Abstract
Nonalchoholic fatty liver disease (NAFLD) is the most common cause of liver dysfunction and is associated with metabolic diseases, including obesity, insulin resistance, and type 2 diabetes. We mapped a quantitative trait locus (QTL) for NAFLD to chromosome 17 in a cross between C57BL/6 (B6) and BTBR mouse strains made genetically obese with the Lepob/ob mutation. We identified Tsc2 as a gene underlying the chromosome 17 NAFLD QTL. Tsc2 functions as an inhibitor of mammalian target of rapamycin, which is involved in many physiological processes, including cell growth, proliferation, and metabolism. We found that Tsc2+/− mice have increased lipogenic gene expression in the liver in an insulin-dependent manner. The coding single nucleotide polymorphism between the B6 and BTBR strains leads to a change in the ability to inhibit the expression of lipogenic genes and de novo lipogenesis in AML12 cells and to promote the proliferation of Ins1 cells. This difference is due to a different affinity of binding to Tsc1, which affects the stability of Tsc2.
Keywords: diabetes, fatty acid/metabolism; genetics; triglycerides; cell proliferation; nonalcoholic fatty liver disease
Nonalcoholic fatty liver disease (NAFLD), the accumulation of triglyceride (TG) in hepatocytes, is the most common cause of liver dysfunction and is associated with several metabolic diseases, including obesity, insulin resistance, type 2 diabetes, dyslipidemia, and cardiovascular diseases (1). It affects ∼25% of the US population. Twelve to 20% of NAFLD patients develop an inflammatory response and nonalcoholic steatohepatitis, and 5 to 15% progress to fibrosis and cirrhosis as the final stage of the disease, which is a great risk factor for hepatocellular carcinoma (2–4). Ten percent of liver transplants in the USA are done due to cirrhosis related to NAFLD (5).
Genetic background has been shown to play an important role in NAFLD, and the prevalence of NAFLD differs in various racial and ethnic groups, with African-Americans having the lowest incidence (24%), European-Americans having intermediate incidence (33%), and Hispanics having the highest incidence (45%) (6). Genetic approaches, including genome-wide association studies, have been used to identify DNA sequence variations that contribute to NAFLD. Single nucleotide polymorphisms (SNPs) of several genes, including PNPLA3, PPARGC1A, ADIPONECTIN, and CLOCK, have been shown to be involved in NAFLD (7–10). In spite of this progress, the genetic contribution to the regulation of lipid metabolism, which leads to the development and progression of NAFLD, has yet to be elucidated (11).
The repertoire of available inbred mouse strains represents as much genetic variance as that found in the human population. This is an important resource for the identification of genes and pathways that confer resistance or susceptibility to diseases, including cancer, diabetes, and infectious diseases (12–17). Although several mouse models have been used to study the roles that specific genes play in lipid metabolism in the liver (18–21), there are only a few published genome-wide scans for this disease phenotype (8, 22).
Our laboratory has carried out genetic studies in two inbred mouse strains that differ in diabetes susceptibility. The C57BL/6 (B6) strain, when made obese, is relatively resistant to diabetes, whereas the BTBR strain develops severe diabetes when made obese due to a failure to expand β-cell mass and to a defect in insulin secretion (14, 23, 24). In addition to diabetes, the B6 and BTBR strains differ in their susceptibility to develop hepatic steatosis (25). Based on the differences in liver TG content between the obese B6 and BTBR strains, we sought to map the genetic loci contributing to these differences in a genome-wide scan of an F2 cross of ∼550 animals. We mapped a quantitative trait locus (QTL) for NAFLD to chromosome 17. We investigated the genes on chromosome 17 from 0 to 60 Mb and identified Tsc2, a candidate gene that regulates NAFLD.
MATERIALS AND METHODS
Animals and breeding strategy
The C57BL/6 (B6) and BTBR T+tf (BTBR) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred at the University of Wisconsin-Madison as parental lean animals. The Lepob mutation was introgressed into all strains using Lepob/+ mice as breeders to generate Lepob/ob mice. Tsc2+/− mice in B6 background were gifts from Dr. Avtar Roopra. Wild-type littermates were used as control. All mice were maintained at the Department of Biochemistry, University of Wisconsin-Madison animal care facility on a 12 h dark-light cycle (6 PM to 6 AM). The mice were fed Purina Formulab Chow 5008 and water ad libitum.
Genetic analysis
550 F2ob/ob mice were genotyped with the 5K GeneChip (Affymetrix). Traits were mapped across the genome using R/qtl (26). The significance of high logarithm of the odds (LOD) peaks was evaluated using 1,000 permutations (27). Support intervals are genomic regions within 1.5 units of the peak per chromosome (28).
Fasting and refeeding study
Animals were separated into two groups: fasted for 24 h or fasted for 24 h followed by refeeding for 8 h. Liver tissues were collected after euthanization to harvest RNA for quantitative PCR, protein lysates for immune-blotting, and nuclei for detecting nuclear Srebp1c.
Cell culture and retroviral infection
Tsc2−/− mouse embryonic fibroblasts (MEFs) (a gift from Dr. Brendan Manning) were cultured in DMEM containing 25 mM glucose, 100 units/ml of antibiotic-antimycotic, and 10% FBS. The rat β-cell line Ins1 (832/13, a gift from Dr. Chris Newgard, Duke University) was cultured in RPMI 1640 medium containing 11 mM glucose supplemented with 10% heat-inactivated FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 100 units/ml of antibiotic-antimycotic, and 50 mM β-mercaptoethanol. Exogenous Tsc2 were introduced by MMLV-based retroviral infection. A total of 1 × 105 Ins1 cells expressing GFP, B6, or BTBR alleles of Tsc2 were plated into 60 mm dishes. Cells numbers were counted in a TC10 automated cell counter (BIO-RAD) 4, 10, and 14 days after plating to measure the growth rate.
Isolation and quantification of total RNA
RNA from liver and cell lines was extracted using the QIAGEN RNeasy Plus Kit. After extraction, RNA was used for cDNA synthesis (Applied Biosystems). The mRNA abundance was determined by quantitative PCR using FastStart SYBR Green (Roche), and gene expression was represented by comparative delta CT method.
Triglyceride quantification
Triglyceride from 20 to ∼40 mg of liver tissue was extracted by chloroform and quantified by a Wako L-Type TG H measurement kit (Wako Chemicals). The amount of TG was normalized to the amount of protein in the sample and presented as μg TG/mg protein.
Immunoprecipitation and immunoblotting
Cells were lysed in 1% Triton-X lysis buffer with a protease and phosphatase inhibitor cocktail (Roche). Immunoprecipitation was performed as described in the Cell Signaling Technology immunoprecipitaion protocol with Tsc1 or Tsc2 antibody. Proteins were detected by immunoblotting following the Cell Signaling Technology immunoblotting protocol with desired antibodies. Tsc1, Tsc2, phospho-S6, total Akt, phospho-Akt, and Actin antibodies were purchased from Cell Signaling Technology. Srebp1 antibody was purchased from BD Pharmingen. CREB antibody was purchased from Santa Cruz Biotechnology.
De novo lipogenesis assay
AML12 cells expressing GFP or different alleles of Tsc2 were starved from insulin for 8 h, followed by a 24-h insulin treatment. Three hours before harvest, cells were incubated in the media with 0.2 mCi/ml 3H2O. Cells were harvested in 2.5 M KOH/EtOH solution and saponified at 85°C for 2 h. Saponifiable lipids were extracted with hexane and separated on TLC plates. The amount of 3H incorporation into fatty acid pool was counted as a measurement of de novo lipogenesis.
Measurement of protein turnover
Tsc2−/− MEFs expressing B6 or BTBR alleles of Tsc2 were treated with 100 μg/ml cycloheximide. Cells were harvested in 1% Triton-X lysis buffer as described above 2, 4, and 8 h after treatment. Protein concentration was measured with a Pierce BCA protein assay kit (Thermo Scientific). From each sample, 25 μg of lysate was loaded to perform immunoblotting to determine Tsc2 protein level.
Statistical analysis
Data were expressed as means ± standard error of means. The statistical comparisons were made using Student's t-test at P < 0.05.
RESULTS
Strain effect of liver TG content
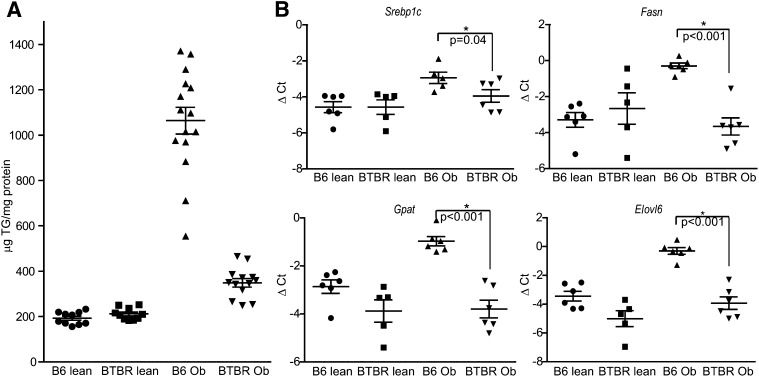
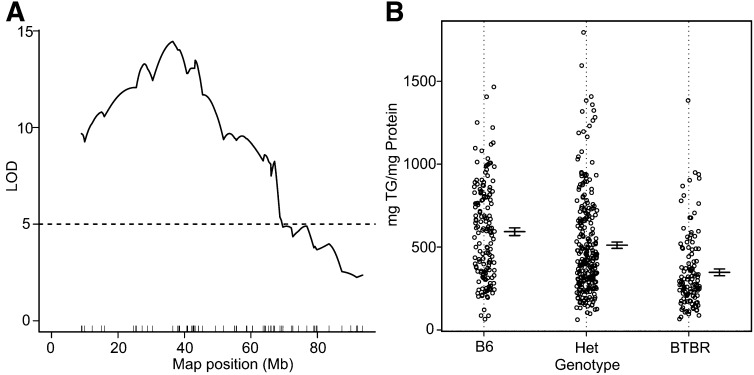
We measured liver TG in obese (Leptinob/ob) B6 and BTBR mice at 10 weeks of age. The B6 livers contain about three times more TG than livers from obese BTBR mice (Fig. 1A). Obese B6 mice also have higher transcript levels for lipogenic genes, including Srebp1c, Fasn, Gpat, and Elovl6 (Fig. 1B), as measured by real-time PCR, indicating that a potential increase in de novo lipogenesis contributes to the higher amount of TG in B6 livers compared with BTBR livers. Based on this large strain difference in liver TG content, we mapped the gene loci contributing to these differences in an F2 cross (sample size ∼550) and detected strong linkage on chromosome 17 with a LOD peak of 15 at ∼42 Mb (Fig. 2A). F2 mice with the B6/B6 genotype at the peak marker on chromosome 17 had an ∼2-fold higher liver TG content than mice with the BTBR/BTBR genotype (P < 0.0001; Fig. 2B). The heterozygotes had an intermediate phenotype.
Fig. 1.
Strain effect of liver TG content. A: TG content in liver tissue was measured in B6 and BTBR mice at 10 weeks of age. The animals were fasted for 4 h before being euthanized. B6 mice had about three times the amount of liver TG as the BTBR mice when they were made genetically obese with Lep ob/ob mutation (n > 10 in each group; P < 0.0001). B: Mouse cDNA was made from the livers of previously described animals. The levels of lipogenic gene transcripts were measured by real-time PCR and presented as ΔCt after normalized to β actin as an internal control (n = 5 in each group).
Fig. 2.
Liver TG QTL on chromosome 17. A: LOD profile for liver triglyceride on chromosome 17; peak LOD = 15 at ∼42.5 Mb, from a F2 cross of 550 mice. B: Liver TG content according to genotype at the coding SNP of LOD peak marker across the F2 population.
Differential Srebp1c and downstream lipogenic gene expression in WT and Tsc2+/− mouse livers
Although positional cloning ultimately identifies specific genes that are causal for a physiological phenotype, this process is slow due to the low resolution inherent in an F2 intercross and the time required to refine QTL intervals by generating congenic mice. We therefore investigated candidate genes on chromosome 17. We focused our attention on the genes reside within 10 to 50 Mb (LOD score greater than 8) with nonsynonymous coding SNPs between B6 and BTBR and identified a promising gene: Tsc2. Tsc2 encodes Tuberin, which functions as a GTPase-activating protein (GAP) toward Rheb to regulate the GTP-binding status of Rheb, which serves as an inhibitor of mammalian target of rapamycin complex (mTORC)1 (29–33).
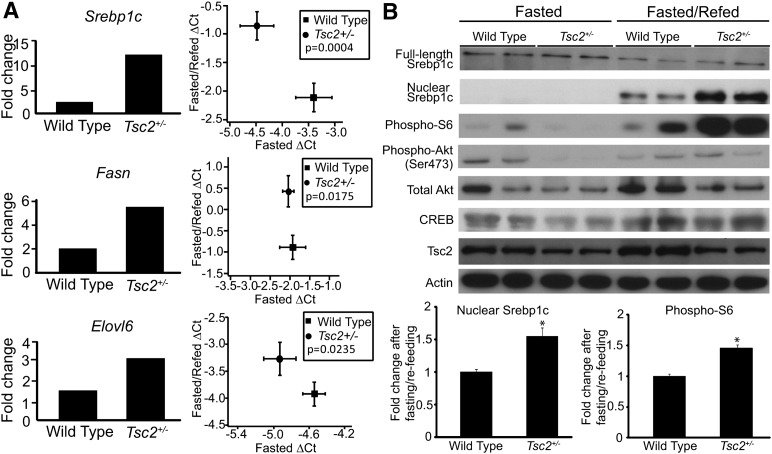
The role of mTORC1 in regulating metabolism led us to hypothesize that Tsc2 is a gene underlying the chromosome 17 QTL that regulates the accumulation of TG in the liver. Tsc2-null mice are embryonic lethal. However, Tsc2+/− animals are viable (34). To understand the role of Tsc2 in the regulation of insulin-dependent lipogenesis in vivo, we obtained Tsc2+/− mice in the B6 background and performed fasting/refeeding studies. We observed a 12.3-fold increase of Srebp1c expression after refeeding in the livers of Tsc2+/− mice, whereas their wild-type littermates showed only a 2.4-fold induction. The transcript levels of downstream lipogenic genes (Fasn and Elovl6) follow the same trend (Fig. 3A). The higher level of phosphorylated S6 ribosomal protein in Tsc2+/− mice indicates that it has a higher level of mTORC1 activation compared with wild-type, independent of the phosphorylation status of upstream Akt (Fig. 3B). We also measured the protein level of processed nuclear Srebp1c and found that Tsc2+/− mice have about 50% more nuclear Srebp1c protein after refeeding compared with wild-type littermates (Fig. 3B).
Fig. 3.
Lipogenic gene expression in wild-type and Tsc2+/− mouse livers. Wild-type and Tsc2+/− mice were fasted for 24 h or fasted for 24 h and refed for 8 h before being euthanized (n > 6 in each group). A: The amount of Srebp1c, Fasn, and Elovl6 mRNA was measured by real time-PCR. The difference in the induction of gene expression after fasting and refeeding procedure between wild-type and Tsc2+/− animals is presented in two ways: (1) mRNA levels from fasted animals were set to be one, and mRNA levels from fasted and refed animals were compared with the fasted group to obtain the fold difference in expression; and (2) the ΔCt value of mRNA level from fasted animals was plotted against the ΔCt value from fasted and refed animals. B: Western blotting was performed on the liver tissues described in (A) to detect Srebp1c and phosphorylated S6 ribosomal protein, phosphorylated Akt (Ser473), and total Akt. The Akt levels in three more WT mice were measured and compared with the first mouse (Supplementary Figure I). The levels of phospho-S6 ribosomal protein were measured in 25 more animals on four separate blots (Supplementary Figure II). The intensity of nuclear Srebp1c and phospho-S6 blots (including the blots not shown in the figure) was measured, and the protein level after fasting and refeeding was compared with the protein level in fasted state. The fold change is expressed as a bar graph. CREB and actin blots serve as loading controls.
Tsc2 allelic differences in regulation of lipogenic gene expression and de novo lipogenesis
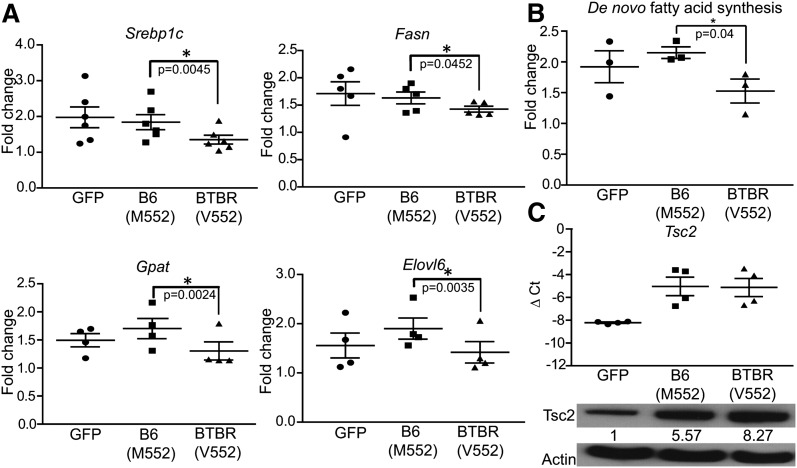
We sequenced the B6 and BTBR alleles of the Tsc2 gene and found a nonsynonymous coding SNP (M552V) located at an unexplored region of the protein. We hypothesized that this coding difference causes the B6 form of Tsc2 to inhibit mTORC1 less efficiently than the BTBR form and therefore causes an increase in lipogenic gene expression in the liver through greater mTORC1 activation. We tested this hypothesis by introducing either the B6 (M552) or the BTBR (V552) forms of Tsc2 into AML12 (a mouse hepatoma cell line) through retroviral infection. The insulin-stimulated expression of lipogenic genes (Srebp1c, Fasn, Gpat, and Elovl6) was greater in cells expressing the B6 (M552) allele of Tsc2 than in cells expressing the BTBR allele (V552) (Fig. 4A). In these experiments, both allelic forms of ectopic Tsc2 were expressed at similar mRNA levels (Fig. 4C).
Fig. 4.
Allelic difference in regulation of lipogenesis. AML12 cells expressing GFP, B6 (M552), or BTBR (V552) alleles of Tsc2 were treated with insulin after insulin starve for 16 h. A: The expression levels of lipogenic genes were measured by real-time PCR and are presented as fold change after insulin induction. B: The amount of tritium incorporation into fatty acids from 3H2O was used as a measurement of de novo lipogenesis after insulin treatment. C: The amount of Tsc2 transcript was measured by real-time PCR in each cell population to confirm that they were expressed to comparable levels. Western blotting was performed on these cells, and the level of Tsc2 was quantified. The numbers underneath Tsc2 blot represent the relative level after correcting for Actin loading control.
De novo fatty acid synthesis in these AML12 cells was measured by treating the cells with 3H2O for 2 h before harvest and measuring the amount of incorporation of 3H into fatty acids. Cells expressing the B6 (M552) allele of Tsc2 have a higher level of de novo lipogenesis than those expressing the BTBR (V552) allele (Fig. 4B). This allelic difference in regulation of lipogenesis provides one mechanism for the difference in hepatic TG accumulation between the two mouse strains.
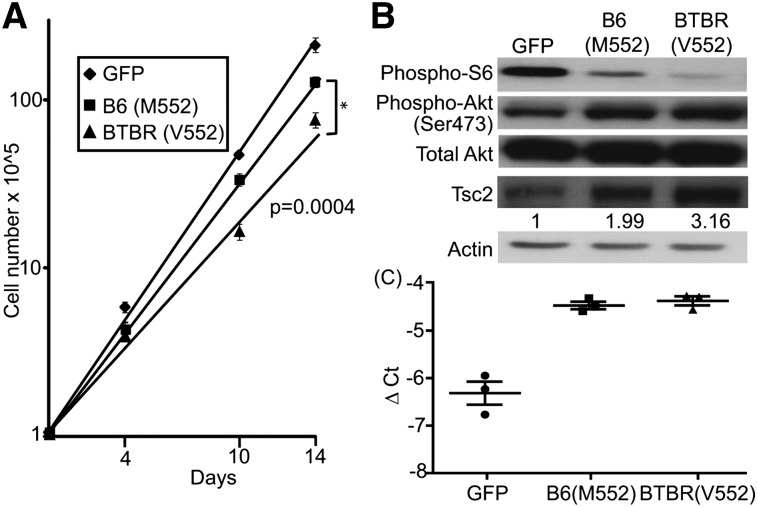
Allelic difference in regulation of β-cell proliferation
Previous work has established Tsc2 as a key regulator of β-cell proliferation (35). Given the fact that BTBR mice are severely diabetic when made obese due to the failure to expand β-cell mass, we hypothesized that the genetic variation of Tsc2 causes insufficient β-cell proliferation through the mTORC1 pathway and contributes to the diabetic phenotype in BTBR animals. To test this hypothesis, we introduced GFP, the B6 (M552), or the BTBR (V552) alleles of Tsc2 into the β-cell line, Ins1 cells, via retroviral infection. Cell proliferation was inhibited by the introduction of either allele of Tsc2. However, the BTBR (M552) Tsc2 allele inhibited cell growth to a greater extent than the B6 (V552) allele (doubling times 38.5 vs. 29.7 h; P = 0.0004; Fig. 5A), whereas the ectopic Tsc2 alleles were expressed at comparable mRNA levels (Fig. 5C). The Tsc2-mediated inhibition of mTORC1 activity was independent of Akt activation, as judged by the phosphorylation status of Akt and S6 ribosomal protein (Fig. 5B). These data confirm prior studies showing that Tsc2 functions as an inhibitor of β-cell proliferation (35, 36) and show that the coding variation of Tsc2 alters the potency of this inhibition, which may contribute to the difference in the proliferation capacity of β cells in B6 versus BTBR islets. We suggest that the polymorphism in the Tsc2 gene affects two distinct functions in two different tissues: TG accumulation in the liver and β-cell proliferation in the pancreas (35).
Fig. 5.
Allelic difference in regulation of β-cell proliferation. GFP, B6 (M552), or BTBR (V552) alleles of Tsc2 were introduced into Ins1 cells via retroviral infection. A: A total of 105 infected cells were plated into 6-well plates 2 days after infection. Cell growth was monitored by counting cell numbers over time. B: The amount of phosphorylation status of S6 ribosomal protein, Akt, and Tsc2 was detected by Western blotting. The numbers underneath the Tsc2 blot represent the relative protein levels. C: The amount of Tsc2 transcript was measured by real-time PCR in each cell population to confirm that they were expressed to comparable levels.
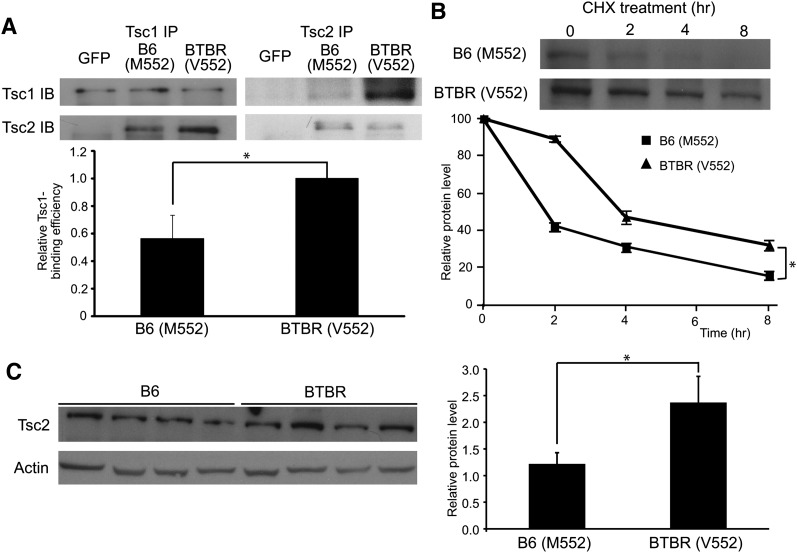
Allelic difference in binding Tsc1 and protein turnover
Binding of Tsc1 to Tsc2 prevents the proteasomal degradation of Tsc2 (37–42). Although the coding variation of Tsc2 between our two mouse strains is located in an unexplored region of the protein, it is near the Tsc1-binding domain (43). We hypothesized that the polymorphism of Tsc2 affects its binding to Tsc1. To test this hypothesis, the B6 (M552) or BTBR (V552) forms of Tsc2 were introduced into Tsc2−/− MEFs, and the Tsc complexes were immunoprecipitated and immunoblotted with Tsc1 or Tsc2 antibodies. When immunoprecipitated with Tsc1, more of the BTBR (V552) form of Tsc2 is coimmunoprecipitated than the B6 (M552) form (Fig. 6A, left panel). In contrast, when the Tsc2 antibody was used to perform the immunoprecipitation, more Tsc1 was pulled down with the BTBR (V552) form than with the B6 (M552) form of Tsc2 (Fig. 6A, right panel). The amounts of Tsc1 and Tsc2 in the complexes were quantified, and the binding efficiencies of Tsc1 and Tsc2 were determined by measuring the amount of binding partner being immunoprecipitated (Fig. 6A). The B6 (M552) form of Tsc2 binds to Tsc1 38% less efficiently than the BTBR (V552) form of Tsc1 on a per-molecule basis.
Fig. 6.
A: Allelic difference in binding of Tsc2 to Tsc1 and turnover of Tsc2 protein B6 (M552) or BTBR (V552) forms of Tsc2 were introduced into Tsc2−/− MEFs. TSC complexes were immunoprecipitated and immunoblotted with Tsc1 or Tsc2 antibody. The amounts of Tsc1 and Tsc2 in the complexes were quantified, and the binding efficiency of Tsc1 and Tsc2 is shown in a bar graph. The amount of binding of the BTBR (V552) form of Tsc2 to Tsc1 is set to one (n > 4; P = 0.013). B: Tsc2-expressing cells described above were treated with 100 μg/ml cycloheximide. Cells were harvested 2, 4, and 8 h after treatment. Western blotting was performed to measure the degradation of Tsc2 over time. The quantified Tsc2 signal from three independent experiments is plotted over time (n > 4; P < 0.001). C: Tsc2 protein from the parental mouse livers was detected by Western blotting (four mice from each strain). The levels of Tsc2 were quantified and expressed as a bar graph after correcting for Actin loading control (P = 0.03).
We asked if the allelic difference in the affinity of Tsc2 for Tsc1 is correlated with a different half-life of the Tsc2 protein. The MEFs described above were treated with cycloheximide to arrest protein synthesis. Protein lysates were harvested at 2, 4, and 8 h after treatment and subjected to immunoblotting to monitor protein degradation. The results indicate that the B6 allelic form of Tsc2 is degraded more rapidly than the BTBR form (t1/2 = 2.3 vs. 3.7 h; P < 0.001). The levels of B6 and BTBR Tsc2 proteins in AML12, Ins1 cells, and the parental strains support these results (Fig. 4C, 5B, and 6C). This difference in turnover of Tsc2 provides a mechanism by which the coding variation between the B6 and BTBR strains causes differential mTORC1 activation, lipogenesis in the liver, and β-cell proliferation in the pancreas (35).
DISCUSSION
Tsc2 is physically located on chromosome 17 at 24.7 Mb. It encodes Tuberin and was first identified as a tumor suppressor gene that causes Tuberous Sclerosis Complex (TSC) when mutated (44, 45). TSC2 requires heterodimerization with TSC1 to stabilize the protein by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase (37). TSC2 functions as a GAP toward Rheb to stimulate the internal GTPase activity of Rheb (29–31, 37). GTP-bound Rheb stimulates the phosphorylation and activation of mTORC (32, 33). mTORC1 signaling is involved in many physiological processes, including cell growth, proliferation, and metabolism (46). Because the TSC1/TSC2 complex is a target of a variety of different signaling molecules, including PI3K, Akt, AMPK, ERK, and IKKβ, to regulate its activity via phosphorylation, it serves as a hub to regulate the physiological outcomes downstream of the mTORC1 pathway (46, 47).
mTORC1 was recently found to be involved in several different aspects of metabolism (48–51). It is required for the stimulation of lipogenesis in rat liver in an insulin-dependent manner. Thus, the insulin induction of lipogenic genes (Srebp1c, Fasn, and Scd1) can be inhibited by the mTORC1 inhibitor rapamycin (50). Tsc2−/− MEFs have a rapamycin-sensitive increase in de novo lipogenesis compared with their wild-type counterparts (49). mTORC1 promotes the function of SRE-binding protein by controlling the phosphorylation and localization of Lipin1 (52). These observations led us to hypothesize that Tsc2 is a gene underlying the chromosome 17 QTL that regulates the accumulation of TG in the liver.
We performed fasting and refeeding studies and observed greater activation of Srebp1c and induction of lipogenic genes (Fasn and Elovl6) after refeeding in the livers of Tsc2+/− mice vs. their wild-type littermates. This is the first evidence that Tsc2 plays a direct role in the regulation of lipogenesis in vivo.
We sequenced the B6 and BTBR alleles of the Tsc2 gene and found a nonsynonymous coding SNP (M552V), located at an unexplored region of the protein. We show that this coding difference causes the B6 form of Tsc2 to inhibit mTORC1 less efficiently than the BTBR form and therefore causes increased de novo lipogenesis in the liver.
In the presence of insulin, PI3K is recruited to phospho-IRS1 and activated, resulting in the phosphorylation and activation of Akt. Activated Akt is able to phosphorylate and inactivate Tsc2 (53), which allows Rheb to remain in the GTP-bound form and activate mTORC1 (54), leading to the regulation of downstream pathways, including lipid metabolism. However, surprising results from a recent study show that Akt is able to stimulate lipogenesis through the suppression of Insig2a in an mTORC1-independent manner. This pathway seems to be more pronounced when the regulation of mTORC1 by the TSC complex is absent (55). Other than the canonical Akt-mTORC1 pathway, Akt2 was also recently reported to be an independent signaling molecule regulating postprandial hepatic lipid metabolism (56).
In this study, we have focused on Tsc2’s function in regulating de novo lipogenesis through the mTORC1 pathway. However, decreasing β-oxidation in the liver, increasing free fatty acid uptake from peripheral blood, and decreasing VLDL secretion into circulation can be potential pathways that also lead to TG accumulation in the liver. Preliminary data from our lab showing that Cpt1 is expressed at a higher level in BTBR livers than in B6 (data not shown) indicates that de novo lipogenesis might not be the only pathway that accounts for the strain difference in hepatic TG accumulation.
In our model, we only observed the strain difference in steatosis susceptibility when the animals were made genetically obese. Leptin-deficiency serves as a challenge for the steatosis to develop by providing excess energy from over-eating. It is also possible that the lack of leptin signaling decreases AMPK activity, which has been shown to be an inhibitor of mTORC in the liver (57, 58).
In addition to de novo lipogenesis, Tsc2 has been shown to regulate β-cell proliferation through the mTORC1 pathway. Tissue-specific disruption of Tsc2 induces β-cell proliferation and improved glucose tolerance in a mTORC1-dependent manner (35). Knockdown of Tsc2 increased the number of cells in S/G2-M phase in β-cell lines and was reversed by rapamycin treatment (36). Given that obese BTBR mice develop severe dia betes due to the failure to expand β-cell mass (23), we hypothesized that the genetic variation of Tsc2 causes insufficient β-cell proliferation through the mTORC1 pathway, contributing in part to the diabetic phenotype in BTBR mice. We found that when introduced into Ins1 cells, an insulinoma cell line, the BTBR allele of Tsc2 inhibited cell growth more effectively than the B6 allele. This difference between B6 and BTBR Tsc2 may reflect differential binding to Tsc1, which affects the stability of the Tsc2 protein.
The inhibition of mTORC1 activation by Tsc2 can be regulated on many different levels. The interaction of Tsc2 with Tsc1 affects the half-life of Tsc2. The phosphorylation status of Tsc2 affects its GAP enzymatic activity. We have shown that the coding SNP in Tsc2 affects the interaction between Tsc2 and Tsc1 and results in different half-lives of the Tsc2 protein. The polymorphism between the B6 and BTBR alleles of Tsc2 is located close to its known phosphorylation sites (S540 and S664). Thus, it is possible that the polymorphism affects the phosphorylation status of Tsc2.
NAFLD is a general term used to describe a range of liver disorders, all characterized by the accumulation of excess TG in the liver. In some cases, hepatic steatosis progresses to nonalcoholic steatohepatitis and the devel opment of hepatocellular carcinoma (59–61). Genetic predisposition affects the susceptibility to NAFLD hepatocellular carcinoma (59). Given that Tsc2+/− mice spontaneously develop liver tumors without steatosis (62), the dual function of Tsc2 in regulating lipogenesis and cell proliferation through the mTORC1 pathway leads us to postulate that loss of function of Tsc2 can contribute to hepatic steatosis and hepatocellular carcinoma.
The obese B6 mice are diabetes resistant compared with the obese BTBR mice. Our previous studies showed that the obese B6 mice exhibit greater β-cell mass due to an induction of β-cell proliferation. In contrast, this obesity induction of β-cell mass does not occur in BTBR mice (23). It is likely that multiple genetic loci coordinately regulate β-cell proliferation. The present study suggests that the genetic variation within a single gene, Tsc2, can simultaneously affect hepatic TG and influence β-cell replication.
Supplementary Material
Acknowledgments
The authors thank Merck for the DNA extraction and genotyping of the F2 mice, Dr. Avtar Roopra for providing the Tsc2+/− animals, and Dr. Brendan Manning for providing us Tsc2−/− MEFs and for discussion and comments regarding this study.
Footnotes
Abbreviations:
- GAP
- GTPase-activating protein
- LOD
- logarithm of the odds
- MEF
- mouse embryonic fibroblast
- mTORC
- mammalian target of rapamycin complex
- NAFLD
- nonalchoholic fatty liver disease
- QTL
- quantitative trait locus
- SNP
- single-nucleotide polymorphism
- TG
- triglyceride
- TSC
- Tuberous Sclerosis Complex
This work was supported by the NIH: DK066369, DK058037, and American Diabetes Association Mentor-Based Fellowship 1-11-MN-03.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
References
- 1.Schwimmer J. B., Deutsch R., Kahen T., Lavine J. E., Stanley C., Behling C. 2006. Prevalence of fatty liver in children and adolescents. Pediatrics. 118: 1388–1393 [DOI] [PubMed] [Google Scholar]
- 2.Adams L. A., Angulo P. 2005. Recent concepts in non- alcoholic fatty liver disease. Diabet. Med. 22: 1129–1133 [DOI] [PubMed] [Google Scholar]
- 3.Adams L. A., Angulo P., Lindor K. D. 2005. Nonalcoholic fatty liver disease. Can. Med. Assoc. J. 172: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams L. A., Lymp J. F., St Sauver J., Sanderson S. O., Lindor K. D., Feldstein A., Angulo P. 2005. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 129: 113–121 [DOI] [PubMed] [Google Scholar]
- 5.McCullough A. J. 2004. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin. Liver Dis. 8: 521–533 (viii.) [DOI] [PubMed] [Google Scholar]
- 6.Browning J. D., Szczepaniak L. S., Dobbins R., Nuremberg P., Horton J. D., Cohen J. C., Grundy S. M., Hobbs H. H. 2004. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 40: 1387–1395 [DOI] [PubMed] [Google Scholar]
- 7.Musso G., Gambino R., De Michieli F., Durazzo M., Pagano G., Cassader M. 2008. Adiponectin gene polymorphisms modulate acute adiponectin response to dietary fat: Possible pathogenetic role in NASH. Hepatology. 47: 1167–1177 [DOI] [PubMed] [Google Scholar]
- 8.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L. A., Boerwinkle E., Cohen J. C., Hobbs H. H. 2008. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40: 1461–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sookoian S., Castano G., Gemma C., Gianotti T. F., Pirola C. J. 2007. Common genetic variations in CLOCK transcription factor are associated with nonalcoholic fatty liver disease. World J. Gastroenterol. 13: 4242–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoneda M., Hotta K., Nozaki Y., Endo H., Uchiyama T., Mawatari H., Iida H., Kato S., Hosono K., Fujita K., et al. 2008. Association between PPARGC1A polymorphisms and the occurrence of nonalcoholic fatty liver disease (NAFLD). BMC Gastro enterol. 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day C. P. 2006. Non-alcoholic fatty liver disease: current concepts and management strategies. Clin. Med. 6: 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilger A., Bennett L. M., Carabeo R. A., Chiaverotti T. A., Dvorak C., Liss K. M., Schadewald S. A., Pitot H. C., Drinkwater N. R. 2004. A potent modifier of liver cancer risk on distal mouse chromosome 1: linkage analysis and characterization of congenic lines. Genetics. 167: 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilger A., Sullivan R., Prunuske A. J., Clipson L., Drinkwater N. R., Dove W. F. 2008. Widespread hyperplasia induced by transgenic TGFalpha in ApcMin mice is associated with only regional effects on tumorigenesis. Carcinogenesis. 29: 1825–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clee S. M., Attie A. D. 2007. The genetic landscape of type 2 diabetes in mice. Endocr. Rev. 28: 48–83 (PubMed) [DOI] [PubMed] [Google Scholar]
- 15.Clee S. M., Yandell B. S., Schueler K. M., Rabaglia M. E., Richards O. C., Raines S. M., Kabara E. A., Klass D. M., Mui E. T., Stapleton D. S., et al. 2006. Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat. Genet. 38: 688–693 [DOI] [PubMed] [Google Scholar]
- 16.Kotb M., Fathey N., Aziz R., Rowe S., Williams R. W., Lu L. 2008. Unbiased forward genetics and systems biology approaches to understanding how gene-environment interactions work to predict susceptibility and outcomes of infections. Novartis Found. Symp. 293: 156–165; discussion 165–157, 181–153 [DOI] [PubMed] [Google Scholar]
- 17.Marquis J. F., Gros P. 2008. Genetic analysis of resistance to infections in mice: A/J meets C57BL/6J. Curr. Top. Microbiol. Immunol. 321: 27–57 [DOI] [PubMed] [Google Scholar]
- 18.Reddy J. K., Hashimoto T. 2001. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21: 193–230 [DOI] [PubMed] [Google Scholar]
- 19.Smith S. J., Cases S., Jensen D. R., Chen H. C., Sande E., Tow B., Sanan D. A., Raber J., Eckel R. H., Farese R. V., Jr 2000. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat. Genet. 25: 87–90 [DOI] [PubMed] [Google Scholar]
- 20.Tolwani R. J., Hamm D. A., Tian L., Sharer J. D., Vockley J., Rinaldo P., Matern D., Schoeb T. R., Wood P. A. 2005. Medium-chain acyl-CoA dehydrogenase deficiency in gene-targeted mice. PLoS Genet. 1: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X. X., Murray S. F., Pandey S. K., Booten S. L., Bao D., Song X. Z., Kelly S., Chen S., McKay R., Monia B. P., et al. 2005. Antisense oligonucleotide reduction of DGAT2 expression improves hepatic steatosis and hyperlipidemia in obese mice. Hepatology. 42: 362–371 [DOI] [PubMed] [Google Scholar]
- 22.Chalasani N., Guo X., Loomba R., Goodarzi M. O., Haritunians T., Kwon S., Cui J., Taylor K. D., Wilson L., Cummings O. W., et al. 2010. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology. 139: 1567–1576, e1561–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller M. P., Choi Y., Wang P., Davis D. B., Rabaglia M. E., Oler A. T., Stapleton D. S., Argmann C., Schueler K. L., Edwards S., et al. 2008. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 18: 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatnagar S., Oler A. T., Rabaglia M. E., Stapleton D. S., Schueler K. L., Truchan N. A., Worzella S. L., Stoehr J. P., Clee S. M., Yandell B. S., et al. 2011. Positional cloning of a type 2 diabetes quantitative trait locus; tomosyn-2, a negative regulator of insulin secretion. PLoS Genet. 7: e1002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan H., Rabaglia M. E., Stoehr J. P., Nadler S. T., Schueler K. L., Zou F., Yandell B. S., Attie A. D. 2003. Gene expression profiles of nondiabetic and diabetic obese mice suggest a role of hepatic lipogenic capacity in diabetes susceptibility. Diabetes. 52: 688–700 [DOI] [PubMed] [Google Scholar]
- 26.Broman K. W., Wu H., Sen S., Churchill G. A. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 19: 889–890 [DOI] [PubMed] [Google Scholar]
- 27.Churchill G. A., Doerge R. W. 1994. Empirical threshold values for quantitative trait mapping. Genetics. 138: 963–971 (PubMed) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manichaikul A., Dupuis J., Sen S., Broman K. W. 2006. Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus. Genetics. 174: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benvenuto G., Li S., Brown S. J., Braverman R., Vass W. C., Cheadle J. P., Halley D. J., Sampson J. R., Wienecke R., DeClue J. E. 2000. The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene. 19: 6306–6316 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A., Pan D. 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5: 578–581 [DOI] [PubMed] [Google Scholar]
- 31.Inoki K., Li Y., Xu T., Guan K. L. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17: 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tee A. R., Blenis J., Proud C. G. 2005. Analysis of mTOR signaling by the small G-proteins, Rheb and RhebL1. FEBS Lett. 579: 4763–4768 [DOI] [PubMed] [Google Scholar]
- 33.Tabancay A. P., Jr, Gau C. L., Machado I. M., Uhlmann E. J., Gutmann D. H., Guo L., Tamanoi F. 2003. Identification of dominant negative mutants of Rheb GTPase and their use to implicate the involvement of human Rheb in the activation of p70S6K. J. Biol. Chem. 278: 39921–39930 [DOI] [PubMed] [Google Scholar]
- 34.Onda H., Lueck A., Marks P. W., Warren H. B., Kwiatkowski D. J. 1999. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J. Clin. Invest. 104: 687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rachdi L., Balcazar N., Osorio-Duque F., Elghazi L., Weiss A., Gould A., Chang-Chen K. J., Gambello M. J., Bernal-Mizrachi E. 2008. Disruption of Tsc2 in pancreatic beta cells induces beta cell mass expansion and improved glucose tolerance in a TORC1-dependent manner. Proc. Natl. Acad. Sci. USA. 105: 9250–9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartolome A., Guillen C., Benito M. 2010. Role of the TSC1–TSC2 complex in the integration of insulin and glucose signaling involved in pancreatic beta-cell proliferation. Endocrinology. 151: 3084–3094 [DOI] [PubMed] [Google Scholar]
- 37.Chong-Kopera H., Inoki K., Li Y., Zhu T., Garcia-Gonzalo F. R., Rosa J. L., Guan K. L. 2006. TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J. Biol. Chem. 281: 8313–8316 [DOI] [PubMed] [Google Scholar]
- 38.Inoki K., Li Y., Zhu T., Wu J., Guan K. L. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4: 648–657 [DOI] [PubMed] [Google Scholar]
- 39.Kwiatkowski D. J. 2003. Rhebbing up mTOR: new insights on TSC1 and TSC2, and the pathogenesis of tuberous sclerosis. Cancer Biol. Ther. 2: 471–476 [DOI] [PubMed] [Google Scholar]
- 40.Kwiatkowski D. J. 2003. Tuberous sclerosis: from tubers to mTOR. Ann. Hum. Genet. 67: 87–96 (PubMed) [DOI] [PubMed] [Google Scholar]
- 41.Nellist M., Sancak O., Goedbloed M. A., Rohe C., van Netten D., Mayer K., Tucker-Williams A., van den Ouweland A. M., Halley D. J. 2005. Distinct effects of single amino-acid changes to tuberin on the function of the tuberin-hamartin complex. Eur. J. Hum. Genet. 13: 59–68 [DOI] [PubMed] [Google Scholar]
- 42.Nellist M., Sancak O., Goedbloed M. A., van Veghel-Plandsoen M., Maat-Kievit A., Lindhout D., Eussen B. H., de Klein A., Halley D. J., van den Ouweland A. M. 2005. Large deletion at the TSC1 locus in a family with tuberous sclerosis complex. Genet. Test. 9: 226–230 [DOI] [PubMed] [Google Scholar]
- 43.Hodges A. K., Li S., Maynard J., Parry L., Braverman R., Cheadle J. P., DeClue J. E., Sampson J. R. 2001. Pathological mutations in TSC1 and TSC2 disrupt the interaction between hamartin and tuberin. Hum. Mol. Genet. 10: 2899–2905 [DOI] [PubMed] [Google Scholar]
- 44.Kandt R. S., Haines J. L., Smith M., Northrup H., Gardner R. J., Short M. P., Dumars K., Roach E. S., Steingold S., Wall S., et al. 1992. Linkage of an important gene locus for tuberous sclerosis to a chromosome 16 marker for polycystic kidney disease. Nat. Genet. 2: 37–41 [DOI] [PubMed] [Google Scholar]
- 45.van Slegtenhorst M., de Hoogt R., Hermans C., Nellist M., Janssen B., Verhoef S., Lindhout D., van den Ouweland A., Halley D., Young J., et al. 1997. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 277: 805–808 [DOI] [PubMed] [Google Scholar]
- 46.Huang J., Manning B. D. 2008. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 412: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoki K., Corradetti M. N., Guan K. L. 2005. Dysregulation of the TSC-mTOR pathway in human disease. Nat. Genet. 37: 19–24 [DOI] [PubMed] [Google Scholar]
- 48.Zhang H. H., Huang J., Duvel K., Boback B., Wu S., Squillace R. M., Wu C. L., Manning B. D. 2009. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS ONE. 4: e6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duvel K., Yecies J. L., Menon S., Raman P., Lipovsky A. I., Souza A. L., Triantafellow E., Ma Q., Gorski R., Cleaver S., et al. 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 39: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S., Brown M. S., Goldstein J. L. 2010. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. USA. 107: 3441–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakrabarti P., English T., Shi J., Smas C. M., Kandror K. V. 2010. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 59: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., Balderas E., Guertin D. A., Madden K. L., Carpenter A. E., Finck B. N., et al. 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 146: 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manning B. D. 2004. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J. Cell Biol. 167: 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell. 25: 903–915 [DOI] [PubMed] [Google Scholar]
- 55.Yecies J. L., Zhang H. H., Menon S., Liu S., Yecies D., Lipovsky A. I., Gorgun C., Kwiatkowski D. J., Hotamisligil G. S., Lee C. H., et al. 2011. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 14: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan M., Leavens K. F., Saleh D., Easton R. M., Guertin D. A., Peterson T. R., Kaestner K. H., Sabatini D. M., Birnbaum M. J. 2011. Postprandial hepatic lipid metabolism requires signaling through Akt2 independent of the transcription factors FoxA2, FoxO1, and SREBP1c. Cell Metab. 14: 516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reiter A. K., Bolster D. R., Crozier S. J., Kimball S. R., Jefferson L. S. 2005. Repression of protein synthesis and mTOR signaling in rat liver mediated by the AMPK activator aminoimidazole carboxamide ribonucleoside. Am. J. Physiol. Endocrinol. Metab. 288: E980–E988 [DOI] [PubMed] [Google Scholar]
- 58.Reiter A. K., Bolster D. R., Crozier S. J., Kimball S. R., Jefferson L. S. 2008. AMPK represses TOP mRNA translation but not global protein synthesis in liver. Biochem. Biophys. Res. Commun. 374: 345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hill-Baskin A. E., Markiewski M. M., Buchner D. A., Shao H., DeSantis D., Hsiao G., Subramaniam S., Berger N. A., Croniger C., Lambris J. D., et al. 2009. Diet-induced hepatocellular carcinoma in genetically predisposed mice. Hum. Mol. Genet. 18: 2975–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang S., Lin H. Z., Hwang J., Chacko V. P., Diehl A. M. 2001. Hepatic hyperplasia in noncirrhotic fatty livers: is obesity-related hepatic steatosis a premalignant condition? Cancer Res. 61: 5016–5023 [PubMed] [Google Scholar]
- 61.Bugianesi E. 2007. Non-alcoholic steatohepatitis and cancer. Clin. Liver Dis. 11: 191–207 (x–xi.) [DOI] [PubMed] [Google Scholar]
- 62.Finlay G. A., Malhowski A. J., Polizzi K., Malinowska-Kolodziej I., Kwiatkowski D. J. 2009. Renal and liver tumors in Tsc2(+/−) mice, a model of tuberous sclerosis complex, do not respond to treatment with atorvastatin, a 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor. Mol. Cancer Ther. 8: 1799–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.