated in autophagy through Ypt31/32.
Figure 7. Ypt31, but not Ypt1, suppresses the GFP-Atg8 transport defect in trs130ts and trs65ts mutant cells.
The indicated Ypt1 and Ypt31 plasmids (Ypt31S for the GDP-bound form of Ypt31 and Ypt31Q for the GTP-bound form of Ypt31, all in pRS425 vectors) were transformed into trs130ts, trs65ts and WT strains. (A) WT and the GTP-bound Ypt31 suppressed the autophagy defect in trs130ts mutant cells (left) and trs65ts mutant cells (right) containing GFP-Atg8. Cells were grown as in Figure 1A for starvation and stained with FM4-64 for the vacuole during the last hour. Cells were washed and examined under a fluorescence microscope. Different plasmids in WT (TRS130 or TRS65) had similar GFP-Atg8 phenotypes and only the empty vector pRS425 (Ø) in WT is shown. Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A. (B) WT and GTP-bound Ypt31 suppressed Pho8Δ60 activity defect in trs130ts mutant cells (in the TN 1 2 4 background). For high-temperature treatment, cells were grown as in Figure 1A except at 36.5°C, where TN124 cells are more temperature sensitive than in the TRS130 background. Pho8Δ60 assay is described in Materials and Methods. Average activity of TRS130 samples from SD-N at 36.5°C was set to 100%. Data presented are average activities (with standard deviation) of four repeats from two independent experiments. Asterisks indicate P < 0.001 as highly significant. (C) WT and GTP-bound Ypt31 suppressed GFP-Atg8 degradation in trs130ts mutant cells (left) and trs65ts mutant cells (right). Immunoblots were used to examine GFP-Atg8 degradation using anti-GFP for cells in (A). Overexpression of Ypt1 or Ypt31 was confirmed with anti-Ypt1 or anti-Ypt31. G6PDH served as a loading control. White asterisks below Ypt31 bands mark traces of Ypt1 from incomplete stripping during the immunoblot assay. Blots were quantified and presented as in Figure 1B.
Discussion
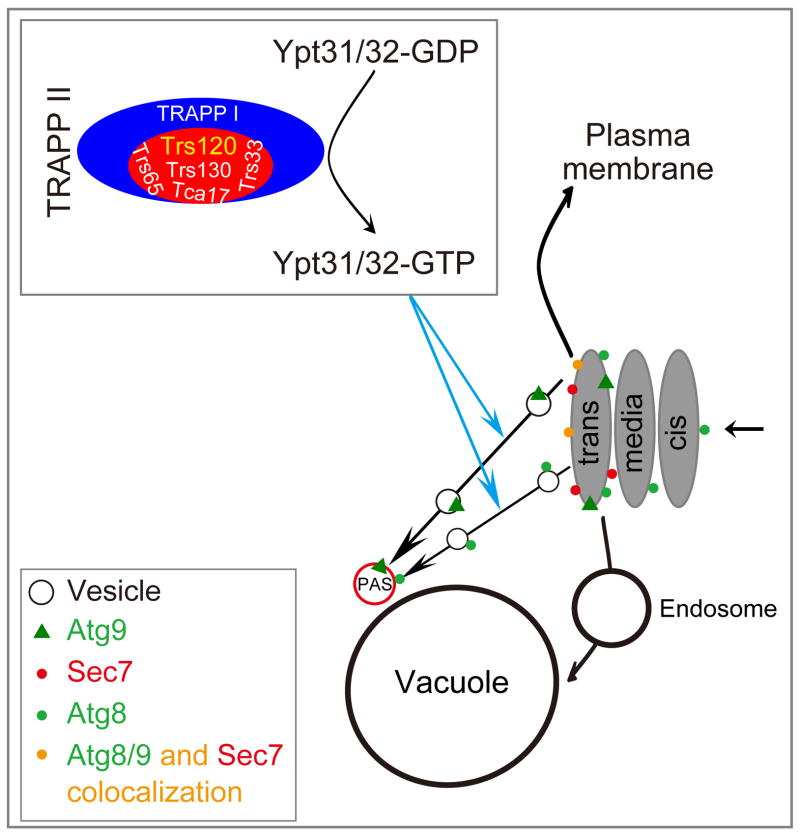
Trs130 is a TRAPP II-specific subunit; and TRAPP II is the GEF of Ypt31/32 (10). In this study, we found that the Cvt pathway and starvation-induced autophagy were impaired in trs130ts mutants at the restrictive temperature. Mislocalized Atg8 and Atg9 proteins in trs130ts mutants at the restrictive temperature did not assemble at the PAS, but showed some accumulation in the trans-Golgi. Most importantly, the defective autophagy in trs130ts (and trs65ts) mutants was rescued by Ypt31(WT), Ypt31(GTP-bound form) or Ypt32, but not by Ypt1 or the GDP-bound form of Ypt31. Therefore, TRAPP II and its downstream GTPases Ypt31/32 appear to regulate the trafficking of Atg proteins from the trans-Golgi to the PAS (Figure 8).
Figure 8. Schematic model for interpreting the effect of Trs130 and other TRAPP II-specific subunits on autophagy.
Trs130 and other TRAPP II-specific subunits assemble together with TRAPP I to form the TRAPP II complex, which switches the small GTPases Ypt31 and Ypt32 from inactive form to active form, modulating intra-Golgi protein trafficking through and traffic from the Golgi to the plasma membrane (54), and autophagy (20). Mutation of TRS130 or related TRAPP II-specific subunits interferes with autophagy, including partial blocking of Atg8/Atg9 at the trans-Golgi in trs130ts. Transports of Atg8 and Atg9 from the trans-Golgi to the PAS are shown separately because the proteins have different characteristics. No evidence indicates that Atg8 and Atg9 move together from the trans-Golgi to the PAS. The GFP-Atg8 transport defect in TRAPP II-specific subunit mutants can be suppressed with Ypt31/32 but not Ypt1. We propose that Trs130/TRAPP II regulates autophagy at least partially through Ypt31/32 with regulation of autophagy proteins.
Trs130 and Trs65 localize at the trans-Golgi (9, 10), while Ypt31/32 work between the Golgi and the plasma membrane. Therefore, we hypothesized that these proteins might participate in autophagy via regulation of vesicular trafficking of autophagic components. Some Atg proteins, particularly integral membrane proteins such as Atg9, must follow the secretory pathway to travel via the trans-Golgi. They cannot exit the trans-Golgi properly when Trs130 is impaired alone or together with Trs65. We observed multiple Atg9 dots outside the PAS in trs130ts and trs65ts mutants (Figures 4 and S3) with some presented at the trans-Golgi (Figure 5). Our finding of some Atg9 protein at the trans-Golgi is consistent with recent reports of the Golgi apparatus as an Atg9 vesicle source (44) and Golgi-related secretory pathways as a source of Atg9-containing structures (45, 46). Atg8 is not a transmembrane protein, but needs to conjugate with PE during the autophagy process to attach to membranes (22). Our data showed that mutation of Trs130 partially blocked Atg8 traffic at the trans-Golgi (Figure 5); these Atg8 molecules are likely conjugated to PE. The precise identities of the many mislocalized Atg8 and Atg9 dots in trs130ts mutant cells are still unknown (Figure 5).
The TRAPP II and TRAPP III complexes are involved in autophagy through their respective GTPases Ypt31/32 and Ypt1 ((6, 14) and this study). However, the step at which TRAPP II is involved in autophagy is likely to be distinct from TRAPP III involvement. We found multiple GFP-Atg8 dots in TRAPP II mutants at the restrictive temperature. This is the opposite of the observation that GFP-Atg8 dots are reduced in a TRS85 deletion mutant or Ypt1 mutant (6, 14, 47, 48). TRAPP III/Trs85 is on the PAS, where it activates Ypt1 (6, 14), while TRAPP II/Trs130/Trs65 is not on the PAS (6). No evidence suggests that TRAPP III activates Ypt1 before TRAPP II activates Ypt31/32 in the autophagic pathway. Our GFP-Atg8 phenotype of the trs130ts trs85Δ double mutants at a restrictive temperature was between the trs130ts and the trs85Δ single mutants, suggesting that TRAPP III and TRAPP II complexes did not have a strict upstream-downstream relationship, but a parallel relationship in the autphagic pathway (Figure 1A). This parallel function of Trs85 and Trs130 in the autophagic pathway synergistically inhibited the autophagy process, resulting in more severe inhibition of Atg8 degradation and preApe1 maturation in trs130ts trs85Δ double mutants than in trs130ts or trs85Δ single mutants alone (Figure 1B). The interplay between these two TRAPP complexes in autophagy will be a focus of our future research.
Previous genetic and biochemical data on Trs130-containing TRAPP II shows that TRAPP II acts as a GEF for Ypt1 and Ypt31/32 (10, 41–43, 49), although there are results indicate that Trs130-containing TRAPP II complex does not function as a GEF for Ypt31/32, but is a GEF for Ypt1 (8, 50). The suppression of defective autophagy in trs130ts and trs65ts mutants by Ypt31/32 (Figures 7, S4C-D and S5) was consistent with TRAPP II being a GEF for Ypt31/23. These data support the view that TRAPP II-specific subunits function in the autophagic pathway by being part of the TRAPP complex that serves as the GEF for Ypt31/32. However, the roles of Trs130 and Ypt31/32 in the autophagic pathway appear not to be identical because trs130ts and ypt31Δ/32ts cells present slightly different autophagic defects. For example, in an atg1Δ background, Atg9 forms multiple dots in trs130ts, but not in ypt31Δ/32ts (our results and (20)). Either Trs130 or Ypt31/32 might have additional independent roles in the autophagic pathway. Similarly, the mutant effects of each TRAPP II-specific subunit in autophagy were also variable (Figures 1 and S1). We hypothesize that any mutation of the TRAPP complex subunit that affects its GEF activity to a Ypt/Rab protein autophagy.
Taken together, the deletion or mutation of TRAPP II-specific subunit resulted in severe defects in the Cvt pathway and autophagy. Our results found that some Atg proteins accumulated at the trans-Golgi in trs130ts mutants, which in turn impaired normal trafficking to the PAS. Our genetic analysis suggested that autophagy defects in TRAPP II-specific subunit mutants were partially caused by the reduced activity of Ypt31/32. This suggests that the GEF-GTPase relationship between TRAPP II and Ypt31/32 is involved in Golgi traffic and autophagy (Figure 8).
Materials and Methods
Strains, plasmids and reagents
Yeast strains and plasmids used in this study are summarized in Table S1. The main TRAPP II-specific subunit mutants are trs130ts (10) and trs65ts (9). For other TRAPP II-specific subunit mutants, trs120 mutants were made by adding Ypt31 and deleting TRS120; these strains were designated as trs120Δ (trs120Δ+Ypt31) (43). A trs33 mutant was made by deleting TRS33 from a Trs130-HA background strain and designating resulting strain trs33Δ (trs33Δ Trs130-HA) because a TRS33 deletion in a Trs130-HA Trs120-myc background is lethal (7). A tca17ts mutant was made by deleting TCA17 from a Trs130-HA Trs120-myc background strain and designating the resulting strain as tca17ts (tca17Δ Trs130-HA Trs120-myc). Desired genes from WT and trs130ts/ trs65ts background were PCR amplified and transferred to strain TN124 for Pho8Δ60 activity assays. Integration plasmids for GFP-ATG8 (URA3), RFP-APE1 (LEU2), ATG9-3XGFP (URA3), GFP-SNC1 (URA3) were used to generate strains. Chromosomal gene deletion was through PCR amplification of a drug-resistance cassette with gene flanking regions for recombination. For genetic interaction experiments, the open reading frame (ORF) of YPT1 or YPT31 with 1000bp of the 5′-promoter region was cloned into the pRS425 vector (2′, LEU2) (51) and mutated to different nucleotide-bound forms of Ypt31 using overlap PCR (52). The cloned YPT1 or YPT31 gene in pRS425 was moved into pRS426 (2′, URA3) YPT32, TRS130, TRS65, and TCA17 were into YEp351 (41) using the same procedure as cloning YPT31 into pRS425. All yeast transformations used the lithium acetate method (53). Escherichia. coli transformation was by electroporation.
Antibodies used were mouse anti-GFP (IgG fraction; Molecular Probes, Eugene, OR), rabbit anti-Ape1 (a gift from Y. Ohsumi), affinity-purified rabbit anti-Ypt31 (54), affinity-purified rabbit anti-Ypt1 (21), rabbit anti-glucose-6-phosphate dehydrogenase (G6PDH, Sigma-Aldrich, MO). Goat-anti-rabbit IgG-horseradish peroxidase linked and an ECL kit were from Millipore Corporation (Billerica, MA).
All chemical reagents were from Amersco (Fair Lawn, NJ), unless otherwise noted. SynaptoRed, also known as FM4-64, was from Molecular Probes (Eugene, OR). Geneticin was from Gibco Laboratories (Grand Island, NY). Restriction enzymes and buffers were from Takara Biotechnology (Dalian) (Dalian, China).
Yeast culture conditions and induction of autophagy
For live-cell fluorescence microscopy, yeast cultures were grown at permissive temperature (26°C) in rich medium (YPD) or selection medium (when plasmid was used or as indicated in growth conditions) to mid-log phase, and grown at 26°C or switched to restrictive temperature (37°C) for 1.5 hours. If the cells were subjected to nitrogen starvation, they were washed and transferred to SD-N at 26°C for 2 hours, or pretreated for 40 min at 37°C before washing and transferring to SD-N at 37°C for 2 hours. If necessary, FM4-64 was added to a final concentration of 1.6 μM to stain the vacuole in the last hour of incubation. Cells on slides were examined with a Nikon inverted research microscope Eclipse Ti (Tokyo, Japan). More than five fields were collected for each sample. Each experiment was repeated at least twice. Fluorescence microscopy data were quantitated as indicated in the figure legends.
Immunoblot Assay
Immunoblot assays were conducted as previously described (9) and repeated at least twice. Blots were immunoblotted with anti-Ape1 to determine the degree of the conversion of Ape1 from preApe1 to mApe1, with anti-GFP to determine GFP-Atg8 degradation, and anti-G6PDH as a loading control. Blots from two independent experiments were quantified with ImageJ software (National Institute of Health) for band density. The percentage of free GFP (% GFP) was calculated as GFP/(GFP-Atg8 + GFP) X 100% and mature Ape1 (% mApe1) was calculated as mApe1/(preApe1 + mApe1) X 100%.
Pho8Δ60 assay
Pho8Δ60 assay was performed using a spectrophotometric method as previously described (31). Briefly, cells were grown to mid-log phase in YPD, and then half the cells were grown in rich medium for 1.5 hours at either 26°C or 36.5°C. The other half was transferred to SD-N medium to starve at 26°C for 2 hours or pretreated at 36.5°C for 40 mins before transferring to SD-N medium to starve at 36.5°C for 2 hours. Protein extracts were prepared and the Pho8Δ60 assay was conducted as previously described (31).
Supplementary Material
Table S1. Yeast strains and plasmids used in this study
Figure S1. Cvt pathway and starvation-induced autophagy are impaired in other TRAPP II-specific subunit mutants at the restrictive temperature. (A) Atg8 and Ape1 localization in trs65ts and tca17ts mutant cells. WT-II (for indicating TRAPP II-specific subunits Trs130 and Trs120 are tagged as Trs130-HA Trs120-myc) and mutant cells were tagged using GFP-Atg8 and RFP-Ape1 integration plasmids. Cells were grown and treated as in Figure 1A. GFP-Atg8 morphology of WT-II and tca17ts cells starved for 0.5 hour at 37°C is at right. Arrowheads indicate colocalization of Atg8 and Ape1. Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A. (B) Ape1 maturation was blocked under non-starvation conditions and GFP-Atg8 degradation was reduced under starvation conditions in trs65ts mutant cells at 37°C. Cells grown as in (A) but in YPD were subjected to immunoblot assay, quantitated and presented as in Figure 1B. II for WT-II as in (A). (C)-(F): GFP-Atg8 localization in the trs120Δ mutant (trs120Δ+Ypt31) (C) and the trs33Δ mutant (trs33Δ in Trs130-HA background) (Trs33 was a candidate for TRAPP II-specific subunit in this study) (E); immunoblot assay for autophagy defects in trs120Δ mutant (D) and trs33Δ mutant (F). Experiments were done as in (A) and (B).
Figure S2. Localization of Atg8 in trs130ts mutants with deletion of ATG genes without starvation. (A) Multiple GFP-Atg8 dots were formed in trs130ts atg1Δ without starvation at the restrictive temperature; this was rescued by Trs130. Cells grown in rich medium (YPD or SD-Leu) to mid-log phase at 26°C were treated as in Figure 1A for non-starvation and stained with FM4-64 during the last hour to detect the vacuole. Bar, 5 μm. (B) Yeast strains were grown and treated as described in Figure 3B except that incubation was in SD-Ura for 1.5 hours at 37°C without starvation. Cells were examined by live microscopy imaging for changes in GFP-Atg8 and RFP-Ape1. Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A.
Figure S3. Anterograde transport of Atg9 to the PAS is altered in trs65ts mutant cells at the restrictive temperature. WT and trs65ts mutant cells tagged with Atg9-3XGFP and RFP-Ape1 in an atg1Δ background were incubated as described in Figure 4 and examined for fluorescence. (A) Non-starvation. (B) Starvation. Bar, 7 μm. Data are presented as in Figure 4. Asterisks indicate P < 0.001 as highly significant.
Figure S4. Changes in autophagic flux in some TRAPP II-specific subunit mutants and suppression of growth defects in mutants by Ypt31. (A) Pho8Δ60 activity in trs130ts and trs65ts mutants. WT (TN124) and the atg1Δ (in TRS130 background) strains were used as positive and negative controls. All cells were treated as in Figures 1B except that 36.5°C was used to guarantee that the trs130ts cells would be viable after treatment since the mutants in the TN124 background were more temperature sensitive than the strains in Figures 1A and S1A. The trs130ts and trs65ts mutants were transferred from 36.5°C in SD-N to 26°C in SD-N for 2 hours recovery and designated SD-NR (2h). Pho8Δ60 assay data are presented as in Figure 7B except with absolute values because of slightly lower Pho8Δ60 activity in TRS130 than in TN124. Asterisks indicate P < 0.001 as highly significant. (B) The trs130ts and trs65ts mutants were viable after treatment at 36.5°C in SD-N for 2 hours. WT and the mutants incubated in SD-N at 26°C and 36.5°C for 2 hours were plated on YPD plates with tenfold serial dilution from top to bottom and grown at 26°C for 2 days. (C)–(E) Cells as in Figures 7 and S1 but without RFP-Ape1 tagging were transformed with the indicated plasmids to examine growth. Cells were spotted onto SD-Leu medium with tenfold serial dilution from top to bottom and grown at various temperatures. Ypt31 (WT and GTP-bound form) but not Ypt1 suppressed the growth defect of trs130ts mutant cells (C), trs65ts and tca17ts mutant cells (D), and trs33Δ mutant cells (E) at 37°C. (F) The Ypt1 plasmid in (C)-(E) was used to overexpress Ypt1 in ypt1ts to confirm expression and function. Ypt31 served as a negative control. Growth was checked as in (C) and protein expression was examined as in Figure 7C.
Figure S5. Ypt32 fully suppresses growth and autophagy defects in trs130ts mutant cells, but TRAPP II subunits suppress ypt31Δ/32ts growth defects weakly. (A) Trs130 and Ypt32 suppress the growth defect in trs130ts mutant cells at the restrictive temperature. Trs130 and Ypt32 in YEp351 were transformed into WT and trs130ts mutant cells tagged with GFP-Atg8 and grown as in Figure S4C. (B) Trs130 and Ypt32 suppressed the GFP-Atg8 transport defect in trs130ts cells. Yeast transformants as in (A) were investigated to determine whether Trs130 and Ypt32 suppressed the GFP-Atg8 transport defect in trs130ts cells in synthetic minimal medium (SD-Leu) or starvation medium (SD-N) at the restrictive temperature. Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A. (C) TRAPP II-specific subunits (Trs130, Trs65 and Tca17) weakly suppressed the growth defect in ypt31Δ/32ts mutant cells at the non-permissive temperature (34°C). Trs130, Trs65 and Tca17 in YEp351 (all are functional) were transformed and grown as in Figure S4C. (D) TRAPP II-specific subunits (Trs130, Trs65 and Tca17) did not obviously suppress the GFP-Atg8 transport defect in ypt31Δ/32ts cells. Yeast transformants as in (C) were investigated to determine whether TRAPP II-specific subunits suppressed the GFP-Atg8 transport defect in ypt31Δ/32ts cells in synthetic minimal medium (SD-Leu) at a growth suppression temperature (34°C) or in starvation medium (SD-N) at a temperature at which GFP-Atg8 transport starts to show defects (36°C). Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A.
Acknowledgments
We thank Y. Ohsumi for providing anti-Ape1 antibody, R. Schekman for providing the sec7-4ts mutant and F. Reggiori for providing Cu-Cherry-ATG8-415 plasmid. We thank H Ghiradella for help with language. This work was supported by grants from the Natural Science Foundation of China (31271520 to Y. Liang; 30971441 and 31171285 to ZX; 31070242 to SL); the National Key Basic Research Program of China (2011CB910100 to ZX); the Research Fund for the Doctoral Program of Higher Education of China (20090097120039), the Project-sponsored by Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry ([2011]508), Nanjing Agricultural University (680-804094-521), the Fundamental Research Funds for the Central Universities (KYT201001 and KYZ201123) to Y. Liang; the National Institutes of Health (GM-45444 to NS). SZ was supported by the Program for Scientific Innovation Research of College Graduate in Jiangsu Province (CXLX11_0663).
Footnotes
The authors have no conflicts of interest.
References
- 1.Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol. 2001;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 2.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 3.Segev N. Ypt/Rab GTPases: regulators of protein trafficking. Sci STKE. 2001:RE11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- 4.Yu S, Liang Y. A trapper keeper for TRAPP, its structures and functions. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-00012-01024-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yip CK, Berscheminski J, Walz T. Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nat Struct Mol Biol. 2010;17:1298–1304. doi: 10.1038/nsmb.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tokarev AA, Taussig D, Sundaram G, Lipatova Z, Liang Y, Mulholland JW, Segev N. TRAPP II complex assembly requires Trs33 or Trs65. Traffic. 2009;10:1831–1844. doi: 10.1111/j.1600-0854.2009.00988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y, Chin HF, Lazarova D, Menon S, Fu C, Cai H, Sclafani A, Rodgers DW, De La Cruz EM, Ferro-Novick S, Reinisch KM. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Y, Morozova N, Tokarev AA, Mulholland JW, Segev N. The role of Trs65 in the Ypt/Rab guanine nucleotide exchange factor function of the TRAPP II complex. Mol Biol Cell. 2007;18:2533–2541. doi: 10.1091/mbc.E07-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morozova N, Liang Y, Tokarev AA, Chen SH, Cox R, Andrejic J, Lipatova Z, Sciorra VA, Emr SD, Segev N. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat Cell Biol. 2006;8:1263–1269. doi: 10.1038/ncb1489. [DOI] [PubMed] [Google Scholar]
- 11.Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR, 3rd, Abeliovich H, Ferro-Novick S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, Ferro-Novick S. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7:433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- 14.Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci U S A. 2012;109:6981–6986. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi C, Davey M, Schluter C, Pandher P, Fang Y, Foster LJ, Conibear E. Organization and assembly of the TRAPPII complex. Traffic. 2011;12:715–725. doi: 10.1111/j.1600-0854.2011.01181.x. [DOI] [PubMed] [Google Scholar]
- 16.Montpetit B, Conibear E. Identification of the novel TRAPP associated protein Tca17. Traffic. 2009;10:713–723. doi: 10.1111/j.1600-0854.2009.00895.x. [DOI] [PubMed] [Google Scholar]
- 17.Sclafani A, Chen S, Rivera-Molina F, Reinisch K, Novick P, Ferro-Novick S. Establishing a role for the GTPase Ypt1p at the late Golgi. Traffic. 2010;11:520–532. doi: 10.1111/j.1600-0854.2010.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat Rev Mol Cell Biol. 2010;11:759–763. doi: 10.1038/nrm2999. [DOI] [PubMed] [Google Scholar]
- 19.Bridges D, Fisher K, Zolov SN, Xiong T, Inoki K, Weisman LS, Saltiel AR. Rab5 proteins regulate activation and localization of target of rapamycin complex 1. J Biol Chem. 2012;287:20913–20921. doi: 10.1074/jbc.M111.334060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- 22.Yu ZQ, Ni T, Hong B, Wang HY, Jiang FJ, Zou S, Chen Y, Zheng XL, Klionsky DJ, Liang Y, Xie Z. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8:883–892. doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair U, Yen WL, Mari M, Cao Y, Xie Z, Baba M, Reggiori F, Klionsky DJ. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy. 2012;8:780–793. doi: 10.4161/auto.19385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He C, Klionsky DJ. Atg9 trafficking in autophagy-related pathways. Autophagy. 2007;3:271–274. doi: 10.4161/auto.3912. [DOI] [PubMed] [Google Scholar]
- 28.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 29.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci U S A. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noda T, Klionsky DJ. The quantitative Pho8Δ60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 35.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 37.Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Yang Z, Nair U, Mao K, Liu X, Klionsky DJ. Phosphatatidylinositol 4-kinases are required for autophagic membrane trafficking. J Biol Chem. 2012 doi: 10.1074/jbc.M112.371591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Vaart A, Reggiori F. The Golgi complex as a source for yeast autophagosomal membranes. Autophagy. 2010;6:800–801. doi: 10.1091/mbc.E09-04-0345. [DOI] [PubMed] [Google Scholar]
- 40.van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto K, Jigami Y. Mutation of TRS130, which encodes a component of the TRAPP II complex, activates transcription of OCH1 in Saccharomyces cerevisiae. Curr Genet. 2002;42:85–93. doi: 10.1007/s00294-002-0336-5. [DOI] [PubMed] [Google Scholar]
- 42.Zhang CJ, Bowzard JB, Greene M, Anido A, Stearns K, Kahn RA. Genetic interactions link ARF1, YPT31/32 and TRS130. Yeast. 2002;19:1075–1086. doi: 10.1002/yea.903. [DOI] [PubMed] [Google Scholar]
- 43.Sciorra VA, Audhya A, Parsons AB, Segev N, Boone C, Emr SD. Synthetic genetic array analysis of the PtdIns 4-kinase Pik1p identifies components in a Golgi-specific Ypt31/rab-GTPase signaling pathway. Mol Biol Cell. 2005;16:776–793. doi: 10.1091/mbc.E04-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy. 2005;1:37–45. doi: 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, Eskelinen EL, Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- 49.Zou S, Liu Y, Zhang XQ, Chen Y, Ye M, Zhu X, Yang S, Lipatova Z, Liang Y, Segev N. Modular TRAPP Complexes Regulate Intracellular Protein Trafficking Through Multiple Ypt/Rab GTPases in Saccharomyces cerevisiae. Genetics. 2012;191:451–460. doi: 10.1534/genetics.112.139378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W, Ferro-Novick S. A Ypt32p exchange factor is a putative effector of Ypt1p. Mol Biol Cell. 2002;13:3336–3343. doi: 10.1091/mbc.01-12-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 52.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 53.Gietz D, Stjean A, Woods RA, Schiestl RH. Improved method for high-efficiency transformation of intact yeast-cells. Nucleic Acids Res. 1992;20:1425–1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J Cell Biol. 1997;137:563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Yeast strains and plasmids used in this study
Figure S1. Cvt pathway and starvation-induced autophagy are impaired in other TRAPP II-specific subunit mutants at the restrictive temperature. (A) Atg8 and Ape1 localization in trs65ts and tca17ts mutant cells. WT-II (for indicating TRAPP II-specific subunits Trs130 and Trs120 are tagged as Trs130-HA Trs120-myc) and mutant cells were tagged using GFP-Atg8 and RFP-Ape1 integration plasmids. Cells were grown and treated as in Figure 1A. GFP-Atg8 morphology of WT-II and tca17ts cells starved for 0.5 hour at 37°C is at right. Arrowheads indicate colocalization of Atg8 and Ape1. Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A. (B) Ape1 maturation was blocked under non-starvation conditions and GFP-Atg8 degradation was reduced under starvation conditions in trs65ts mutant cells at 37°C. Cells grown as in (A) but in YPD were subjected to immunoblot assay, quantitated and presented as in Figure 1B. II for WT-II as in (A). (C)-(F): GFP-Atg8 localization in the trs120Δ mutant (trs120Δ+Ypt31) (C) and the trs33Δ mutant (trs33Δ in Trs130-HA background) (Trs33 was a candidate for TRAPP II-specific subunit in this study) (E); immunoblot assay for autophagy defects in trs120Δ mutant (D) and trs33Δ mutant (F). Experiments were done as in (A) and (B).
Figure S2. Localization of Atg8 in trs130ts mutants with deletion of ATG genes without starvation. (A) Multiple GFP-Atg8 dots were formed in trs130ts atg1Δ without starvation at the restrictive temperature; this was rescued by Trs130. Cells grown in rich medium (YPD or SD-Leu) to mid-log phase at 26°C were treated as in Figure 1A for non-starvation and stained with FM4-64 during the last hour to detect the vacuole. Bar, 5 μm. (B) Yeast strains were grown and treated as described in Figure 3B except that incubation was in SD-Ura for 1.5 hours at 37°C without starvation. Cells were examined by live microscopy imaging for changes in GFP-Atg8 and RFP-Ape1. Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A.
Figure S3. Anterograde transport of Atg9 to the PAS is altered in trs65ts mutant cells at the restrictive temperature. WT and trs65ts mutant cells tagged with Atg9-3XGFP and RFP-Ape1 in an atg1Δ background were incubated as described in Figure 4 and examined for fluorescence. (A) Non-starvation. (B) Starvation. Bar, 7 μm. Data are presented as in Figure 4. Asterisks indicate P < 0.001 as highly significant.
Figure S4. Changes in autophagic flux in some TRAPP II-specific subunit mutants and suppression of growth defects in mutants by Ypt31. (A) Pho8Δ60 activity in trs130ts and trs65ts mutants. WT (TN124) and the atg1Δ (in TRS130 background) strains were used as positive and negative controls. All cells were treated as in Figures 1B except that 36.5°C was used to guarantee that the trs130ts cells would be viable after treatment since the mutants in the TN124 background were more temperature sensitive than the strains in Figures 1A and S1A. The trs130ts and trs65ts mutants were transferred from 36.5°C in SD-N to 26°C in SD-N for 2 hours recovery and designated SD-NR (2h). Pho8Δ60 assay data are presented as in Figure 7B except with absolute values because of slightly lower Pho8Δ60 activity in TRS130 than in TN124. Asterisks indicate P < 0.001 as highly significant. (B) The trs130ts and trs65ts mutants were viable after treatment at 36.5°C in SD-N for 2 hours. WT and the mutants incubated in SD-N at 26°C and 36.5°C for 2 hours were plated on YPD plates with tenfold serial dilution from top to bottom and grown at 26°C for 2 days. (C)–(E) Cells as in Figures 7 and S1 but without RFP-Ape1 tagging were transformed with the indicated plasmids to examine growth. Cells were spotted onto SD-Leu medium with tenfold serial dilution from top to bottom and grown at various temperatures. Ypt31 (WT and GTP-bound form) but not Ypt1 suppressed the growth defect of trs130ts mutant cells (C), trs65ts and tca17ts mutant cells (D), and trs33Δ mutant cells (E) at 37°C. (F) The Ypt1 plasmid in (C)-(E) was used to overexpress Ypt1 in ypt1ts to confirm expression and function. Ypt31 served as a negative control. Growth was checked as in (C) and protein expression was examined as in Figure 7C.
Figure S5. Ypt32 fully suppresses growth and autophagy defects in trs130ts mutant cells, but TRAPP II subunits suppress ypt31Δ/32ts growth defects weakly. (A) Trs130 and Ypt32 suppress the growth defect in trs130ts mutant cells at the restrictive temperature. Trs130 and Ypt32 in YEp351 were transformed into WT and trs130ts mutant cells tagged with GFP-Atg8 and grown as in Figure S4C. (B) Trs130 and Ypt32 suppressed the GFP-Atg8 transport defect in trs130ts cells. Yeast transformants as in (A) were investigated to determine whether Trs130 and Ypt32 suppressed the GFP-Atg8 transport defect in trs130ts cells in synthetic minimal medium (SD-Leu) or starvation medium (SD-N) at the restrictive temperature. Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A. (C) TRAPP II-specific subunits (Trs130, Trs65 and Tca17) weakly suppressed the growth defect in ypt31Δ/32ts mutant cells at the non-permissive temperature (34°C). Trs130, Trs65 and Tca17 in YEp351 (all are functional) were transformed and grown as in Figure S4C. (D) TRAPP II-specific subunits (Trs130, Trs65 and Tca17) did not obviously suppress the GFP-Atg8 transport defect in ypt31Δ/32ts cells. Yeast transformants as in (C) were investigated to determine whether TRAPP II-specific subunits suppressed the GFP-Atg8 transport defect in ypt31Δ/32ts cells in synthetic minimal medium (SD-Leu) at a growth suppression temperature (34°C) or in starvation medium (SD-N) at a temperature at which GFP-Atg8 transport starts to show defects (36°C). Bar, 5 μm. The percentage of cells with Atg8 dots was quantitated and presented as in Figure 1A.