Abstract
Amyloid-β (Aβ) oligomers destabilize cellular ionic homeostasis, mediating Alzheimer's disease (AD). It is still unclear whether the mechanism (i) is mediated by cell surface receptors; (ii) is direct, with Aβ oligomers interacting with membrane lipids; or (iii) both mechanisms take place. Recent studies indicate that Aβ oligomers may act by either of the last two. Little is known about the oligomers’ structures and how they spontaneously insert into the membrane. Using explicit solvent molecular dynamics (MD) simulations, we show that fibril-like Aβ17-42 (p3) oligomer is capable of penetrating the membrane. Insertion is similar to that observed for protegrin-1 (PG-1), a cytolytic β-sheet-rich antimicrobial peptide (AMP). Both Aβ and PG-1 favor the amphipathic interface of the lipid bilayer in the early stage of interaction with the membrane. U-shaped Aβ oligomers are observed in solution and in the membrane, suggesting that the preformed seeds can be shared by amyloid fibrils in the growth phase and membrane toxicity. Here we provide sequential events in possible Aβ oligomer membrane-insertion pathways. We speculate that for the U-shaped motif, a trimer is the minimal oligomer size to insert effectively. We propose that monomers and dimers may insert in (apparently on-pathway) aggregation-intermediate β-hairpin state, and may (or may not) convert to a U-shape in the bilayer. Together with earlier observations, our results point to a non-specific, broadly heterogeneous landscape of membrane-inserting oligomer conformations, pathways, and membrane-mediated toxicity of β-rich oligomers.
Keywords: Toxic amyloid oligomer, β-sheet conformation, cytolytic activity, membrane insertion, molecular dynamics simulation
INTRODUCTION
The β-amyloid (Aβ) peptide is the primary component of extracellular fibrillar deposits, termed amyloid plaques, found in postmortem brains of patients with Alzheimer's disease (AD).1 In the lag phase, Aβ monomers associate to form small Aβ oligomer seeds; in the next growth phase, they are extended into fibrils. The highly ordered Aβ fibrillar oligomers were believed to be a non-toxic pathway state, whereas the small soluble Aβ oligomers a toxic state.2-4 However, more recent studies identified the small soluble Aβ oligomers as Aβ fibrillar oligomers (FOs) that contain 3-10 Aβ monomers and exhibit fibril-like morphology rich in β-sheet structure.5 Aβ FOs do not show thioflavin-T positive fibril formation, but nucleate monomer conversion into newly replicated FOs. Analysis of atomic force microscopy (AFM) images indicated that Aβ FOs are protofilaments5 which fit the U-shaped β-strand-turn-β-strand peptide motif.6-8 Aβ oligomers with this motif expose the hydrophobic surface of the C-terminal β-strands, which are able to diffuse into cell membranes forming toxic pores. A less toxic Aβ oligomer is deficient in β-sheet conformation, while a more toxic Aβ oligomer has a solvent exposed hydrophobic face.9
Aβ neurotoxicity is triggered by the small fibril-like Aβ oligomer intermediates, predominantly parallel β-sheet structures.10 Aβ toxicity is related to the loss of ionic homeostasis via ion channel formation mediated by the small Aβ oligomers, which is supported by loss of calcium homeostasis, electrophysiological recordings, and AFM images.11-16 Molecular dynamics (MD) simulations revealed channel conformations with truncated Aβ peptides (Aβ17-42 (p3) and Aβ9-42 (N9))17-22 and full-length Aβ1-42 peptides with mutations.23-26 These studies suggested that Aβ channels are dynamic assemblies of 3-6 mobile β-sheet subunits in the lipid bilayers. The size of β-sheet subunits, which contain 2-6 Aβ monomers, indicates that they are small fibril-like Aβ oligomers with morphological similarity to the preformed seeds in amyloid aggregation. This suggests that the same seeds can be shared in amyloid fibrils in solution and channels in cell membrane. If small fibril-like Aβ oligomers like the preformed seeds are the predominant toxic species in AD, the oligomers can be expected to be capable of penetrating into the cell membrane. However, it is still unclear how these small oligomers are spontaneously insert into the bilayer.
Several theoretical efforts addressed the mechanism of Aβ insertion into cell membranes. In early studies, modeling of Aβ monomer insertion by Aβ1-40/42 and FAD mutants proposed that monomers adopt a partially inserted conformation in one leaflet of the bilayer.27 Those studies employed implicit simulations and did not account for β-sheet formation and channel organization. In studies of Aβ1-42 monomers on the surfaces of zwitterionic and anionic bilayers, it was found that in the early stages of Aβ aggregation, Aβ monomers near the bilayer surface do not conserve any significant β-structure, but form unstable β-hairpin on the anionic bilayer.28 In the same studies on the anionic bilayer, Aβ dimerization via β-sheet was promoted by strong protein-protein interactions.29 In recent studies of aggregation of three α-helical Aβ monomers on a mixed lipid bilayer containing cholesterols, a conversion of the secondary structure from α-helices to β-strands was observed for a short segment of parallel β-sheet.30 This suggests that β-sheet formation is catalyzed by the presence of bilayer membranes.31 Although such studies which illustrate that Aβ monomers bound to the bilayer surface could provide putative models for Aβ insertion and lipid-induced Aβ oligomerization, Aβ monomers and fibrils are less cytotoxic than Aβ oligomer intermediates.32 It was reported that Aβ oligomers bind more strongly to the membrane causing Aβ-induced permeation as compared to fibrillar Aβ.33 In recent studies, interactions of Aβ9-40 oligomers in a protofibril architecture with the lipid bilayer indicated that strong electrostatic attraction between charged sidechains and the lipid headgroups, and hydrogen bonding with the lipid heagroups drive the loss of the initial fibril structure, resulting in the penetration of C-terminal residues into the hydrophobic region of the bilayer.34 However, the initial structure of octameric Aβ9-40 protofibrils revealed that the fibrils formed by two layers of U-shaped peptides bury the hydrophobic faces between the sheets, indicating the less toxic oligomer motif.9
Here, we show interactions of fibril-like Aβ oligomers formed by the N-terminal-truncated p3 peptides with the lipid bilayer. The peptide is found in amyloid plaques of AD and in preamyloid lesions of the Down's syndrome (DS).35 Although, the p3 fragment may be involved in Aβ fibrillation, the full-length Aβ peptide alone is generally believed to be nontoxic, as recently also validated in a comprehensive study.36 In aqueous environment, p3 peptides do not grow to mature fibrils at room temperature,37 but the presence of small oligomeric p3 protofibrils is predicted in MD simulations.38 Using AFM, we illustrate that aggregation phases of p3 largely depend on hydrophobicity provided by the surrounding environments. The peptide is highly hydrophobic and hence enhances its ordered states on the hydrophobic graphite surface and in the hydrophobic membrane core. However, on the membrane-water interface, rather than assembling into large macromolecular aggregates, p3 peptides favor formation of small oligomers, which are easily inserted into the membrane. The different aggregation behaviors of p3 point to the importance of examining its lag phase, and how it adjusts its aggregation pathway in the key phase of membrane insertion. Using MD simulations, we test preassembled fibril-like p3 pentamers in five different configurations on/in the lipid bilayer. These configurations represent possible sequential states along the membrane insertion pathway, and as such may provide a mechanism for Aβ oligomer insertion. Based on these, our proposed mechanism is similar to that of the cytolytic antimicrobial peptide (AMP) protegrin-1 (PG-1). In the early stage of the membrane attack by PG-1, positively charged residues are mostly responsible for disruption/thinning effects and ultimately channel formation.39,40 PG-1 interacts with the lipids with the β-sheet plane lying obliquely to the bilayer surface. This oblique attack may increase the pressure on the bilayer surface, resulting in some void created by the evading lipids.41,42 Recently, it was demonstrated that the Aβ peptide possesses antimicrobial properties with characteristics similar to those of host defense peptides (HDPs).43-46
The p3 peptide used here has the β-strand-turn-β-strand motif. The β-sheet-rich morphology with the exposed hydrophobic face provides the features of toxic Aβ oligomers.5,9,10 Our major result is that the U-shaped p3 pentamer is able to insert into the lipid bilayer, and that a trimer is likely the minimal oligomer size to insert effectively. Smaller species may insert as β-hairpin intermediates. This leads us to propose a role for small β-hairpin aggregate-intermediates in toxicity.
MATERIALS AND METHODS
AFM Imaging
Experiments were conducted as described earlier.37 Briefly, a Multimode AFM equipped with a Nanoscope IVa controller (Bruker, Santa Barbara, CA) was used. Oxide sharpened cantilevers with spring constants of 0.12 N/m or 0.58 N/m were employed. A fluid cell (Bruker) was utilized. Oligomers were observed in tapping and contact modes. For tapping mode, oscillation frequencies of ~9 kHz and drive amplitudes between 10 - 20 mV were used. In contact mode, the force was minimized by adjusting the setpoint to the lowest possible value before the tip came out of contact with the surface. The width of oligomers was measured as the full width at half maximum (FWHM) in the height profile. All experiments were performed at room temperature. Highly Oriented Pyrolitic Graphite (grade SPI-1) was purchased from SPI supplies (West Chester, PA). p3 peptides (purchased from Anaspec, Santa Ana, Ca) were aliquoted in 1% NH4OH (Sigma-Aldrich, St. Louis, MO) aqueous solutions and further diluted (1:10 by vol) in PBS without nominal Ca2+ or Mg2+ content (Fisher Scientific, Pittsburgh, PA). Concentrations of 7.5 μM, peptides were diluted in PBS solutions with 1% NH4OH solutions and sonicated for 20 min in an ice water bath at temperatures of ~4 °C. Graphite surfaces were freshly cleaved before each experiment and a region of the sample was imaged for ~1 hr as reference before injecting p3 peptides into the AFM fluid cell. DOPC (Dioleoyl, 1,2-dioleoyl-sn-glycero-3-phosphocholine) was purchased from Avanti Polar Lipids (Alabaster, Al). Supported lipid bilayers were prepared by rupture and fusion of liposomes. These were deposited on mica at lipid concentrations of 0.1 - 1 mg/mL, allowed to adsorb for ~1 min and rinsed with PBS buffer.
Modeling p3 Oligomers
Initial coordinates of the p3 (Aβ17-42) monomer were extracted from the Aβ1-42 fibril, the structure obtained from a combination of hydrogen/deuterium-exchange NMR data, side-chain packing constraints from pair-wise mutagenesis, ssNMR and EM (PDB code: 2BEG).6 This ssNMR-based structure provided the coordinates for the central and C-terminal portions of Aβ1-42. The N-terminal coordinates (residues 1-16) were missing due to disorder. The structure formed by residues 17-42 (Aβ17-42) has the U-shaped β-strand-turn-β-strand motif.7,8 The Aβ17-42 peptide corresponds to that of p3 observed in vivo following cleavage by α-secretase. Using the U-shaped p3 monomer with both N- and C-termini blocked by acetyl and amine groups, respectively, we generated fibril-like pentamer and dimer by stacking each monomer along the fibril axis, with intermolecular hydrogen bonds (H-bonds). We initially set both p3 pentamer and dimer at various locations/orientations on the lipid bilayer containing DOPC lipid. Throughout the paper, we mainly discuss the p3 pentamer simulations, and mention the p3 dimer explicitly when it is presented. We consider 5 different initial configurations (Fig. 1), to represent sequential stages of p3 pentamers during the membrane insertion.
FIGURE 1.
Schematic diagrams representing five different initial configurations for the MD simulations for the p3 (Aβ17-42) pentamers interacting with the DOPC bilayer. The pentamers at various locations in the lipid bilayer imply the sequential events in the membrane insertion pathway. For the U-shaped pentamers containing two parallel β-sheets, the N- and C-termini are marked in the diagram of configuration 1. Red pentagons attached to the pentamers represent the negatively charged Glu22 sidechains. The p3 dimer simulations also employed the same initial configurations as used in the p3 pentamer simulations.
Five Different Initial Configurations
Configuration 1 mimics a possible ‘initial touch’, where the p3 pentamer is sitting on top of the bilayer, with its hydrophobic C-terminal strands facing the bilayer and barely interacting with the lipids. It has been shown that AMPs bind to the interface and subsequently penetrate the bilayer if their hydrophobic faces, and not their positively charged faces, are directed toward the interface.47 Configuration 2 denotes ‘partial deposition’, where the p3 pentamer is further inserted into the interior of the bilayer. The hydrophobic C-terminal strands interact with the lipid tails, while the negatively charged N-terminal strands are still located at the bulk water area. In both configurations 3 and 4, which are denoted ‘oblique insertion’, the p3 pentamers are inserted obliquely into the lipid bilayer, representing the state that the pentamers are in the midst of membrane crossing via rotation of their long molecular axis. In configuration 3, the termini of the pentamer slip across the hydrophobic core of the lipid bilayer, while the pentamer's turns slip in configuration 4. Finally, the p3 pentamer fully crosses the lipid bilayer in configuration 5 denoted as ‘embedded’.
MD Simulations of Explicit Lipid Bilayer
The CHARMM program48 using the revised CHARMM27 (C27r) force field for lipids49 and the modified TIP3P water model50 were used to construct the set of starting points and to relax the systems to a production-ready stage. For each configuration with p3 pentamer and dimer, a unit cell containing two layers of DOPC lipids is constructed. TIP3P waters were added at both sides with lipid/water with ratios of ~1/160 and ~1/120 for the pentamer and dimer simulations, respectively. The p3 monomer has a net negative charge. To neutralize the system, five (5 Na+) and two (2 Na+) counterions for the pentamer and dimer simulations, respectively, were added to the bilayer system. In addition, to obtain a physiological salt concentration near 100 mM, additional 50 Na+ and 50 Cl- for the pentamer and 18 Na+ and 18 Cl- for the dimer systems were added.
Production Runs
We generated five different initial configurations for each p3 pentamer and dimer simulation. A total of ten independent initial systems within explicit solvents were constructed and gradually relaxed, with the peptide held rigid. A series of minimizations was performed for the initial systems to remove overlaps of the alkane chains in the lipids and to gradually relax the solvents around the p3 peptides. These initial configurations were gradually relaxed through dynamic cycles with electrostatic cutoffs (12 Å). In the subsequent preequilibrium stages, a series of dynamic cycles was performed with the harmonically restrained p3 peptides, and then the harmonic restraints were gradually diminished until gone with the full Ewald electrostatics calculation. Our simulation employed the NPAT (constant number of atoms, pressure, surface area, and temperature) ensemble with a constant normal pressure applied in the direction perpendicular to the membrane. A Nosé-Hoover thermostat/barostat was used to maintain a constant temperature of 300 K. For both p3 pentamer and dimer simulations with configuration 1, production runs of 500 ns were performed, while for the simulations with configuration 2 to 5, production runs of 100 ns were performed. The longer timescale simulations for configuration 1 ensure that the p3 peptides are able to diffuse into the amphipathic interface of the lipid bilayer. However, for those configurations with pre-inserted p3 peptides into the lipid bilayer, the 100 ns simulations were enough to capture relaxed peptide conformations in the lipid environment and detailed peptide–lipid interactions. Total production runs of 1.8 Ps for the starting points with the NAMD code51 were performed on a Biowulf cluster at the NIH.
RESULTS
The Aβ Peptides Favor the Formation of Small Oligomers Rather Than Fibrils on the Surface of the Lipid Bilayer
A mixed population of globular and linear p3 (Aβ17-42) oligomers, along with amorphous areas, was observed in regions of p3 films formed on graphite surfaces (Fig. 2A). The diameter of the globular oligomers was ~3 nm and the lengths of the linear oligomers were typically 5 - 10 nm. These structures coexisted with the more common linear filaments and criss-cross patterns described previously, although even those filaments possessed a globular substructure when observed in detail.37 The hydrophobic graphite surface supports linear elongation of the β-sheet as compared to annular extension of the β-sheet in the Aβ channels embedded in the hydrophobic core of the lipid bilayers.17-22 p3 fibrils were not observed on the surface of DOPC bilayers, although some AFM images were suggestive of the presence of globular oligomers on the bilayer surface (Fig. 2B).37 It is noted that the supported lipid bilayers on the mica surface do not allow large oligomers to insert into the hydrophobic core of the lipid bilayer, while small oligomers might be able to insert. We hypothesize that these small Aβ oligomers within β-sheets are responsible for the membrane insertion and the toxic ion channel formation. However, it is not clear whether these oligomers possess the β-sheet or not, since the AFM resolution does not allow visualization of the secondary structures of these oligomers. Even though these oligomers possess the β-sheet and insert into the hydrophobic core of the lipid bilayer, AFM is not able to detect the oligomers when they are embedded in the bilayer.
FIGURE 2.
(A) p3 (Aβ17-42) film with 1.1 nm thickness formed after prolonged contact (~450 min) of 7.5 μM aqueous solutions of p3 peptides with a graphite surface. Some globular oligomers are encircled in yellow and a linear oligomer is encircled in green. An overall parallel alignment of the linear oligomers following crystallographic directions of graphite is still visible, as described previously.37 (B) AFM image of p3 oligomers on the surface of DOPC bilayer. The image was reproduced from the original data.37
Five Different Aβ Oligomers Interact with the Lipid Bilayer along the Possible Insertion Pathways
We generated five different initial configurations for a 5-mer p3 oligomer interacting with the DOPC lipid bilayer. These configurations may represent sequential events during membrane insertion of the Aβ oligomer, which may suggest some possible pathways. We used experiment-based Aβ17-42 coordinates6 and constructed the p3 pentamers on/in the lipid bilayer. In configuration 1, the p3 pentamer is initially located at the bilayer surface in the upper bilayer leaflet, barely interacting with lipids. This configuration illustrates that the fibril-like Aβ oligomer pre-assembled in the extracellular medium touches the cell membrane in the early stage of a cytolytic attack. In configurations 2 to 5, the p3 pentamers partially (confs. 2 to 4) or fully (conf. 5) cross the lipid bilayer, representing membrane embedded states of Aβ oligomers during the insertion process. The p3 pentamer in configuration 1 is referred to as pentamer 1 (with the 500 ns MD simulation), while the p3 pentamers in other configurations are referred to as pentamers 2 to 5 (with the 100 ns simulations). In the longer timescale simulation for pentamer 1, we are able to capture events such as local membrane perturbation by the pentamer; however, spontaneous pentamer insertion into the hydrophobic core of the lipid bilayer is not observed, since this event may occur on much longer simulation timescale. Pentamer 1 slowly reaches equilibration after 100 ns, while the other pentamers in the embedded states quickly reach equilibration after 30 ns (Supporting Information, Fig. S1).
The averaged pentamer structures and the peptide-averaged RMSD values with respect to the starting points are presented in Fig. 3. Partially or fully membrane-bound pentamers 2-5 yield smaller RMSD values compared to membrane-unbound pentamer 1, suggesting that the lipid bilayer supports the fibril-like Aβ oligomer conformation.31 Increases in the peptide-lipid interactions stabilize the U-shaped Aβ conformation when the pentamers further diffuse into the bilayer (Table 1). In contrast, increases in the peptide-water interactions reduce the fibril-like features of the Aβ oligomer, as indicated by the reduced p3 fibrillar structures in solution.37 It can be noted that for pentamer 5 in the fully crossed state, individual peptide interactions with surrounding environments (e.g. lipids, water, and other peptides) are well-balanced (Supporting Information, Fig. S2).
FIGURE 3.
Simulated structures of p3 (Aβ17-42) pentamers in a cartoon representation averaged over the simulations starting from five different initial configurations. Averaged RMSD values calculated for each peptide in the pentamers with respect to the starting points are included in parenthesis. In the cartoons, white meshes around the pentamer represent three-dimensional density maps of water with high probability of 0.2 indicating the solvation sites. In the peptides, red sticks represent the negatively charged Glu22 and Asp23 sidechains, while blue sticks denote the positively charged Lys28 sidechains. Yellow dotted lines indicate the bilayer surfaces.
Table 1.
Calculated properties of p3 (Aβ17-42) pentamers. Interaction energies of the pentamer with DOPC lipids and waters are shown in the first two columns. The deviation of center of mass of the pentamer, Δz, from the upper bilayer leaflet is shown in the third column. In the fourth column, β-strand tilt angle, θ, between the vector connecting the turn to the tail and the membrane normal is shown. The fibril axis tilt angle, δ, between the vector along the hydrogen bonding pairs in the direction from peptide 5 to peptide 1 and the membrane normal is shown in the last column.
| Lipid interaction energy (kcal/mol) | Water interaction energy (kcal/mol) | Center of mass deviation, Δz (Å) | β-Strand tilt angle, θ (deg) | Fibril axis tilt angle, δ (deg) | |
|---|---|---|---|---|---|
| Pentamer 1 | -337.6 ± 104.9 | -3468.9 ± 397.7 | 14.6 ± 1.7 | 53.8 ± 6.4 | 94.3 ± 5.8 |
| Pentamer 2 | -837.0 ± 70.7 | -3261.9 ± 164.1 | -1.5 ± 0.7 | 84.4 ± 2.6 | 98.6 ± 3.6 |
| Pentamer 3 | -664.8 ± 88.4 | -3013.5 ± 138.2 | -1.2 ± 1.0 | 100.6 ± 3.8 | 95.3 ± 4.4 |
| Pentamer 4 | -667.9 ± 77.8 | -3248.9 ± 132.5 | -2.5 ± 1.0 | 75.5 ± 2.2 | 87.0 ± 3.8 |
| Pentamer 5 | -904.6 ± 92.3 | -2884.9 ± 142.8 | -13.2 ± 0.9 | 163.5 ± 2.4 | 94.9 ± 2.5 |
The U-shaped p3 peptides with the β-strand-turn-β-strand motif form two parallel β-sheets. The C-terminal β-sheet is hydrophobic, while the N-terminal β-sheet contains negatively charged residues. Secondary structure analysis with database of secondary structure assignments (DSSP)52 for the p3 pentamers shows that the β-sheet is enhanced as the pentamers are fully inserted into the bilayer (Fig. 4A). The hydrophobic interactions between the C-terminal β-strands and the lipid tails are essential to preserve the fibril-like feature of the Aβ oligomer. To measure the β-sheet planarity, we calculated the peptide order parameter,53 which can be experimentally measured by the polarized attenuated total reflectance Fourier-transform infrared (ATR-FTIR) spectroscopy,54,55 using an equation defined by
| (1) |
where θij is the angle between two vectors of the backbone carbonyl bonds, C=O, in the i and j residues, and N is the total number of the vector pairs. In the calculation, the C=O bonds in the turn region are omitted. In all initial configurations, all C=O vectors in the peptides closely lie on the x-y plane that is perpendicular to the membrane normal. For a perfect planar β-sheet, one can obviously expect that SC=O ≈ 1. The calculated order parameter for the starting conformation of p3 pentamer is ~0.8, indicating a highly planar β-sheet. However, the planarity of β-sheet in the relaxed pentamers is decreased during the simulations (Fig. 4B). We found that the probability distributions curves are located at SC=O = ~0.1, ~0.25, ~0.38, ~0.5, and ~0.42 for the pentamers from 1 to 5, respectively. Lower SC=O value for pentamer 1 reflects that the β-strands are disordered, especially the β-strands in the peptides at both ends of the pentamer. Note that the SC=O value can also decrease due to the typical “β-sheet twist” (or cross β-sheet). We observed that pentamers 2, 3, and 5 present highly cross β-sheets states, while pentamer 4 has a lower β-sheet twist (Fig. 3).
FIGURE 4.
(A) Percent of β-sheet content based on the secondary structure analysis with DSSP52 calculated for the N-terminal (dark gray bars) and C-terminal (light gray bars) β-strands separately as a function of the number of p3 (Aβ17-42) pentamer. (B) Probability distribution of the peptide order parameter, SC=O, for the β-sheets in the pentamers.
During the simulations, the relaxed p3 pentamers alter their orientations and heights in the lipid bilayer deviating from the initial settings. To observe from the geometrical features how the Aβ oligomers evolve in the bilayer, we introduced three parameters: (i) the deviation of the center of mass of the pentamer, Δz, from the upper bilayer leaflet, (ii) β-strand tilt angle, θ, between the vector connecting the turn to the tail and the membrane normal, and (iii) the fibril axis tilt angle, δ, between the vector along the hydrogen bonding pairs in the direction from peptide 5 to peptide 1 and the membrane normal (Table 1). The calculated Δz values for the starting points are 13.2, -2.1, -6.4, -8.8, and -13.2 Å for the pentamers from 1 to 5, respectively. Except for the obliquely inserted pentamers in configurations 3 and 4, pentamers 1, 2, and 5 seem to retain their initial heights in the lipid bilayer (the differences are less than 2 Å). Pentamer 1 slightly increases the Δz value due to the rotation and the β-sheet twist, generating the spectrum Δz values for individual peptides in the pentamer (Supporting Information, Fig. S3). The rotation of the pentamer can be explained by the change in θ from a right angle. Pentamer 2 mostly retains its initial bilayer setting during the simulation with a very small change in the center of mass deviation of 0.6 Å and the β-strand tilt angle of 84.4°, which is very close to a right angle. For the obliquely inserted pentamers, we observed that pentamers 3 and 4 bounce to the bilayer surface as soon as the simulations started (Supporting Information, Fig. S4). This indicates that the charged atoms in the termini or turns of the pentamers are unfavorably located in the middle of the hydrophobic core of the lipid bilayer. As a result, the strong repulsive forces exerted on the termini or turns relocate the pentamers to the amphipathic interface of the lipid bilayer. At the starting points of both pentamers, the insertion angle, α, which can be defined by the angle between the long molecular axis of the pentamer and the bilayer surface, is ~25°. However, after the simulations, we observed that the insertion angles, which can be calculated from the averaged β-strand tilt angles, θ = 100.6° and 75.5° for pentamers 3 and 4, decreased to 10.6° and 15.5°, respectively. In the fully embedded state, pentamer 5 stays at the same bilayer height, but slightly alters the tilt angle from the starting point due to the solvated N-terminal strands (see below). All pentamers maintain their initial δ values near δ = 90°, suggesting that the fibril axis of Aβ oligomer always lie on the plane along the bilayer surface during the membrane insertion.
The Cytolytic Activity of the Aβ Oligomer Leads to Spontaneous Diffusion into the Amphipathic Interface
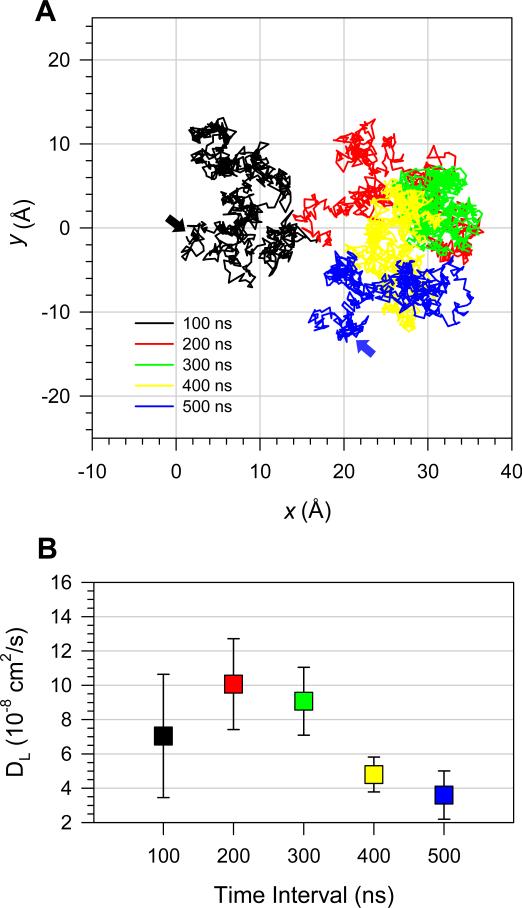
Configuration 1 mimics the event where a small fibril-like Aβ oligomer lands on the membrane surface for the first time and proceeds to a cytolytic attack. To bury its hydrophobic residues in the hydrophobic core of the lipid bilayer, the oligomer at the surface has to move to the amphipathic interface of the lipid bilayer. Configuration 2 represents the partial deposition state where the pentamer buries the C-terminal β-strands in the hydrophobic core and leaves the charged N-terminal β-strands in the bulk water area. To observe the transition event from configuration 1 to 2, which involves the pentamer crossing over a high free energy barrier in a low-dielectric medium, multiscale time simulations ranging over few hundreds of microseconds are required, which is far beyond our simulation timescale. However, our 500 ns simulation could be long enough to monitor the perturbation in the lipid bilayer induced by the pentamer. We observed that, in the beginning of the simulation, pentamer 1 barely contacts the lipid bilayer and moves around the bilayer surface seeking an appropriate spot to dig (Fig. 5A). During the first 300 ns simulation time, our calculated lateral diffusion coefficients for each 100 ns time frame indicate that the pentamer moves as fast as DOPC (Fig. 5B), where the lateral diffusion coefficient for DOPC is known to be DL = ~10 × 10-8 cm2/s at 30 °C.56 After t = 300 ns, when longitudinal diffusion begins to take place, the lateral diffusion of the pentamer is reduced. During this time, the pentamer diffuses slightly from the top of the bilayer surface into the amphipathic interface of the lipid bilayer.
FIGURE 5.
(A) Lateral trajectory of p3 (Aβ17-42) pentamer in configuration 1 plotted on the x-y plane denoting the DOPC bilayer surface. Line colors correspond to each consecutive 100 ns time frame in the order of black, red, green, yellow, and blue. Black and blue arrows on the plane indicate the starting and ending points, respectively. (B) Lateral diffusion coefficients of p3 pentamer in configuration 1 calculated for each 100 ns time interval.
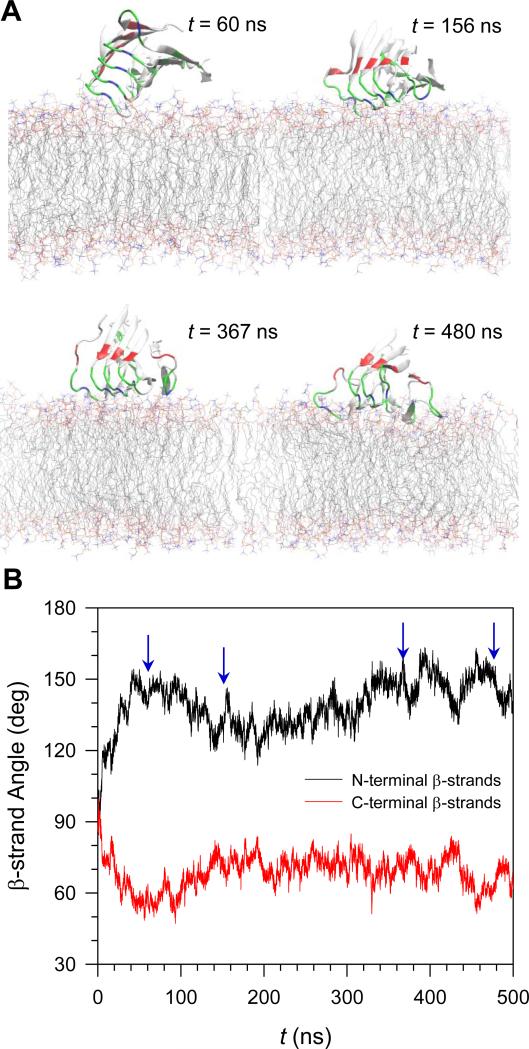
Pentamer 1 initially loses a contact with the bilayer and then comes back to attack the bilayer. Figure 6 shows snapshots taken during the simulations and the time series of the β-strand angles with respect to the membrane normal for the N- and C-terminal β-strands separately. The strands are directly opposite to each other. During the first 100 ns simulation, the β-strand angles for both strands largely deviate from the initial value of 90° due to the elevation of the termini (snapshot at t = 60 ns). In this situation, only the turn of peptide 5 in the pentamer is marginally hinged to the bilayer surface. The strong electrostatic interaction between the positively charged sidechain of Lys28 at the turn and the negatively charged phosphate groups of the lipids prevent the pentamer from falling off. After t = 100 ns, the pentamer starts to reorient, resulting in all peptide turns being able to contact the bilayer surface (snapshot at t = 156 ns). The sidechain of Lys28 at the turn in peptide 1 on the other side of the pentamer interacts with the lipid headgroup, establishing another hinge point. The turn region of the pentamer is now slightly immersed in the amphipathic interface of the lipid bilayer (snapshot at t = 367 ns). As the simulation progresses, the pentamer further moves into the hydrophobic core of the lipid bilayer (snapshot at t = 480 ns). Two loosened Lys28 sidechains from both edges of the pentamer turn region energetically disturb the lipid heads and consequently extract the lipids underneath the turn region. The evaded lipids provide room for the hydrophobic residues nearby, and the peptide turns to slip easily into the hydrophobic core of the lipid bilayer. Thus, overall, during the simulation we observed that the pentamer interaction with the lipids is mainly driven by two Lys28 residues from both edges of the pentamer turn region, indicating that the peptide-lipid interactions largely reflect the Lys28-lipid interactions (Supporting Information, Fig. S5).
FIGURE 6.
(A) Snapshots of p3 (Aβ17-42) pentamer in configuration 1 at selected times, t = 60, 156, 367, and 480 ns, representing the various orientations on the DOPC bilayer. (B) Time series of β-strand angle with respect to the membrane normal for the N-terminal (black line) and C-terminal (red line) β-strands separately for the pentamer in configuration 1.
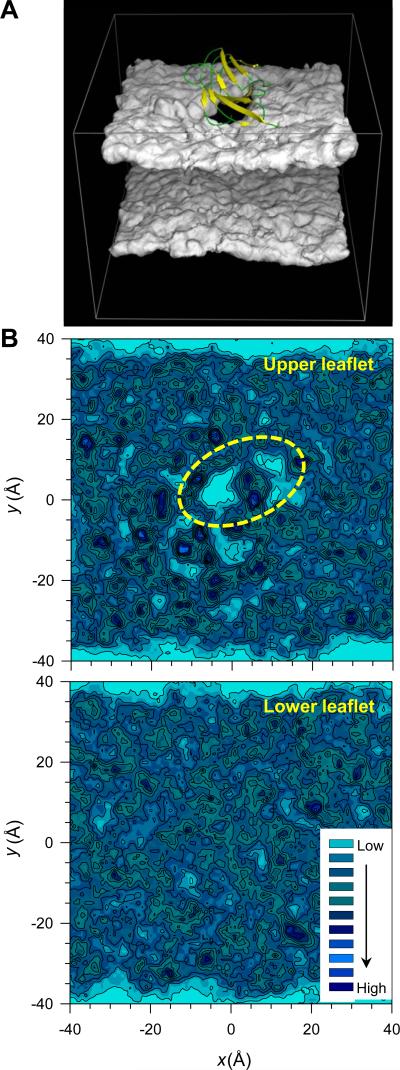
It has been known that cytolytic AMP PG-1 behaves like an amyloid with the capability to form fibrils, and amyloids possess cytotoxic properties with some antimicrobial activity.45,46 They share structural, functional, and biological similarities. Consistent with the PG-1 cytolytic activity, here we observed that the β-sheet of the p3 pentamer also induces membrane disruption effect at the amphipathic interface of the lipid bilayer. The three-dimensional density map of the lipid headgroups clearly shows a void below the pentamer (Fig. 7A). The contour maps of the center of mass of individual lipids reveal reduced contour lines, representing the low probability distribution of lipids on the pentamer-pressured surface at the upper bilayer leaflet (Fig. 7B). In contrast, contour lines of the isotropic probability distribution of lipids at the low bilayer leaflet can be observed. In the early stage of the bilayer attack, the β-sheet of the p3 pentamer lies obliquely to the bilayer surface (Fig. 6), increasing the surface pressure and disturbing the lipid molecules. In the calculations of the lipid order parameters for the heads and tails separately, we observed that lipids which are located in close proximity to the pentamer are less ordered than those located far away (Supporting Information, Fig. S6).
FIGURE 7.
(A) Three-dimensional density map of the lipid headgroup for the DOPC bilayer with embedded p3 (Aβ17-42) pentamer in configuration 1. The dark hole underneath the pentamer representing the low population density of lipid headgroup formed due to the cytolytic activity of pentamer is clearly visible. (B) Mapping of x, y coordinates of the center of mass of each lipid at the upper and lower bilayer leaflets separately onto the x-y plane for the DOPC bilayer. The contour lines enclose highly populated locations of the center of mass of each DOPC lipid. Yellow dotted line enclosed a significant deficiency in the distribution of the locations of the lipids’ center of mass corresponding to the empty space underneath the pentamer in the lipid headgroup density map.
Hydrophobic Core Crossing by the Aβ Oligomer Can Take Place via Two Different Pathways
In the partial deposition state of configuration 2, the p3 pentamer favors preserving the U-shaped conformation since the hydrophobic surface with the C-terminal β-strands faces the lipid tails, while the polar surface with the N-terminal β-strands is exposed to the bulk water. Compared to other pentamers, pentamer 2 strongly interacts with lipids with the strength of the interaction comparable to that of pentamer 5 in the fully embedded state (see Table 1). However, for cytotoxicity through channel formation, pentamer 2 has to slip into the bilayer so that the β-strands can align with the lipids. Core crossing can occur through either rotations of the termini or the turns of configuration 2. The obliquely inserted pentamers in configurations 3 and 4 can be described as states in the middle of the rotation. However, in the simulations, we did not observe any spontaneous crossing by the obliquely inserted state to reach a fully embedded state. Instead, the pentamers bounce to the bilayer surface back to a partial deposition state. In order to cross the bilayer, the pentamers have to cross over the high free energy barrier between two states. The surface plots for the peptide-lipid interaction energy as a function of the insertion angle, α, illustrate that the equilibrium MD simulations with limited samplings provide high and flat energy surfaces spanning over two energy minima representing the states of configurations 2 and 5, where configurations 3 and 4 merge into configuration 2 (Fig. 8). The surface plots do not represent the complete free energy surface for the pentamers. However, it can be assumed that the interaction energy can approximate the free energy, if the internal energy of the pentamer is consistent across the simulations.
FIGURE 8.
Insertion angle, α, dependence of the interaction energies of the pentamers with lipids during the simulations. The energy surface can be calculated from the occupancy probability of visiting each grid point on the plane of the interaction energy and insertion angle. Two sets of three combined configurations were sampled from the equilibrium simulations of p3 (Aβ17-42) pentamer for the pathways connecting the configurations (A) 2 → 3 → 5 and (B) 2 → 4 → 5. The insertion angle can be defined by an angle between the long molecular axis of pentamer and the membrane surface. The initial value of α for both pentamers is ~25°.
To sample events taking place while the p3 pentamers cross the hydrophobic core by rotating on their long molecular axis, we used the implicit generalized Born simple switching model (GBSW).57,58 The pentamers are sampled over the insertion angles, α, ranging from 0° to 90° with respect to the bilayer surface for two different pathways of pentamer rotations (Fig. 9A). The data are collected from a series of the implicit bilayer simulations varying the angles with a bin size of 3°. To mimic the DOPC bilayer, we set the GBSW membrane parameters as follows: 30 Å for the bilayer hydrophobic core thickness, 5 Å for the thickness of both interfaces, and a surface tension coefficient of 0.04 kcal/molÅ2. To sample more accurately at the transitions, the umbrella sampling method employing the miscellaneous mean field potential (MMFP) is used to restrain the pentamers harmonically along the reaction coordinates. However, the biased force is removed by using the weighted histogram analysis method (WHAM)59,60 that generates the unbiased histogram overlap. Our WHAM calculations estimate the potential of mean force (PMF) profiles representing the relative free energy changes along the reaction coordinates for two different pentamer rotations (Fig. 9B). The one-dimensional PMF profile indicates that pathway 1 has a higher free energy barrier than pathway 2, suggesting that rotating the termini across the hydrophobic core does cost more energy than rotation of the turns. Both pathways exhibit relative free energy minima at ~84°, which is close to ~74° obtained from the β-strand tilt angle in the equilibrium simulation. Although the implicit method does not provide information for lipid orientation and water penetration into the bilayer, it provides qualitative estimation how the pentamers are inserted through any specific pathway in the lipid bilayer.
FIGURE 9.
(A) Schematic diagrams describing two different pathways of p3 (Aβ17-42) pentamer rotation by increasing the insertion angle, α, in the hydrophobic core of the lipid bilayer. The insertion angle can be defined by an angle between the long molecular axis of pentamer and the membrane surface. Pathway 1 denotes the rotation of termini, while pathway 2 indicates the turn rotation. Red pentagons attached to the pentamers represent the negatively charged Glu22 sidechains. (B) The potential of mean force (PMF) profiles representing the relative free energy changes as a function of the insertion angle, α, for two different rotation pathways.
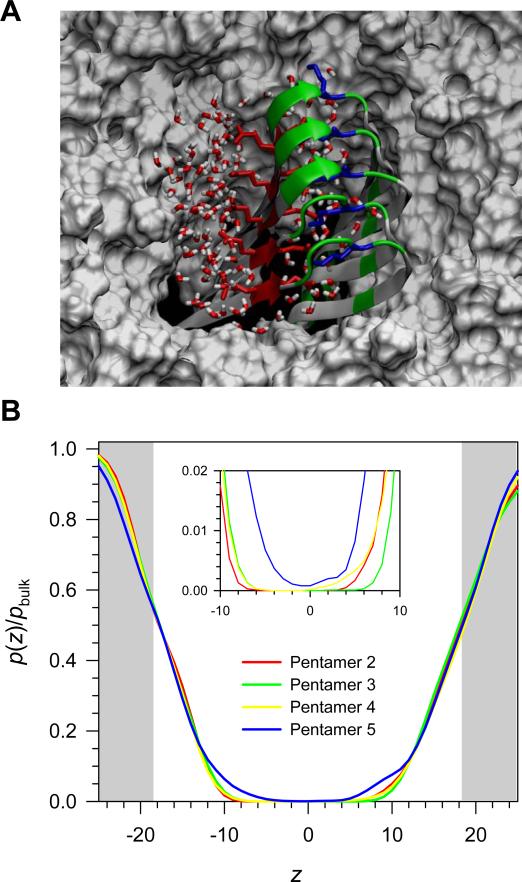
After crossing the hydrophobic core, the p3 pentamer is in the fully embedded state as in configuration 5. In this stage, the pentamer preserves the U-shaped conformation and strongly interacts with lipids. In the bilayer core, the hydrophobic interactions between the C-terminal β-strands and the lipid tails stabilize the β-sheet structure, while the electrostatic interactions of the charged atoms in both termini or in the turns with the lipid heads at the hinge at both ends in the amphipathic interfaces. However, the N-terminal β-strands containing the negatively charged Glu22 sidechains that point toward the hydrophobic core are unfavorably located in the middle of the bilayer. To avoid the unfavorable interactions with lipid tails, the pentamer brings water molecules to screen the charged sidechains in the hydrophobic core. At the starting point for the pentamer 5 simulation, no waters were initially placed below the glycerol groups inside the lipid bilayer, but they permeated during the course the simulation. A snapshot from the end of simulation clearly illustrates the screening effect at the solvation site (Fig. 10A). In configuration 5, water molecules deeply penetrate into the hydrophobic core compared to the other configurations (Fig. 10B). The water molecules brought by the pentamer are important, since these molecules can directly serve as water molecules inside the pore if, or when, few subunits containing 2 - 6 peptides align to form a channel.
FIGURE 10.
(A) Snapshot of p3 (Aβ17-42) pentamer in configuration 5 embedded in the DOPC bilayer taken from the 100 ns simulation in the top view. In the peptides, hydrophobic residues are shown in white, polar and Gly residues are shown in green, negatively charged residues in red, and positively charged residues in blue. Red sticks represent the negatively charged Glu22 and Asp23 sidechains, while blue sticks denote the positively charged Lys28 sidechains. Water molecules are shown in tiny red (oxygen) and white sticks (hydrogen), and the gray surface represents the DOPC bilayer. (B) A ratio of probability distribution functions for water in the DOPC bilayer as a function of distance from the bilayer center. The white area in the graph denotes the lipid bilayer, whiles gray areas at both sides indicate the bulk water. Inset highlights the hydrophobic core of the lipid bilayer.
Membrane Insertion of Aβ Dimer
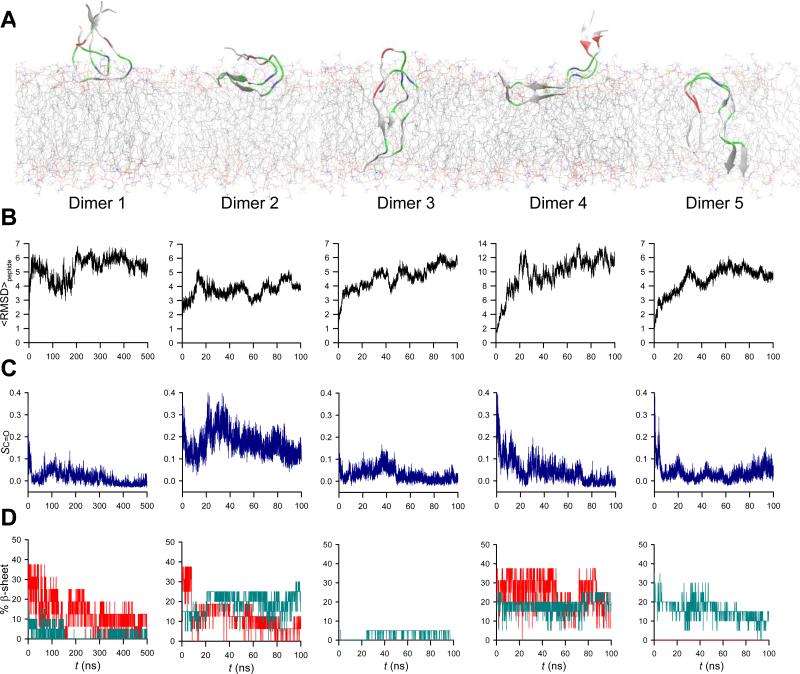
We repeated the simulations for the p3 dimer in the five different configurations, which are the same set of configurations used for the 5-mer p3 oligomer simulations. Our simulations showed that the p3 dimers are less stable in the lipid bilayer than the p3 pentamers (Fig. 11). Among them, dimers 2 and 5 are relatively stable with low RMSD values, marginally preserving the U-shaped conformation. Dimer 1 slightly diffuses into the amphipathic interface of the lipid bilayer as observed in the 5-mer case, but significantly loses the ordered conformation with decreasing in the β-sheet content. In contrast, dimer 2 retains the U-shaped conformation with relatively high values of the peptide order parameter and β-sheet content. In the obliquely inserted states, the dimers dismiss their initial configurations. Dimer 3 seems to cross spontaneously the hydrophobic core of the lipid bilayer, but loses its ordered conformation and the β-sheet. We observed that a small portion of an intermolecular antiparallel β-sheet between the N- and C-terminal strands barely holds the dimer interface. Although dimer 4 retains the β-sheet during the rotation on the bilayer surface, its U-shaped conformation deviates significantly due to the tumbled N-terminal strands in the bulk water area. In dimer 5, the C-terminal strands preserve the β-sheet, while the N-terminal strands lose the intermolecular H-bonds due to the separation. Dimer 5 contains the U-shaped structure in the lipid bilayer, but the overall structure is slightly different from that of dimer 2.
FIGURE 11.
(A) Simulated structures of p3 (Aβ17-42) dimers in a cartoon representation averaged over the simulations starting from five different initial configurations. (B) The root-mean-squared deviation (RMSD) from the starting point for backbone heavy atoms averaged over the peptides in the dimers. (C) Time series of the peptide order parameter, SC=O, for the β-sheets in the dimers. (D) Time series of the percent of β-sheet content based on the secondary structure analysis with DSSP52 calculated for the N-terminal (red lines) and C-terminal (blue lines) β-strands separately in the dimers.
DISCUSSION
We have simulated p3 (Aβ17-42) pentamers in different positions and states in the lipid bilayer. The p3 pentamer conformation derived from experiment-based coordinates of Aβ1-42 pentamer6 (where the N-terminal coordinates, residues 1-16 were missing due to disorder) is the U-shaped structure with the β-strand-turn-β-strand motif. It is still difficult to determine high resolution structures of toxic Aβ oligomers in solution, since the oligomer conformations are highly polymorphic. Our AFM image shows globular-like p3 oligomers on the DOPC bilayer. However, the exact secondary structures of these oligomers are still unknown. The NMR-based Aβ1-42 pentamer is a small piece of fibril, which can be determined because of the highly populated structure in solution, which can easily propagate into fibrils. We assume that the small piece of fibril in the oligomer size can similarly be populated in the Aβ oligomer conformational space in solution. A recent NMR study reported partial-helical structures of Aβ peptides in solution, but the early helical intermediates can convert into β structures upon binding to membrane.61 Thus, in our simulations the fibril-like p3 pentamer represents the Aβ oligomers in the aggregation stage in the presence of membrane. The U-shaped p3 peptide has been shown to be capable of forming a toxic ion channel in the lipid bilayer.17,18,20-22 Compared to full length Aβ peptides (Aβ1-40 and Aβ1-42), the N-terminal truncated p3 peptide is highly hydrophobic, which may facilitate its membrane insertion. Recent implicit membrane simulations of Aβ1-42 oligomers indicated that the hydrophobic core-spanning part of the Aβ peptide involves the residues 17 – 42, and that the transmembrane structures of Aβ1-42 oligomers are highly populated by β-sheets containing several β-hairpin intermediates.62
The Aβ oligomers were initially located on/in the lipid bilayer with five different configurations that represent sequential events in the Aβ oligomer membrane insertion pathways. Our simulations are based on the postulate that a possible insertion pathway would follow the sequence of events of ‘initial touch’ → ‘partial deposition’ → ‘oblique insertion’ → ‘embedded’ oligomer. A prerequisite to support such hypothesis is that Aβ peptides form an oligomer with fibril-like features including the β-sheet structure. The morphology of the p3 pentamer exhibits features of toxic Aβ oligomers, since it is fibril-like with parallel β-sheets and has an exposed hydrophobic face.5,9,10 The p3 peptides form fibrils on hydrophobic graphite surfaces, but not on hydrophilic mica or lipid bilayers.37 Instead, small p3 oligomers are adsorbed on the bilayer surface. p3 peptides do not form fibrils in solution at room temperature. However, short p3 protofibrils can exist in solution, serving as seeds for amyloid fibrils in appropriate environments,38 and mature p3 fibrils are able to form in solution at 37 °C.63 Preformed small fibril-like Aβ oligomers can also serve as seeds for amyloid channels, since asymmetric hydrophobic and polar faces are suitable for describing the subunits assembly architecture of amyloid channels.
A recent study demonstrates that the mechanism of membrane disruption by Aβ1-40 is a two-step process: the initial formation of ion-selective channels and the nonspecific membrane fragmentation during fiber formation which are enhanced in the presence of ganglioside.64 In our simulation, the mechanism of p3 pentamer insertion is similar to AMP PG-1's for several reasons:41,42 (i) the pentamer favors to locate at the amphipathic interface of the lipid bilayer in the early stage of membrane attack; (ii) the hydrophobic face of pentamer is toward the bilayer surface; (iii) positively charged residues initially drive the bilayer perturbation; and finally, (iv) after crossing into the bilayer, toxic β-sheet channel is formed in both peptide cases. The p3 pentamer in the U-shape morphology containing the hydrophobic-polar faces searches for a more energetically favorable environment inside the lipid bilayer. In order to insert, the Aβ oligomer contacts the lipid bilayer with its hydrophobic surface facing the bilayer surface, a mechanism common for AMPs.47 In the early events of membrane attack, the p3 pentamer also rotates the β-sheet plane obliquely to the bilayer surface. As a result, the turn region of the pentamer exerts pressure on the bilayer surface inducing bilayer perturbation. Two positively charged Lys28 sidechains which project out from the turns at both ends are important in the interactions with lipids and promoting the initial perturbation. Local thinning of the bilayer followed by membrane disruption facilitates the pentamer insertion, suggesting that the cytolytic activity of Aβ on the bilayer surface mimics the diffusion mechanism of AMPs.
The Aβ oligomer can translocate into the amphipathic interface of the lipid bilayer in the early stage of the membrane attack. It induces cytolytic activity with membrane disruption effects, increasing the surface pressure to insert into the hydrophobic core of the lipid bilayer. Once embedded in the bilayer, lipids support the U-shaped β-sheet motif. The fibril-like Aβ oligomer conformation is further stabilized when the oligomer fully crosses the hydrophobic core.31 The fibril axis of the Aβ oligomer is almost on the plane parallel to the bilayer surface, suggesting that additional oligomer interactions can occur in the lateral direction of the lipid bilayer. The Aβ dimer with the U-shaped motif is not effective in the membrane insertion, since the dimer loses its β-sheet structure in the lipid bilayer. We speculate that a trimeric Aβ oligomer with the U-shaped motif appears the smallest oligomer containing β-sheet core, which is able to effectively propagate through the membrane insertion pathway. For Aβ dimer and monomer, the formation of a β-hairpin intermediate may facilitate membrane insertion, similar to AMP PG-1, which has a β-hairpin structure. In this case, conversion to the U-shaped structure followed by interaction with monomers or small oligomers in the bilayer may take place. Recent experimental and theoretical studies support monomeric Aβ β-hairpin intermediates in solution.65-67 As in the PG-1, the β-hairpin can also form amyloid channels, extending the heterogeneous populated Aβ channel conformations.
To summarize, we propose a membrane insertion pathway for Aβ oligomers with a fibril-like β-sheet structure and provide the morphology of toxic Aβ oligomer in AD. Amyloid fibrils and channels can share the preformed seed of the U-shaped Aβ oligomer. Amyloid fibrils are highly polymorphic, resulting from growth of an ensemble of broad and heterogeneous population of preformed seeds. This suggests that other membrane insertion pathways are possible with different amyloid seeds. Among them, the populated Aβ oligomers with the β-sheet structure which are capable of inducing cytolytic activity, is a mechanism shared with the membrane inserting and pore forming AMPs.
Supplementary Material
ACKNOWLEDGMENTS
This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. This research was supported by the National Institutes of Health (National Institute on Aging AG028709 to RL). All simulations had been performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Time series of peptide averaged RMSD; averaged interaction energies of each monomer; time series of the center of mass deviation for selected residues; time series of interaction energies of each monomer in pentamer 1; probability distribution of the angle between the P-N vector and the bilayer normal, and deuterium order parameters for the tails of DOPC lipids. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Irvine GB, El-Agnaf OM, Shankar GM, Walsh DM. Mol Med. 2008;14:451–464. doi: 10.2119/2007-00100.Irvine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 3.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 4.Walsh DM, Selkoe DJ. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 5.Wu JW, Breydo L, Isas JM, Lee J, Kuznetsov YG, Langen R, Glabe C. J Biol Chem. 2010;285:6071–6079. doi: 10.1074/jbc.M109.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. Proc. Natl. Acad. Sci. USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petkova AT, Yau WM, Tycko R. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. J Am Chem Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 9.Ladiwala AR, Litt J, Kane RS, Aucoin DS, Smith SO, Ranjan S, Davis J, Vannostrand WE, Tessier PM. J Biol Chem. 2012;287:24765–24773. doi: 10.1074/jbc.M111.329763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Nat Struct Mol Biol. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 11.Arispe N, Rojas E, Pollard HB. Proc. Natl. Acad. Sci. USA. 1993;90:567–571. doi: 10.1073/pnas.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arispe N, Pollard HB, Rojas E. Proc. Natl. Acad. Sci. USA. 1996;93:1710–1715. doi: 10.1073/pnas.93.4.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagan BL, Azimov R, Azimova R. J. Membr. Biol. 2004;202:1–10. doi: 10.1007/s00232-004-0709-4. [DOI] [PubMed] [Google Scholar]
- 14.Kourie J, Henry C, Farrelly P. Cell Mol. Neurobiol. 2001;21:255–284. doi: 10.1023/A:1010995121153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin HAI, Bhatia R, Lal R. FASEB J. 2001;15:2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 16.Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R. Proc. Natl. Acad. Sci. USA. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang H, Zheng J, Nussinov R. Biophys J. 2007;93:1938–1949. doi: 10.1529/biophysj.107.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang H, Zheng J, Lal R, Nussinov R. Trends Biochem Sci. 2008;33:91–100. doi: 10.1016/j.tibs.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Jang H, Arce FT, Capone R, Ramachandran S, Lal R, Nussinov R. Biophys. J. 2009;97:3029–3037. doi: 10.1016/j.bpj.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL, Nussinov R, Lal R. Proc Natl Acad Sci U S A. 2010;107:6538–6543. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang H, Arce FT, Ramachandran S, Capone R, Lal R, Nussinov R. J Phys Chem B. 2010;114:9445–9451. doi: 10.1021/jp104073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang H, Arce FT, Ramachandran S, Capone R, Lal R, Nussinov R. J. Mol. Biol. 2010;404:917–934. doi: 10.1016/j.jmb.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capone R, Jang H, Kotler SA, Connelly L, Teran Arce F, Ramachandran S, Kagan BL, Nussinov R, Lal R. J Chem Theory Comput. 2012;8:1143–1152. doi: 10.1021/ct200885r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capone R, Jang H, Kotler SA, Kagan BL, Nussinov R, Lal R. Biochemistry. 2012;51:776–785. doi: 10.1021/bi2017427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connelly L, Jang H, Arce FT, Capone R, Kotler SA, Ramachandran S, Kagan BL, Nussinov R, Lal R. J Phys Chem B. 2012;116:1728–1735. doi: 10.1021/jp2108126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Connelly L, Jang H, Arce FT, Ramachandran S, Kagan BL, Nussinov R, Lal R. Biochemistry. 2012;51:3031–3038. doi: 10.1021/bi300257e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mobley DL, Cox DL, Singh RR, Maddox MW, Longo ML. Biophys J. 2004;86:3585–3597. doi: 10.1529/biophysj.103.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis CH, Berkowitz ML. J Phys Chem B. 2009;113:14480–14486. doi: 10.1021/jp905889z. [DOI] [PubMed] [Google Scholar]
- 29.Davis CH, Berkowitz ML. Proteins. 2010;78:2533–2545. doi: 10.1002/prot.22763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao LN, Chiu SW, Benoit J, Chew LY, Mu Y. J Phys Chem B. 2011;115:12247–12256. doi: 10.1021/jp2065985. [DOI] [PubMed] [Google Scholar]
- 31.Kagan BL, Thundimadathil J. Adv Exp Med Biol. 2010;677:150–167. doi: 10.1007/978-1-4419-6327-7_13. [DOI] [PubMed] [Google Scholar]
- 32.Pike CJ, Burdick D, Walencewicz AJ, Glabe CG, Cotman CW. J Neurosci. 1993;13:1676–1687. doi: 10.1523/JNEUROSCI.13-04-01676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams TL, Serpell LC. Febs J. 2011;278:3905–3917. doi: 10.1111/j.1742-4658.2011.08228.x. [DOI] [PubMed] [Google Scholar]
- 34.Tofoleanu F, Buchete NV. J Mol Biol. 2012;421:572–586. doi: 10.1016/j.jmb.2011.12.063. [DOI] [PubMed] [Google Scholar]
- 35.Lalowski M, Golabek A, Lemere CA, Selkoe DJ, Wisniewski HM, Beavis RC, Frangione B, Wisniewski T. J Biol Chem. 1996;271:33623–33631. doi: 10.1074/jbc.271.52.33623. [DOI] [PubMed] [Google Scholar]
- 36.Prangkio P, Yusko EC, Sept D, Yang J, Mayer M. PLoS One. 2012;7:e47261. doi: 10.1371/journal.pone.0047261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arce FT, Jang H, Ramachandran S, Landon PB, Nussinov R, Lal R. Soft Matter. 2011;7:5267–5273. doi: 10.1039/C1SM05162H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng J, Jang H, Ma B, Tsai CJ, Nussinov R. Biophys J. 2007;93:3046–3057. doi: 10.1529/biophysj.107.110700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang H, Ma B, Lal R, Nussinov R. Biophys J. 2008;95:4631–4642. doi: 10.1529/biophysj.108.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capone R, Mustata M, Jang H, Arce FT, Nussinov R, Lal R. Biophys J. 2010;98:2644–2652. doi: 10.1016/j.bpj.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang H, Ma B, Woolf TB, Nussinov R. Biophys J. 2006;91:2848–2859. doi: 10.1529/biophysj.106.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jang H, Ma B, Nussinov R. BMC Struct Biol. 2007;7:21. doi: 10.1186/1472-6807-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soscia SJ, Kirby JE, Washicosky KJ, Tucker SM, Ingelsson M, Hyman B, Burton MA, Goldstein LE, Duong S, Tanzi RE, Moir RD. PLoS One. 2010;5:e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris F, Dennison SR, Phoenix DA. Faseb J. 2012;26:1776–1781. doi: 10.1096/fj.11-199208. [DOI] [PubMed] [Google Scholar]
- 45.Jang H, Arce FT, Mustata M, Ramachandran S, Capone R, Nussinov R, Lal R. Biophys J. 2011;100:1775–1783. doi: 10.1016/j.bpj.2011.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kagan BL, Jang H, Capone R, Teran Arce F, Ramachandran S, Lal R, Nussinov R. Mol Pharm. 2012;9:708–717. doi: 10.1021/mp200419b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shepherd CM, Vogel HJ, Tieleman DP. Biochem J. 2003;370:233–243. doi: 10.1042/BJ20021255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 49.Klauda JB, Brooks BR, MacKerell AD, Jr., Venable RM, Pastor RW. The journal of physical chemistry. 2005;109:5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 50.Durell SR, Brooks BR, Bennaim A. J. Phys. Chem. 1994;98:2198–2202. [Google Scholar]
- 51.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 53.Lague P, Roux B, Pastor RW. J. Mol. Biol. 2005;354:1129–1141. doi: 10.1016/j.jmb.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 54.Han X, Tamm LK. Proc. Natl. Acad. Sci. USA. 2000;97:13097–13102. doi: 10.1073/pnas.230212097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Axelsen PH, Kaufman BK, McElhaney RN, Lewis RN. Biophys. J. 1995;69:2770–2781. doi: 10.1016/S0006-3495(95)80150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Filippov A, Oradd G, Lindblom G. Biophys J. 2003;84:3079–3086. doi: 10.1016/S0006-3495(03)70033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Im W, Feig M, Brooks CL., 3rd Biophys J. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Im W, Lee MS, Brooks CL., 3rd J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 59.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 60.Roux B. Comput Phys Commun. 1995;91:275–282. [Google Scholar]
- 61.Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A. Biochem Biophys Res Commun. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strodel B, Lee JW, Whittleston CS, Wales DJ. J Am Chem Soc. 2010;132:13300–13312. doi: 10.1021/ja103725c. [DOI] [PubMed] [Google Scholar]
- 63.Schmechel A, Zentgraf H, Scheuermann S, Fritz G, Pipkorn R, Reed J, Beyreuther K, Bayer TA, Multhaup G. J Biol Chem. 2003;278:35317–35324. doi: 10.1074/jbc.M303547200. [DOI] [PubMed] [Google Scholar]
- 64.Sciacca MF, Kotler SA, Brender JR, Chen J, Lee DK, Ramamoorthy A. Biophys J. 2012;103:702–710. doi: 10.1016/j.bpj.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoyer W, Gronwall C, Jonsson A, Stahl S, Hard T. Proc Natl Acad Sci U S A. 2008;105:5099–5104. doi: 10.1073/pnas.0711731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sandberg A, Luheshi LM, Sollvander S, Pereira de Barros T, Macao B, Knowles TP, Biverstal H, Lendel C, Ekholm-Petterson F, Dubnovitsky A, Lannfelt L, Dobson CM, Hard T. Proc Natl Acad Sci U S A. 2010;107:15595–15600. doi: 10.1073/pnas.1001740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cruz L, Rao JS, Teplow DB, Urbanc B. J Phys Chem B. 2012;116:6311–6325. doi: 10.1021/jp301619v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.