Abstract
The apical Na+-dependent bile salt transporter (ASBT/SLC10A2) is essential for maintaining the enterohepatic circulation of bile salts. It is not known when Slc10a2 evolved as a bile salt transporter or how it adapted to substantial changes in bile salt structure during evolution. We characterized ASBT orthologs from two primitive vertebrates, the lamprey that utilizes early 5α-bile alcohols and the skate that utilizes structurally different 5β-bile alcohols, and compared substrate specificity with ASBT from humans who utilize modern 5β-bile acids. Everted gut sacs of skate but not the more primitive lamprey transported 3H-taurocholic acid (TCA), a modern 5β-bile acid. However, molecular cloning identified ASBT orthologs from both species. Cell-based assays using recombinant ASBT/Asbt's indicate that lamprey Asbt has high affinity for 5α-bile alcohols, low affinity for 5β-bile alcohols, and lacks affinity for TCA, whereas skate Asbt showed high affinity for 5α- and 5β-bile alcohols but low affinity for TCA. In contrast, human ASBT demonstrated high affinity for all three bile salt types. These findings suggest that ASBT evolved from the earliest vertebrates by gaining affinity for modern bile salts while retaining affinity for older bile salts. Also, our results indicate that the bile salt enterohepatic circulation is conserved throughout vertebrate evolution.
Keywords: sodium-dependent transporter, enterohepatic circulation, bile acids, bile alcohols, taurocholic acid, lamprey, petromyzon marinus, skate, leucoraja erinacea
Bile alcohols and bile acids are the end products of cholesterol metabolism. Most bile alcohols and bile acids are conjugated with sulfate, taurine, or glycine at the terminal carbon of the side chain and are isolated as bile salts (1). Bile salts play critical physiological roles in vertebrates. They facilitate lipid absorption, inhibit microbe growth in the biliary tract and intestine, and function as signaling molecules that regulate energy expenditure and carbohydrate and lipid metabolism (2). The bile salt pool is maintained in an enterohepatic circulation by bile salt transporters in the distal ileum and the liver (3). Central to this process is the apical sodium (Na+)-dependent bile salt transporter (ASBT/SLC10A2) located on the luminal membrane in the distal ileum and proximal tubule of the kidney in humans and rodents (4). ASBT maintains the enterohepatic and renal-hepatic circulation of bile salts by facilitating their reabsorption from the intestinal lumen and renal tubules. Dysfunction of ASBT/Asbt interrupts the bile salt enterohepatic circulation, reduces the bile salt pool size by 80% in mice, and leads to bile salt malabsorption, diarrhea, and steatorrhea in humans, where reduced plasma levels of cholesterol are also observed (5, 6). Thus, there is considerable pharmaceutical interest in ASBT inhibition as a potential target for drug discovery for the treatment of hypercholesterolemia and diabetes mellitus type 2 (7, 8). Furthermore, an ASBT inhibitor has been demonstrated to be beneficial for patients with chronic idiopathic constipation (9, 10). In contrast to what is known for mammalian species, very little is known about the presence or function of Asbt in other vertebrates.
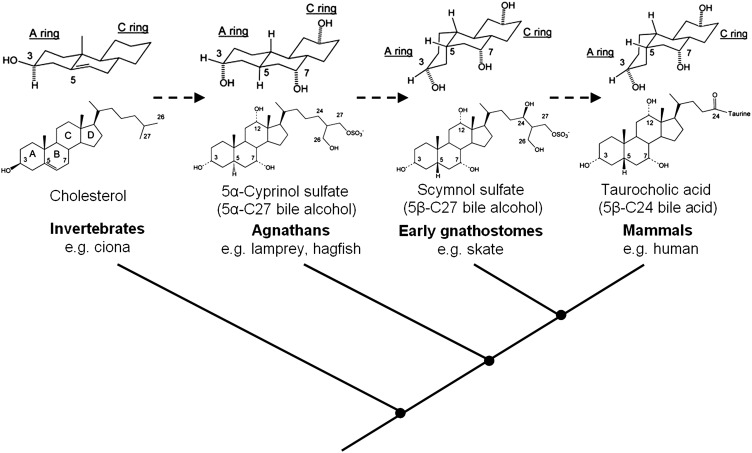
Bile salts demonstrate considerable structural variation across vertebrate classes (1, 11) (Fig. 1). The enzymatic pathway that converts cholesterol into bile salts is complex and requires a minimum of five enzymes in primitive vertebrates and up to 16 enzymes in humans (12–14). The most primitive vertebrates (agnathans or jawless fish) use early evolving C27 sulfated bile alcohols with a C-5 hydrogen at α configuration (i.e., 5α), which is an overall planar structure to the four conjoined steroid rings, one that is similar to that of cholesterol (1, 11). Evolutionarily “intermediate” 5β-C27 sulfated bile alcohols in which the four rings of the steroid possess a tilted structure are mostly detected in jawed cartilaginous fishes (early gnathostomes). More complex vertebrates including mammals primarily use later evolving bile salts, which are 5β-C24 bile acid conjugates with a bent ring juncture and a shortened side chain containing a carboxylic acid. Because of these structural changes, bile salt composition has been proposed as a complementing biochemical trait to characterize evolutionary relationships among species (1).
Fig. 1.
The structural change of bile salts during vertebrate evolution. A simplified phylogenetic tree of vertebrate classes is shown, with respective major bile salt class indicated above, where dashed lines represent the molecular evolution of bile salts. Bile salts are metabolites of cholesterol, a versatile molecule that is already seen in invertebrates that do not use bile salts. Agnathans (jawless fish) are the earliest vertebrate class that primarily use “ancient” 5α-C27 sulfated bile alcohols with an overall planar structure as cholesterol, such as 5α-cyprinol sulfate. Of note, the bile salt spectrum of lamprey comprises a mixture of C27- and C24-5α sulfated bile alcohols that includes C24 5α-Petromyzonol sulfate (5α-PZS), a bile alcohol similar to 5α-cyprinol sulfate but with a shortened side-chain. The evolutionarily more advanced early gnathostomes (jawed cartilaginous fish) mostly use 5β-C27 sulfated bile alcohols with a tilted structure of the steroid rings, such as scymnol sulfate, the major bile salt of skate. As evolution further progressed, more complex vertebrates including mammals mostly use 5β-C24 bile acids with a shortened side-chain containing a carboxylic acid, such as taurocholic acid, a major bile salt in humans.
The major structural variation of bile salts seen over the course of vertebrate evolution raises the question as to how the various transporters that move bile salts into and out of cells located in the intestine and liver have adapted to these changes. Specifically, do nonmammalian Slc10a2’s in more primitive species function as Na+-dependent bile salt transporters and how have they adapted structurally and functionally with the changing shapes of bile salts? Insight into the structure/function relationship of SLC10A/Slc10a members has been advanced by solving the crystal structure of a distant ASBT homolog from the bacterium Neisseria meningitidis (15). Still, the structural determinants of ASBT/Asbt for its bile salt substrates remain unclear because of low homology in the substrate binding pocket between ASBT/Asbt and this bacterial homolog. In particular, it is not known which residues directly bind bile salts when ASBT is configured in an outward direction to accept substrates for uptake.
In this report, we have characterized two ASBT orthologs at the molecular and functional level that represent early stages in the vertebrate lineage. First, we identified an ASBT ortholog in the sea lamprey (Petromyzon marinus), representing agnathans, the most primitive vertebrate class that diverged from a more complex lineage ∼500 million years ago and whose bile consists of some of the earliest 5α-C27 bile alcohols. Second, we identified an ASBT ortholog in the little skate (Leucoraja erinacea), an early gnathostome that diverged ∼300 million years ago, whose bile salts are 5β-C27 bile alcohols (“intermediate” bile salts). Finally, we compared our findings with the structure and substrate specificity of ASBT from humans, whose bile contains the later evolving 5β-C24 bile acids. Our phylogenetic and experimental findings support the concept that ASBT emerged at the very beginning of vertebrate evolution with a limited ability to transport bile salts. As vertebrate evolution progressed, the substrate specificity of ASBT/Asbt for bile salts expanded while at the same time retaining its ability to transport the earlier evolved forms. These findings also indicate that the enterohepatic circulation of bile salts is a conserved function throughout vertebrate evolution.
MATERIALS AND METHODS
Chemicals
Unless otherwise stated, all chemicals were from Sigma (St. Louis, MO). 3H-taurocholic acid (TCA, activity 5.0 Ci/mmol) and 3H-estrone-3-sulfate (activity 57.3 Ci/mmol) were purchased from PerkinElmer (Waltham, MA). 5α-petromyzonol sulfate (PZS) was from Toronto Research Chemicals. 5α-cyprinol sulfate was kindly provided by Dr. Lee Hagey (University of California at San Diego, San Diego, CA). Taurodehydrocholic acid was from Calbiochem (San Diego, CA) and bilirubin-ditaurate was from Frontier Scientific (Logan, UT). Oligonucleotides and DNA sequencing were provided by the Keck Biotechnology Resource Laboratory at Yale University. Fluorescence dye labeled DNA probes were made by Integrated DNA Technologies (Coralville, IA).
Animals
All animal experiments were performed at the Mount Desert Island Biological Laboratory (MDIBL) in Salisbury Cove, ME. Animal experiments were approved by the Institutional Animal Care and Use Committee and in concordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals. Larval lampreys were acquired from Acme Lamprey Co. (Harrison, ME). Adult lampreys were caught while migrating upstream in the Kennebunk River, Maine, in May–June 2011. Larval and adult lampreys were kept in dark-adapted freshwater tanks at 11°C. Skates were collected in June–July 2010 from the Gulf of Maine, off Biddeford, and maintained in seawater tanks at 15°C.
Everted gut sac 3H-TCA uptake assay
All animals were anesthetized with Tricaine before euthanasia. The intestine was removed proximally at the liver or bile duct junction and distally at the start of the rectum. Proximal and distal everted gut sacs were prepared as described by Lack and Weiner (16). Gut sacs were washed four times in either lamprey Ringer's solution (130 mM NaCl, 2.1 mM KCl, 1.8 mM MgCl2, 2.6 mM CaCl2, 1 mM NaHCO3, 4 mM D-glucose, 4 mM HEPES, tetramethyl ammonium hydroxide to pH 7.4) or elasmobranch Ringer's solution, prepared as described previously (17). Next, all sacs were submerged in their respective solutions supplemented with 50 µM 3H-TCA (1 mCi/mmol), lightly gassed with ambient air, and incubated at 15°C water bath for 30, 60, or 120 min with gentle manual agitation. Following incubation, sacs were washed four times in ice-cold solution, homogenized, and lysed in 0.5% Triton X-100 PBS. Lysate was centrifuged at 18,000 g for 10 min and supernatant collected for measurement of protein and radioactivity. Protein concentration was determined according to the Bradford method using a commercial kit (Bio-Rad). Radioactivity was measured in a Tri-Carb 2100TR liquid scintillation counter (Packard) and data was normalized to total cell protein.
RNA extraction and quantification
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) and purified using a kit (RNeasy Clean-up Kit, Qiagen, Valencia, CA). Two micrograms of total RNA from each sample was reverse transcribed into cDNA using a kit from Roche (Indianapolis, IN). TaqMan real-time RT-PCR was performed on an ABI 7500 Sequence Detection System (Applied Biosystems, Carlsbad, CA). The specific primers and probes are listed in supplementary Table I. Because the expression of the housekeeping gene β-actin varied significantly in the broad range of tissues that were examined, we normalized mRNA expression to 1 µg of total RNA. Results were expressed in copy number, where the cloned constructs were used to establish a standard curve.
Molecular cloning
To clone lamprey Asbt (lpAsbt), we first retrieved sequence fragments from the lamprey genome by ortholog searching using human ASBT (hASBT) protein sequence as query. We then designed primers and amplified a 600 bp fragment from lamprey intestine using RT-PCR. DNA sequencing and phylogenetic analysis confirmed that this fragment encoded a portion of lpAsbt. A full-length lpAsbt was obtained by 5′- and 3′- RACE PCR using a kit from Clontech. To clone skate Asbt (skAsbt), we first acquired a DNA fragment by RT-PCR using degenerate primers that matched two conserved regions of ASBT/Asbts. The full-length skAsbt was also obtained by RACE PCR. To obtain hASBT, we directly amplified the coding region from Caco-2 cells and inserted it into a pcDNA3 vector, and confirmed sequence identity with GenBank data. To functionally characterize lpAsbt and skAsbt, both were also subcloned into pcDNA3 vectors. For both lpAsbt and skAsbt, at least six full-length clones were sequenced and one clone with identical sequence to the lamprey genome or original RACE-PCR products was picked for further experiments. In addition, we made lpAsbt-FLAG and skAsbt-FLAG constructs in pcDNA3 vectors to tag these two proteins at the C terminus for purposes of Western blotting and immunofluorescent labeling. The primers are listed in supplementary Table I.
Phylogenetic analysis
Sequence alignment was performed with the ClustalW2 algorithm assuming the Gonnet replacement matrix (18). We inferred phylogeny with Bayesian Markov Chain Monte Carlo (MCMC) analysis using MrBayes v3.2.0 software (19). The analysis was performed assuming the Jones model for amino acid replacement, an equal rates γ distribution with four categories and run for 300,000 generations with one cold chain and three heated chains. Posterior probabilities were calculated by sampling every 100 generations and discarding the first 500 samples as “burn-in”. The phylogenetic tree was rooted using SLC17A5 as an out-group and visualized with FigTree v1.3.1.
COS-7 cell based 3H-TCA uptake assay
COS-7 cells were maintained at low passage number in growth medium (DMEM with 10% FBS, 50 U/ml penicillin and 50 µg/ml streptomycin, all from Invitrogen). When cells reached 80% confluence, they were transfected with either pcDNA3 (control), pcDNA3-lpAsbt, pcDNA3-skAsbt or pcDNA3-hASBT using Fugene HD or X-tremeGENE 9 transfection reagent (Roche). Forty hours posttransfection, cells were subjected to the uptake assay as previously described (20). Transport activity was normalized to total cell protein. Kinetic constants are expressed as value ± SE and were calculated by nonlinear fitting of data to the Michaelis-Menten equation using least squares (Graphpad Prism 5, Graphpad Software).
ASBT-farnesoid X receptor α (FXR/NR1H4) luciferase reporter assay for bile salt transport
A dual-luciferase gene reporter assay (Promega, Madison, WI) was utilized to assess the ability of conjugated bile salts to be transported into cells transfected with ASBT/Asbt's. HEK293T cells were maintained in growth medium. When cells reached 80% confluence, the culture medium was changed to DMEM supplemented with 0.5% charcoal-stripped FBS and cotransfected with 50 ng pcDNA3 (control) or 4 ng pcDNA3-lpAsbt and 46 ng pcDNA3 or 50 ng pcDNA3-skAsbt or 50 ng pcDNA3-hASBT, along with 50 ng pCMX-hFXRα, 37.5 ng pCMX-hRXRα, 125 ng pGL3-hIBABP, and 1.5 ng phRL-CMV in triplicates, using 3μl Lipofectamine 2000 (Invitrogen) for each well in 24-well plates. Twenty-four hours posttransfection, cells were treated with bile salts for an additional 24 h in 0.5% charcoal-stripped FBS DMEM. Passive lysis buffer (Promega) was used to prepare cell lysate and luminescence was detected in a Synergy2 Microplate Reader (BioTek). Firefly luciferase readings were normalized to Renila luciferase, the internal control. To validate the bile salt transport function of lpAsbt, cells were treated with lipid extract (1:1000 dilution) isolated from adult lamprey liver.
Statistical analysis
Statistical analysis was performed with Graphpad Prism 5 (Graphpad Software). Unpaired two-tailed t-test was used to detect differences between two groups and one-way ANOVA was used to detect differences between more than two groups, followed by Tukey's post hoc test for pairwise comparison. P < 0.05 was considered to be statistically significant.
RESULTS
An intestinal Na+-dependent transport system for the modern bile salt TCA is absent in lamprey in contrast to the little skate
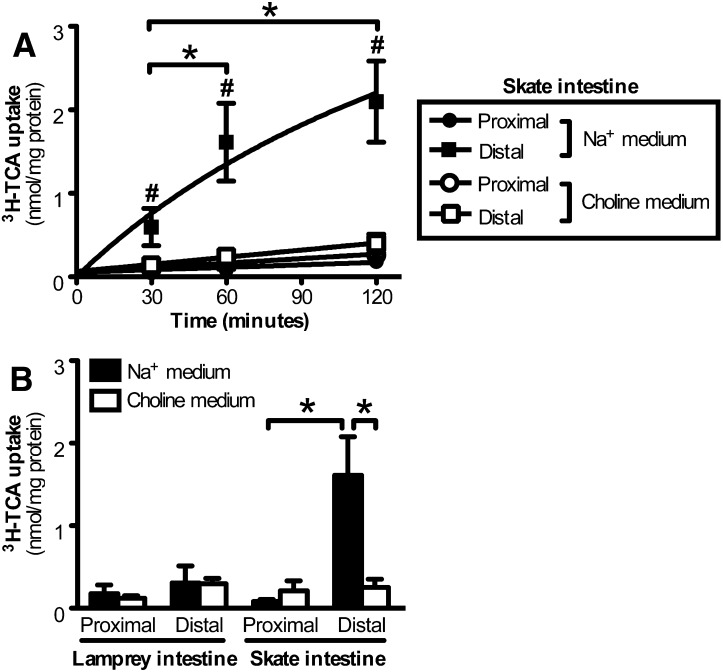
Previous work by our group demonstrated that bile salts are reabsorbed from the intestine of the little skate, suggesting that an active transport system is present for bile salts (21). However, it was not known if this transport system is Na+-dependent, as is human and rodent ASBT/Asbt. To address this question, 3H-TCA uptake was assessed in everted gut sacs isolated from the little skate (16). As demonstrated in Fig. 2A, a significant time-dependent uptake of 3H-TCA was observed in the distal but not proximal intestine. When Na+ was replaced by choline in the medium, uptake of 3H-TCA was abolished, indicating that a Na+-dependent bile salt transporter was present in the distal intestine of skate. A similar transport experiment was then carried out in adult lamprey, and in contrast to skate intestine, neither the proximal nor distal intestine of the lamprey showed significant Na+-dependent uptake of 3H-TCA (Fig. 2B). This negative result suggests that either an ASBT ortholog has not evolved in lamprey or its substrate specificity is limited and does not accommodate the modern bile salt TCA.
Fig. 2.
Uptake of 3H-TCA into skate and lamprey intestine. A: Everted gut sacs prepared from the proximal and distal skate intestine were incubated for 30, 60, or 120 min in medium supplemented with 50 μM 3H-TCA containing Na+ or choline (Na+-void). Values represent means (n = 3–4) ± SD. *, P <0.05; #, P < 0.01 versus all other groups at the same time point. B: Uptake of 50 μM 3H-TCA in everted gut sacs prepared from proximal or distal intestine of skate and lamprey, incubated for 60 min in Na+ or choline (Na+-void) medium. Values represent means (n = 3–4) ± SD. *, P < 0.05.
ASBT orthologs are identified in the distal intestine of both lamprey and skate
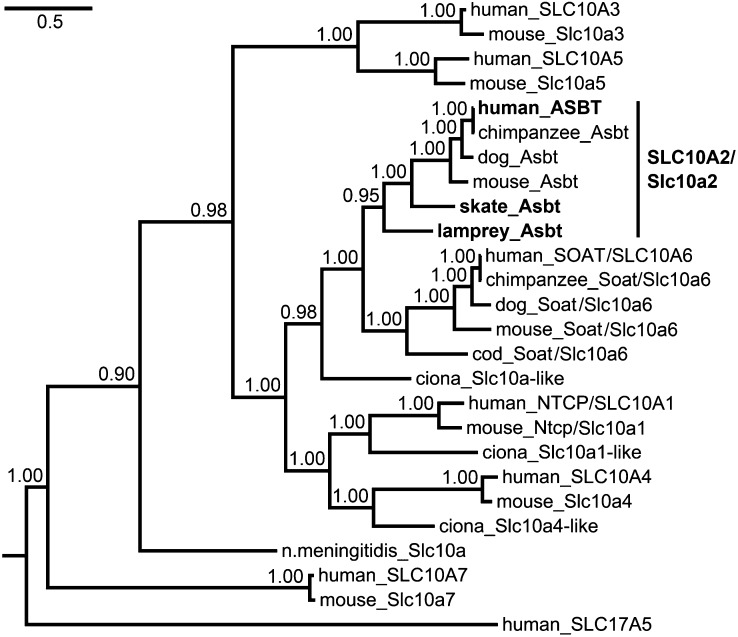
A search of the lamprey genome revealed DNA sequences with the potential to encode portions of an ASBT ortholog. Subsequent RT-PCR and RACE PCR identified a full-length lpAsbt transcript which encodes 363 amino acids with a 132 bp 5′-untranslated region (UTR) and 1.7 kb 3′-UTR (GenBank accession number JX014266). To identify skAsbt, we performed RT-PCR using degenerate primers, followed by RACE PCR. We amplified a full-length skAsbt transcript 2.3 kb in size, encoding for 393 amino acids with 189 bp at the 5′-UTR and 918 bp at the 3′-UTR (GenBank accession number JX014267). LpAsbt and skAsbt share 58% and 64% amino acid identity to hASBT, respectively (supplementary Table II). Phylogenetic analysis placed lpAsbt and skAsbt as the most primitive of known ASBT/SLC10A2 orthologs (Fig. 3). BLAST search of genome of the sea squirt (Ciona intestinalis), an invertebrate that is believed to share the last common ancestor with vertebrates, produced 16 putative protein sequences that could be members of the Slc10 protein family. However, phylogenetic analysis did not identify a potential ortholog of SLC10A2 (Fig. 3 and supplementary Figure I). The ciona sequence that was most closely related was placed on the common branch of Slc10a2 and Slc10a6 subfamilies. Based on these findings, we propose that a distinct Asbt/Slc10a2 first emerged from an ancient Slc10a2/a6-like gene near the beginning of vertebrate evolution, presumably from a gene duplication between ciona and lamprey evolution, at a time that coincides with the extensive utilization of cholesterol, the emergence of bile salts, the development of a biliary system and the ability to produce bile.
Fig. 3.
Phylogeny of the Slc10a family shows that ortholog genes identified in lamprey and skate are the most primitive members of the ASBT/SLC10A2 subfamily. Phylogeny was inferred using Bayesian MCMC analysis. Posterior probabilities are indicated at nodes. Branch length is expressed as number of expected substitutions per site. Accession numbers of protein sequences used are listed in supplementary Table III.
Alignment of experimentally verified sequences of ASBT/Asbt's revealed that the proposed transmembrane domains are well conserved, including the Na+-binding core and the substrate binding pocket based on the recently solved crystal structure of a distant ASBT homolog in N. meningitidis (15) (supplementary Figs. II and III). In contrast, the sequences of the N-terminus and C terminus show great interspecies variation. For example, while a N-glycosylation site in hASBT Asn-10 is well conserved (22), computer software predicts an additional N-glycosylation site at Asn-22 in skAsbt (supplementary Fig. II).
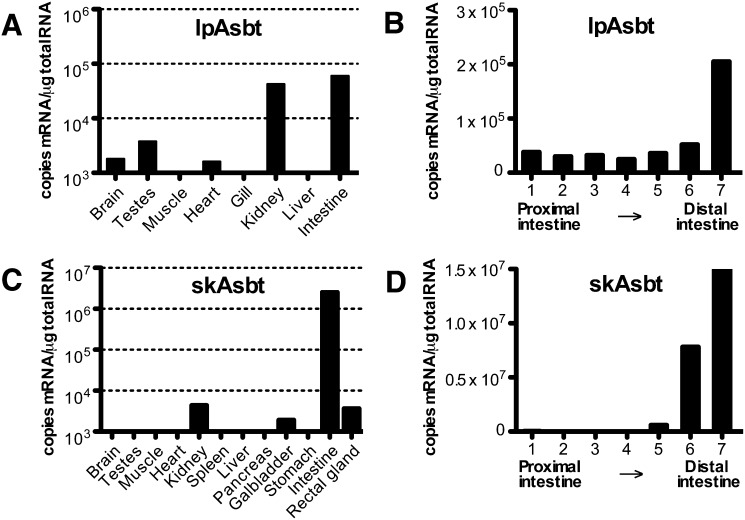
To determine the tissue distribution of lpAsbt and skAsbt, we performed real-time RT-PCR. As shown in Fig. 4A, lpAsbt mRNA was expressed primarily in the kidney and intestine; less in testes, heart, and brain; and was undetectable in muscle, gill, and liver. A similar expression pattern was found in lamprey larva. SkAsbt mRNA was highly expressed in the intestine with lower levels in the kidney, gallbladder, and rectal gland; it was absent from other tissues (Fig. 4C). In addition, we analyzed mRNA distribution along segments of intestine, which demonstrated that both lpAsbt and skAsbt are most abundant in the distal intestine (Fig. 4B, D). This was particularly evident in skate with a greater than 300-fold expression in the distal compared with the proximal intestine. These expression profiles are similar in human and rodents and correspond with the known bile salt recycling function of ASBT.
Fig. 4.
Tissue distribution of lpAsbt and skAsbt mRNA by quantitative real-time RT-PCR. A broad variety of tissues was tested for mRNA expression of lpAsbt (A) and skAsbt (C). Expression along the intestine was determined by dividing the intestine in 7 segments of equal length (B and D). Segment 1 starts at the intestinal connection to the bile duct/liver and segment 7 ends at the start of the rectum. Expression below 1,000 copies mRNA/μg total RNA was considered background and is not shown. All values represent one animal and are expressed as means from triplicate measurements.
Affinity for the modern bile salt TCA is absent in lpAsbt, low in skAsbt, and high in hASBT
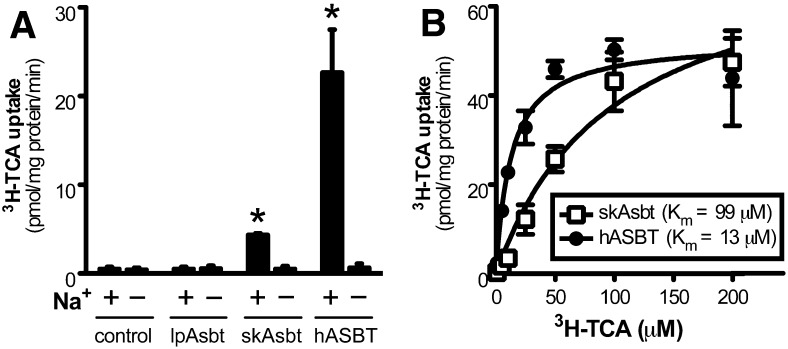
To functionally characterize lpAsbt and skAsbt and to compare their substrate specificity to hASBT, we subcloned these three genes into a pcDNA3 plasmid vector and transfected them into COS-7 cells for expression. As shown in Fig. 5A, skAsbt and hASBT demonstrated Na+-dependent uptake of 10 µM 3H-TCA that increased significantly over vector control. In contrast, lpAsbt did not show any specific transport activity for 3H-TCA (10–100 µM) with or without Na+ in the media, despite confirmation by Western blot and immunofluorescent labeling that the lpAsbt-FLAG fusion protein was expressed on the plasma membrane of transfected cells (supplementary Fig. IV). These results are consistent with the negative results of 3H-TCA transport in the everted gut sac experiments (Fig. 2B) and suggest that either lpAsbt is not a functional bile salt transporter or it is functionally inactive for the modern 5β-C24 bile salt TCA.
Fig. 5.
3H-TCA uptake assay in transfected COS-7 cells. A: Uptake of 10 μM 3H-TCA in cells transiently transfected with vector without insert (control), skAsbt, lpAsbt or hASBT. Cells were incubated for 10 min in medium with Na+ (+) or medium where Na+ was substituted with choline (–). *, P < 0.05 versus choline. B: Kinetics of 3H-TCA uptake in cells transfected with skAsbt or hASBT. Cells were incubated for 10 min and background uptake levels derived from cells transfected with vector control were subtracted. lpAsbt did not show increased uptake over background (vector control) at 10 μM and 100 μM and was excluded. Data was normalized to total cell protein. Kinetic parameters were obtained by nonlinear curve fitting using the Michaelis-Menten equation. All values represent at least three independent experiments and are expressed as means ± SD.
Michaelis-Menten analysis of 3H-TCA transport kinetics indicated that skAsbt has a Vmax of 75 ± 9 pmol/mg protein/min with a Km of 99 ± 24 µM, whereas hASBT has a Vmax of 51 ± 3 pmol/mg protein/min with a significantly lower Km of 13 ± 2 µM (Fig. 5B). Of note, the Vmax was derived from data normalized to total protein in transfected cells, not to transporter protein, which prevents direct comparison between absolute transport rates and subsequently comparison of Vmax. However, Km is independent of absolute transport rate, and can be compared to determine differences in affinity. The findings of lack of TCA affinity for lpAsbt and low affinity for skAsbt, albeit lower than for hASBT, suggest that structural changes in Asbt have evolved that allowed accommodation of this new bile acid structure sometime between the development of agnathans and early gnathostomes, whereas TCA affinity was optimized further between early gnathostome and mammalian evolution.
Bile salt substrate affinity is limited to early evolving bile salts for lpAsbt, early and “intermediate” bile salts for skAsbt, whereas hASBT has high affinity for all bile salt forms
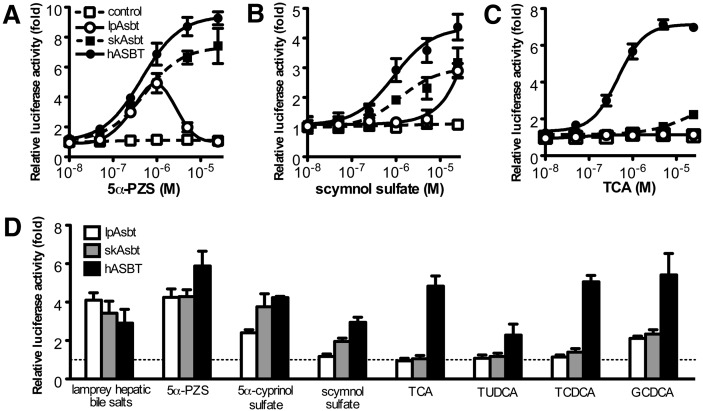
To test if lpAsbt transports bile salts, we utilized a luciferase-based ASBT-FXR reporter assay. Because conjugated bile salts require a specific transporter (e.g., ASBT) to cross the cell membrane and they are ligands for human FXR, transactivation of FXR reflects the ability of ASBT to transport a given bile salt into the transfected cells. In this experiment, expression constructs of lpAsbt, skAsbt, hASBT, or empty vector were cotransfected with FXR reporter constructs into HEK293T cells. The transfected cells were then treated with 5α-PZS, scymnol sulfate, and TCA, the three major bile salts in lamprey, skate, and human, respectively. Interestingly, lpAsbt cotransfection transactivated FXR after 5α-PZS treatment, indicating lpAsbt does transport this endogenous bile salt (Fig. 6A). SkAsbt and hASBT also demonstrated transport activity for 5α-PZS. When 5β-C27 scymnol sulfate was tested, lpAsbt showed significant activity only at 25 µM or higher concentrations whereas skAsbt and hASBT demonstrated activity with nanomolar concentrations (Fig. 6B). When cells were treated with 5β-C24 TCA, lpAsbt did not show any transport activity, skAsbt showed low activity, whereas hASBT showed high activity (Fig. 6C). To further confirm that lpAsbt, skAsbt, and hASBT have differential selectivity for bile salts, we tested additional early and late evolving bile salts with the ASBT-FXR reporter assay. As indicated by Fig. 6D, lpAsbt can transport lamprey bile salts from its liver lipid extract and 5α-cyprinol sulfate (1 μM), another 5α-C27 bile alcohol, but not modern 5β-C24 bile salts, including tauroursodeoxycholic acid (TUDCA), taurochenodeoxycholic acid (TCDCA), TCA, glycocholic acid, and glycoursodeoxycholic acid. Interestingly, glycochenodeoxycholic acid (GCDCA) is an exception and apparently was transported by lpAsbt. skAsbt transported scymnol sulfate in addition to all the lpAsbts substrates, but again demonstrated low affinity for most modern bile salts (Fig. 6D). In contrast, all tested bile salts were effectively transported by hASBT. Collectively, these findings indicate that lpAsbt's affinity for bile salts is largely confined to the 5α-C27 early evolving “ancient” bile salts, skAsbt can effectively transport both the “ancient” and evolutionarily “intermediate” bile salts whereas hASBT effectively transports bile salts with the entire spectrum of structures.
Fig. 6.
ASBT-FXR luciferase reporter assay in cotransfected HEK293T cells. Cells were cotransfected with FXR reporter constructs plus either vector without insert (control), lpAsbt, skAsbt or hASBT for 24 h and subjected to bile salt treatment for additional 24 h. Dose-response curve of cells treated with 5α-petromyzonol sulfate (5α-PZS), the endogenous bile salt of lamprey (A); with 5β-scymnol sulfate, the endogenous bile salt of skate (B); and with taurocholic acid (TCA), the endogenous bile salt of humans(C). Luciferase readings of dose-response curves are relative to cells transfected with vector control treated with 10−8 M bile salts. (D) Cells transfected with vector control, lpAsbt, skAsbt or hASBT were treated with 1 μM indicated bile salt or ∼1 μM bile salts from adult lamprey liver. Luciferase readings of this analysis are relative to cells transfected with vector control. All values represent at least three independent experiments and are expressed as means ± SD.
Substrate specificity of skAsbt is confined to bile salts, similar to hASBT
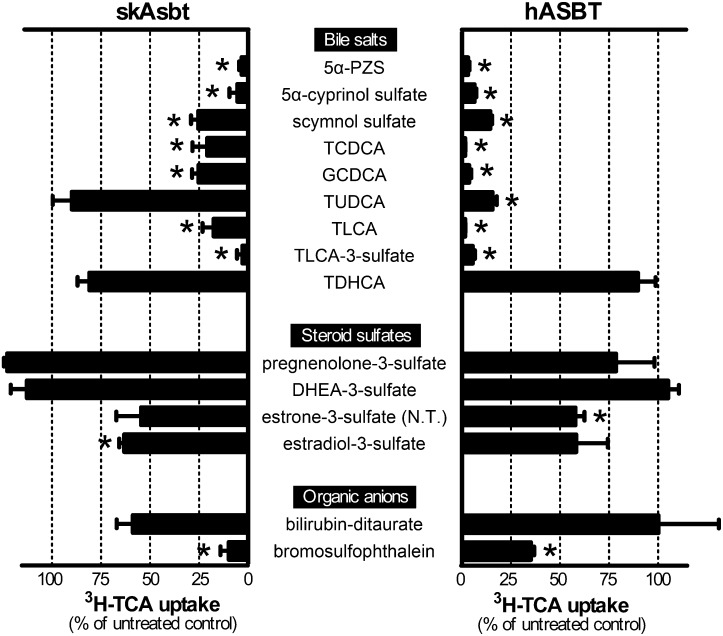
To further assess skAsbt substrate specificity, we tested an array of bile salts, steroid sulfates, and organic anions in assays competing with uptake of 3H-TCA. In general, more bile salt structures inhibited hASBT 3H-TCA uptake more effectively than with skAsbt (Fig. 7). Specifically, the early evolved bile salts (5α-PZS, 5α-cyprinol sulfate), “intermediate” bile salts (5β-C27 scymnol sulfate) and modern 5β-C24 bile acids (TCDCA, GCDCA, taurolithocholic acid, and taurolithocholic acid -3-sulfate) significantly reduced 3H-TCA transport activity in both skAsbt and hASBT. Interestingly, TUDCA effectively reduced 3H-TCA transport activity in hASBT but not in skAsbt, consistent with results from the ASBT-FXR reporter assay. Taurodehydrocholic acid had no effect on 3H-TCA transport activity with either skAsbt or hASBT. We then determined the affinity of skAsbt for 5β-C27 scymnol sulfate, the major endogenous bile salt in skate, in a competition assay for 3H-TCA uptake. Scymnol sulfate has a Ki of 42 ± 12 µM, whereas the Km for 3H-TCA was calculated as 87 ± 38 µM (supplementary Fig. V).
Fig. 7.
Competition assay of 3H-TCA transport. COS-7 cells transfected with skAsbt or hASBT were incubated for 10 min in Na+ medium supplemented with 10 μM 3H-TCA without competitor (untreated control) or with 100 μM competitor. Background levels derived from cells transfected with vector control were subtracted and uptake measurements were normalized for protein concentration of the lysate.*, P < 0.05 for untreated control versus competitor. All values represent at least three independent experiments and are expressed as means ± SD.
In addition, we tested whether or not steroid sulfates could inhibit hASBT and skAsbt transport of 3H-TCA. These molecules are structurally similar to bile salts and are transported by SOAT/SLC10A6 (23), a close paralog of ASBT (Fig. 3). Neither pregnenolone-3-sulfate nor dehydroepiandrosterone-3-sulfate inhibited uptake activity of hASBT and skAsbt for 3H-TCA. Estradiol-3-sulfate weakly inhibited skAsbt but not hASBT, whereas estrone-3-sulfate weakly inhibited hASBT but not skAsbt. However, 3H-estrone-3-sulfate (25 μM) was not transported by lpAsbt, skAsbt, or hASBT. Also, both skAsbt and hASBT did not show significant affinity for the organic anion bilirubin-ditaurate, whereas bromosulfophtalein significantly inhibited both transporters. Taken together, these competition experiments suggest that even as the substrate specificity of these transporters expanded for bile salts, this expansion did not extend to the closely related family of steroid sulfates.
DISCUSSION
In order to explore the evolutionary development of ASBT/SLC10A2 and its substrate specificity, we functionally characterized this intestinal bile salt transport system in the sea lamprey and the little skate, two species that represent early evolving members of vertebrate evolution, and compared these with the human system. Our results demonstrate that primitive ASBT/SLC10A2 orthologs in lamprey and skate have the ability to transport bile salts (Figs. 2, 5, and 6). Like mammalian ASBT/Asbt, the substrate specificity of lpAsbt and skAsbt appears confined to bile salts, as they did not demonstrate affinity for structurally related steroid sulfates and other organic anions (Fig. 7). The mRNA tissue distribution of both lpAsbt and skAsbt was most abundant in the distal intestine and kidney (Fig. 4), consistent with the development of transporters to reabsorb bile salts from the intestinal lumen, an essential function for establishing an enterohepatic circulation. This molecular evidence indicates that the enterohepatic circulation of bile salts was present at the earliest beginnings of vertebrate evolution. Because an ASBT ortholog was not identified in the sea squirt (Fig. 2 and supplementary Fig. I), a late nonvertebrate, we propose that ASBT/Asbt is a bile salt transporter that evolved at the beginning of vertebrate evolution.
We also demonstrated that skAsbt transports bile salts in a Na+-dependent manner. At this time, we were not able to directly determine whether bile salt transport by lpAsbt is Na+-dependent since appropriate radiolabeled bile salt substrates are not available. However, protein sequence alignment reveals that amino acids directly involved in Na+-binding for N. meningitidis Slc10a (Asbt-like) are completely conserved in lpAsbt, skAsbt, mammalian ASBT/Asbt's and even Na+-dependent paralogs NTCP and SOAT (supplementary Fig. II) (15), suggesting that lpAsbt is likely to be a Na+-dependent transporter.
In this study, we assessed transport activity and substrate specificity with 3H-TCA uptake assays in intact intestinal tissue (Fig. 2) and cell-based assays as well as in ASBT-FXR reporter assays using recombinant expressed transporters (Figs. 5 and 6). Collectively, these assays demonstrate that lpAsbt has a bile salt substrate specificity with high affinity only for 5α-PZS (the endogenous bile salt of lamprey) and 5α-cyprinol sulfate, two early evolving “ancient” 5α bile alcohols, whereas the more evolutionarily advanced “intermediate” 5β-C27 bile alcohol scymnol sulfate was a low affinity substrate. In contrast, the modern 5β-C24 bile acids TCA and TCDCA were not transported. In the case of skAsbt, not only was 5β-C27 scymnol sulfate (its endogenous bile salt) a high affinity substrate but so were the “ancient” 5α-bile alcohols. The later evolving 5β-C24 bile acids such as TCA and TCDCA showed only low affinity for skAsbt. Surprisingly, all the structural forms of bile salts were high affinity substrates for hASBT, even though the major endogenous bile salts in humans are 5β-C24 bile acids. Altogether, these results suggest that ASBT expanded its substrate specificity whenever a novel class of bile salts emerged in evolution (1, 11).
This broad substrate affinity for human ASBT is unusual, as most orthologs generally lose affinity for their earlier substrates when they acquire affinity for novel substrates during evolution (24–26), a phenomenon termed “ligand-receptor” coevolution. ASBT/Asbt has apparently retained affinity for old substrates even as it gained affinity for more modern bile salts. We speculate that modern ASBT/Asbt retains its broad substrate specificity in order to recover as many different species of bile salts as possible. In mammals, 16 different enzymatic reactions are required to convert cholesterol to 5β-C24 bile acids. As each step is not 100% efficient, small amounts of intermediate bile salts will be produced and excreted in to bile (27). In order to most efficiently maintain the bile salt pool size, these “by-products of bile salt synthesis” are also reclaimed. We speculate that other transporters involved in the enterohepatic circulation of bile salts might also retain specificity for a wide variety of bile salt substrates structures, although this hypothesis has not yet been tested.
Our phylogenetic analysis indicates that the recently solved crystal structure of N. meningitidis Slc10a (formally called nmASBT) is not a member of the SLC10A2 gene family (Fig. 3). Although it transports TCA, it is not known whether it can transport other bile salts or other molecules. In addition, N. meningitidis Slc10a shares lower identity to hASBT than does SLC10A3, SLC10A4, SLC10A5, and SOAT/SLC10A6 (supplementary Table II). Because the latter four solute carriers do not transport TCA, this raises a question as to the specificity of the substrate binding pocket of N. meningitidis Slc10a for bile salts. Because lpAsbt, skAsbt, and hASBT share much higher sequence identity and demonstrate differential selectivity for bile salts, it should be possible to identify specific amino acids that are responsible for the expansion of mammalian ASBT/Asbt substrate specificity using further sequence analyses and mutation experiments. Doing so will help to determine ASBT/Asbt's evolutionary path while providing insight into the structure-function determinants of hASBT.
In conclusion, the present study demonstrates that Asbt is a functional bile salt transporter in the most primitive vertebrates, occurring together with the emergence of bile salts and the ability of organisms to form bile. This study establishes that ASBT/Asbt and the enterohepatic circulation of bile salts were present at the beginning of vertebrate evolution, giving vertebrates the transport ability essential to regulate both bile salt and lipid homeostasis. Our findings also suggest that as ASBT evolved, it gained substrate specificity to novel bile salt structures while retaining affinity for its old substrates.
Supplementary Material
Acknowledgments
We thank Dr. Lee Hagey (University of California at San Diego, San Diego, CA) for generously providing 5α-cyprinol sulfate and for critical review and editing of this manuscript. Dr. Carol Soroka and Shuhua Xu in our laboratory have provided technical support. Also, we are grateful to Professor Ulrich Beuers (University of Amsterdam) for advice and constructive discussions.
Footnotes
Abbreviations:
- ASBT/Asbt
- apical sodium-dependent bile salt transporter
- FXR/Fxr
- farnesoid X receptor
- hASBT
- human ASBT
- lpAsbt
- lamprey Asbt
- skAsbt
- skate Asbt
- TCA
- taurocholic acid
- UTR
- untranslated region
This work was supported by the National Institutes of Health grants P30 DK34989 and R37 DK25636 (J.L.B.) and student fellowship grant 10-06s from the Dutch Digestive Foundation (D.A.L.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables, five figures, supplementary Materials and Methods, data, and references.
REFERENCES
- 1.Hofmann A. F., Hagey L. R., Krasowski M. D. 2010. Bile salts of vertebrates: structural variation and possible evolutionary significance. J. Lipid Res. 51: 226–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trauner M., Claudel T., Fickert P., Moustafa T., Wagner M. 2010. Bile acids as regulators of hepatic lipid and glucose metabolism. Dig. Dis. 28: 220–224 [DOI] [PubMed] [Google Scholar]
- 3.Dawson P. A., Lan T., Rao A. 2009. Bile acid transporters. J. Lipid Res. 50: 2340–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson P. A. 2011. Role of the intestinal bile acid transporters in bile acid and drug disposition. Handb. Exp. Pharmacol. 201: 169–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson P. A., Haywood J., Craddock A. L., Wilson M., Tietjen M., Kluckman K., Maeda N., Parks J. S. 2003. Targeted deletion of the ileal bile acid transporter eliminates enterohepatic cycling of bile acids in mice. J. Biol. Chem. 278: 33920–33927 [DOI] [PubMed] [Google Scholar]
- 6.Oelkers P., Kirby L. C., Heubi J. E., Dawson P. A. 1997. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2). J. Clin. Invest. 99: 1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis M. C., Brieaddy L. E., Root C. 1995. Effects of 2164U90 on ileal bile acid absorption and serum cholesterol in rats and mice. J. Lipid Res. 36: 1098–1105 [PubMed] [Google Scholar]
- 8.Bhat B. G., Rapp S. R., Beaudry J. A., Napawan N., Butteiger D. N., Hall K. A., Null C. L., Luo Y., Keller B. T. 2003. Inhibition of ileal bile acid transport and reduced atherosclerosis in apoE2/2 mice by SC-435. J. Lipid Res. 44: 1614–1621 [DOI] [PubMed] [Google Scholar]
- 9.Chey W. D., Camilleri M., Chang L., Rikner L., Graffner H. 2011. A randomized placebo-controlled phase IIb trial of a3309, a bile acid transporter inhibitor, for chronic idiopathic constipation. Am. J. Gastroenterol. 106: 1803–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simren M., Bajor A., Gillberg P. G., Rudling M., Abrahamsson H. 2011. Randomised clinical trial: the ileal bile acid transporter inhibitor A3309 vs. placebo in patients with chronic idiopathic constipation–a double-blind study. Aliment. Pharmacol. Ther. 34: 41–50 [DOI] [PubMed] [Google Scholar]
- 11.Haslewood G. A. 1967. Bile salt evolution. J. Lipid Res. 8: 535–550 [PubMed] [Google Scholar]
- 12.Russell D. W. 2009. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50(Suppl): S120–S125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norlin M., Wikvall K. 2007. Enzymes in the conversion of cholesterol into bile acids. Curr. Mol. Med. 7: 199–218 [DOI] [PubMed] [Google Scholar]
- 14.Hagey L. R., Moller P. R., Hofmann A. F., Krasowski M. D. 2010. Diversity of bile salts in fish and amphibians: evolution of a complex biochemical pathway. Physiol. Biochem. Zool. 83: 308–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu N. J., Iwata S., Cameron A. D., Drew D. 2011. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 478: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lack L., Weiner I. M. 1961. In vitro absorption of bile salts by small intestine of rats and guinea pigs. Am. J. Physiol. 200: 313–317 [DOI] [PubMed] [Google Scholar]
- 17.Fricker G., Hugentobler G., Meier P. J., Kurz G., Boyer J. L. 1987. Identification of a single sinusoidal bile salt uptake system in skate liver. Am. J. Physiol. 253: G816–G822 [DOI] [PubMed] [Google Scholar]
- 18.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- 19.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- 20.Wong M. H., Oelkers P., Craddock A. L., Dawson P. A. 1994. Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J. Biol. Chem. 269: 1340–1347 [PubMed] [Google Scholar]
- 21.Fricker G., Wossner R., Drewe J., Fricker R., Boyer J. L. 1997. Enterohepatic circulation of scymnol sulfate in an elasmobranch, the little skate (Raja erinacea). Am. J. Physiol. 273: G1023–G1030 [DOI] [PubMed] [Google Scholar]
- 22.Zhang E. Y., Phelps M. A., Banerjee A., Khantwal C. M., Chang C., Helsper F., Swaan P. W. 2004. Topology scanning and putative three-dimensional structure of the extracellular binding domains of the apical sodium-dependent bile acid transporter (SLC10A2). Biochemistry. 43: 11380–11392 [DOI] [PubMed] [Google Scholar]
- 23.Geyer J., Doring B., Meerkamp K., Ugele B., Bakhiya N., Fernandes C. F., Godoy J. R., Glatt H., Petzinger E. 2007. Cloning and functional characterization of human sodium-dependent organic anion transporter (SLC10A6). J. Biol. Chem. 282: 19728–19741 [DOI] [PubMed] [Google Scholar]
- 24.Moyle W. R., Campbell R. K., Myers R. V., Bernard M. P., Han Y., Wang X. 1994. Co-evolution of ligand-receptor pairs. Nature. 368: 251–255 [DOI] [PubMed] [Google Scholar]
- 25.Park Y., Kim Y. J., Adams M. E. 2002. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc. Natl. Acad. Sci. USA. 99: 11423–11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker M. E. 2011. Origin and diversification of steroids: co-evolution of enzymes and nuclear receptors. Mol. Cell. Endocrinol. 334: 14–20 [DOI] [PubMed] [Google Scholar]
- 27.Griffiths W. J., Sjovall J. 2010. Bile acids: analysis in biological fluids and tissues. J. Lipid Res. 51: 23–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.