Abstract
Cytosine hydroxymethylation is an epigenetic control factor in higher organisms. New discoveries of the biological roles of hydroxymethylation serve to raise questions about how this epigenetic modification exerts its functions and how organisms discriminate cytosine hydroxymethylation from methylation. Here, we report investigations that reveal an effect of cytosine hydroxymethylation on mechanical properties of DNA under load. The findings are based on molecular force assay measurements and steered molecular dynamics simulations. Molecular force assay experiments identified significant effects of hydroxymethylation on stretching-induced strand separation; the underlying physical mechanism has been revealed by steered molecular dynamics simulations. We find that hydroxymethylation can either upregulate or downregulate DNA’s strand separation propensity, suggesting that hydroxymethylation can control gene expression by facilitating or obstructing the action of transcription machinery or the access to chromosomal DNA.
Introduction
Genetic information, which determines the development of an organism, is encoded in the DNA sequence. Many studies showed that so-called epigenetic mechanisms such as DNA methylation and histone deacetylation change gene expression in the cell. Additional information may thus be stored and read without altering the DNA sequence (1,2). DNA methylation is found to exist widely in the CpG sites (where a cytosine base exists next to guanine base), and it plays an important role in silencing genes by impeding transcription factors (3–5). DNA methylation arises also in noncoding regions of DNA and is linked to gene control at the chromosomal level (6). Cytosine hydroxymethylation was recently discovered as another important epigenetic modification on DNA in mammalian cells (7,8). Similar to methylation, hydroxymethylation replaces, at the C5 position in cytosine, the hydrogen atom by a hydroxymethyl group. It has been demonstrated that cytosine hydroxymethylation is also involved in gene regulation (7–10). For example, the hydroxymethylation level has been found to be associated with pluripotency of stem cells (7). Disturbed hydroxymethylation of DNA cytosine can result in disordered cell functions, causing different types of cancers, e.g., myeloid cancers (11).
Prior studies suggest that hydroxymethylated cytosine is an intermediate in a pathway of DNA demethylation in the mammalian zygote (12), or even may be the final product of genome-wide demethylation (13). Recently, the TET (ten-eleven translocation) proteins were identified as converting methylcytosine to hydroxymethylcytosine in stem cells, inducing active demethylation (7,14,15). Although many studies could identify the biological roles of DNA hydroxymethylation, the mechanisms explaining how these roles come about are still unclear. Unlike DNA methylation, to our knowledge, no protein has been found yet that binds to hydroxymethylation sites on DNA to, thereby, possibly influence gene expression. It is speculated that DNA hydroxymethylation excludes the binding of methylcytosine-binding proteins to influence gene activities associated with DNA methylation (16,17).
Cytosine hydroxymethylation was observed to be a stable DNA modification in mammalian tissues and relatively abundant in the central nervous system (9,18–20). Some groups have quantified the amount of hydroxymethylated cytosines in some mammalian tissues (9,19,21), but the specific positions of hydroxymethylated cytosines are still unknown. The technical challenge of discrimination of methylated cytosine and hydroxymethylated cytosine is due to the similarity between them. Wanunu et al. (22) utilized solid-state nanopores to discriminate methylcytosine from hydroxymethylcytosine in DNA molecules, suggesting that hydroxymethylation may affect the flexibility and stability of DNA duplexes differently than methylation does.
Prior studies have shown that methylation has only a small effect on DNA melting temperature, while the shape of the melting curve of DNA changes more significantly (23,24). These results indicate that methylation leaves DNA thermodynamic stability nearly unaltered, but alters DNA mechanical cooperativity. In the case of hydroxymethylation, one expects similar effects.
In the cell, DNA is involved in many mechanical processes, in which molecular machines exert mechanical forces on DNA (25,26). For example, RNA polymerase promotes DNA strand separation to access genetic information stored in the DNA sequence (27). In addition, protein binding usually induces local DNA deformation, such as elongation, twist and bend; sometimes it even causes strand separation at the binding site (28,29). Hence, a change of the mechanical stability of DNA could alter (facilitate or inhibit) a related biological process. Chemical modifications on DNA, e.g., methylation and hydroxymethylation, may change the mechanical properties of DNA and, thereby, affect gene expression. Recent studies showed that DNA methylation, in particular, affects alternative splicing (an important regulatory mechanism of gene expression to generate protein diversity), possibly by changing elongation rates of RNA polymerase (15,30,31). Indeed, we have demonstrated in a recent experimental-computational study (32) that methylation influences double-stranded DNA’s propensity for strand separation, which might contribute to epigenetic regulation in cells. Because DNA replication and transcription all involve strand separation, an in-depth characterization of hydroxymethylation effects on strand separation of DNA should be critical for a better understanding of epigenetic mechanisms.
In this study, we investigated the effect of hydroxymethylation on mechanical properties of DNA using molecular force assay (MFA) and molecular dynamics (MD) simulation. MFA measurements were conducted to compare the stabilities of different hydroxymethylated DNA duplexes when stretched in zipper and shear geometry. DNA exhibits different stabilities in the two pulling geometries. It is observed that DNA with more hydroxymethylcytosine (hmC) sites is more stable than DNA with fewer hmC sites, when it is pulled in zipper geometry; in the case of shear geometry pulling, the stability of DNA not only associates with the number of hmC sites, but also their positions and context, giving rise to a possible environmental effect. MD simulations provide a detailed description of the pathway of DNA separation stretched in zipper and shear geometry.
Materials and Methods
MFA-chip and PDMS-stamp
The molecular setup of the MFA, shown in Fig. 1, has been assembled as described previously except for some modifications (32,33). A molecular force probe (MFP) consists of DNA oligomers labeled 1 and 2, which form the bottom duplex, and oligomers 2 and 3, which form the top duplex.
Figure 1.
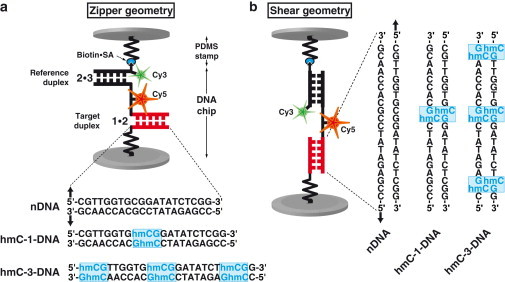
Schematic representation of MFP. Each MFP is comprised of DNA strands 1, 2 and 3, which form two DNA duplexes that are coupled in series. DNA strand 1 is anchored to the substrate (lower surface) and strand 3 is modified with a biotin for coupling to streptavidin on the PDMS-stamp (upper surface). The target duplex 1 ⋅ 2 exhibits three different variants with none (nDNA), one (hmC-1-DNA), or three (hmC-3-DNA) 5-hydroxymethylated CpG steps, while the reference duplex 2 ⋅ 3 is the same for all three cases. Furthermore, DNA strand 2 carries a Cy5 (red) and strand 3 a Cy3 (green) fluorescent marker. Depending on the sequences of the DNA strands, the DNA duplexes are oriented such that they are loaded in shear or zipper geometry as shown in panels a and b.
The MFPs are built up on the bottom surface (MFA-chip) as follows: DNA oligomer 1 is amine-modified for covalent linkage to aldehyde-functionalized glass slides. Approximately 104 duplicates of MFPs are built up in parallel per μm2. The experiments are conducted for three hydroxymethylation levels with the bottom duplex 1 • 2, containing zero, one, or three 5-hydroxymethylcytosine (hmC) per strand. Depending on the direction of the DNA sequences employed, the MFPs are realized in zipper (Fig. 1 a) or shear geometry (Fig. 1 b). Artifacts in the force measurements, which could be caused by structural changes of the DNA duplex or unwanted hybridizations of the strands, are kept to a minimum by using a well-characterized DNA sequence with minimal self-complementarity and, hence, minimal hairpin-formation (34).
The second part of the MFA is the top surface of the chip. The top surface is a polydimethylsiloxane (PDMS) stamp, fabricated and functionalized as described previously (35,36). DNA oligomer 3 is biotinylated and forms a link to the stamp upon contact. The sequences of the DNA oligomers are provided in the Supporting Material. All experiments are carried out in 1 × phosphate buffered saline at room temperature.
MFA contact process, readout, and analysis
A detailed description of the measurement process can be found in a previous article (33). In brief, a fluorescence microscope is combined with a custom-built contact device. The contact device controls the distance between PDMS-stamp and MFA-chip via a closed-loop piezoelectric actuator.
To begin, the MFA-chip and the soft PDMS-stamp are separated and the fluorescence signal of the MFA-chip is measured twice: First, Cy5 is excited and the fluorescence signal is measured. Second, Cy3 is excited and the fluorescence signal of Cy5 is measured (the fluorophores Cy3 and Cy5 form a FRET pair due to their close proximity). Then, the stamp is lowered until both surfaces are brought into contact, allowing the connection of strand 3 of the MFPs to the streptavidin on the top surface via biotin • streptavidin complexation (Fig. 1). To achieve a sufficient level of MFPs that couple to the PDMS-stamp, the contact is held for 10 min. Then, the surfaces are separated with a retract velocity of 5 μm/s. During the separation process, the force builds up gradually in each MFP until one of the duplexes 1 ⋅ 2 or 2 ⋅ 3 ruptures. At the last step, and are read out for the second time.
From the analysis of the four fluorescence images ( and before contact and after separation), the normalized fluorescence intensity (NF) can be determined (33) as
| (1) |
where is the background- and bleaching-corrected fluorescence image (after separation) divided by the background- and bleaching-corrected image (before contact). The NF is interpreted as the ratio between broken reference bonds (2 ⋅ 3) and total amount of MFPs that have been under load. The NF reflects the relative mechanical stability between the target duplex 1 ⋅ 2 and the reference duplex 2 ⋅ 3 of a MFP. Higher values for the NF denote an increased mechanical stability of the target duplex over the reference duplex and an NF value of 0.5 represents equally stable duplexes.
Molecular dynamics simulations
Oligonucleotides employed in the MD simulations were all of the same sequence (CGTTGGTGCGGATATCTCGG) and involved the same hydroxymethylation pattern, namely nDNA, hmC-1-DNA, and hmC-3-DNA, as used in the experiments. A double-stranded helix of nDNA was built with the program X3DNA (37); hmC-1-DNA and hmC-3-DNA were obtained through mutating cytosines in CpG steps to hydroxymethylcytosines. The topology of DNA along with the missing hydrogen atoms were generated using the psfgen plug-in of the program VMD (38) with a topology file corresponding to the CHARMM27 force field (39). Each DNA was placed in a water box with 0.1 mol/l KCl added. In shear geometry stretching simulations, the size of the water box was 61 Å × 61 Å × 275 Å; in unzipping geometry simulations, the size of the water box was 240 Å × 61 Å × 61 Å. The total size of the simulated systems was in the 100,000–150,000 atom range.
Simulations were performed using the program NAMD 2.7 (40) with the CHARMM27 force field for DNA (39) and the TIP3P water model (41). Periodic boundary conditions were assumed and the particle-mesh Ewald summation method was employed for evaluating Coulomb forces. The van der Waals energy was calculated using a smooth cutoff of 12 Å. The integration time step was 1 fs. The temperature was kept at 295 K by applying Langevin forces with a damping coefficient of 0.1 ps−1 (42) only to the oxygen atoms of water molecules. After energy-minimization for 4000 steps and 4 ps heating to 295 K, each simulated system was equilibrated for 500 ps with harmonic restraints applied to all DNA atoms under NPT ensemble conditions using Nosé-Andersen Langevin piston pressure control (42,43). Subsequently, with restraints turned off, each system was subjected to 2 ns equilibration under NVT ensemble conditions. Finally, steered molecular dynamics (SMD) simulations were carried out in the NVT ensemble. Table 1 lists all such simulations carried out in this study.
Table 1.
List of performed simulations
| Name | DNA | Pulling geometry | Velocity | Time (ns) |
|---|---|---|---|---|
| A1–A2 | nDNA | Zipper | 1 Å/ns | 180 ∼ 200 |
| B1–B2 | hmC-1-DNA | Zipper | 1 Å/ns | 180 ∼ 200 |
| C1–C2 | hmC-3-DNA | Zipper | 1 Å/ns | 180 ∼ 200 |
| D1–D6 | nDNA | Shear | 1 Å/ns | 110 ∼ 120 |
| E1–E6 | hmC-1-DNA | Shear | 1 Å/ns | 110 ∼ 120 |
| F1–F6 | hmC-3-DNA | Shear | 1 Å/ns | 110 ∼ 120 |
For each system, we separated DNA strands in two different modes, namely, in zipper geometry and shear geometry. In zipper geometry, a 3′-end of DNA was fixed and a 5′-end was pulled by attaching a harmonic spring to the terminal O3 and pulling the other end of the spring at a constant velocity of 1 Å/ns. Two independent SMD simulations in zipper geometry were conducted for each DNA. For shear geometry, six independent SMD simulations were performed, in which a 5′-end of DNA was fixed, and the other 5′-end was pulled at 1 Å/ns.
Results
Molecular force assay measurements
MFA is a sensitive method to experimentally characterize the unbinding forces of interacting molecules such as DNA-protein interactions or DNA strand separation. Thermal fluctuations of the force sensor limit typically the sensitivity of single-molecule force techniques (e.g., in the case of AFM, optical tweezers or magnetic tweezers) and shrinking of the sensor helps to increase the sensitivity (44). Following this idea, the MFA is designed so that a single molecular bond is utilized as a force sensor, in our case the bonding between the strands of a DNA duplex (the reference duplex). Actually, MFA measurement directly compares the mechanical stability of two DNA duplexes, a target and a reference duplex, against each other like a scale that balances a target weight against a reference weight. Due to the highly parallel format of the assay, MFA permits the examination of different sequences under the same experimental (solvent, force actuator, etc.) conditions in a single experiment, which is crucial for the value of the results.
At the molecular level, MFA consists of molecular force probes (MFPs) that are anchored in parallel on a glass slide with a density of ∼104 MFPs per μm2. Each MFP itself is composed of a target duplex 1 ⋅ 2 and a reference duplex 2 ⋅ 3 that are coupled in series and connected between a glass slide (lower surface) and a PDMS stamp (top surface) (see Materials and Methods). The target DNA duplex is 20-bp long and contains zero (nDNA), one (hmC-1-DNA), or three (hmC-3-DNA) 5-hydroxymethylcytosines (hmC) per strand, while the reference duplex is the same for all three different MFPs. Depending on the attachment points of the modifications on each DNA strand, the MFP is built up either in a zipper (Fig. 1 a) or in a shear geometry (Fig. 1 b). The different MFPs are immobilized as well separated spots on the glass substrate and probed with a single PDMS stamp all at once.
In the measurement process, the PDMS stamp is moved away from the chip at a speed of 5 μm/s. The polymeric anchors of the DNA strands are stretched and a force builds up gradually in each MFP until either the target duplex or the reference duplex ruptures. After complete separation of PDMS stamp and chip, the chip is read out via fluorescence microscopy: if bond 2 ⋅ 3 ruptures, the Cy5 dye is still on the lower surface (chip) and contributes to the signal; if bond 1 ⋅ 2 ruptures, Cy5 is on the PDMS stamp and is not contributing to , because the fluorescence is only read out on the lower surface. From the analysis of the fluorescence images pixel-by-pixel, one obtains an image of normalized fluorescence (NF). The NF is defined as the ratio of broken reference bonds to the total amount of MFPs that have been under load. Accordingly, the NF is a measure for the relative mechanical stability between the target and the reference duplex. A higher NF denotes an increased mechanical stability of the target duplex over that of the reference duplex.
Representative results for a typical experiment in zipper and shear geometry are shown in Fig. 2. The Gaussian fits of the histograms of the NF-images result in the following mean values and standard deviations:
For zipper geometry (Fig. 2 a), NF(nDNAzip) = (0.498 ± 0.011); NF(hmC-1-DNAzip) = (0.505 ± 0.013); and NF(hmC-3-DNAzip) = (0.537 ± 0.011).
For shear geometry (Fig. 2 b), NF(nDNAshear) = (0.502 ± 0.007); NF(hmC-1-DNAshear) = (0.395 ± 0.008); and NF(hmC-3-DNAshear) = (0.657 ± 0.006).
Figure 2.
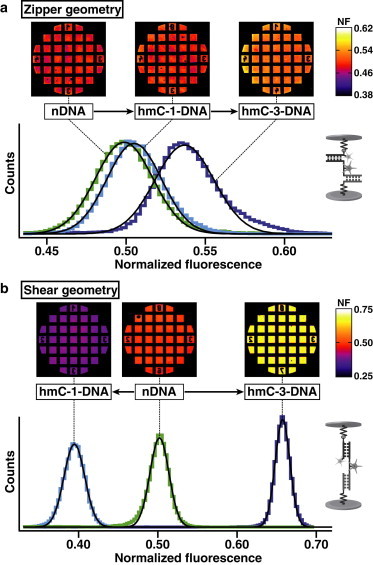
NF images and histograms of one representative experiment. (a) Zipper geometry: the NF images constitute the quantitative result of the relative unbinding forces between target and reference duplex. The visible squares (each 100 μm × 100 μm) in the NF images correspond to the contacted and probed area of the PDMS stamp. Due to the highly parallel measurement format, ∼104 MFPs are probed per μm2. Histograms of the NF images are fitted with a Gaussian. (b) Shear geometry.
The difference in NF reflects a quantitative measure for a difference in mean rupture force between nDNA, hmC-1-DNA and hmC-3-DNA. The width of the histograms does not necessarily reflect the width of the force distribution, because the width is influenced by the exposure time of the fluorescence images and the coupling efficiency. Typically, the coupling efficiency was ∼15–20% lower for zipper than for shear geometry. This might be due to steric conformation differences of the MFPs in zipper and shear geometry, which might influence the accessibility of the biotin on strand 3 to the streptavidin on the PDMS stamp.
Summarizing all pads of all experiments, we determined the following mean values and standard errors:
For zipper geometry (Fig. 3 a), NF(nDNAzip) = (0.504 ± 0.002); NF(hmC-1-DNAzip) = (0.521 ± 0.004); and NF(hmC-3-DNAzip) = (0.538 ± 0.003).
Figure 3.
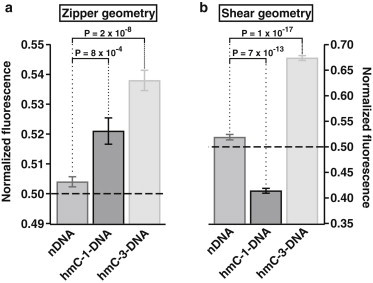
(a) Analysis of 42 pads from four different experiments in zipper geometry. The mean rupture force measured in NF rises with increasing number of 5-hydroxymethylated CpG steps. (b) Analysis of 39 pads from four different experiments in shear geometry. In contrast to the case of zipper geometry, hmC-1-DNA shows a lower and hmC-3-DNA a higher mean rupture force compared to nDNA. This effect was also observed for 5-methylated CpG steps, but the hydroxymethylated DNA exhibits an even more enhanced effect on the mechanical stability than does methylated DNA, namely a stronger destabilization for hmC-1-DNA than mC-1-DNA and an increased stabilization for hmC-3-DNA than mC-3-DNA in shear geometry.
For shear geometry (Fig. 3 b), NF(nDNAshear) = (0.519 ± 0.005); NF(hmC-1-DNAshear) = (0.414 ± 0.005); and NF(hmC-3-DNAshear) = (0.674 ± 0.005).
In zipper geometry, the P-value between nDNA and hmC-1-DNA is 8 × 10−4, and for nDNA and hmC-3-DNA it is 2 × 10−8. In the case of shear geometry, the P-value between nDNA and hmC-1-DNA is 7 × 10−13, and for nDNA and hmC-3-DNA it is 1 × 10−17.
Clearly, the MFA experiments prove a significant influence of hydroxymethylcytosine on the mechanical properties of DNA. The strength of the influence on the mechanical stability depends on the direction of the applied force (zipper versus shear geometry). In zipper geometry, we found that the mechanical stability increases with the number of hmC bases. In shear geometry, hmC-1-DNA exhibits a lower mechanical stability than nDNA, and hmC-3-DNA exhibits a higher stability. Our results indicate that cytosine hydroxymethylation of DNA can both enhance and decrease the propensity for strand separation. All reported cases show a significant change in mechanical stability of the DNA.
Molecular dynamics simulations
In order to demonstrate, at the atomic level, how hydroxymethylation affects DNA strand separation, SMD simulations were performed to stretch DNA with the same sequence and hydroxymethylation patterns as used in experiments.
When dsDNA is stretched in zipper geometry, the DNA helix unwinds and the basepairs break one by one, forming eventually two extended single-stranded DNA molecules, as shown in Fig. 4 a. A movie (see Movie S1 in the Supporting Material) showing the dynamics of unzipping DNA is provided. The rupture force of separating DNA in zipper geometry mainly arises from three aspects: (1) Unwinding of the DNA helix; (2) Breaking of hydrogen bonds between basepairs; (3) Extending of the single-stranded DNA. Fig. 4 b shows a typical force profile of unzipping DNA, which contains many small force peaks. During stretching, the number of DNA basepairs decreases one-by-one. Fig. S1 in the Supporting Material shows the time-evolution of force and number of still intact basepairs in six unzipping simulations. In all unzipping simulations, the frequency of appearance of force peaks is close to the frequency of basepair breaking, indicating that almost every basepair breaking event gives rise to a small force peak. After averaging all force values over time (each data point is obtained from two independent SMD simulations), we obtained mean rupture forces for nDNA, hmC-1-DNA, and hmC-3-DNA, as shown in Fig. 4 c. Consistent with MFA results, hmC-3-DNA exhibits strongest mechanical stability and requires the strongest rupture force (∼62 pN) in comparison with nDNA (∼53 pN) and hmC-1-DNA (∼53 pN). A difference between hmC-1-DNA and nDNA is not seen in simulations, probably due to small sampling. It is notable that the error bar of the mean rupture force of hmC-3-DNA is smaller than the error bars of hmC-1-DNA and nDNA, indicating that hmC-3-DNA, indeed, is more stable than hmC-1-DNA and nDNA.
Figure 4.
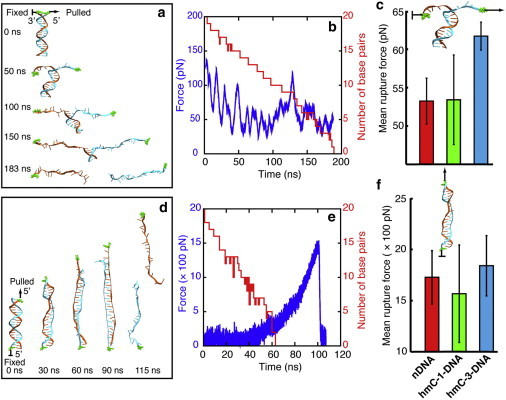
Analysis of MD simulations. (a) Snapshots of dsDNA unzipping from a steered molecular dynamics simulation (simulation A1). The top 3′-end (highlighted in green) of DNA strands is subject to constraint force and the adjacent 5′-end (highlighted also in green) is stretched. (b) Typical force profile (blue) of dsDNA unzipping and corresponding time evolution of the number of still intact basepairs (red) (simulation C1). (c) Histogram of the mean rupture force of separating DNA strands in zipper geometry. In comparison to hmC-1-DNA and nDNA, rupturing hmC-3-DNA requires the strongest force (∼62 pN). Due to small sampling (only two independent simulations), the small difference observed between hmC-1-DNA and nDNA can be reconciled as a random event. (d) Snapshots of dsDNA shearing from a steered molecular dynamics simulation (simulation D1). The bottom 5′-end (highlighted in green) of the DNA strands is subject to constraint force and the top 5′-end (highlighted also in green) is stretched. (e) Typical force profile (blue) of dsDNA shearing and the corresponding time evolution of the number of still intact basepairs (red) (simulation F1). (f) Histogram of mean rupture force of separating DNA strands in shear geometry. Consistent with experimental results, the order of the mean rupture force, F, is hydroxymethylation-dependent: FhmC-3-DNA > FnDNA > FhmC-1-DNA. Sampling involved six independent simulations for each DNA.
Different from the case of unzipping DNA, stretching dsDNA in shear geometry causes DNA strand separation in two steps (an example is shown in Movie S2):
In the first step, DNA elongates and unwinds as can be seen in the 30-ns and 60-ns snapshots in Fig. 4 d. In the meantime, DNA Watson-Crick basepairs begin to break and bases of the two separate strands begin to stack on top of each other, forming the so-called zipperlike DNA (45), as seen in the 90-ns snapshot in Fig. 4 d. The force increases slowly during the first step (see Fig. 4 e).
In the second step, the acting force increases rapidly. When the pulling force reaches a certain peak value, the two strands of DNA separate (see the 115 ns snapshot in Fig. 4 d), after which moment the force drops very rapidly to zero, as seen in Fig. 4 e. Accordingly, a typical rupture force profile for shearing DNA (Fig. 4 e) exhibits a peak rupture force, reflecting the DNA stability against strand separation by shearing. Fig. 4 f shows the mean peak rupture force of nDNA, hmC-1-DNA, and hmC-3-DNA (each obtained from six independent SMD simulations); hmC-3-DNA requires the strongest rupture force for strand separation; the mean peak force of nDNA is stronger than that of hmC-1-DNA, which is also consistent with MFA results. However, we note that the error bar for the peak force of hmC-1-DNA is very large.
To understand why different DNAs exhibit different peak rupture forces, we compared the conformation of DNA at the moment when its rupture force reaches the peak value, as shown in Fig. S2. In general, hmC-3-DNA assumes a more ordered DNA form with intercalated bases (fewer bubbles inside the DNA zipper and more compact ends) than do hmC-1-DNA and nDNA; hmC-1-DNA assumes the most disordered conformations, which reduces its mean peak rupture force. Similar to the findings in a previous study on effects of methylation on strand separation of methylated DNA (32), hydroxymethylation also affects bubble formation during DNA stretching, which controls the propensity for strand separation.
Discussion
We demonstrated in a previous study that methylcytosine might exert a biological function by itself, through influencing the mechanical stability of the DNA double helix (32). It was shown that the propensity of DNA strand separation is changed upon methylation of CpG, which might have a biological effect, possibly on transcription machinery or helicases (32). Because methylcytosine (mC) can alter, in a pronounced fashion, the mechanical properties of DNA, the question arises whether hydroxymethylation can also change the mechanical properties of DNA.
In the MFA experiments reported here, the relative stability of a target DNA duplex is measured against a reference DNA duplex. The MFA experiments were conducted in shear and zipper geometry. It should be noted that during forced strand separation in the MFA, individual basepairs in zipper geometry may unbind and rebind driven by thermal fluctuations, averaging out the well pronounced peaks resolved in the much faster MD simulations. In the case of shear geometry, we found that hmC-3-DNA is more stable compared to nDNA, and hmC-1-DNA is less stable compared to nDNA, as reflected by respective differences in NF values of 0.155 and 0.105, respectively. This means that the probability of strand separation is lower for hmC-3-DNA and higher for hmC-1-DNA than for nDNA. In zipper geometry, the relative stability for both hmC-1-DNA and hmC-3-DNA are higher than for nDNA, as reflected in NF value differences of 0.017 and 0.034, respectively. Because the experiments with hmC-3-DNA, hmC-1-DNA, and nDNA were conducted in parallel, with spatially separated spots in the same well, their measurement conditions were identical. Hence, the measured differences in the strand separation probability among hmC-3-DNA, hmC-1-DNA, and nDNA in zipper and shear geometry are highly reliable. The effect of hydroxymethylation on the mechanical stability is more pronounced in shear than in zipper geometry (by a factor of 5).
Consistent with MFA results, SMD simulations of stretching DNA in zipper and shear geometry showed that hydroxymethylation indeed affects the stability of DNA. Simulations demonstrated at the atomic level the DNA strand separation pathways, explaining why the effect of hydroxymethylation on DNA stability is dependent on pulling fashions. Stretched in zipper geometry, DNA basepairs break one-by-one. Hence, unzipping DNA mainly measures the strength of single basepairs. As observed in simulations, hmCG basepairs are relatively more stable than CG basepairs during unzipping and, therefore, the more hmCG basepairs are contained in DNA, the more stable the DNA. When DNA is stretched in shear geometry, the B-form double helix unwinds, followed by breaking of Watson-Crick basepairs, and bases from two strands slip past each other and intercalate. With an increase of shearing force, bubbles accumulating in this DNA begin to form and, when they reach a critical concentration, strand separation occurs. Observed from MD simulations, hydroxymethylation seems to change the likelihood of DNA strand separation by affecting bubble formation during stretching. Shearing DNA measures the stability of the entire DNA duplex and, therefore, not only the number of hmC, but also the context and position of hmC, influence the stability of DNA.
MD simulations provide a microscopic picture of DNA strand separation that complements MFA measurements. However, different from MFA experiments that conduct millions of independent measurements at one time, SMD simulations can only be performed with fast pulling and small sampling on a very few dsDNA specimens, due to limitation of computational resources. The pulling velocity employed in our SMD simulation is 1 Å/ns, much faster than the experimental pulling velocity of 5 × 10−5 Å/ns. Despite the very fast pulling, the total simulation time of our study still involves ∼3 μs, requiring three months of computation on 720 CPUs. The fast pulling velocity increases the rupture force for strand separation as more bonds are broken in a short time. Despite the fast rupture, the actual rupture pathway seen in the MFA experiments should still be revealed by SMD simulations (46,47). In the case of rupturing DNA, we observed the same strand separation pathway as reported by prior SMD studies (48–50) and DNA force-extension curves show the same shape as obtained from experiments (except magnitude) (51,52). Hence, effects of hydroxymethylation on strand separation observed in MD simulations should offer a reliable explanation of the experimental data.
In a previous study, we investigated the influence of mC on the mechanical stability of DNA strand separation (32). Because we use in this study the same DNA sequence and same cytosine position for hydroxymethylation as we used for methylation before, it is possible to compare the effects of hmC with mC. First of all, the tendencies to stabilize and destabilize DNA in shear and zipper geometry are the same: hmC-1-DNA destabilizes the DNA in shear geometry like mC-1-DNA, while hmC-3-DNA and mC-3-DNA stabilize the DNA in shear geometry. In zipper geometry, hmC-1-DNA and hmC-3-DNA as well as mC-1-DNA and mC-3-DNA stabilize the DNA. For mC-1-DNA, NF was ΔNF = 0.063 lower and for mC-3-DNA it was ΔNF = 0.104 higher than for nDNA (32). Therefore, the relative difference ((ΔNF (hmC-1-DNA)-ΔNF (mC-1-DNA))/ΔNF(mC-1-DNA)) is ∼70% and for hmC-3-DNA it is 50%, i.e., we observe qualitatively the same stabilization and destabilization for hmC as for mC. Previously, we had found that the effect of cytosine methylation does not only depend on the methylation level, but also on the sequence context of methylated sites. Because we observed the same effects in stabilization and destabilization for hmC as for mC, we assume a comparable environmental effect exists for hmC as for mC.
The changes of hydroxymethylation and methylation effects as seen typically in thermal stability measurements are subtle in comparison to the pronounced differences in mechanical stability, which arise from methylation and hydroxymethylation (22). This prompts the question if the effect reported here and in Severin et al. (32) has a biological function. On the one hand, processes like the mechanical manipulation of DNA in transcription initiation or the processivity of helicases exert forces on DNA to separate the DNA duplex into two single strands. On the other hand, these processes can also be influenced, e.g., in regard to the rate or processivity, through the mechanical stability of the DNA double strand (53–55).
The direction of force, which acts in vivo on the DNA double helix to separate the two DNA strands, depends on the molecular machinery. For DNA helicases, a DNA unzipping geometry might be more representative (55). The mechanical manipulation of DNA in transcription initiation happens in the confined setting of highly structured polynucleosomes. Hence, the shear geometry motion, which takes place more or less within the volume of nonstretched DNA, is relevant in the rather compact, structured polynucleosome setting found in the cell nucleus. Our measurements and simulations of DNA stretching in shear and zipper geometry should cover the different force directions that actually arise in the cell. Our findings might also be relevant to explain a chromosomal level role of methylation and hydroxymethylation on noncoding DNA (6).
In summary, we demonstrated through experiment and simulation a pronounced effect of hydroxymethylation on the propensity of DNA strand separation. Even though we identified this effect, we could not uncover fully the associated molecular mechanism. Therefore, further investigations are needed. The effect of hydroxymethylation exceeds in strength what was previously demonstrated for the effect of methylation. Our study reveals that hydroxymethylation could regulate gene expression through altering mechanical properties of DNA.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, the Nanosystems Initiative Munich, National Institutes of Health grants 9P41GM104601, R01-GM073655, and National Science Foundation grant PHY0822613. P.M.D.S. is grateful to the Elite Network of Bavaria for a doctoral fellowship. K.S. acknowledges an award from the Alexander von Humboldt Foundation. Supercomputer time was provided by the Texas Advanced Computing Center and the National Center for Supercomputing Applications via XSEDE Resource Allocation Committee grant MCA93S028.
Contributor Information
Klaus Schulten, Email: kschulte@ks.uiuc.edu.
Hermann E. Gaub, Email: gaub@physik.uni-muenchen.de.
Supporting Material
References
- 1.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Jones P.A. DNA methylation and cancer. Oncogene. 2002;21:5358–5360. doi: 10.1038/sj.onc.1205597. [DOI] [PubMed] [Google Scholar]
- 4.Straussman R., Nejman D., Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 5.Rottach A., Leonhardt H., Spada F. DNA methylation-mediated epigenetic control. J. Cell. Biochem. 2009;108:43–51. doi: 10.1002/jcb.22253. [DOI] [PubMed] [Google Scholar]
- 6.Thurman R.E., Rynes E., Stamatoyannopoulos J.A. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M., Koh K.P., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Münzel M., Globisch D., Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Chem. Int. Ed. Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- 10.Münzel M., Globisch D., Carell T. 5-Hydroxymethylcytosine, the sixth base of the genome. Angew. Chem. Int. Ed. Engl. 2011;50:6460–6468. doi: 10.1002/anie.201101547. [DOI] [PubMed] [Google Scholar]
- 11.Ko M., Huang Y., Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito S., D’Alessio A.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iqbal K., Jin S.-G., Szabó P.E. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H., Zhang Y. Tet1 and 5-hydroxymethylation: a genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011;10:2428–2436. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schübeler D. Molecular biology. Epigenetic islands in a genetic ocean. Science. 2012;338:756–757. doi: 10.1126/science.1227243. [DOI] [PubMed] [Google Scholar]
- 16.Valinluck V., Tsai H.-H., Sowers L.C. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin S.G., Kadam S., Pfeifer G.P. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penn N.W., Suwalski R., Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szwagierczak A., Bultmann S., Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng J., Zhou Y., Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson A.B., Dahl J.A., Klungland A. A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2011;39:e55. doi: 10.1093/nar/gkr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wanunu M., Cohen-Karni D., Drndic M. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J. Am. Chem. Soc. 2011;133:486–492. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefebvre A., Mauffret O., Fermandjian S. Sequence dependent effects of CpG cytosine methylation. A joint 1H-NMR and 31P-NMR study. Eur. J. Biochem. 1995;229:445–454. doi: 10.1111/j.1432-1033.1995.0445k.x. [DOI] [PubMed] [Google Scholar]
- 24.Wojdacz T.K., Dobrovic A., Hansen L.L. Methylation-sensitive high-resolution melting. Nat. Protoc. 2008;3:1903–1908. doi: 10.1038/nprot.2008.191. [DOI] [PubMed] [Google Scholar]
- 25.Bustamante C., Chemla Y.R., Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 26.Prévost C., Takahashi M., Lavery R. Deforming DNA: from physics to biology. ChemPhysChem. 2009;10:1399–1404. doi: 10.1002/cphc.200900253. [DOI] [PubMed] [Google Scholar]
- 27.Cheetham G.M.T., Steitz T.A. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2000;10:117–123. doi: 10.1016/s0959-440x(99)00058-5. [DOI] [PubMed] [Google Scholar]
- 28.Hogan M.E., Austin R.H. Importance of DNA stiffness in protein-DNA binding specificity. Nature. 1987;329:263–266. doi: 10.1038/329263a0. [DOI] [PubMed] [Google Scholar]
- 29.Young M.A., Ravishanker G., Berman H.M. Analysis of local helix bending in crystal structures of DNA oligonucleotides and DNA-protein complexes. Biophys. J. 1995;68:2454–2468. doi: 10.1016/S0006-3495(95)80427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de la Mata M., Alonso C.R., Kornblihtt A.R. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Shukla S., Kavak E., Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Severin P.M., Zou X., Schulten K. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Res. 2011;39:8740–8751. doi: 10.1093/nar/gkr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Severin P.M., Ho D., Gaub H.E. A high throughput molecular force assay for protein-DNA interactions. Lab Chip. 2011;11:856–862. doi: 10.1039/c0lc00302f. [DOI] [PubMed] [Google Scholar]
- 34.Strunz T., Oroszlan K., Güntherodt H.J. Dynamic force spectroscopy of single DNA molecules. Proc. Natl. Acad. Sci. USA. 1999;96:11277–11282. doi: 10.1073/pnas.96.20.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albrecht C.H., Clausen-Schaumann H., Gaub H.E. Differential analysis of biomolecular rupture forces. J. Phys. Condens. Matter. 2006;18:81–99. [Google Scholar]
- 36.Ho D., Dose C., Gaub H.E. Quantitative detection of small molecule/DNA complexes employing a force-based and label-free DNA-microarray. Biophys. J. 2009;96:4661–4671. doi: 10.1016/j.bpj.2009.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu X.J., Olson W.K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 39.Foloppe N., MacKerell A.D., Jr. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000;21:86–104. [Google Scholar]
- 40.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 42.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 43.Feller S.E., Zhang Y., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 44.Viani M.B., Schäffer T.E., Hansma P.K. Small cantilevers for force spectroscopy of single molecules. J. Appl. Physiol. 1999;86:2258–2262. [Google Scholar]
- 45.Chou S.-H., Chin K.-H. Zipper-like Watson-Crick base-pairs. J. Mol. Biol. 2001;312:753–768. doi: 10.1006/jmbi.2001.4962. [DOI] [PubMed] [Google Scholar]
- 46.Sotomayor M., Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 47.Lee E.H., Hsin J., Schulten K. Discovery through the computational microscope. Structure. 2009;17:1295–1306. doi: 10.1016/j.str.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lohikoski R., Timonen J., Laaksonen A. Molecular dynamics simulation of single DNA stretching reveals a novel structure. Chem. Phys. Lett. 2005;407:23–29. [Google Scholar]
- 49.Santosh M., Maiti P.K. Force-induced DNA melting. J. Phys. Condens. Matter. 2009;21:034113. doi: 10.1088/0953-8984/21/3/034113. [DOI] [PubMed] [Google Scholar]
- 50.Balaeff A., Craig S.L., Beratan D.N. B-DNA to zip-DNA: simulating a DNA transition to a novel structure with enhanced charge-transport characteristics. J. Phys. Chem. A. 2011;115:9377–9391. doi: 10.1021/jp110871g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith S.B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 52.Bustamante C., Smith S.B., Smith D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 53.Cheng W., Dumont S., Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc. Natl. Acad. Sci. USA. 2007;104:13954–13959. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dumont S., Cheng W., Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson D.S., Bai L., Wang M.D. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


