Abstract
Sulfur, a key contributor to biological reactivity, is not amendable to investigations by biological NMR spectroscopy. To utilize selenium as a surrogate, we have developed a generally applicable 77Se isotopic enrichment method for heterologous proteins expressed in E. coli. We demonstrate 77Se NMR spectroscopy of multiple selenocysteine and selenomethionine residues in the sulfhydryl oxidase augmenter of liver regeneration (ALR). The resonances of the active site residues were assigned by comparing the NMR spectra of ALR bound to oxidized and reduced FAD. An additional resonance appears only in the presence of the reducing agent and disappears readily upon exposure to air and subsequent reoxidation of the flavin. Hence, 77Se NMR spectroscopy can be used to report the local electronic environment of reactive and structural sulfur sites, as well as changes taking place in those locations during catalysis.
Keywords: 77Se NMR, selenium NMR, selenocysteine, selenoproteins, augmenter of liver regeneration, ALR, structure
While detection of carbon, nitrogen and proton resonances is routine in biological NMR, the only NMR-active isotope of sulfur, 33S, is a low-sensitivity quadrupolar nucleus unsuited for detection in biological systems. To gain insight into the multifaceted roles of sulfur in biology, the NMR-active isotope, 77Se, may be used as a surrogate. Selenium is located below sulfur in the Periodic Table and shares many physicochemical properties, including electronegativity, van der Waals radius and redox states.1 Substitution of cysteine’s sulfur by selenium, generating selenocysteine (Sec), occurs in nature,2 does not generally compromise function, and can even generate enzymes with new properties.3
The full potential of selenium NMR in biology has yet to be realized. While selenium is easily detected with conventional hardware and experiments, its large chemical shielding response brings about efficient relaxation routes resulting in short transverse relaxation rates that broaden 77Se lines in solution.4This problem is not pronounced for L-selenomethionine (Sem) whose chemical shift tensor span is Ω=580 ppm5 and can be detected with 77Se at natural abundance (7.5%).6 However, isotopic enrichment is necessary for Sec whose resonance widths are reported to be broader in biological systems (see below).4 The span of selenium chemical shift tensors in diselenide bonds of organic molecules range between 500–900 ppm7 and the span of the dimeric form of Sec, L-selenocystine, isΩ=601.5 ppm (unpublished measurements). However, the span of the chemical shift tensor of selenylsulfide bonds is expected to be larger.4 While the chemical shift tensor of Sec has not been reported, measurements in our group and others suggest it is indeed a more difficult target to detect (the resonance line widths at half-height are reported to be 150 to 500 Hz at a magnetic field of 14.1 T).
Earlier NMR studies were hindered by the availability of selenium-rich proteins (specifically Sec enriched proteins) and by low sensitivity in the absence of isotopic enrichment. Seleno proteins were prepared by chemical reactions with small seleno-compounds,8 or by using an animal diet enriched with 77Se.10 These earlier studies described direct detection of specific biological species such as selenenic (ESeO2H), selenenylsulfide (ESeSR) and the selenolate form (ESe−) in selenosubtilisin,9 denatured ribonuclease A, lysozyme8 and glutathione peroxide.10 Relaxation properties were described for 77Se introduced by reacting free thiol groups of proteins with the selenium analog of Ellman’s reagent (5,5’-dithiobis-(2-nitrobenzoate)).4 More recently, 77Sec-containing peptides were prepared by solid-phase peptide synthesis allowing assignment of diselenide connectivity in a 37-residue spider toxin11 and determination of pKa of individual Sec residues in a bioactive peptide hormone and neurotransmitter.12 These studies reaffirmed the sensitivity of 77Se NMR to its environment and established its ability to provide structural information for small peptides.
As the paucity of previous biological selenium NMR data testifies, current methods for preparation of 77Sec-rich proteins are not broadly applicable. Solid-state synthesis allows labeling of residues at specific sites but has a molecular weight limit, and along with chemical ligation requires specialized equipment and expertise. Incorporation of Sec in proteins by E. coli’s genetic incorporation machinery offers specificity but usually necessitates introducing mutations in the target protein.13 Depending on the specific system it may also suffer from low yield. Consequently, it has yet to be used for the large scale production necessary for NMR experiments. The enrichment of proteins with seleno-amino acids by supplementing E. coli’s growth media with the respective amino acids is an efficient and high yield method. However, for isotopic labeling with 77Se, the 77Se-enriched amino acids are not commercially available and the cost of their custom synthesis presents anobstacle for their routine utilization. Further, in house synthesis of 77Sec is not generally accessible to the non-specialist given selenium’s toxicity. Hence, none of the current methods to isotopically enrich proteins with 77Se offer an inexpensive, non-specialized procedure generally applicable to proteins irrespective of their amino acid sequence and molecular weight. To gain additional insight into the capabilities of 77Se NMR and EPR in larger biological systems we have developed such a procedure for the enrichment of proteins with 77Se by heterologous expression in E. coli. Here, we describe a broadly applicable, cost effective, selenium enrichment method that circumvents the need for costly (and hazardous) chemical synthesis. Furthermore, the selenium enrichment method described here is not only compatible with 77Se isotopic enrichment but also with incorporation of 13C, 15N, and 2H. It also allows the ratio of sulfur to selenium substitution to be controlled. Therefore, this new method can tailor proteins with different properties by fine-tuning the percent of selenium incorporation: for disulfide-containing proteins, partial substitution of sulfur with selenium generates proteins rich in selenylsulfides (S-Se) bonds, while diselenides (Se-Se) predominate at higher substitution ratios. Since the redox potential of sulfur and selenium differs21 this could potentially lead to subtle differences in their reactivity (vide infra).
Because of the toxicity of selenium, previous investigations of Se uptake and its random incorporation in E. coli proteins focused only on the effect of limited amounts of selenium. We have tested several selenium concentrations and means of supplementation to optimize a defined growth medium compatible with both 77Se labeling of sulfur sites in Met and Cys and the incorporation of 13C, 15N, and 2H. A conspicuous benefit of our method is that it utilizes 77selenite, which is readily obtained from the cost effective elemental 77Se. Hence the cost per liter growth media is comparable to that of 15N. E. coli cultures are grown in the presence of limited amounts of sulfur, so that the sulfur pool is exhausted prior to induction. Once growth is stalled, protein expression is induced, and the requisite mixture of sulfate/selenite is added to a final concentration of 100 µM in two steps, spaced 4 h apart, to achieve the desired sulfur-to-selenium substitution ratio (see details in materials and methods). Table 1 shows that this method affords workable expression levels and reproducible ratios of substitution for two structurally diverse proteins. E. coli thioredoxin contains two cysteine residues, forming a redox-active disulfide bond, and two methionine residues (Figures S1 and S2). The structurally more complex protein augmenter of liver regeneration (ALR) is a FAD-linked sulfhydryl oxidase that is involved in signal transduction and oxidative protein folding in the mitochondrial intermembrane space. The short form of this protein contains two catalytic disulfides, four structural disulfides, and four methionine residues per homodimer of 32 kDa (Figure S2). Interestingly, we have not detected a noticeable bias in selenium incorporation at specific sites even though the incorporation of selenium at different positions may influence the folding path in E. coli.
Table 1.
Sulfur-to-selenium substitution ratio and protein yield for growth media with varying ratios of sulfate and selenite.
| Protein | Sulfate in growth media (µM) |
Selenite in growth media (µM) |
Yield (mg/L) |
Selenium incorporation (% Se) |
|---|---|---|---|---|
| Trx | 5000 | 0 | 67±3 | 0* |
| Trx | 70 | 30 | 47±7 | 12±3 |
| Trx | 50 | 50 | 46±8 | 22±5 |
| Trx | 30 | 70 | 35±4 | 25±1 |
| Trx | 10 | 90 | 28±2 | 68±1 |
| Trx | 0 | 100 | 10±4 | 78±2 |
| ALR | 100 | 0 | 23±3 | 0* |
| ALR | 20 | 80 | 12.5±1 | 56±6 |
| ALR | 0 | 100 | 4±0.5 | 87±7 |
within the measurement accuracy
error was estimated based on the range of two independent measurements of yield and at least 3 independent measurements of selenium incorporation from different batches.
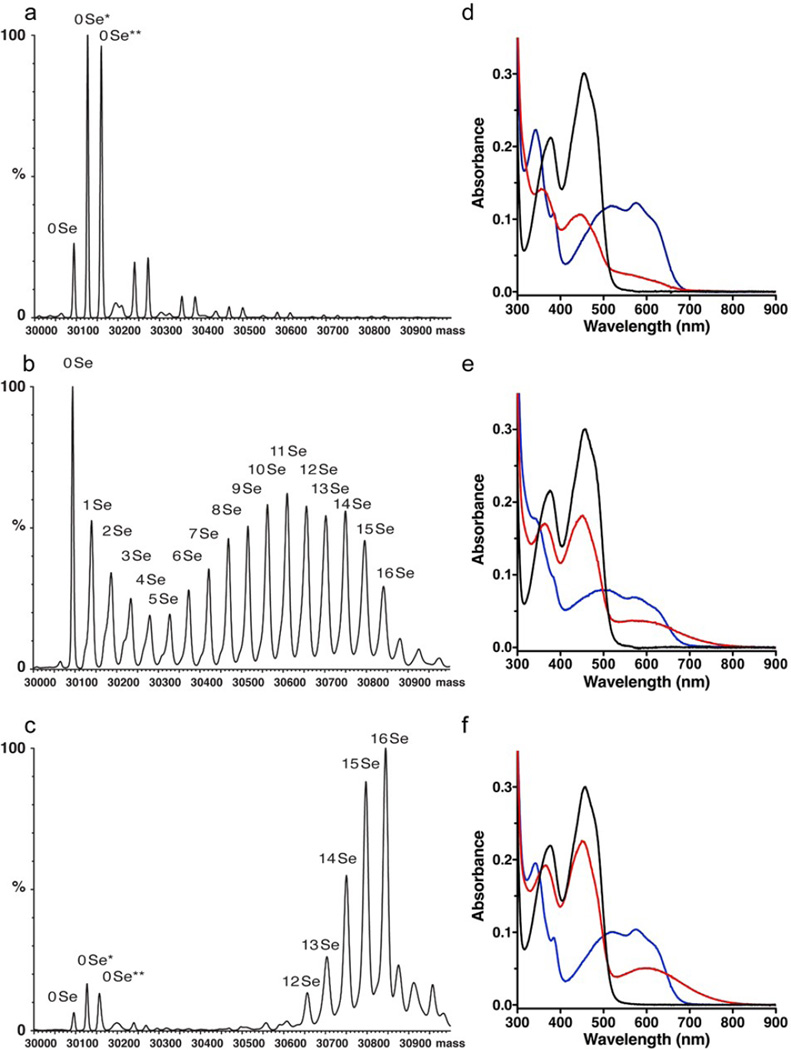
We use the cysteine-rich ALR to evaluate the effect of selenium substitution on the enzymatic activity, stability, and 3D structure of this cytokine-like protein. Incorporation of up to ~90% selenium into ALR (Figures 1a–c and S3), still yields an active flavo enzyme and generates the expected blue neutral flavo semiquinone upon aerobic incubation with the model substrate DTT (Figure 1d–f).25 Further, replacement of the active site disulfide by a diselenide forms a new charge-transfer intermediate with the flavin prosthetic group (visualized 5 sec after mixing), with an absorbance band extending > 750 nm (Figure 1f).
Figure 1.
Mass spectroscopy and UV-visible spectra of ALR as a function of selenite concentration in the growth media. Panels a–c: Mass spectroscopy of ALR as a function of selenite concentration in the growth media. (a) ALR grown solely on sulfate. The star denotes oxidation. Unlabeled peaks at increasing molecular weight are trifluoroacetic acid adducts. (b) ALR grown in a 1:4 sulfate/selenite molar ratio mixture yielding a selenium incorporation of ~60%. (c) ALR grown solely on selenite, where ~90% of the sulfur atoms are replaced by selenium. Panels d–f: DTT treatment of ALR followed by UV-VIS spectroscopy (black line: before addition of DTT to the enzyme, red line: 5 s and blue line: 5 min after adding DTT). (d) The spectra at 5 min show native and selenium-substituted enzymes form the blue flavo semiquinone. This species is developing at 5 sec in panel d for native ALR, whereas panels e and f reveal a featureless long-wavelength charge-transfer band with the oxidized flavin. (e) ALR with ~60% selenium incorporation (~48% of the selenium atoms are in selenylsulfide bonds) has a charge transfer band, easily discerned at wavelengths >550 nm. (f) Selenium-rich ALR with ~90% selenium incorporation (~81% of the selenium atoms are in diselenide bonds) displays the highest levels of charge transfer complex.
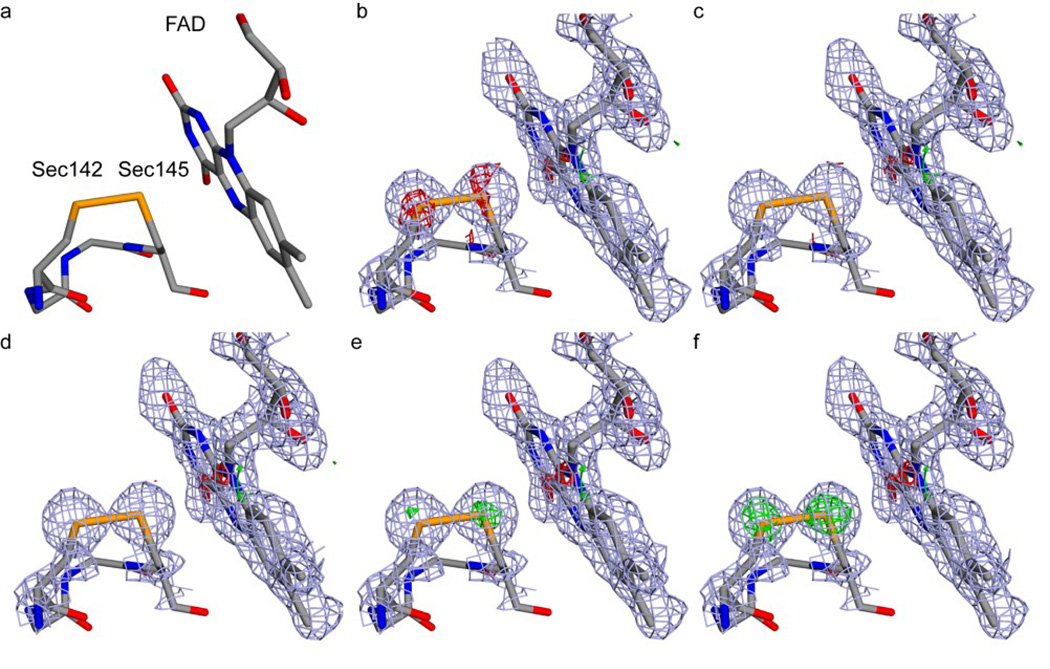
In addition to the role of selenium in modulating the behavior of the redox active cysteines, ALR contains both intra- and inter-chain structural disulfides (Figure S2). It was therefore of interest to determine the impact of selenium substitution on both the global thermal stability of ALR and on the rate of flavin release rate from the enzyme (Figure S4). Both approaches show that the stability of selenium-based ALR is only slightly lower than that of the native ALR.29 A more detailed picture of the effect of substitution emerges from the structure of the selenium-rich ALR solved by X-ray crystallography (resolution of 1.5 Å, Rfree of 0.221, PDB code 3U5S, Table S1). The high resolution data provided the first opportunity to refine a protein X-ray structure with occupancy of sulfur and selenium using the substitution ratio determined by mass spectrometry and inductively coupled plasma spectroscopy (Figure 2). Alignment of the native ALR structure against selenium-rich ALR shows an RMSD of 0.305 Å for all atoms. The only significant structural changes reflect the longer Se-Se and Se-C bonds and the corresponding adjustments in dihedral angles (Figure S5 and Table S2). For selenium-rich ALR, these torsion angles are 2–4° larger than those of the native sulfur-containing protein.
Figure 2.
Electron density difference maps of the disulfide/diselenide and flavin in the active site of ALR refined with different ratios of selenium and sulfur occupancy. (a) A view of the active site residues near the flavin in CPK coloring. Panels b–f: the difference electron density maps computed following refinement with 60–100% selenium occupancy are shown for the redox diselenide (Sec142-Sec145) and flavin group of selenium-rich ALR. The 2Fo-Fc map is drawn in grey contoured at 1.2 σ. The Fo-Fc maps were contoured at 3.0 σ, with positive difference density shown in green and negative difference density shown in red. (b) 100% Se, 0% S. (c) 90% Se, 10% S. (d) 80% Se, 20% S. (e) 70% Se, 30% S. (f) 60% Se, 40% S.
An important facet of this selenium enrichment method is the ability to incorporate 77Se into all the sulfur-containing residues, hence allowing detection by NMR spectroscopy. 77Se, a spin I=1/2 nucleus, has a pronounced chemical shielding response.30 The latter renders it highly sensitive to changes in the electronic environment, such as modification in bonding and conformation. Consequently, 77Se NMR measurements, in combination with theoretical calculations of the magnetic shielding tensor, will provide new approaches to probe the role of the local environment in shaping the reactivity of cysteines. Further, 77Se NMR could be used to investigate oxidative damage to Sec and selenomethionine (Sem) residues; address the binding of metals, ligands and proteins and detect changes in the local environment during catalysis, folding and conformational changes.
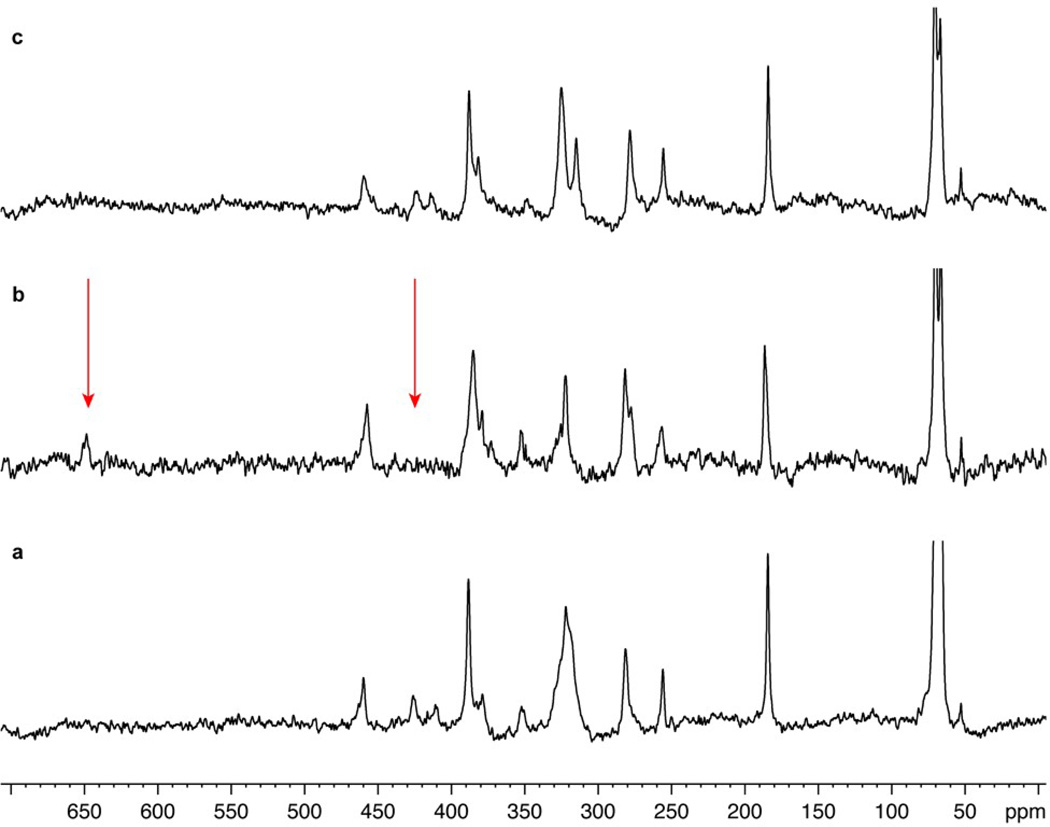
To demonstrate our ability to detect and resolve multiple selenium sites in proteins at positions normally occupied by sulfur, Figure 3 presents solution-state NMR detection of selenium in ALR at a magnetic field of 14.1 T (1H frequency of 600 MHz, 77Se frequency of 114.493 MHz). Here, ALR was enriched to ~50% selenium to generate a combination of diselenide and selenylsulfide bonds. Based on the count of sulfur-containing amino acids in ALR – 6 Cys and 3 Met – we expect resonances of selenium in 6 selenylsulfide, 6 diselenide and 3 seleno-ether bonds. As shown, the resonances of most species are readily observed by direct detection of selenium. The half-height line width of selenium resonances ranges from 180 to 320 Hz, due to chemical shift anisotropy driven relaxation. The differences in line width suggest that the T2 relaxation time may be influenced by local protein dynamics. The isotropic chemical shifts of selenium in Sem are between 50–70 ppm, a common range for Sem in proteins.6 The isotropic chemical shifts of selenium in Sec range from 185 to 460 ppm, where 10 resonances are resolved. The sample did not have resonances corresponding to oxidized selenium residues (Se-OH, Se-O2H and SeO3H at 1100–1300 ppm10) or reduced Sec (~−220 ppm9). This wide range for selenium resonances in diselenide and selenylsulfide bonds in the same protein is at first glance surprising, considering the fact that ALR’s three selenylsulfide/diselenide bonds are all at water-exposed positions. However, studies in our laboratory indicate that even for a single Sec within a given redox motif (such as Cys-Gly-Ala-Sec), it is possible to detect resonances that differ in chemical shifts by about 135 ppm and by a seven-fold difference in line width. Similar differences in chemical shift values were recently reported for Sec positioned in different locations in a peptide.12 The isotropic chemical shifts are likely to depend on the dihedral angle at which the selenium is located, nearby charges, etc. Assignment of specific resonances is however not straight forward since information on how these factors affect Sec resonances in proteins is sparse. While the chemical shifts of ALR have been reported and are available via the Biological Magnetic Resonance Data Bank (BMRB),33 correlating Se resonances with other nucleivia 2D experiments is complicated by the fast T2 relaxation time. (The line width of individual resonances reported here range from 50 to 300 Hz). In parallel, the assignment and interpretation of ALR selenium NMR spectra is continuing via genetic incorporation of Sec in unique positions using E. coli’s innate insertion machinery34 as well as site-specific mutagenesis. It would be interesting to employ chemical shielding calculations to further study the local environment of individual Sec residues using the high-resolution structural information obtained by the X-ray refinement described above (see Figure 2 and Table S2).
Figure 3.
77Se NMR spectroscopic characterization of flavin reduction in 77Se-labeled ALR. Proton-decoupled 77Se spectra of ALR with 50% selenium enrichment acquired at 14.1 T. (a) Spectra of 77Se-labeled ALR with oxidized FAD (Sem resonances are truncated for clarity). (b) Spectra of 77Se-labeled ALR with reduced FAD under anaerobic conditions. Upon addition of a reducing agent, the resonance peaks at 412, and 426 disappears, while a new resonance peak at 651 ppm appears (highlighted with an arrow). (c) Same sample shown in panel b following exposure to air and removal of the reducing agent.
Further assignment of the resonances and an additional assessment for 77Se NMR sensitivity is possible by comparing the spectra of ALR when bound to oxidized and reduced flavin (Figure 3). When the FAD is reduced with sodium dithionite, two resonances at 412 and 426 ppm disappear. These two resonances were assigned to be selenylsulfide bonds by comparing the NMR spectra of ALR with different selenium incorporation levels (data not shown). The resonances reappear upon reoxidation of FAD and removal of the reducing agent (Figure 3c). Hence, we conclude that the resonances are likely to be the selenylsulfide bonds in the active site residues proximal to the flavin prosthetic group (Figure 2a). In addition to the changes in the 412–426 ppm region, a new resonance is detected at 651 ppm. The modification is reversible, as discerned from the disappearance of the 651 ppm resonance upon removal of the reducing agent and reoxidation of FAD (Figure 3c).
In sum, the reported substitution method provides a robust, flexible and general platform that can be used to enrich heterologous proteins in E. coli with selenium, without the need to modify expression systems, synthesize selenium compounds or calibrate sulfur/selenium consumption. We demonstrate the ability to acquire spectra of multiple selenium atoms in a mid–sized protein in a time-effective fashion. It is clear that selenium NMR can be used as a reporter for multiple sulfur/selenium locations without the need to introduce selenium in a site-specific fashion. This allows us to directly probe the environment, conformation and interactions of the site of interest and the capacity to directly detect modifications in Sec residues and changes in the local environment taking place during catalysis. Hence, 77Se provides a powerful spectroscopic probe for the multiple roles of cysteine and methionine in protein structure, stability and function.
Materials and Methods
Materials
Elemental 77selenium (99.20%) was purchased from Isoflex USA (San Francisco, CA). Enzymes used for molecular biology were acquired from New England Biolabs (Ipswich, MA). The pMHT Delta 238 plasmid expressing Tobacco Etch Virus (TEV) protease fused to the cytoplasmic maltose binding protein35 was purchased from the Protein Structure Initiative-Material Repository. Chromatography media was supplied by GE Healthcare Bio-Sciences Corporation (Pittsburgh, PA). Crystallization reagents were from Hampton research (Aliso Viejo, CA). All other chemicals and reagents were supplied by Sigma-Aldrich (St. Louis, MO), Acros Organics (Geel, Belgium) and GoldBio (St. Louis, MO). All reagents and solvents were at least analytical grade and were used as supplied.
Expression of ALR and Trx
The pTrcHisA (Life Technologies, NY) expression vector containing the short form of human ALR (residues 81–205) gene with mutations C154A and C165A was as previously described.25 For this study a TEV cleavage site was introduced between the N-terminal hexahistidine tag and the first Met. Following cleavage with TEV protease a Ser is present before residues 81–205. E. coli Trx fused to a C-terminal hexahistidine tag was expressed in pET32a (Addgene plasmid 11516). The two proteins were expressed in BL21(DE3).
Cells were grown in a defined media adapted from Studier36 C-750501 minimal media designed for 13C labeling and abbreviated as MC-750501. The modified medium contained: 50 mM Na2HPO4, 50 mM KH2PO4, 10 mM NaCl, 50 mM NH4Cl, 2 mM MgCl2, 0.2× metals, 1× vitamins, 0.4% glucose, 200 µM CaCl2, supplemented with antibiotics. 1× concentration trace metals solution contained 50 µM FeCl3, 20 µM CaCl2, 10 µM MnCl2, 10 µM ZnCl2, 2 µM CoCl2, 2 µM CuCl2, 2 µM NiCl2, 2 µM Na2MoO4 and 2 µM H3BO3. The recipe for 1000× vitamins solution was as detailed by Studier.36
A 20 mL starter culture of MC-750501 supplemented with 5 mM Na2SO4 and antibiotics selection was grown for about 9 h. A 1 mL of starter culture was used to innoculate 1 L growth media supplemented with 50 μM Na2SO4 in 2.8 L baffled flasks. For constructs that required the use of kanamycin sulfate we typically reduce the concentration of the kanamycin to 50 μM and use the antibiotics as the sole source of sulfur in the main incubation. Cells were grown at 37 °C, with good aeration and an antibiotics selection until the cell density no longer increased. Typically, this value is reached after 14 h with an OD close to 0.8 at 600 nm. Note that the doubling time of the cells in the MC-7504501 growth media is about 60 min. At this point, protein expression was induced with 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG). At induction the cells were supplied with 50 µM mixture of Na2SO4/Na2SeO3 at the desired ratio. Following 4 h of growth, a second 50 µM aliquot was added. Cells were harvested 9–12 h past induction by centrifuging at 5,000 g for 10 min at 4 °C. In general, the optimal expression time is approximately double that of the expression time in LB, due to the longer doubling times in the relatively spartan growth media. The cell paste was resuspended in 50 mM potassium phosphate buffer, pH 7.5, containing 500 mM NaCl (IMAC buffer), flash frozen using liquid nitrogen and stored at −80 °C.
For NMR experiments, the appropriate amount of elemental 77Se was oxidized to 77selenite in a minimal volume of nitric acid.37 The expected selenite was confirmed by solution 77Se NMR. The volume was kept to a minimum in order to reduce the amount of acid added to the growth media.
Purification of ALR and Trx
The cell paste was thawed and lysed in IMAC buffer supplemented with 1 mM PMSF and 0.5 mM benzamidine using a high pressure homogenizer (EmulsiFlex-C5, Avestin, Ottawa, Canada). Cell debris was removed by centrifugation at 15,000 g for 1 h at 4 °C. Clear supernatant was loaded onto a 5 mL HisTrap FF column and the column was washed with IMAC buffer, supplemented with 20 mM imidazole. ALR was eluted using 5 mL fractions of IMAC buffer supplemented with 50 mM, 200 mM, 500 mM and 1 M imidazole. Yellow fractions containing ALR were combined and the buffer was exchanged into 20 mM Tris (pH 7.5), 1 mM EDTA by dialysis and the hexahistidine tag cleaved with TEV protease, at 4°C for 18 h. The resulting protein was then dialyzed against 20 mM Tris, 1 mM EDTA pH 7.5 at 4 °C. The protein was applied to a 5 mL HiTrap SP HP column to remove the TEV and then loaded on a Source 15Q column and eluted with a salt gradient between 0–100 mM NaCl over 20 column volumes. Purification of Trx was by identical IMAC chromatography using IMAC buffer with 400 mM imidazole. The buffer was then exchanged to 20 mM Tris (pH 8.0), 1 mM EDTA and the protein loaded on a 5 mL HiTrap Q HP and eluted with a salt gradient between 0–300 mM NaCl over 20 column volumes. For both ALR and Trx the fractions containing proteins were combined. The protein was dialyzed against 20 mM Tris (pH 7.5), 1 mM EDTA, then concentrated to 20 mg/mL and flash frozen using liquid N2 and stored at −80 °C until further use. Protein purity, as determined by 15% SDS-PAGE Tris-Glycine gels, was higher than 99%. Protein concentration was determined using the molar extinction coefficients for FAD bound ALR of 11.6 mM−1 cm −1 at 456 nm.25 The ratio of sulfur and selenium was determined by LC-ESI-TOF mass spectrometry and ICP.
While we have optimized the protocol for a final concentration of 100 μM selenite we have found for 77Se incorporation that a lower concentration of selenium (70 μM) yields identical selenium incorporation ratio but a higher yield (sometimes by a factor of 2). For simultaneous incorporation of 13C, regarded to be the most costly isotopes discussed here, we have tested the Marley method in which an initial culture is grown to exponential phase in LB, washed with saline, and resuspended in growth media containing isotopically labeled metabolic precursors.39 We found that this method yields identical results as long as the cells are allowed to equilibrate after suspension and cell density monitored to ensure that the internal sulfur has been depleted. If 13C and 15N is not considered then it is also possible to enrich the growth media with additional amino acids.36
Mass spectroscopy
Mass spectra were obtained using a QTOF Ultima (Waters, MA) operating under positive electrospray ionization (+ESI) mode connected to a LC-20AD (Shimadzu, Kyoto, Japan). ALR samples were separated from small molecules by reverse phase chromatography on a C4 column (Waters XBridge BEH300) using an acetonitrile gradient from 30–71.4% with 0.1% TFA as the mobile phase in 25 min. Data were acquired from m/z 350 to 25,000 at a rate of 1 sec/scan.
UV-Vis spectroscopy
UV-VIS spectra were recorded using a HP8453 diode array spectrophotometer (Hewlett-Packard, CA). DTT and ALR were prepared in 20 mM Tris, 1 mM EDTA, pH 7.5 and mixed to give final concentrations of 20 µM ALR, 5 mM DTT. DTT solutions were standardized with DTNB.
Thermal and chemically induced unfolding transitions
Thermal unfolding transitions of 10 μM ALR in 10 mM phosphate, pH 7.5 were followed by circular dichroism using a J-810 circular dichroism spectropolarimeter (Jasco, Essex, UK). Spectra were collected between 190–260 nm, in 2 °C increments from 40 to 96 °C using a 10 mm cell. The data was analyzed using the mean residue ellipticities (deg cm2 dmol−1) at 222 nm as a function of temperature using a two-state model. Data were an average of two independent measurements.
Flavin dissociation was monitored by recording the decrease in absorbance at 496 nm in the presence of 5 M guanidine hydrochloride. Spectra were recorded at room temperature using 10 µM ALR in 20 mM Tris buffer, 1 mM EDTA, 5 M guanidine hydrochloride, pH 7.5.
Crystallization and data collection
Protein crystals were obtained by hanging drop vapor diffusion at 22 °C. The selenium-rich ALR reproducibly crystallized in a broad range of protein and precipitant concentrations. The best crystals formed in a reservoir solution of 20 mM MES, pH 6.5, 16% PEG 8000. Crystallization drops were prepared by mixing 1 μL of selenium rich ALR (10 mg/mL protein in 20 mM MES, pH 6.5) with 1 µL of the reservoir solution and placing over 250 μL of the reservoir solution. Crystals appeared within hours and reached their maximum size in 2 weeks. Crystals were flash-cooled in liquid nitrogen following incubation of the crystal in a cryo solution made from the reservoir well solution, which was 25% saturated with the cryo protectant xylitol.
Diffraction data was collected using an in-house Rigaku-RUH3R rotating anode generator with a RAXIS IV image plate area detector. X-ray diffraction data was collected at −180 °C with a total of 180 15 min 1° oscillations from one crystal. The diffraction data was indexed and scaled with the program HKL2000.40
Crystal structure solution and refinement
The crystal of the selenium-rich ALR was indexed in the space group C2221 with cell dimensions a, b, c of 50.9, 76.9, 63.5 Å, respectively. There was one subunit of protein in the asymmetric unit and the solvent content was 34.7%. The crystal structure was solved by molecular replacement using the program MOLREP of CCP441 using as a search model the structure of human ALR (PDB code 3MBG).29 During model building, refinement was carried out using the program REFMAC5 of CCP4. Model modifications were performed using the graphics program COOT.42 In contrast to the search model ALR in which the electron density was clear only starting with residue Asp94, the entire N-terminus was built in the selenium-rich model, starting with Ser80. (Residue S80 is an extra residue at the N-terminus that remains after protease cleavage of the hexahistidine tag.) To facilitate the refinement of 90% Se and 10% S partial occupancy for Cys and Met residues, the program Phenix43 was used for the final rounds of refinement. Water molecules were placed during successive cycles of model building and refinement. The final model contains 126 amino acid residues, one FAD molecule, and 112 water molecules. A final 2Fo - Fc difference electron density map (Figure S5) for the model confirmed the validity of the final model. The final Rworking and Rfree values were 0.200 and 0.221, respectively.
NMR spectroscopy
Solution state NMR spectra were acquired on a 14.1 T Bruker AV600 spectrometer equipped with a Bruker 5 mm broad-band outer coil liquid-state probe operating at a frequency of 114.493 MHz. Proton-decoupled 77Se spectra of selenium was recorded using spectral width of 96.1 KHz, an acquisition time of 0.085 s, with 250,000 scans, pulse delay of 0.5 s, a 90° pulse width of 15 µs, and WALTZ decoupling with a 2.78 kHz field. While the spin-lattice relaxation time, T1, was not measured for ALR, values for Sec in a variety of other selenoproteins in our lab range between 0.9–1.5 s at a field of 14.1 T. NMR spectra were processed using backwards linear prediction of the first 4 points of the FID and 100 Hz exponential apodization. 77Se chemical shifts are reported with respect to diphenyl diselenide used as an external secondary chemical shift reference standard, set at 463.0 ppm (dimethyl selenide as primary reference at 0 ppm).44 Sample temperature was 25 °C, controlled to within 0.1 °C. The NMR sample contained 2 mM ALR in 50 mM phosphate, 1 mM EDTA, 10% D2O, pH 7.5 in a volume of 350 µL in a Shigemi NMR tube (Shigemi Inc., PA). ALR was enriched to 50% 77selenium. The sample was reduced by adding sodium dithionite to the NMR tube to a final concentration of 6 mM in a gas-tight Shigemi NMR tube (Wilmad Lab Glass, NJ).
Accession numbers
The atomic coordinates for the structure of selenium-substituted ALR have been deposited in the PDB (PDB code 3U5S).
Highlights.
The NMR-active isotope 77Se is introduced into proteins by heterologous expression in Escherichia coli.
The method is cost effective and requires no synthesis or modifications to existing expression systems.
The method permits control over the ratio of sulfur to selenium substitution in proteins allowing the manipulation of protein populations for medical and biochemical research.
It is possible to resolve multiple selenomethionine and selenocysteine residues in the 32 kDa dimer of augmenter of liver regeneration by conventional NMR spectroscopy.
Using the protein augmenter of liver regeneration, we further demonstrate the feasibility of routine biological Se-NMR spectroscopy as a probe of structure and function.
Acknowledgments
We thank Shawn Gannon, Dr. Steve Bai and Dr. Mike Geckle for technical assistance and helpful discussions. C.T. and S.S. were supported by NIH GM26643 and 1-T32-GM08550. R.P.R. thank the David A. Plastino Scholars Alumni Undergraduate Research Fellowship Program. W.W. was supported by the University of Delaware Research Foundation Research Experience for Undergraduates Program. S.R. and B.B. acknowledge support from the University of Delaware Strategic Initiative Grant (10000431) and grants from the National Center for Research Resources (5P30RR031160-03) and the National Institute of General Medical Sciences (8 P30 GM103519-03). S.R. acknowledges support from NSF MRI-R2 Grant 0959496.This material is based upon work supported by the National Science Foundation under Grant No. MCB-1054447 “CAREER: Reactivity of Selenoproteins” (S.R.).
Abbreviations
- ALR
augmenter of liver regeneration
- Trx
thioredoxin
- Sec
selenocysteine
- Sem
selenomethionine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:
References
- 1.Wessjohann LA, Schneider A, Abbas M, Brandt W. Selenium in chemistry and biochemistry in comparison to sulfur. Biological Chemistry. 2007;388:997–1006. doi: 10.1515/BC.2007.138. [DOI] [PubMed] [Google Scholar]
- 2.Lobanov AV, Hatfield DL, Gladyshev VN. Eukaryotic selenoproteins and selenoproteomes. Biochimica Et Biophysica Acta-General Subjects. 2009;1790:1424–1428. doi: 10.1016/j.bbagen.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansson L, Gafvelin G, Arner ESJ. Selenocysteine in proteins - properties and biotechnological use. Biochimica et Biophysica Acta-General Subjects. 2005;1726:1–13. doi: 10.1016/j.bbagen.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Gettins P, Wardlaw SA. NMR relaxation properties of Se-77-labeled proteins. Journal of Biological Chemistry. 1991;266:3422–3426. [PubMed] [Google Scholar]
- 5.Potrzebowski MJ, Katarzynski R, Ciesielski W. Selenium-77 and carbon-13 high-resolution solid-state NMR studies of selenomethionine. Magnetic Resonance in Chemistry. 1999;37:173–181. [Google Scholar]
- 6.Zhang MJ, Vogel HJ. 2-dimensional NMR-studies of selenomethionyl calmodulin. Journal of Molecular Biology. 1994;239:545–554. doi: 10.1006/jmbi.1994.1393. [DOI] [PubMed] [Google Scholar]
- 7.Demko BA, Wasylishen RE. Solid-state selenium-77 NMR. Progress in Nuclear Magnetic Resonance Spectroscopy. 2009;54:208–238. [Google Scholar]
- 8.Luthra NP, Costello RC, Odom JD, Dunlap RB. Demonstration of the feasibility of observing nuclear magnetic-resonance signals of Se-77 covalently attached to proteins. Journal of Biological Chemistry. 1982;257:1142–1144. [PubMed] [Google Scholar]
- 9.House KL, Dunlap RB, Odom JD, Wu ZP, Hilvert D. Structural characterization of selenosubtilisin by Se-77 NMR-spectroscopy. Journal of the American Chemical Society. 1992;114:8573–8579. [Google Scholar]
- 10.Gettins P, Crews BC. Se-77 NMR characterization of Se-77-labeled ovine erythrocyte glutathione-peroxidase. Journal of Biological Chemistry. 1991;266:4804–4809. [PubMed] [Google Scholar]
- 11.Mobli M, de Araujo AD, Lambert LK, Pierens GK, Windley MJ, Nicholson GM, Alewood PF, King GE. Direct visualization of disulfide bonds through diselenide proxies using Se-77 NMR spectroscopy. Angewandte Chemie-International Edition. 2009;48:9312–9314. doi: 10.1002/anie.200905206. [DOI] [PubMed] [Google Scholar]
- 12.Mobli M, Morgenstern D, King GF, Alewood PF, Muttenthaler M. Site-specific pKa determination of selenocysteine residues in selenovasopressin by using 77Se NMR spectroscopy. Angewandte Chemie-International Edition. 2011;50:11952–11955. doi: 10.1002/anie.201104169. [DOI] [PubMed] [Google Scholar]
- 13.Johansson L, Chen CY, Thorell JO, Fredriksson A, Stone-Elander S, Gafvelin G, Arner ESJ. Exploiting the 21st amino acid - purifying and labeling proteins by selenolate targeting. Nature Methods. 2004;1:61–66. doi: 10.1038/nmeth707. [DOI] [PubMed] [Google Scholar]
- 14.Bar-Noy S, Moskovitz J. Mouse methionine sulfoxide reductase B: effect of selenocysteine incorporation on its activity and expression of the seleno-containing enzyme in bacterial and mammalian cells. Biochemical and Biophysical Research Communications. 2002;297:956–961. doi: 10.1016/s0006-291x(02)02314-8. [DOI] [PubMed] [Google Scholar]
- 15.Su D, Li YH, Gladyshev VN. Selenocysteine insertion directed by the 3'-UTR SECIS element in Escherichia coli. Nucleic Acids Research. 2005;33:2486–2492. doi: 10.1093/nar/gki547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strub MP, Hoh F, Sanchez JF, Strub JM, Bock A, Aumelas A, Dumas C. Selenomethionine and selenocysteine double labeling strategy for crystallographic phasing. Structure. 2003;11:1359–1367. doi: 10.1016/j.str.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Salgado PS, Taylor JD, Cota E, Matthews SJ. Extending the usability of the phasing power of diselenide bonds: SeCys SAD phasing of CsgC using a non-auxotrophic strain. Acta Crystallographica Section D-Biological Crystallography. 2011;67:8–13. doi: 10.1107/S0907444910042022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moroder L. Isosteric replacement of sulfur with other chalcogens in peptides and proteins. Journal of Peptide Science. 2005;11:187–214. doi: 10.1002/psc.654. [DOI] [PubMed] [Google Scholar]
- 19.Wessjohann LA, Schneider A. Synthesis of selenocysteine and its derivatives with an emphasis on selenenylsulfide (-Se-S-) formation. Chemistry & Biodiversity. 2008;5:375–388. doi: 10.1002/cbdv.200890038. [DOI] [PubMed] [Google Scholar]
- 20.Stocking EM, Schwarz JN, Senn H, Salzmann M, Silks LA. Synthesis of L-selenocystine, L- Se-77 selenocystine and L-tellurocystine. Journal of the Chemical Society-Perkin Transactions. 1997;1:2443–2447. [Google Scholar]
- 21.Metanis N, Keinan E, Dawson PE. Synthetic seleno-glutaredoxin 3 analogues are highly reducing oxidoreductases with enhanced catalytic efficiency. Journal of the American Chemical Society. 2006;128:16684–16691. doi: 10.1021/ja0661414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller S, Heider J, Bock A. The path of unspecific incorporation of selenium in Escherichia coli. Archives of Microbiology. 1997;168:421–427. doi: 10.1007/s002030050517. [DOI] [PubMed] [Google Scholar]
- 23.Cowie DB, Cohen GN. Biosynthesis by escherichia-coli of active altered proteins containing selenium instead of sulfur. Biochimica Et Biophysica Acta. 1957;26:252–261. doi: 10.1016/0006-3002(57)90003-3. [DOI] [PubMed] [Google Scholar]
- 24.Tuve T, Williams HH. Metabolism of selenium by escherichia coli - biosynthesis of selenomethionine. Journal of Biological Chemistry. 1961;236:597–601. [PubMed] [Google Scholar]
- 25.Farrell SR, Thorpe C. Augmenter of liver regeneration: A flavin-dependent sulfhydryl oxidase with cytochrome c reductase activity. Biochemistry. 2005;44:1532–1541. doi: 10.1021/bi0479555. [DOI] [PubMed] [Google Scholar]
- 26.Daithankar VN, Farrell SR, Thorpe C. Augmenter of liver regeneration: Substrate specificity of a flavin-dependent oxidoreductase from the mitochondrial intermembrane space. Biochemistry. 2009;48:4828–4837. doi: 10.1021/bi900347v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metanis N, Hilvert D. Strategic use of non-native diselenide bridges to steer oxidative protein folding. Angewandte Chemie-International Edition. 2012;51:5585–5588. doi: 10.1002/anie.201109129. [DOI] [PubMed] [Google Scholar]
- 28.Lees WJ. Going Non-native To Improve Oxidative Protein Folding. ChemBioChem. 2012;13:1725–1727. doi: 10.1002/cbic.201200288. [DOI] [PubMed] [Google Scholar]
- 29.Daithankar VN, Schaefer SA, Dong M, Bahnson BJ, Thorpe C. Structure of the human sulfhydryl oxidase augmenter of liver regeneration and characterization of a human mutation causing an autosomal recessive myopathy. Biochemistry. 2010;49:6737–6745. doi: 10.1021/bi100912m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duddeck H. Se-77 NMR spectroscopy and its applications in chemistry. Annual Reports on NMR Spectroscopy. 2004;52:105–166. [Google Scholar]
- 31.Tanioku A, Hayashi S, Nakanishi W. Analysis of one-bond Se-Se nuclear couplings in diselenides and 1,2-diselenoles on the basis of molecular orbital theory: torsional angular dependence, electron density influence, and origin in (1)J(Se, Se) Bioinorganic Chemistry and Applications. 2009:1–9. doi: 10.1155/2009/381925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potrzebowski MJ, Michalska M, Blaszczyk J, Wieczorek MW, Ciesielski W, Kazmierski S, Pluskowski J. Molecular modeling, X-ray-diffraction, and C-13, Se-77 CP/MAS NMR-studies of bis(2,3,4,6-tetra-O-acetyl-beta-D-glucopyranosyl) diselenide and disulfide. Journal of Organic Chemistry. 1995;60:3139–3148. [Google Scholar]
- 33.Banci L, Bertini I, Calderone V, Cefaro C, Ciofi-Baffoni S, Gallo A, Kallergi E, Lionaki E, Pozidis C, Tokatlidis K. Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4811–4816. doi: 10.1073/pnas.1014542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arner ESJ, Sarioglu H, Lottspeich F, Holmgren A, Bock A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. Journal of Molecular Biology. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 35.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expression and Purification. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expression and Purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Gladyshev VN, Khangulov SV, Stadtman TC. Nicotinic acid hydroxylase from Clostridium barkeri: Electron paramagnetic resonance studies show that selenium is coordinated with molybdenum in the catalytically active selenium-dependent enzyme. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:232–236. doi: 10.1073/pnas.91.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Engineering. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 39.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. Journal of Biomolecular NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Bailey S. The CCP4 suite - Programs for protein crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duddeck H. Se-77 nuclear magnetic resonance spectroscopy. Progress in Nuclear Magnetic Resonance Spectroscopy. 1995:1–323. [Google Scholar]