Abstract
Variants of human carbonic anhydrase II (HCA II) with amino-acid replacements at residues in contact with water molecules in the active-site cavity have provided insights into the proton transfer rates in this protein environment. X-ray crystallography and 18O exchange measured by membrane inlet mass spectrometry have been used to investigate structural and catalytic properties of variants of HCA II containing the replacements of Tyr7 with Phe (Y7F) and Asn67 with Gln (N67Q). The rate constants for proton transfer from His64 to the zinc-bound hydroxide in catalysis were 4 μs-1 and 9 μs-1 for Y7F and Y7F-N67Q, respectively, compared with a value of 0.8 μs-1 for wild-type HCA II. These higher values observed for Y7F and Y7F-N67Q HCA II could not be explained by differences in the values of the pKa of the proton donor (His64) and acceptor (zinc-bound hydroxide) or by orientation of the side chain of the proton shuttle residue His64. They appeared to be associated with reduced branching in the networks of hydrogen-bonded water molecules between the proton shuttle residue His64 and the zinc-bound solvent molecule as observed in crystal structures at 1.5 – 1.6 Å resolution. Moreover, Y7F-N67Q HCA II is unique among the variants studied in having a direct, hydrogen-bonded chain of water molecules between the zinc-bound solvent and Nδ of His64. This study provides the clearest example to date of the relevance of ordered water structure to rate constants for proton transfer in catalysis by carbonic anhydrase.
The carbonic anhydrases (CAs) are primarily zinc metalloenzymes that rapidly hydrate carbon dioxide to form bicarbonate and a proton. The well studied α class includes the mammalian CAs with roles in acid-base balance, fluid secretion, respiration, and other physiological processes (1). Humans are known to express many CA isozymes throughout the body (1, 2), including the structurally and kinetically well characterized human carbonic anhydrase II (HCA II) (3-5). The first stage of the catalysis is the binding of bicarbonate to zinc followed by the dehydration of bicarbonate (eq 1). The second stage is the transfer of a proton from buffer in solvent BH+ to the proton shuttle His64 and subsequently to zinc-bound hydroxide to regenerate the catalytic zinc-bound water (eq 2).
| (1) |
| (2) |
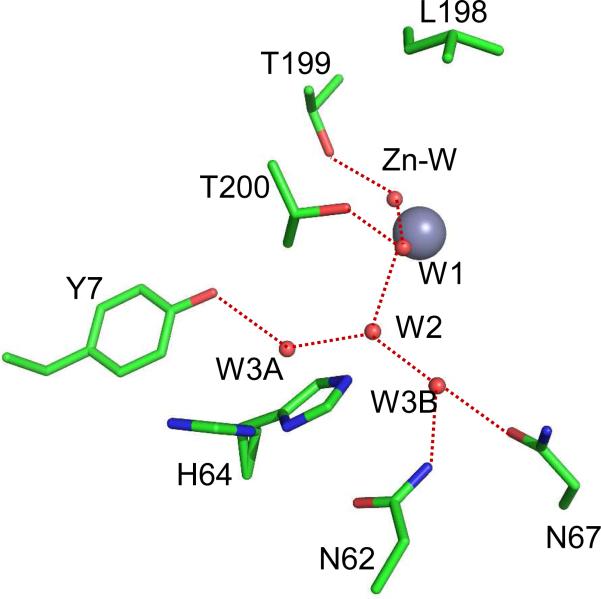
The intramolecular proton transfer from His64 to the zinc-bound hydroxide in the second stage of the catalysis (eq 2) is as fast as 1 μs-1 and is rate limiting for the maximal velocity of HCA II (6, 7). The position of the side-chain of His64 is observed in inward and outward orientations (Figure 1); in the inward orientation the imidazole ring of His64 is about 7.5 Å from the zinc (8-10). An ordered, hydrogen-bonded network of water molecules extending between the zinc-bond water and His64 is observed in crystal structures of HCA II (Figure 1) and is believed to offer clues to the pathway for intramolecular proton transfer in catalysis (3-5).
Figure 1.
Active site of wild type HCA II from the crystal structure at pH 8.0 (PDB code 2ili (8). The zinc ion and the oxygen of water molecules are shown as gray and red spheres, respectively. The water network of the active-site is labeled W1, W2, etc. Dashed red lines are assumed hydrogen bonds. The side chain of the proton shuttle His64 is shown in both the inward and outward orientations. This figure was generated and rendered with PyMOL (www.pymol.org).
The water network in HCA II appears to form hydrogen bonds with hydrophilic residues lining the active-site cavity and is branched at the water molecule labeled W2 in Figure 1. A number of studies have replaced residues in the active-site cavity altering the structure of the water network as well as changes in the orientation of His64 and the pKa of its imidazole ring (11-13) in order to determine the relevance of the observed water structure to the rate constants for proton transfer. One significant observation is that the variant of HCA II with Tyr7 replaced with Phe (Y7F) has an enhanced rate constant of intramolecular proton transfer, five-fold greater than wild type, and less branching of the water network connecting His64 and the zinc-bound solvent (11). Computational studies indicate that the rate of proton transfer is expected to be faster in unbranched water chains (14, 15).
In this work we have constructed the variant Y7F-N67Q HCA II that has a water network showing less branching than in wild type. It also has a completed hydrogen-bonded water chain and a shorter distance between His64 and the zinc-bound solvent as determined from the crystal structure at 1.6 Å resolution. The appropriate single mutants Y7F and N67Q HCA II are also examined. For the double mutant the rate constant for intramolecular proton transfer at 9.0 μs-1 is ten-fold greater than wild type measured by 18O exchange. This allows a more complete understanding of the factors that influence proton transfer through water chains in a protein environment. HCA II serves as a simple model for examining proton transfers in more complex systems such as the photosynthetic reaction center, bacteriorhodopsin, and cytochrome c oxidase.
METHODS
Enzymes
Variants of HCA II were made with the QuickChange II Site-Directed Mutagenesis Kit (Agilent) on the expression vector coding the full-length wild-type HCA II. The entire region coding HCA II was determined for each mutant to confirm the correct DNA sequence. Plasmids were transformed for expression into Escherichia coli BL21(DE3)pLysS cells (Agilent). The transformed cells were grown in LB media to which was added 1.0 mM ZnSO4 and induced with 1.0 mM IPTG when the OD600 nm reached 0.6. Each variant was purified by affinity chromatography using p-(aminomethyl)benezene-sulfonamide coupled to agarose beads (Sigma)(16). The concentration of HCA II and variants was determined by titration of active sites by the tight-binding inhibitor ethoxzolamide while measuring activity by the 18O method.
Oxygen-18 exchange
The catalysis of HCA II has been well studied by measuring the exchange of 18O from labeled bicarbonate and CO2 species to water using membrane inlet mass spectrometry (17). The method is based on the depletion of 18O from CO2 that passes across the membrane inlet into a mass spectrometer (Extrel EXM-200). The apparatus allows a continuous measurement of isotopic content of CO2 under controlled conditions in solution. The initial step of the catalysis by HCA II is the dehydration of labeled bicarbonate with a probability of leaving a 18O-labeled hydroxide at the zinc (eq 3). The second step involves the protonation of the zinc-bound hydroxide subsequently releasing H218O which is very greatly diluted by H216O in solution (eq 4).
| (3) |
| (4) |
The 18O-exchange method allows a rate for both steps of the catalysis to be determined (17). The rate of the first step, exchange between CO2 and HCO3- at chemical equilibrium, is designated R1 and described by eq 5. The maximal rate constant of the conversion between CO2 and HCO3- is kcatex, the apparent binding constant of the substrate to enzyme is KeffS, and [S] is the concentration of the substrate either CO2 or bicarbonate for the hydration or dehydration direction (18). The ratio kcatex/KeffS is in principle and practice equivalent to kcat/Km determined in steady state experiments (18).
| (5) |
The rate of the second step, the transfer of a proton and release of water in eq 4, is termed RH2O. The RH2O is interpreted in terms of a rate constant kB for proton transfer to the zinc-bound hydroxide (eq 6). The ionization constants of the proton donor and the zinc-bound water molecule are given in eq 6 as (Ka)donor, and (Ka)ZnH2O.
| (6) |
The uncatalyzed and carbonic anhydrase catalyzed 18O exchange at chemical equilibrium was measured in the absence of buffer at a total substrate concentration (all CO2 species) of 25 mM using membrane-inlet mass spectrometry (17). Reactions were carried out at 10 °C and 25 °C as noted, and total ionic strength of solution was maintained at 0.2 M by the addition of Na2SO4. The determination of the kinetic and ionization constants of eqs 5 and 6 were carried out by nonlinear least-squares methods (Enzfitter, Biosoft).
Crystallography
Crystals of the mutants N67Q and Y7F-N67Q HCA II were obtained using the hanging drop method (19). The crystallization drops were prepared by mixing 5 μL of protein [concentration ~15 mg/mL in 100 mM Tris-HCl (pH 8.0)] with 5 μL of the precipitant solution [1.25 M sodium citrate, 100 mM Tris-Cl (pH 8.0)] against a well of 1000 μL precipitant solution. Crystals were observed within a week at 293K.
The N67Q and Y7F-N67Q HCAII crystals were flash cooled after a soak in cryo-protectant, 30% glycerol plus precipitant solution, before mounting. The x-ray data were obtained at 100K using an R-AXIS V++ optic system from Viarimax HR a Rigaku RU-H3R Cu rotating anode operating at 50 kV and 100 mA. The detector to crystal distance was set to 80 mm. The oscillation steps were 1° with a 6 min exposure per image for 360 degrees. Data set statistics for the crystals are given in Table 1.
Table 1.
Crystal Structure Data and Refinement Statistics for N67Q and Y7F+N67Q HCA II
| Data-collection statistics | N67Q | Y7F-N67Q |
| Wavelength (Å) | 1.5418 | 1.5418 |
| Space group | P 21 | P 21 |
| Unit-cell parameters (Å,°) a, b, c (Å); β (°) | 42.0, 41.2, 72.1; 104.3 | 42.1, 41.2, 71.9; 104.5 |
| Total number of reflections | 35820 | 31729 |
| Redundancy | 3.6 (2.5)* | 3.8(3.6) |
| Completion % | 97.7 (80.5) | 99.7 (99.8) |
| Resolution (Å) | 50.0-1.50 (1.55-1.50) | 50.0-1.60 (1.66-1.60) |
| aRsym | 7.3 (38.9) | 8.4 (47.3) |
| I/σ(I) | 14.5 (4.1) | 13.1 (2.8) |
| bRcryst (%) | 18.5 | 16.6 |
| cRfree(%) | 21.4 | 20.5 |
| Amino acid residues | 3-261 | 3-261 |
| No. of protein atoms | 2180 | 2199 |
| No. of H2O molecules | 376 | 319 |
| R.m.s.d. for bond lengths (Å), angles (°) | 0.006 1.086 |
0.006 1.019 |
| Ramachandran statistics (%) Most favored, allowed, outlier | 87.6, 12.4, 0.0 | 86.6, 13.4, 0.0 |
| Average B factors (Å2) main-, side-chain, Zn, solvent | 18.2, 21.4, 11.5, 29.6 | 19.8, 22.0, 10.5, 28.7 |
Rsym = Σ |I - <I>|/ Σ <I>.
Rcryst = (Σ |Fo| - |Fc|/ Σ |Fobs|) × 100.
Rfree is calculated in same manner as bRcryst, except that it uses 5% of the reflection data omitted from refinement.
Values in parenthesis represent highest resolution bin.
The model building was done manually with the program Coot (20) and refinement was carried out with PHENIX suite (21). The starting phasing model was the wild-type HCA II structure of Fisher et al. (22) (PDB code: 1tbt) with the waters removed and the mutated residues as well as His64 changed to Ala to reduce model bias.
RESULTS
Catalysis
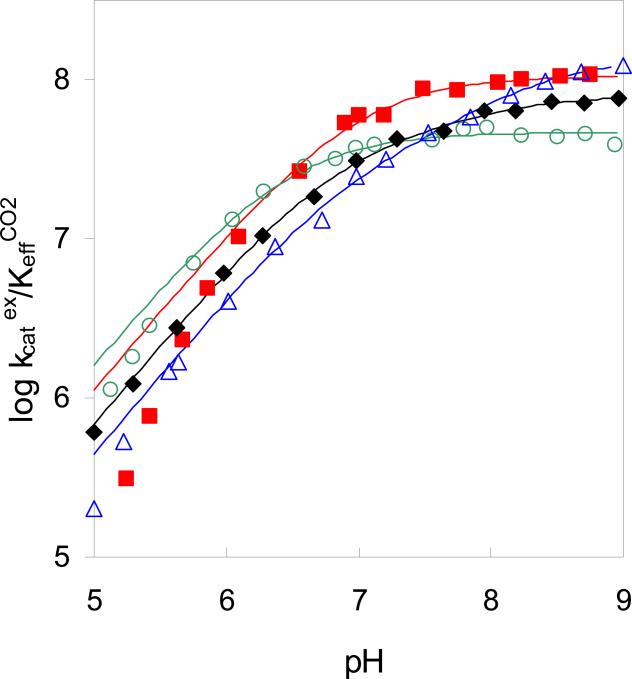
The pH dependence of catalysis of the hydration of CO2 determined by 18O exchange was determined for each of the variants Y7F, N67Q, and Y7F-N67Q HCA II. The data for Y7F HCA II were reported previously (11). Profiles of R1/[E] over a range of pH were obtained, and eq 5 was used to determine the catalytic constants kcatex/KeffCO2 for hydration of CO2 (Figure 2). Ionic strength was maintained at 0.2 M using Na2SO4 and the substrate was NaHCO3; pH was adjusted with dilute H2SO4 and no buffers were added. Fits to a single ionization appear satisfactory except at very low pH < 6 where the data showed decreased values for kcatex/KeffCO2 possibly due to additional ionizations or denaturation. This was particularly apparent in the variants containing the replacement Y7F, as already noted for Y7F HCA II (11). The maximal pH independent values of kcatex/KeffCO2 are compared in Table 2; these are similar with values between 50 and 120 μM-1s-1. The pKa values of the zinc-bound water, pKaZnH2O, determined from the fits to a single ionization obtained from the pH profiles of kcatex/KeffCO2 (Table 3, column 2) are also quite similar. These similarities reflect the distances of these residues from the zinc: 7.0 Å between the hydroxyl oxygen of Tyr7 and the zinc, and 8.4 Å between the carboxamide carbon of Asn67 and the zinc.
Figure 2.
The pH profiles for kcatex/KeffCO2 (M-1s-1) for the hydration of CO2 catalyzed by (◆) wild-type HCA II; (○) N67Q HCA II; (Δ) Y7F HCA II; and (■) Y7F-N67Q HCA II. Data were obtained by 18O exchange between CO2 and water using solutions containing 25 mM of all species of CO2 and sufficient Na2SO4 to maintain 0.2 M ionic strength.
Table 2.
Maximal Values of Rate Constants for Hydration of CO2 and Proton Transfer in Dehydration Catalyzed by HCA II and Variants.
| Enzyme | kcatexch/KeffCO2 CO2 hydration (μM-1s-1)a | kB proton transfer (μs-1)b |
|---|---|---|
| Wild type | 120 | 0.80 |
| Y7Fc | 120 | 3.9 |
| N67Q | 50 | 1.7 |
| Y7F-N67Qd | 80 | 9.0 |
Measured from the exchange of 18O between CO2 and water in the hydration direction (eq 5). Derived for each variant by a fit of the data (as in Figure 2) to a single ionization. The standard errors for these rate constants are generally 15% or less.
Measured from the exchange of 18O between CO2 and water (as in Figure 3) using eq 6 in the dehydration direction. The standard errors are 22% or less.
Data at 10°C from Fisher et al. (11).
Data at 10°C.
Table 3.
Values of Apparent pKa Obtained by Kinetic Measurements of Catalysis by HCA II and Variants
| Enzyme | pKa ZnH2Oa | pKa ZnH2Ob | pKa His64b |
|---|---|---|---|
| wild type | 6.9 | 6.8 | 7.2 |
| Y7Fc | 7.1 | 7.0 | 6.0 |
| N67Q | 6.5 | 6.7 | 6.6 |
| Y7F-N67Qd | 6.9 | 6.3 | 6.2 |
Measured from a fit of the data of Figure 2 to a single ionization. The standard errors in pKa are mostly ± 0.1 and no greater than ± 0.2.
Measured from the fits of eq 6 to the data of Figure 3. The values of pKa have standard errors no greater than ± 0.2.
These data at 10 °C from Fisher et al. (11).
These data at 10 °C.
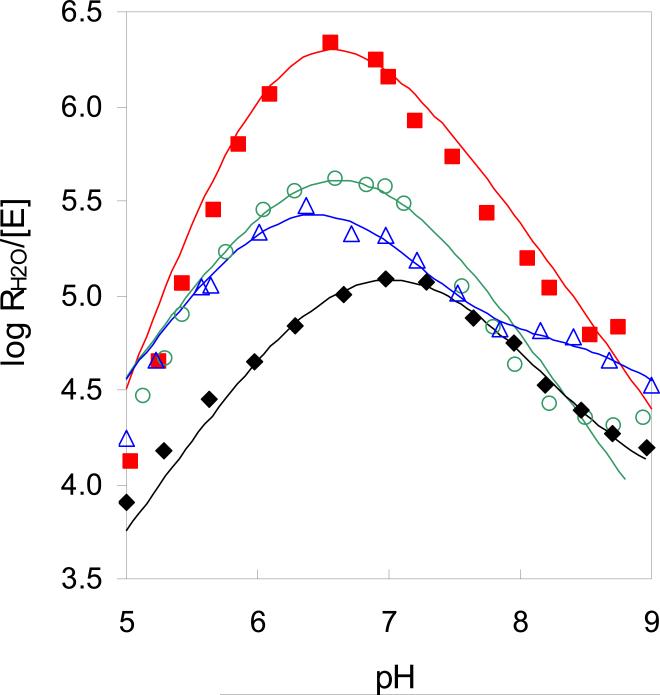
The advantage of the 18O exchange method is that catalysis by variants of CA can be examined in solutions without added buffer to determine rates of intermolecular proton transfer. Experiments carried out at steady state do not have this advantage and must use external buffers to control pH. The rate constant RH2O/[E] measures the second stage of catalysis in which rate-limiting intermolecular proton transfer releases H218O from the active site (eq 4). The pH profiles of RH2O/[E] for each mutant were predominantly bell-shaped (Figure 3), reflecting the proton transfer from the imidazolium side chain of His64 to the zinc-bound hydroxide (23). The bell-shaped curves were fit to eq 6 to yield a rate constant of proton transfer kB. The range in values of kB was approximately ten-fold (Table 2). Since the values of pKa were nearly identical for the proton donor (His64) and acceptor, there was no issue in assignment of these values. The exception is Y7F HCA II in which the pKa of the zinc-bound water was confirmed by measurement of the esterase activity (11).
Figure 3.
The pH profiles for RH2O/[E] (s-1) the proton-transfer dependent rate of release of 18O-labeled water catalyzed by (◆) wild-type HCA II; (○) N67Q HCA II; (Δ) Y7F HCA II; and (■) Y7F-N67Q HCA II. Conditions were as described in Figure 2.
Crystallography
The crystal structures of the variants N67Q and Y7F-N67Q of HCAII were solved to 1.5-1.6Å resolution using data that had completeness greater than 92% (Figures 4, 5). Figure S1 (Supporting information) shows the omit maps with electron densities. The crystal structure data and refinement statistics are listed in Table 1. Overall, no major structural perturbations were observed; the RMSD for Cα atoms was 0.1 Å for both variants when compared to the wild-type HCA II (PDB code 2ili (8)). The proton shuttle residue His64 has been shown to occupy two conformations in wild-type HCA II, inward and outward with respect to orientation in the active-site cavity (8, 10). The outward conformation was dominant in N67Q HCA II while the inward conformation was observed in Y7F-N67Q HCA II, similar to the Y7F variant previously published (11). Coordinates for N67Q HCA II and Y7F-N67Q HCA II have been deposited in the Protein Data Bank with code numbers 3TVN and 3TVO, respectively.
Figure 4.
The active-site structure for N67Q HCA II crystallized at pH 8.0. The three histidine residues (His94, His96, His119) coordinating the zinc (gray sphere) are not labeled. The oxygen atoms of water molecules identified in the active-site cavity are shown as red spheres and labeled W1, W2 .... Presumed hydrogen bonds are represented as dashed red lines. This figure was generated and rendered using Pymol.
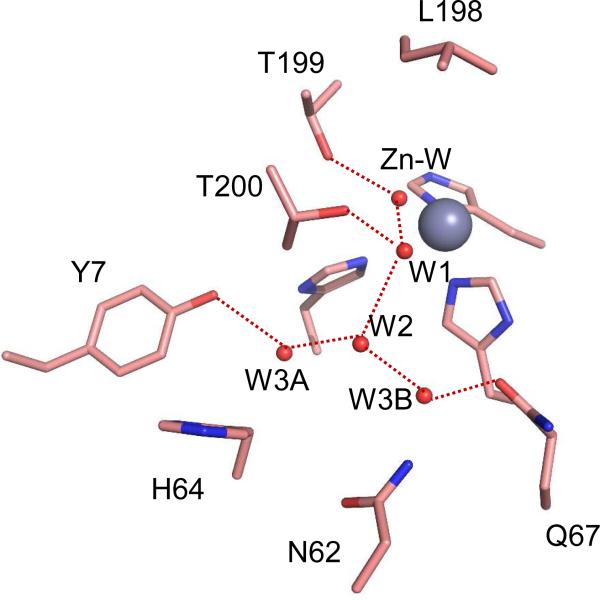
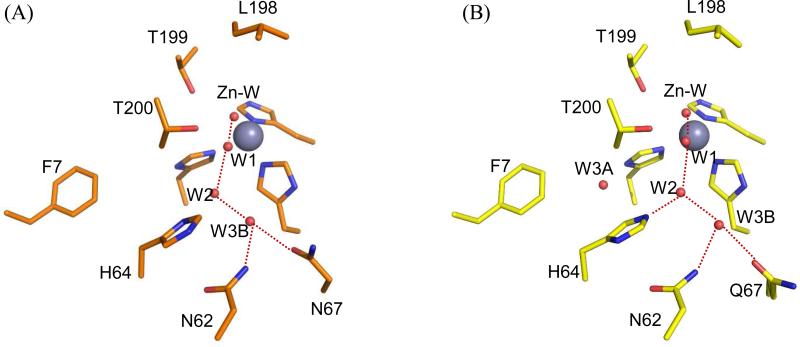
Figure 5.
Comparison of active-site structures for variants of HCA II crystallized at pH 8.0. (A) Y7F HCA II (PDB code 2nxt, (11)); and (B) Y7F-N67Q HCA II. This diagram was constructed as described as in Figure 4.
Compared with wild-type HCA II, the hydrogen-bonded solvent network in the active site cavity was mostly conserved for the three variants of HCA II depicted in Figures 1, 4 and 5. However, there were distinct differences. In the case of Y7F, the water molecule labeled W3A was not observed (Figure 5 (11)). For Y7F-N67Q HCA II, W3A was observed but was not within hydrogen bonding distance to W2; moreover, the distance between W3B and W2 (3.1 Å) was longer than in wild type (2.7 Å)(Table 4). In addition, in Y7F-N67Q the distance between W2 and Nδ of His64 (2.8 Å) is shorter, and hence indicates a stronger hydrogen bond than in wild type for which His64 is inward (3.2 Å) (Table 4). This is also reflected in the respective distances between the zinc-bound solvent and Nδ of His64 (Table 4). The carboxamide group of the Gln substitution at residue 67 extended further into the active site cavity than the Asn residue it replaced. For N67Q, an associated effect was an increase in the distance from the zinc-bound solvent to the Nδ of His64 (10.5 Å, outward orientation) compared with wild type (10.0 Å, outward, Table 4).
Table 4.
Comparison of Distances (Å) in the Proposed Hydrogen Bond Network Determined from the Crystal Structures of Variants of HCA II.
| WT | Y7Fa | Y7F-N67Q | N67Q | |
|---|---|---|---|---|
| ZnSolvent-W1 | 2.7 | 2.7 | 2.7 | 2.7 |
| W1-W2 | 2.7 | 2.6 | 2.6 | 2.9 |
| W2-W3A | 2.8 | n/a | 4.3 | 2.9 |
| W2-W3B | 2.7 | 2.6 | 3.1 | 2.4 |
| W2-H64(Nδ) | 3.2/6.3 | 3.2 | 2.9 | 6.7 |
| ZnSolvent-H64(Nδ) | 7.2/10.0 | 7.1 | 6.7 | 10.5 |
| H64 | in/out | In | in | out |
These data from (11)
DISCUSSION
These catalytic and structural results on variants of HCA II with amino-acid replacements at residues in contact with water molecules in the active-site cavity provide insight into the proton transfer rates in this protein environment. The rate constant kB of 9 μs-1 for proton transfer from His64 to the zinc-bound hydroxide in catalysis by N67Q-Y7F HCA II (Table 2) is the fastest measured for a variant of this isozyme. The previously studied Y7F HCA II was also found to have rapid proton transfer at kB of 4 μs-1 (11). The value for wild-type HCA II is kB = 0.8 μs-1 (Table 2)(11).
These high kB values observed for Y7F and Y7F-N67Q HCA II were not explained by differences in the values of the pKa between the proton donor (His64) and acceptor (zinc-bound hydroxide). This is shown in Table 3 in which the values of pKa are similar for these variants. Moreover, previous reports showed that the pH dependence of kB is rather flat in the vicinity of ΔpKa (pKaZnH2O – pKa His64) near zero (24, 25). We can comment, however, that the increments in the rate constant for proton transfer kB caused by the replacements at residues 7 and 67 are additive in nature, as illustrated in a double mutant cycle (Scheme S1, Supporting Information)(26).
In the cases of Y7F and N67Q-Y7F HCA II, the enhanced rate constants are associated with a predominant inward orientation of the His64 side chain in the crystal structures (Figure 5). However, the orientation of His64 in crystal structures has not been shown to affect the rate constant for proton transfer according to the following data. The variant N62L has His64 predominantly inward and N67L predominantly outward with other aspects of their protein structure nearly identical, yet their values of kB are the same at 0.2 μs-1 (11, 12, 27). The same conclusion that the inward and outward conformations in crystal structures of His64 do not influence the proton transfer rate was reached using a mutant with the substitution of Thr200 with Ser (28). This is supported by computational studies which suggest that the orientation of His64 need not influence this intramolecular proton transfer rate (29, 30).
It remains then to examine the solvent structure observed in the active site. Although these low energy networks of ordered water are observed in crystal structures and are taken as significant clues of the proton transfer pathway, clusters of hydrogen bonded water in the active-site cavity in solution have lifetimes typically in the picosecond range (31). Comparison of the crystal structures of the variants of Table 2 and other reports of variants of HCA II (11, 12, 32) shows that the two most efficient enzymes in proton transfer Y7F and Y7F-N67Q HCA II have less branched water structure, specifically water molecule W3A is not observed in Y7F and is not within hydrogen bond distance of the water chain in Y7F-N67Q (Figure 5; Table 4). Furthermore, in Y7F-N67Q there is a long and weak hydrogen bond between W2 and W3B compared with wild type (Figure 5, Table 4). We also point out that this double mutant is unique among the variants of HCA II we have examined in that it shows a normal hydrogen bond length between W2 and the Nδ of His64, other variants show this as a very weak hydrogen bond at best (Table 4). Thus, Y7F-N67Q has the most completely formed hydrogen-bonded chain of unbranched water molecules between the zinc-bound solvent and Nδ of His64 among the variants examined (Table 4). Computations show that proton transfer through an unbranched, hydrogen-bonded water network is more rapid than through a branched pathway (14, 15, 33). This feature of a more direct pathway in the structures of Y7F and Y7F-N67Q HCA II appears to be most relevant in explaining the proton transfer efficiencies of these variants.
When catalysis was measured by stopped-flow spectrophotometry, the steady-state constants kcat for dehydration were close in magnitude for wild type (0.24 μs-1) and Y7F-N67Q (0.19 μs-1), as were the values of kcat for hydration (Table S1, Supporting Information). The steady-state constant kcat measures many steps in either the dehydration or hydration directions, among which are conversion of bound substrate to product, product dissociation, intramolecular proton transfer, and possibly proton transfer steps between buffer in solution and zinc-bound hydroxide via His64. At steady-state, enzyme species are not necessarily at equilibrium concentrations, and the enzyme species preceding a rate-limiting step accumulates above equilibrium concentrations. The solvent H/D kinetic isotope effects on kcat for hydration of CO2 catalyzed by wild-type and Y7F-N67Q CA II are near 3.2 for both enzymes measured in the presence of excess buffer (Table S1). This indicates that proton transfer is a predominant rate-contributing step for both the wild type and variant measured at steady state.
The 18O exchange rate for proton transfer RH2O from which we obtain kB is determined at chemical equilibrium and focuses more on proton transfer because it contains many fewer steps of the catalysis than kcat. In the 18O exchange experiment there is no buffer in solution, and since there is relatively little proton transfer between the enzyme and solution there is no need for His64 to change orientation to sustain catalysis. These features provide an explanation why kB is greater than kcat. The results are consistent with the influence of the structure of the specific water chains through which proton transfer occurs. In view of the many potential proton transfer pathways that have been found in CA II (34), the data reflect the different proton transfer pathways involved in the equilibrium and steady-state experiments that result in different rate constants.
This study with the greatly enhanced proton transfer of Y7F-N67Q HCA II provides the clearest example to date of the relevance of the ordered water structure to rate constants for proton transfer in catalysis by carbonic anhydrase. The complement of these studies with the pertinent computational results of rapid proton transfer through unbranched water chains (14, 15, 31) is gratifying. It appears that the arrays of ordered, hydrogen-bonded water molecules as observed in crystal structures provide relevant information to explain efficient intramolecular proton transfer in carbonic anhydrase.
Supplementary Material
ABBREVIATIONS
- N67Q HCA II
the variant of human carbonic anhydrase II containing the replacement of Asn67 by Gln
- AE
anion exchange protein
- PDB
Protein Data Bank
- RMSD
root mean squared deviation
Footnotes
This work was supported by a grant from the National Institutes of Health GM25154.
Supporting Information Available
Supporting information consists of a table of steady-state constants for the hydration of CO2 and dehydration of bicarbonate catalyzed by wild-type and Y7F-N67Q HCA II, and a double mutant cycle for the values of kB for catalysis by the enzymes of Table 2. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Chegwidden WR, Carter ND, Edwards YH. The Carbonic Anhydrases New Horizons. Birkhauser Verlag; Basel: 2000. [Google Scholar]
- 2.Supuran CT, Scozzafava A, Conway J. Carbonic Anhydrase - Its Inhibitors and Activators. CRC Press; Boca Raton: 2004. [Google Scholar]
- 3.Silverman DN, McKenna R. Solvent-Mediated Proton Transfer in Catalysis by Carbonic Anhydrase. Acc Chem Res. 2007;40:669–675. doi: 10.1021/ar7000588. [DOI] [PubMed] [Google Scholar]
- 4.Christianson DW, Fierke CA. Carbonic anhydrase: Evolution of the zinc binding site by nature and by design. Accounts of Chemical Research. 1996;29:331–339. [Google Scholar]
- 5.Lindskog S. Structure and mechanism of carbonic anhydrase. Pharmacol Ther. 1997;74:1–20. doi: 10.1016/s0163-7258(96)00198-2. [DOI] [PubMed] [Google Scholar]
- 6.Khalifah RG. Carbon Dioxide Hydration Activity of Carbonic Anhydrase.1. Stop-Flow Kinetic Studies on Native Human Isoenzyme-B and Isoenzyme-C. Journal of Biological Chemistry. 1971;246:2561–2573. [PubMed] [Google Scholar]
- 7.Steiner H, Jonsson BH, Lindskog S. Catalytic Mechanism of Carbonic-Anhydrase - Hydrogen-Isotope Effects on Kinetic-Parameters of Human C Isoenzyme. European Journal of Biochemistry. 1975;59:253–259. doi: 10.1111/j.1432-1033.1975.tb02449.x. [DOI] [PubMed] [Google Scholar]
- 8.Fisher SZ, Maupin CM, Budayova-Spano M, Govindasamy L, Tu C, Agbandje-McKenna M, Silverman DN, Voth GA, McKenna R. Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: Insights into the proton transfer mechanism. Biochemistry. 2007;46:2930–2937. doi: 10.1021/bi062066y. [DOI] [PubMed] [Google Scholar]
- 9.Liljas A, Lovgren S, Bergsten PC, Carlbom U, Petef M, Waara I, Strandbe B, Fridborg K, Jarup L, Kannan KK. Crystal-Structure of Human Carbonic Anhydrase-C. Nature-New Biology. 1972;235:131–137. doi: 10.1038/newbio235131a0. [DOI] [PubMed] [Google Scholar]
- 10.Nair SK, Christianson DW. Unexpected pH-Dependent Conformation of His-64, the Proton Shuttle of Carbonic Anhydrase-II. Journal of the American Chemical Society. 1991;113:9455–9458. [Google Scholar]
- 11.Fisher SZ, Tu CK, Bhatt D, Govindasamy L, Agbandje-McKenna M, McKenna R, Silverman DN. Speeding up proton transfer in a fast enzyme: kinetic and crystallographic studies on the effect of hydrophobic amino acid substitution in the active site of human carbonic anhydrase II. Biochemistry. 2007;42:3803–3813. doi: 10.1021/bi602620k. [DOI] [PubMed] [Google Scholar]
- 12.Zheng JY, Avvaru BS, Tu C, McKenna R, Silverman DN. Role of Hydrophilic Residues in Proton Transfer during Catalysis by Human Carbonic Anhydrase II. Biochemistry. 2008;47:12028–12036. doi: 10.1021/bi801473w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackman JE, Merz KM, Jr., Fierke CA. Disruption of the active site solvent network in carbonic anhydrase II decreases the efficiency of proton transfer. Biochemistry. 1996;35:16421–16428. doi: 10.1021/bi961786+. [DOI] [PubMed] [Google Scholar]
- 14.Cui Q, Karplus M. Is a “proton wire” concerted or stepwise? A model study of proton transfer in carbonic anhydrase. Journal of Physical Chemistry B. 2003;107:1071–1078. [Google Scholar]
- 15.Maupin CM, Saunders MG, Thorpe IF, McKenna R, Silverman DN, Voth GA. Origins of enhanced proton transport in the Y7F mutant of human carbonic anhydrase II. Journal of the American Chemical Society. 2008;130:11399–11408. doi: 10.1021/ja802264j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalifah RG, Strader DJ, Bryant SH, Gibson SM. C-13 Nuclear Magnetic-Resonance Probe of Active-Site Ionizations in Human Carbonic-Anhydrase B. Biochemistry. 1977;16:2241–2247. doi: 10.1021/bi00629a031. [DOI] [PubMed] [Google Scholar]
- 17.Silverman DN. Carbonic anhydrase: oxygen-18 exchange catalyzed by an enzyme with rate-contributing proton-transfer steps. Methods Enzymol. 1982;87:732–752. doi: 10.1016/s0076-6879(82)87037-7. [DOI] [PubMed] [Google Scholar]
- 18.Simonsson I, Jonsson BH, Lindskog S. C-13 NMR study of carbon dioxide-bicarbonate exchange catalyzed by human carbonic anhydrase-C at chemical-equilibrium. European Journal of Biochemistry. 1979;93:409–417. doi: 10.1111/j.1432-1033.1979.tb12837.x. [DOI] [PubMed] [Google Scholar]
- 19.McPherson A. Preparation and Analysis of Protein Crystals. Wiley; New York: 1982. [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher Z, Hernandez Prada JA, Tu C, Duda D, Yoshioka C, An H, Govindasamy L, Silverman DN, McKenna R. Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II. Biochemistry. 2005;44:1097–1105. doi: 10.1021/bi0480279. [DOI] [PubMed] [Google Scholar]
- 23.Tu CK, Silverman DN, Forsman C, Jonsson BH, Lindskog S. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry. 1989;28:7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 24.An H, Tu C, Duda D, Montanez-Clemente I, Math K, Laipis PJ, McKenna R, Silverman DN. Chemical rescue in catalysis by human carbonic anhydrases II and III. Biochemistry. 2002;41:3235–3242. doi: 10.1021/bi0120695. [DOI] [PubMed] [Google Scholar]
- 25.Silverman DN, Tu C, Chen X, Tanhauser SM, Kresge AJ, Laipis PJ. Rate-equilibria relationships in intramolecular proton transfer in human carbonic anhydrase III. Biochemistry. 1993;32:10757–10762. doi: 10.1021/bi00091a029. [DOI] [PubMed] [Google Scholar]
- 26.Mildvan AS, Weber DJ, Kuliopulos A. Quantitative Interpretations of Double Mutations of Enzymes. Archives of Biochemistry and Biophysics. 1992;294:327–340. doi: 10.1016/0003-9861(92)90692-p. [DOI] [PubMed] [Google Scholar]
- 27.Mikulski RL, Silverman DN. Proton transfer in catalysis and the role of proton shuttles in carbonic anhydrase. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2010;1804:422–426. doi: 10.1016/j.bbapap.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krebs JF, Fierke CA, Alexander RS, Christianson DW. Conformational Mobility of His-64 in the Thr200Ser Mutant of Human Carbonic Anhydrase-II. Biochemistry. 1991;30:9153–9160. doi: 10.1021/bi00102a005. [DOI] [PubMed] [Google Scholar]
- 29.Riccardi D, Konig P, Guo H, Cui Q. Proton transfer in carbonic anhydrase is controlled by electrostatics rather than the orientation of the acceptor. Biochemistry. 2008;47:2369–2378. doi: 10.1021/bi701950j. [DOI] [PubMed] [Google Scholar]
- 30.Shimahara H, Yoshida T, Shibata Y, Shimizu M, Kyogoku Y, Sakiyama F, Nakazawa T, Tate S, Ohki SY, Kato T, Moriyama H, Kishida K, Tano Y, Ohkubo T, Kobayashi Y. Tautomerism of histidine 64 associated with proton transfer in catalysis of carbonic anhydrase. J Biol Chem. 2007;282:9646–9656. doi: 10.1074/jbc.M609679200. [DOI] [PubMed] [Google Scholar]
- 31.Maupin CM, McKenna R, Silverman DN, Voth GA. Elucidation of the Proton Transport Mechanism in Human Carbonic Anhydrase II. Journal of the American Chemical Society. 2009;131:7598–7608. doi: 10.1021/ja8091938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domsic JF, Williams W, Fisher SZ, Tu C, Agbandje-McKenna M, Silverman DN, McKenna R. Structural and Kinetic Study of the Extended Active Site for Proton Transfer in Human Carbonic Anhydrase II. Biochemistry. 2010;49:6394–6399. doi: 10.1021/bi1007645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, Voth GA. A computer simulation study of the hydrated proton in a synthetic proton channel. Biophys J. 2003;85:864–875. doi: 10.1016/S0006-3495(03)74526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy A, Taraphder S. Role of protein motions on proton transfer pathways in human carbonic anhydrase II. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2010;1804:352–361. doi: 10.1016/j.bbapap.2009.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.