Abstract
We have previously shown that elevated expression of mitotic kinase aurora kinase A (AURKA) in cancer cells promotes the development of metastatic phenotypes and is associated clinically with adverse prognosis. Here, we first revealed a clinically positive correlation between AURKA and autophagy-associated protein SQSTM1 in breast cancer and further demonstrated that AURKA regulated SQSTM1 through autophagy. Indeed, depletion by siRNA or chemical inhibition of AURKA by the small molecule VX-680 increased both the level of microtubule-associated protein 1 light chain 3-II (LC3-II) and the number of autophagosomes, along with decreased SQSTM1. Conversely, overexpression of AURKA inhibited autophagy, as assessed by decreased LC3-II and increased SQSTM1 either upon nutrient deprivation or normal conditions. In addition, phosphorylated forms of both RPS6KB1 and mechanistic target of rapamycin (MTOR) were elevated by overexpression of AURKA whereas they were suppressed by depletion or inhibition of AURKA. Moreover, inhibition of MTOR by PP242, an inhibitor of MTOR complex1/2, abrogated the changes in both LC3-II and SQSTM1 in AURKA-overexpressing BT-549 cells, suggesting that AURKA-suppressed autophagy might be associated with MTOR activation. Lastly, repression of autophagy by depletion of either LC3 or ATG5, sensitized breast cancer cells to VX-680-induced apoptosis. Similar findings were observed in cells treated with the autophagy inhibitors chloroquine (CQ) and bafilomycin A1 (BAF). Our data thus revealed a novel role of AURKA as a negative regulator of autophagy, showing that AURKA inhibition induced autophagy, which may represent a novel mechanism of drug resistance in apoptosis-aimed therapy for breast cancer.
Keywords: AURKA, SQSTM1, autophagy, breast cancer, MTOR, apoptosis
Introduction
The aurora serine/threonine kinase family, including aurora kinases A, B, and C, are key players in ensuing genetic stability in cell division.1 Aurora kinase A (AURKA) is essential in proper timing of mitotic entry and formation of bipolar spindles.2,3 Ectopic overexpression of AURKA transforms NIH3T3 cells, and these transformed cells can give rise to tumors when implanted in nude mice.4 Indeed, overexpression of AURKA has been reported in various cancer types, such as laryngeal, ovarian, breast, colorectal and gastric cancer.5 Our recent studies indicated that overexpression of AURKA is involved in laryngeal, nasopharyngeal and breast cancer cell metastasis.6-8 Moreover, high AURKA expression induces chemoresistance in breast cancer cells and ovarian cancer cells.9,10 Our previous studies suggest that targeting AURKA may be of therapeutic use, leading to apoptosis in acute myeloid leukemia and tongue squamous cell carcinoma.11,12 Therefore, AURKA is considered a promising molecular target for cancer therapy.
Autophagy, an evolutionarily-conserved degradative pathway, is involved in the turnover of long-lived proteins, cytoplasm and intracellular organelles.13 The role of autophagy in tumorgenesis is complex and highly debated: Some reports suggest that this process can function as a tumor-suppression mechanism,14,15 while other data indicates that tumors may require autophagy to survive under nutrient-limited and low-oxygen conditions.16,17 Autophagy also has a complex relationship with apoptotic cell death in tumor cell lines. Several reports have shown that autophagy enhances caspase-dependent and -independent cell death.18,19 In contrast, autophagy also plays a key role in supporting cell survival against apoptosis.18,19 These findings have shed light from different directions on the role of autophagy in cancer and on the potential of manipulating autophagy as a novel therapeutic strategy in cancer.
Apoptosis-associated proteins have a complex relationship with autophagy.20-22 For example, antiapoptotic proteins such as BCL2, BCL2L2 and MCL1 can block autophagy in cancer.23,24 However, proapoptotic proteins such as BAD, PMAIP1 and BBC3 stimulate autophagy.25,26 Overexpression of AURKA can inhibit apoptosis in human cancer.8,11 Therefore, we investigated the relationship between AURKA with autophagy in breast cancer. In the present study, SQSTM1 was used as a marker for autophagy in breast cancer cells. AURKA was found to positively correlate with SQSTM1. Moreover, we provided evidence that AURKA regulated autophagy associated with the MTOR signaling pathway. Furthermore, we explored the relationship between cell death and autophagy induced by targeting AURKA. We found that enhancement of cell death by targeting AURKA, was significantly increased by inhibition of autophagy. These results suggested that autophagy, induced by targeting AURKA, revealed a major mechanism for resistance to AURKA inhibitors in the treatment of breast cancer.
Results
AURKA correlated with autophagy-associated protein SQSTM1 expression in breast cancer
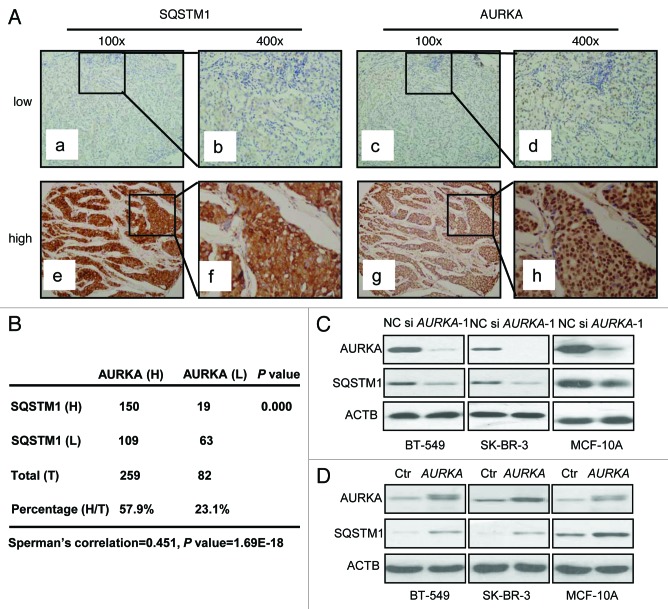
A total of 341 breast cancer tissue specimens were included in this study. Immunohistochemical staining was used to analyze AURKA and SQSTM1 expression in breast carcinoma specimens. Both AURKA and SQSTM1 immunoreactivity were mainly detected in cytoplasm and occasionally in nucleus (Fig. 1A). Among the 341 specimens, high expression of AURKA was detected in 76% of cases (n = 259), and high SQSTM1 expression was found in 49.6% cases (n = 169) (Fig. 1B). In addition, among the 259 high AURKA-expressing specimens, high SQSTM1 expression was found in 57.9% cases (n = 150). Among the 82 low AURKA-expressing specimens, high SQSTM1 expression was found in 23.1% cases (n = 19) (Fig. 1B). As shown in Figure 1B, we observed statistically significant positive correlations between AURKA and SQSTM1 expression (Spearman’s correlation coefficient 0.451, p < 0.001). Similarly, aurora kinase B (AURKB) and SQSTM1 expression were detected in a total of 219 breast cancer tissue specimens by immunohistochemical staining. Results indicated there was no correlation between AURKB and SQSTM1 (Fig. S1A).
Figure 1. AURKA correlated with autophagy-associated protein SQSTM1 expression in breast cancer. (A) Both low and high expression levels of SQSTM1 immunoreactivity were mainly detected in cytoplasm and occasionally in nucleus (a, b, e and f). Both low and high expression levels of AURKA immunoreactivity were seen mainly in cytoplasm of breast cancer tissue (c, d, g and h). 100×, original magnification × 100; 400×, original magnification × 400. (B) The results were calculated on the basis of analyses performed using 341 breast cancer tissue samples. The relationship between AURKA and SQSTM1 expression was compared using Spearman’s correlation coefficient. (C) Three different human cell lines BT-549, SK-BR-3 and MCF-10A were treated with 100 nM AURKA-1 siRNA (si AURKA-1) and negative control (NC) siRNA for 24 h respectively. Cell lysates were subjected to western blot analysis with the indicated antibodies. (D) Control (Ctr) and AURKA-overexpressing cells were lysed and subjected to western blot analysis similarly.
We next tested whether AURKA regulated the expression of SQSTM1 in breast cancer cell lines. We used AURKA siRNA (100 nM) to transfect three breast cancer cell lines, namely SK-BR-3, BT-549 and ZR-75-1 cells and a noncancerous breast epithelial cell line MCF-10A cells for 24 h. For the effects of AURKA overexpression and downregulation to be displayed clearly, it took a much longer exposure in the western blot of AURKA and SQSTM1 in Figure 1C and a much lighter exposure for the western blot of AURKA and SQSTM1 in Figure 1D. In all three cell lines, western blot analysis revealed that reduction of AURKA also decreased SQSTM1 expression (Fig. 1C; Fig. S2A). On the contrary, stable overexpression of AURKA in BT-549, SK-BR-3, MCF-10A and ZR-75-1 cells, using lentiviral vectors, caused an increase of SQSTM1 expression (Fig. 1D; Fig. S2B). As shown in Figure S3A, the SQSTM1 mRNA levels were not decreased by AURKA siRNA in BT-549 and SK-BR-3 cells. Similarly, SQSTM1 mRNA levels were not increased by stable overexpression of AURKA (Fig. S3B). Additionally, we used AURKB siRNA (100 nM) to transfect BT-549 cells for 24 h. As shown in Figure S1B, the SQSTM1 levels were not decreased by AURKB siRNA in BT-549 cells.
To elucidate the relationship between SQSTM1 and AURKA expression in breast cancer cells, we suppressed SQSTM1, using siRNA and found that SQSTM1 siRNA did not change the expression of AURKA in BT-549 cells (Fig. S4A). Conversely, stable overexpression of SQSTM1 using a lentiviral vector failed to change the expression of AURKA in BT-549 cells (Fig. S4B).
Depletion or inhibition of AURKA downregulated SQSTM1 through autophagy
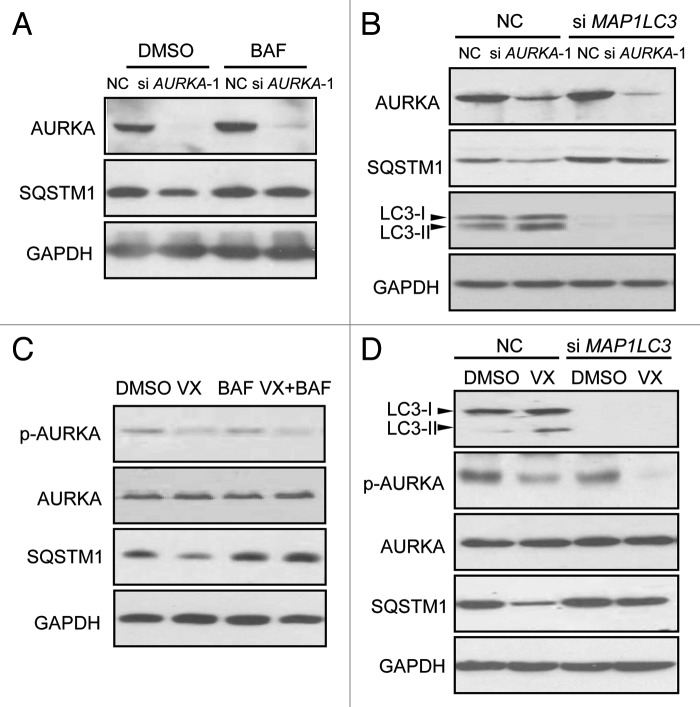
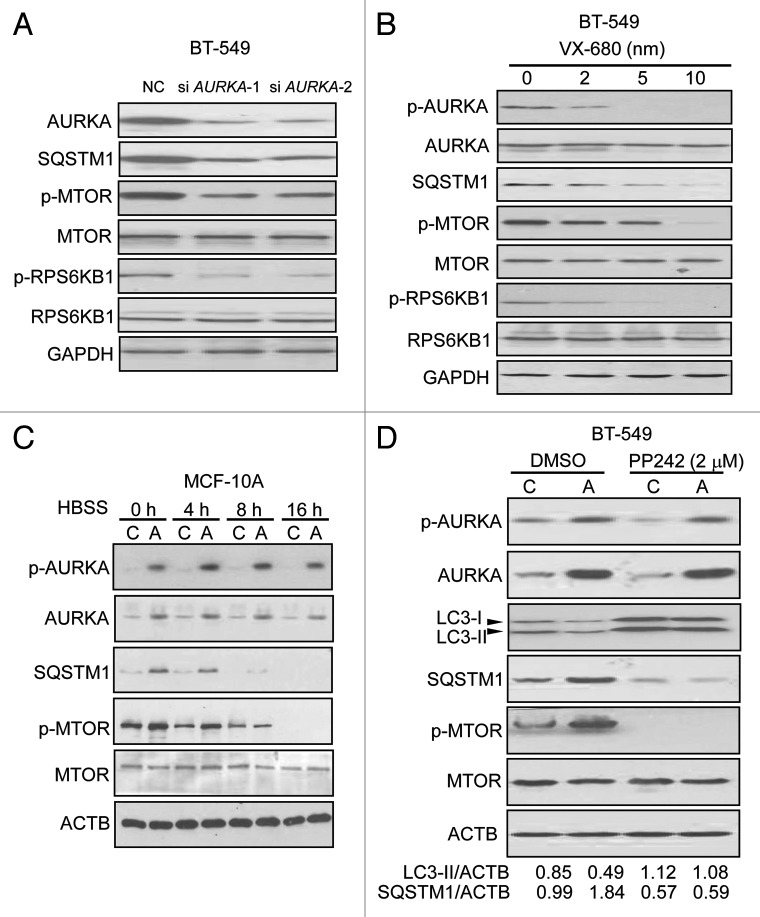
SQSTM1 has been shown to be degraded by autophagy.19 To investigate whether the depletion or inhibition of SQSTM1 in response to depletion of AURKA is mediated by autophagy, BT-549 cells transfected with AURKA siRNA were next treated with the autophagy inhibitor bafilomycin A1 (BAF, 200 nM). BAF inhibits the vacuolar ATPase and blocks the fusion of autophagosomes with lysosomes.20 Western blot analysis showed that BAF reversed the decrease of SQSTM1 induced by AURKA siRNA (Fig. 2A). Additionally, we used siRNA against microtubule-associated protein 1 light chain 3 (LC3) (MAP1LC3 siRNA), to inhibit autophagosomal formation. BT-549 cells transfected with MAP1LC3 siRNA were treated with AURKA siRNA. Figure 2B showed MAP1LC3 siRNA restored the downregulation of SQSTM1 by AURKA siRNA. As shown in Figure 2C and D, VX-680, a selective AURKA kinase inhibitor,27 decreased the expression of SQSTM1. Both BAF and MAP1LC3 siRNA effectively prevented the VX-680-induced downregulation of SQSTM1. These data indicated that regulation of SQSTM1 by AURKA was associated with autophagy. However, the proteosomal inhibitor MG-132 did not prevent the decrease of SQSTM1 induced by depletion or inhibition of AURKA (data not shown), indicating that regulation of SQSTM1 by AURKA was independent of the proteosomal pathway.
Figure 2. Depletion or inhibition of AURKA downregulated SQSTM1 through autophagy. (A) BT-549 cells were treated with 100 nM AURKA siRNA and NC siRNA for 24 h respectively, and then treated with the autophagy inhibitor bafilomycin A1 (BAF, 200 nM) for 10 h. Cell lysates were subjected to western blot analysis with the indicated antibodies. (B) BT-549 cells exposed to MAP1LC3 siRNA. Twenty-four hours after siRNA transfections, cells were transfected with AURKA siRNA for an additional 24 h. Cell lysates were analyzed by western blot. (C) BT-549 cells were pretreated with 200 nM BAF for 1 h, and then treated with 10 nM VX-680 (VX) for an additional 24 h. Cell lysates were subjected to western blot analysis with the indicated antibodies. (D) BT-549 cells exposed to MAP1LC3 siRNA for 24 h, and cells were treated with 10 nM VX-680 (VX) for an additional 24 h. Cell lysates were assayed by western blot analysis similarly.
Depletion or inhibition of AURKA induced autophagy
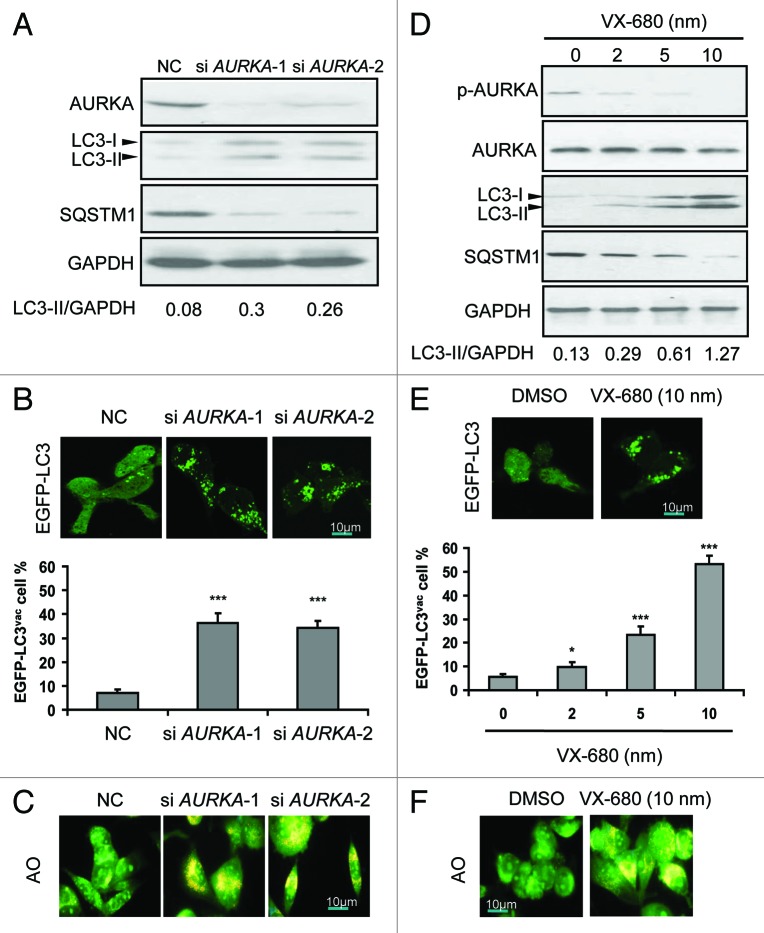
To determine if AURKA regulates autophagy, we examined the protein levels of LC3-II, which is an LC3-phosphatidyl-ethanolamine conjugate and an autophagosomal membrane protein.28 As shown in Figure 3A, a significant increase in endogenous LC3-II accumulation along with reduction of SQSTM1 was found after 24 h of treatment with AURKA siRNA. Besides, BT-549 cells were transfected with a plasmid-expressing EGFP fused with LC3 (EGFP-LC3), for 24 h and AURKA siRNA for an additional 24 h, thereafter the intracellular localization of LC3 in autophagic vacuoles was examined. In the majority of control BT-549 cells, EGFP-LC3 was diffusely distributed in the cytosol and nucleus, and only about 8% of the cells showed a dotted staining pattern of EGFP-LC3, indicative of the accumulation of autophagosomes (Fig. 3B). AURKA siRNA induced the redistribution of EGFP-LC3 fusion protein from a diffuse pattern toward an autophagosomal puncta-staining pattern. The numbers of cells with EGFP-LC3 puncta increased to approximately 40% (Fig. 3B). Moreover, the BT-549 cells were treated with AURKA siRNA for 24 h and then LC3 was detected by immunofluorescence. We found that the puncta of LC3 were detectable in the cytoplasm (Fig. S5D). In addition, BT-549 cells transfected with AURKA siRNA were stained with acridine orange (AO) to visualize acidic vesicular organelles (AVOs). Inhibition of AURKA expression by siRNA increased the number of cells with AVOs significantly (Fig. 3C).
Figure 3. Depletion or inhibition of AURKA induced autophagy. (A) BT-549 cells were treated with 100 nM AURKA-1 siRNA (si AURKA-1), AURKA-2 siRNA (si AURKA-2) and NC siRNA for 24 h respectively, and then cell lysates were subjected to western blot analysis with the indicated antibodies. LC3-II and GAPDH were quantified using Image J software. (B) BT-549 cells were transfected with 2 μg of EGFP-LC3 construct. At 24 h post-transfection, cells were transfected with AURKA-1 and AURKA-2 siRNA (100 nM). After additional 24 h post-transfection, the localization of LC3 in transfected cells was examined by confocal microscopy (magnification × 1000). The percentage of cells showing accumulation of EGFP-LC3 in puncta (EGFP-LC3vac) was quantified. A minimum of 100 EGFP-LC3-transfected cells were counted. Results are the mean ± SD of triplicates. ***p < 0.001, compared with controls. (C) BT-549 cells were treated with 100 nM AURKA-1 siRNA, AURKA-2 siRNA and NC siRNA for 24 h respectively. Cells were stained with acridine orange (AO) and examined by fluorescence microscopy (magnification × 400). Large orange puncta were considered to be acidic vesicular organelles (AVOs). (D) BT-549 cells were treated with increasing doses of VX-680 for 24 h, and then cell lysates were subjected to western blot analysis with the indicated antibodies. LC3-II and GAPDH were quantified using Image J software. (E) BT-549 cells were transfected with EGFP-LC3. At 24 h post-transfection, cells were treated with increasing doses of VX-680 for 24 h. Localization of LC3 in transfected cells was examined by confocal microscopy (magnification × 1000), the percentage of cells showing accumulation of EGFP-LC3 in puncta (EGFP-LC3vac) was quantified. A minimum of 100 EGFP-LC3-transfected cells were counted. Results are the mean ± SD of triplicates. *p < 0.05; ***p < 0.001, compared with control. (F) BT-549 cells were treated with 10 nM VX-680 for 24 h. Cells were stained with acridine orange (AO) and examined by fluorescence microscopy (magnification × 400). Large orange puncta were considered to be acidic vesicular organelles (AVOs).
Additionally, VX-680 resulted in the elevation of LC3-II levels, along with a decline of SQSTM1 in a dose-dependent manner, in BT-549 cells (Fig. 3D). Furthermore, the number of cells with EGFP-LC3 puncta was distinctly enhanced with increasing doses of VX-680 (0–10 nM) (Fig. 3E). Meanwhile, the number of cells with AVOs was clearly increased with 10 nM of VX-680 (Fig. 3F). Similar results were found in SK-BR-3 and MCF-10A cells (Figs. S5 and S6). Therefore, our data showed that depletion or inhibition of AURKA enhanced autophagic vacuolization, indicating an accumulation of autophagosomes and autolysosomes.
The accumulation of autophagosomes and autolysosomes could involve an enhanced autophagic sequestration or a reduced degradation of autophagic material. To distinguish between these possibilities, we detected LC3-II production in the presence of lysosomal proteases pepstatin A plus E64-d. As shown in Figure S7A, pepstatin A plus E64-d increased the presence of LC3-II induced by AURKA siRNA in BT-549 cells. Similarly, pepstatin A plus E64-d enhanced LC3-II levels induced with 5 nM of VX-680 (Fig. S7B). Furthermore, we assessed autophagic vacuolization induced by knockdown of AURKA or VX-680, by monitoring colocalization of LC3 with the lysosomal marker LAMP2. As shown in Figure S7C, AURKA siRNA induced colocalization of LC3 with LAMP2. Similarly, VX-680 increased colocalization of LC3 with LAMP2 (Fig. S7D). These results indicated that depletion or inhibition of AURKA may not block autophagosome-lysosome fusion but enhanced autophagic sequestration, at least in cultured breast cancer cells.
Autophagy may occur via a canonical pathway dependent of BECN1 and ATG5.29,30 To determine whether autophagy regulated by AURKA was involved in such a pathway, we used siRNA as an approach to suppress either BECN1 or ATG5 respectively, and quantified EGFP-LC3 puncta formation. Downregulation of either of these two canonical pathway dependent proteins in AURKA-inhibited cells resulted in a significant reduction of EGFP-LC3 puncta (Fig. S8), indicating that autophagy induction by AURKA inhibition might follow a canonical autophagic pathway.
Overexpression of AURKA inhibited autophagy under conditions of nutrient deprivation
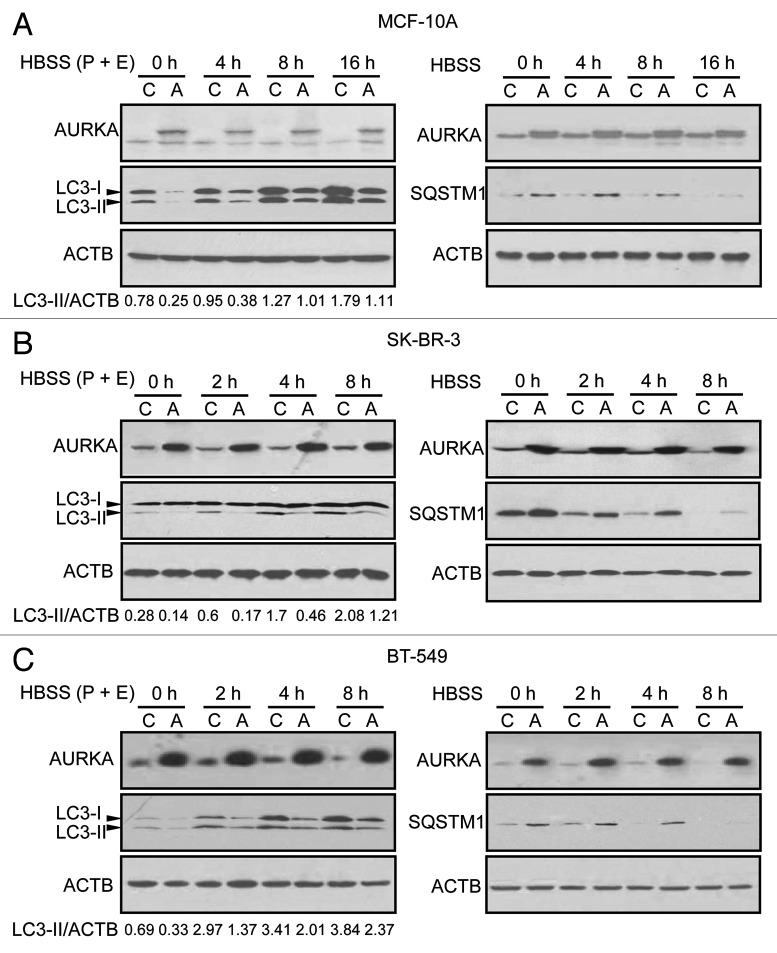
To assess whether overexpression of AURKA inhibits autophagy, MCF-10A cells stably overexpressing AURKA or vector were cultured in Hank's Balanced Salt Solution (HBSS) for different time intervals in the presence of the lysosomal cathepsin inhibitors pepstatin A and E64-d. We showed that HBSS increased LC3-II in a time-dependent manner (Fig. 4A). However, as shown in Figure 4A, overexpression of AURKA suppressed the increase of LC3-II at the time points indicated. Moreover, MCF-10A cells stably overexpressing AURKA or vector in HBSS were treated for different time intervals in the absence of pepstatin A and E64-d, and found that overexpression of AURKA suppressed the reduction of SQSTM1 expression at the time points as indicated (Fig. 4A). In SK-BR-3 and BT-549 cells, overexpression of AURKA also inhibited the production of LC3-II and increased the level of SQSTM1 under HBSS-induced nutrient starvation or normal growth conditions (Fig. 4B and C). These data suggested that overexpression of AURKA inhibited autophagy under starvation conditions. In contrast, AURKA siRNA increased LC3-II production and reduced SQSTM1 level under starvation (HBSS) in BT-549 cells (Fig. S9).
Figure 4. Overexpression of AURKA inhibited autophagy under conditions of nutrient deprivation. (A–C) Control (C) and overexpressed AURKA (A) MCF-10A, SK-BR-3 and BT-549 cells were subjected to HBSS starvation for the indicated time intervals. Pepstatin A (P, 10 μg/ml) and E64-d (E, 10 μg/ml) were added directly to the culture 4 h before lysis (left panel). Cells were not treated with P and E before lysis (shown in right panel). Cell lysates were subjected to western blot analysis. Densitometry analysis of LC3-II levels relative to ACTB was performed using Image J software.
AURKA-suppressed autophagy was associated with MTOR activation
Autophagy is regulated by a number of kinases including MTOR, which is generally thought to be involved in the negative control of mammalian autophagy.31 Here, we examined the effect of depletion of AURKA on the MTOR pathway using western blotting in BT-549 and MCF-10A cells. As shown in Figure 5A and Figure S10A, after a 24 h treatment with AURKA siRNA in BT-549 and MCF-10A cells, we found significant decreases in the levels of phosphorylated MTOR and its downstream ribosomal protein S6 kinase 1, 70 kDa, polypeptide 1 (RPS6KB1). Moreover, the levels of phosphorylated MTOR and RPS6KB1 were decreased in cells treated with increasing concentrations of VX-680 (Fig. 5B; Fig. S10B). Next, we treated MCF-10A cells stably overexpressing AURKA and control cells with HBSS and found that AURKA upregulated the expression of phosphorylated MTOR (Fig. 5C). In contrast, AURKA siRNA decreased phosphorylated MTOR expression under starvation (HBSS) in BT-549 cells (Fig. S11). Additionally, we found that inhibition of MTOR with PP242, an inihibitor of MTOR complex1/2 (MTORC1/2),32 abrogated the differences in LC3-II and SQSTM1 levels between AURKA-overexpressing and control BT-549 cells (Fig. 5D), suggesting that AURKA-inhibited autophagy was reversed by suppressing the MTOR. These results indicated that AURKA-inhibited autophagy was associated with MTOR activation.
Figure 5. AURKA-suppressed autophagy was associated with MTOR activation. (A) BT-549 cells were treated with 100 nM AURKA-1 siRNA, AURKA-2 siRNA and NC siRNA for 24 h respectively, and then cell lysates were analyzed by western blot. (B) BT-549 cells were treated with increasing dose of VX-680 for 24 h, and then cell lysates were subjected to western blot analysis. (C) Control (C) and overexpressed AURKA (A) MCF-10A cells were subjected to HBSS starvation for the indicated time intervals. Cell lysates were subjected to western blot asssays similarly. (D) Control (C) and overexpressed AURKA (A) BT-549 cells were treated with DMSO and 2 μM PP242 for 24 h, and then cell lysates were subjected to western blot analysis similarly. Densitometry analysis of LC3-II and SQSTM1 levels relative to ACTB was performed using Image J software.
Our previous study shows that AURKA regulates AKT1 activity by PtdIns3K.7 To assess whether AURKA increased MTOR activity dependent on the PtdIns3K-AKT1 pathway, the SK-BR-3 cells stably overexpressing AURKA and control cells were treated with the irreversible PtdIns3K inhibitor wortmanin (10 μM) and AKT1 signaling inhibitor-2 (API-2, 15 μM) for 24 h respectively. Both wortmannin and API-2 failed to prevent the increase in phosphorylated MTOR expression caused by AURKA overexpression (Fig. S12), suggesting that AURKA induced MTOR activity independently of the PtdIns3K-AKT1 pathway.
Inhibition of autophagy potentiated cytotoxicity by depletion or inhibition of AURKA in breast cancer cells
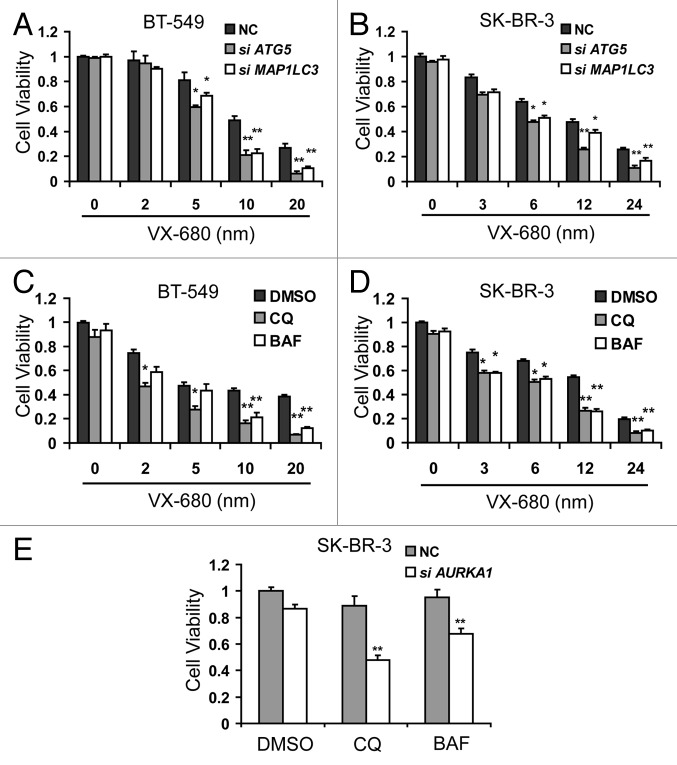
Autophagy may have an anticancer effect or a cytoprotective function against cytotoxic reagents.19 To assess the role of VX-680-induced autophagy in breast cancer cells, we attempted to block the autophagic process induced by VX-680, using siRNAs against ATG5 or MAP1LC3. Suppression of ATG5 or MAP1LC3 resulted in a significant enhancement of cytotoxicity induced by VX-680 in BT-549 and SK-BR-3 cells (Fig. 6A and B). Additionally, inhibition of autophagy by use of pharmacological inhibitors BAF (200 nM) or chloroquine (CQ, an inhibitor of lysosomal acidification29) (100 μM) resulted in increased cytotoxicity by VX-680 in both cell lines (Fig. 6C and D). Similar results were obtained using another autophagy inhibitor 3-methyladenine (3-MA), which blocks the early stages of autophagy (data not shown). These results indicated that VX-680-induced autophagy reduced cytotoxicity in response to VX-680 in breast cancer cells. Furthermore, SK-BR-3 cells were transfected with AURKA siRNA for 24 h and treated with CQ and BAF for an additional 24 h. As shown in Figure 6E, CQ and BAF enhanced the cytotoxicity by AURKA siRNA. Together, our data showed that inhibition of autophagy potentiates cytotoxicity by depletion or inhibition of AURKA in breast cancer cells.
Figure 6. Inhibition of autophagy potentiated cytotoxicity by depletion or inhibition of AURKA in breast cancer cells. (A and B) BT-549 and SK-BR-3 cells were transfected with 100 nM NC siRNA, MAP1LC3 siRNA and ATG5 siRNA respectively. Twenty-four hours later, they were treated with increasing doses of VX-680 for 24 h, cell viability was measured using MTT assay. (C and D) BT-549 and SK-BR-3 cells were pretreated with 100 μM chloroquine (CQ) and 200 nM bafilomycin A1 (BAF) for 1 h, and then cells were treated with increasing dose VX-680 for additional 24 h, cell viability was measured using MTT assay. (E) SK-BR-3 cells were transfected with 100nM NC siRNA and AURKA siRNA respectively. Twenty-four hours later, cells were treated with 100 μM CQ and 200 nM BAF for 24 h, and then cell viability was measured using the MTT assay. Data are the means of triplicate experiments. *p < 0.05; **p < 0.01.
Inhibition of autophagy enhanced VX-680-induced apoptosis in breast cancer cells
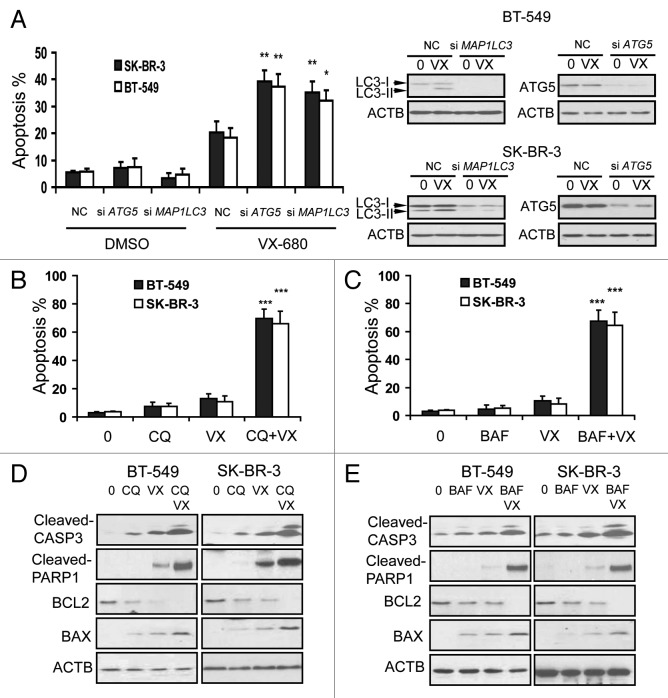
The data shown above indicated that inhibition of autophagy could potentiate the cytotoxicity by VX-680 in BT-549 and SK-BR-3 cells. We then asked if inhibition of autophagy could enhance VX-680-induced apoptosis in breast cancer cells. We knocked down expression of ATG5 or LC3 in BT-549 and SK-BR-3 cells by using specific siRNA, and analyzed VX-680-induced cell apoptosis by Annexin V-FITC/PI staining. As shown in Figure 7A, ATG5 or MAP1LC3 siRNA increased VX-680-induced cell apoptosis. Subsequently, Annexin V-FITC staining assays were performed after a 24 h treatment with VX-680, CQ, or the combination of VX-680 and CQ. Treatment with CQ or VX-680 alone had weak effects on apoptosis in these cells (Fig. 7B). However, there was a significantly higher apoptosis rate found upon treatment with the combination of VX-680 and CQ (Fig. 7B). Similar results were found using other autophagy inhibitors, BAF (Fig. 7C) or 3-MA (data not shown).
Figure 7. Inhibition of autophagy enhanced VX-680-induced apoptosis in breast cancer cells. (A) BT-549 and SK-BR-3 cells were transfected with 100nM NC siRNA, MAP1LC3 siRNA and ATG5 siRNA respectively. Twenty-four hours later, BT-549 and SK-BR-3 were treated with 10 nM and 12 nM VX-680 for 24 h respectively, cell apoptosis was measured using an Annexin V-FITC/PI staining assay. The knockdown effects on LC3 and ATG5 were confirmed by western blot analysis (right panel). (B and C) BT-549 and SK-BR-3 cells were pretreated with 100 μM chloroquine (CQ) and 200 nM bafilomycin A1 (BAF) for 1 h, and then BT-549 and SK-BR-3 cells were treated with 10 nM and 12 nM VX-680 (VX) for additional 24 h respectively, cell apoptosis was measured using an Annexin V-FITC/PI staining assay. (D and E) BT-549 and SK-BR-3 cells were pretreated with 100 μM CQ and 200 nM BAF for 1 h, and then BT-549 and SK-BR-3 cells were treated with 10 nM and 12 nM VX-680 (VX) for additional 24 h respectively, and then cell lysates were assayed by western blot analysis. Data are the means of triplicate experiments. *p < 0.05; **p < 0.01.
Next, the proapoptotic factor BAX and the antiapoptotic protein BCL2 were studied in BT-549 and SK-BR-3 cells after treatment with VX-680, CQ or their combination by western blot analysis. As shown in Figure 7D, CQ increased the expression of BAX induced by VX-680. Conversely, BCL2 expression decreased by VX-680 was considerably diminished by CQ. In addition, upregulation of cleaved CASP3 and cleaved PARP1 expression were also observed (Fig. 7D). Similar results were also generated using BAF or 3-MA combined with VX-680 (Fig. 7E and data not shown). These results revealed that the mitochondrial apoptotic pathway was involved in the apoptosis induced by VX-680, combined with inhibition of autophagy, in breast cancer cells.
Discussion
In this study, we found a significant positive correlation of AURKA with autophagy-associated SQSTM1 protein expression in breast cancer specimens. Similar results were confirmed in breast cancer cell lines. However, the transcription levels of SQSTM1 were not decreased by AURKA siRNA in breast cancer cell lines. Similarly, transcriptional upregulation of SQSTM1 was not found in AURKA-overexpressing breast cancer cell lines. Furthermore, the prevention of SQSTM1 downregulation by both chemical and genetic inhibition of autophagy strongly suggested the involvement of autophagy in the regulation of SQSTM1 by AURKA, in breast cancer cell lines. We further showed that depletion or inhibition of AURKA enhanced autophagy in the cell lines. However, the possibility of a positive correlation between AURKA and defective autophagy in breast cancer could not be excluded. In addition, transcriptional levels of SQSTM1 can be induced by oxidative stress.33 Therefore, there was a possibility of transcriptional upregulation of SQSTM1 in advanced breast cancer under oxidative stress. Additionally, we elucidated that AURKA-inhibited autophagy was associated with MTOR activation in breast cancer cell lines. Notably, we revealed a novel mechanism of drug resistance through targeting AURKA as a potential therapy: a cellular protective role of autophagy activation. We then confirmed this by showing that suppression of autophagy increased the sensitivity of cancer cells to apoptotic cell death induced by AURKA inhibition.
AURKA has been well characterized for its role in centrosome maturation and mitotic progression.1-3 Dysregulation of AURKA has been implicated in genetic instability and consequent tumorigenesis.34 In addition, we have recently found that AURKA induced cell survival and migration.7,8,12 In this study, we demonstrated that AURKA inhibited autophagy in breast cancer cells. Autophagy inhibition promoted tumorigenesis that has been reported in ovarian, breast and prostate cancers.35,36 AURKA overexpression, also found in various types of cancer cells, was shown to play key roles in tumorigenesis. Therefore, it is conceivable that inhibition of autophagy could contribute to AURKA-induced tumorigenesis in breast cancer.
Autophagy is negatively regulated by MTOR.31 Regulation of autophagy by MTOR is dependent on MTOR complex 1 (MTORC1) which contains RPTOR/Raptor, MLST8 and AKT1S1/PRAS40. MTORC1 suppresses autophagy through direct regulation of the ULK1-ATG13-RB1CC1/FIP200 complex.37 Elevated phosphorylation of MTOR Ser2448 in AURKA-transfected cells suggests that AURKA could potentially regulate MTOR pathways.4 In this study, we found that overexpression of AURKA increased the phosphorylation of MTOR Ser2448 and RPS6KB1. Conversely, depletion or inhibition of AURKA decreased the phosphorylation of MTOR Ser2448 and RPS6KB1. To ask whether the regulation of autophagy by AURKA was dependent on the MTOR pathway, MTOR was inhibited by using PP242 in BT-549 cells stably overexpressing either AURKA or vector. We found that inhibition of MTOR by PP242 abrogated the difference in autophagic level between cells overexpressing AURKA and vector. These results were consistent with the possibility that AURKA regulated autophagy through MTOR pathways. AKT1 upregulates MTOR activity by phosphorylating tuberous sclerosis complex 2 (TSC2).38 TSC1/2 inhibit the activity of RHEB, a GTPase that upregulates MTOR activation.39 We and others have previously reported that AURKA can upregulate PtdIns3K and AKT1 activation.7,12 However, we found here that AURKA regulated MTOR independently of the PtdIns3K-AKT1 pathway. Additionally, the interaction between AURKA and MTOR was investigated by coimmunoprecipitation (data not shown). No physical binding of AURKA and MTOR was detected. In a previous study, we found that AURKA enhances the activity of mitogen-activated protein kinase (MAPK).6 MAPK can phosphorylate TSC2 directly in vitro and lead to TSC1-TSC2 dissociation. The dissociation of TSC1-TSC2 complexes increases the ability of RHEB to induce phosphorylation of MTOR.40 Therefore, MAPK can enhance the activity of MTOR through a TSC1/2-RHEB pathway.41 This raised a possibility that AURKA regulates autophagy through MAPK-TSC1/2-RHEB-MTOR pathways.
AURKA expression is upregulated in various types of cancer cells, which has been implicated in resistance to stress-induced apoptosis.34 Inhibition of AURKA by small molecular inhibitor or transient transfection with siRNA has been shown to induce apoptosis in cancer cells.11,12 Targeting AURKA has been a potential therapeutic approach in human cancer. In this study, we found that inhibition of AURKA by VX-680 induced autophagy, as well as apoptosis. Autophagy is a process of degrading proteins and organelles, and recycling materials in response to stress.42,43 Studies show that there are links between pathways that are involved in autophagy and apoptosis, and these connections might be influenced by the stimuli to which cells are exposed.18 Here, we found that inhibition of autophagy potentiated VX-680-induced cell apoptosis. These results indicated that autophagy caused by targeting AURKA contributed to breast cancer cells resistance to VX-680. Similar studies, consistent with these findings, show that activated autophagy can protect tumor cells from targeted therapies, such as trastuzumab in breast cancer,44 the imatinib mesylate in Philadelphia chromosome-positive cells,45 and proteasome inhibitors in prostate cancer.46 Thus, our data, along with these reports, supported that autophagy might contribute to a drug-resistance mechanism. However, there are also reports showing that inhibition of autophagy suppresses cancer cell death by anticancer reagents, indicating that autophagy can be a potential contributor to cell death.43,47 Thus, the involvement of autophagy in cancer cell death is still controversial, probably because the molecules which execute cell death in autophagy have not been thoroughly identified.
Here we demonstrated for the first time, that regulation of autophagy by AURKA and inhibition of AURKA, induced autophagy to counteract the antiproliferative and apoptotic effect of AURKA inhibition in breast cancer cells. Our current findings showed that the use of an autophagy inhibitor may further enhance the anticancer effect of AURKA inhibitors in the treatment of breast cancer.
Materials and Methods
Patients and clinical tissue specimens
Tissue microarray samples from 341 patients with breast carcinoma were obtained from the Department of Pathology, the State Key Laboratory of Oncology in Southern China, Sun Yat-sen University Cancer Center, between 1999 and 2008. The cases selected were based on availability of resection tissue, follow-up data and no preoperative radiation or chemotherapy. Patients without known death cause were excluded from our study. Ages of the 341 patients ranged from 26 to 84 y (mean age, 48.0 y). Cancer stages and pertinent clinical data were assessed according to the World Health Organization pathological system (2003) and the Tumor-Node-Metastasis system of International Union Against Cancer (2002), respectively.48 Tumor histology was coded according to World Health Organization criteria in the International Classification of Diseases for Oncology. This study was approved by the Clinical Ethics Review Board at Sun Yat-sen University Cancer Center and written informed consents were from all patients at their recruitment time.
Immunohistochemical staining and statistical analysis
The immunohistochemical staining of SQSTM1 and AURKA was performed using a standard of two-step techniques. The tissue microarray slide was dried overnight at 37°C, dewaxed in xylene, rehydrated through graded alcohol, and immersed in 3% hydrogen peroxide for 20 min to block endogenous peroxidase activity. An antigen retrieval process was accomplished in a microwave oven with 10 mM citrate buffer (pH = 7.0) for 15 min. The slides were incubated with 3% BSA at room temperature (RT) for 10 min to reduce nonspecific reactions. Subsequently, the tissue microarray slides were incubated with antibody against SQSTM1 (Santa Cruz Biotechnology, sc-28359) and AURKA (Upstate, 07-648) for 1 h at RT. After three rinses with 0.01 M PBS (pH = 7.4) for 10 min. The detection of the primary antibody was achieved with a secondary antibody (Envision, K4003) for 30 min at RT, and stained with DAB (3,3-diaminobenzidine) after washing in PBS again. Finally, the sections were counterstained with Mayer’s hematoxylin, dehydrated, and mounted. For negative controls PBS was used instead of the anti-SQSTM1 and anti-AURKA antibody. The expression of SQSTM1 and AURKA was compared using Spearman’s correlation coefficient. Statistical analysis was performed using SPSS version 16.0 (SPSS, Inc.). Receiver operating characteristic (ROC) curve analysis was applied to determine the cutoff scores for high or low expression of SQSTM1, AURKA and AURKB.
Reagents
Reagents used included bafilomycin A1 (Sigma, B1793), chloroquine (Sigma, C6628), 3-methyladenine (Sigma, M9281), E64-d (Sigma, E8640), pepstatin A (Sigma, P5318), DMSO (Sigma, D2650), VX-680 (Kava Tech, 639089-54-6), acridine orange (Invitrogen, A3568), MG132 (Sigma, C2211), PP242 (Sigma, P0037), API-2 (Calbiochem, 124012), wortmannin (Cell Signaling Technology, 9951).
Cell culture
Cell lines were obtained from American Type Culture Collection. MCF-10A cells were cultured as previously described.49 BT-549 and ZR-75-1 cells were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS, Hyclone, SV30160.03) and 0.023 U/ml of human insulin (Sigma, I0516). SK-BR-3 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 incubator.
Gene expression analysis by real-time quantitative PCR
Total RNA was isolated by TRIzol (Invitrogen, 15596-026) according to the manufacturer’s instructions. cDNA was prepared from total RNA using random primers (Promega, C1181) and the Omniscript RT kit (Qiagen, 205111). The relative levels of mRNA were determined by real-time quantitative PCR using an Eppendorf Realplex Mastercycler (Eppendorf) and Quantitect SYBR Green PCR kit (Qiagen, 204141). GAPDH mRNA levels were used for normalization. Primer sequences used were as follows: SQSTM1: 5′ -GTGGTAGGAACCCGCTACAA-3′, 5′-GCGATCTTCCTCATCTGCTC-3′;50 GAPDH: 5′-CCACCCATGGCAAATTCCATGGCA-3′, 5′-TCTAGACGGCAGGTCAGGTCCACC-3′.
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde at RT for 25 min and permeabilized in 0.5% Triton X-100 in PBS for 15 min. Slides were incubated with anti-LC3 (Novus Biologicals, Inc., 2775s) and anti-LAMP2 (Santa Cruz Biotechnology, sc-18822) antibodies at RT for 60 min. The immune complexes were stained with anti-rabbit IgG tagged with Alexa Fluor 488 (Invitrogen, A11008) or anti-mouse IgG tagged with Alexa Fluor 546 (Invitrogen, A11030) for 1 h. Confocal images were taken with Olympus FV-1000 confocal microscope.
RNA interference
siRNA for downregulating gene expression was done by transfection of RNA oligonucleotides with lipofectamine 2000 (Invitrogen, 11668) according to the manufacturer’s instructions. The negative control (NC) siRNA and siRNAs against SQSTM1, AURKA, MAP1LC3, ATG5 and BECN1 were synthesized by Shanghai GenePharma Co. For SQSTM1, two siRNA oligonucleotides were used, SQSTM1-B: 5′-GGACCCATCTGTCTTCAAA-3′, SQSTM1-C: 5′-GCATTGAAGTTGATATCGA-3′;41 For AURKA, two siRNA oligonucleotides were used, AURKA-1: 5′-AUGCCCUGUCUUACUGUCA-3′,8 AURKA-2: 5′-AACGTGTTCTCGTGACTCAGC-3′;51 For AURKB, 5′-AACGCGGCACUUCACAAUUGA-3′52; For MAP1LC3, a pool of three siRNA oligonucleotides was used, (a): 5′-CGGUGAUAAUAGAACGAUA-3′, (b): 5′-GGUUUGUUCUCUAGAUAGU-3′, (c): 5′-CGUACGCUCUUUACAGAUA-3′;53
For ATG5: 5′-TGATATAGCGTGAAACAAG-3′;54 and for BECN1: 5′-CAGTTTGGCACAATCA ATA-3′.55
Quantification of EGFP-LC3 puncta assay
Cells were transfected with EGFP-LC3 constructs (Addgene, 11546)56 in Opti-MEM (Invitrogen, 11058) using Lipofectamine 2000 (Invitrogen, 11668). Afterward, cells were fixed in 4% paraformaldehyde at RT for 20 min, and washed twice with PBS. EGFP-LC3 distribution was subsequently monitored using an Olympus FV-1000 confocal microscope. Autophagy was quantified as described previously.20 Simply, the percentage of cells showing accumulation of EGFP-LC3 in dots or vacuoles (EGFP-LC3vac, of a minimum of 100 cells per preparation in three independent experiments) was counted. Cells representing several intense punctate EGFP-LC3 aggregates with no nuclear EGFP-LC3 were classified as autophagic, on the contrary, cells presenting a mostly diffuse distribution of EGFP-LC3 in the cytoplasm and nucleus were considered as nonautophagic.
Acridine orange staining for acidic vesicular organelles
Acridine orange was added at a final concentration of 1 μg/ml for 15 min. Thereafter, viewed immediately and pictures were obtained with a fluorescence microscope (Olympus IX71) equipped with a 450–490 nm band-pass blue excitation filter, a 505 nm dichroic mirror, a 520 nm long pass-barrier filter, and a digital camera (Nikon DS-5Mc).
Western blot analysis
Cells were lysed on ice in RIPA buffer. The protein concentration was determined by Bradford dye method. Equal amounts (20 to 40 μg) of cell extract were subjected to electrophoresis in 6–12.5% SDS-PAGE and transferred to nitrocellulose membrane (Millipore, HATF00010) for antibody blotting. The antibodies used were as follows: anti-GAPDH (Ambion, AM4300), anti-ACTB (Cell Signaling Technology, 4967), anti-SQSTM1 (Santa Cruz Biotechnology, sc-28359), anti-AURKA (Upstate, 07-648), anti-phosphorylated AURKA (Thr288) (Cell Signaling Technology, 3079s), anti-AURKB (Cell Signaling Technology, 3094), anti-LC3B (Novus Biologicals Inc., 2775s), anti-phosphorylated MTOR (Ser2448) (Cell Signaling Technology, 2971s), anti-MTOR (Epitomic, s0302), anti-RPS6KB1 (Epitomic, 1494-1), anti-phosphorylated RPS6KB1 (Thr389) (Cell Signaling Technology, 9205), anti-phosphorylated AKT1 (Ser473) (Cell Signaling Technology, 9271), anti-AKT1 (Santa Cruz Biotechnology, sc-1618), anti-ATG5 (Cell Signaling Technology, 2630), anti-BECN1 (Cell Signaling Technology, 3738), anti-cleaved-CASP3 (Cell Signaling Technology, 9661L), anti-BCL2 (Santa Cruz Biotechnology, sc-492), anti-BAX (Santa Cruz Biotechnology, sc-20067) and anti-cleaved-PARP1 (Cell Signaling Technology, 9546). Blots were quantified using Image J software (National Institutes of Health). All experiments were repeated at least three times.
Cell viability and apoptosis assays
Cell viability was assessed by standard MTT (Sigma, M2128) assay. The absorbance was determined at a test wavelength of 490 nm on a multiwell plate reader (Microplate Reader; Bio-Rad). Measurement of apoptosis was conducted by Annexin V-FITC analysis as described previously.11
Statistical analysis
Statistical analysis was performed using SPSS version 16.0 (SPSS, Inc.). The χ2 test and Student's t test were used to make a statistical comparison between groups. p < 0.05 was considered statistically significant. Results were representative of at least three experiments under identical conditions.
Supplementary Material
Acknowledgments
We thank the Quentin Liu lab for their critical comments and technical support. This research work was supported by National Natural Science Foundation Key Program of China (81130040 to Q.L.), National Basic Research Program of China (2012CB967000 to Q.L.), National Science Fund for Distinguished Young Scholars (303041389009 to Q.L.) and National Natural Science Foundation of China (81000217 to Z.-J.L.). We thank K. Kirkegaard for EGFP-LC3 plasmid.
Glossary
Abbreviations:
- AURKA
aurora kinase A
- SQSTM1
sequestosome 1
- ACTB
beta actin
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- AURKB
aurora kinase B
- siRNA
small interfering RNA
- BAF
bafilomycin A1
- CQ
chloroquine
- AO
acridine orange
- AVOs
acidic vesicular organelles
- 3-MA
3-methyladenine
- HBSS
Hank's Balanced Salt Solution
- DMSO
dimethyl sulfoxide
- EGFP
enhanced green fluorescent protein
- LC3
microtubule-associated protein 1 light chain 3
- MAP1LC3
microtubule-associated protein 1 light chain 3
- BCL2
B-cell CLL/lymphoma 2
- BCL2L2
BCL2-like 2
- MCL1
myeloid cell leukemia sequence 1
- BAD
BCL2-associated agonist of cell death
- PMAIP1
phorbol-12-myristate-13-acetate-induced protein 1
- BBC3
BCL2 binding component 3
- MTOR
mechanistic target of rapamycin
- RPS6KB1
ribosomal protein S6 kinase 1, 70kDa, polypeptide 1
- MTORC1/2
MTOR complex1/2
- RPTOR/Raptor
regulatory associated protein of MTOR
- MLST8
MTOR associated protein, LST8 homolog
- AKT1S1
AKT1 substrate 1
- PRAS40
40 kDa proline-rich AKT substrate
- ULK1
unc-51-like kinase 1
- ATG13
autophagy-related 13
- RB1CC1
RB1-inducible coiled-coil 1
- FIP200
200 kDa FAK family kinase-interacting protein
- LAMP2
lysosomal-associated membrane protein 2
- ATG5
autophagy-related 5
- BECN1
Beclin 1
- PtdIns3K
phosphatidylinositol-4,5-bisphosphate 3-kinase
- AKT1
v-akt murine thymoma viral oncogene homolog 1
- API-2
AKT1 signaling inhibitor-2
- MAPK
mitogen-activated protein kinase
- TSC2
tuberous sclerosis 2
- RHEB
Ras homolog enriched in brain
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- BAX
BCL2-associated X protein
- CASP3
caspase 3
- PARP1
poly (ADP-ribose) polymerase 1
- ROC
receiver operating characteristic
- DAB
3,3-diaminobenzidine
- PBS
phosphate buffered saline
- BSA
bovine serum albumin
- FBS
fetal bovine serum
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/22110
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/22110
References
- 1.Meraldi P, Honda R, Nigg EA. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr Opin Genet Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Liu Q, Ruderman JV. Aurora A, mitotic entry, and spindle bipolarity. Proc Natl Acad Sci U S A. 2006;103:5811–6. doi: 10.1073/pnas.0601425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, et al. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–58. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 5.Mountzios G, Terpos E, Dimopoulos MA. Aurora kinases as targets for cancer therapy. Cancer Treat Rev. 2008;34:175–82. doi: 10.1016/j.ctrv.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Wan XB, Long ZJ, Yan M, Xu J, Xia LP, Liu L, et al. Inhibition of Aurora-A suppresses epithelial-mesenchymal transition and invasion by downregulating MAPK in nasopharyngeal carcinoma cells. Carcinogenesis. 2008;29:1930–7. doi: 10.1093/carcin/bgn176. [DOI] [PubMed] [Google Scholar]
- 7.Wang LH, Xiang J, Yan M, Zhang Y, Zhao Y, Yue CF, et al. The mitotic kinase Aurora-A induces mammary cell migration and breast cancer metastasis by activating the Cofilin-F-actin pathway. Cancer Res. 2010;70:9118–28. doi: 10.1158/0008-5472.CAN-10-1246. [DOI] [PubMed] [Google Scholar]
- 8.Guan Z, Wang XR, Zhu XF, Huang XF, Xu J, Wang LH, et al. Aurora-A, a negative prognostic marker, increases migration and decreases radiosensitivity in cancer cells. Cancer Res. 2007;67:10436–44. doi: 10.1158/0008-5472.CAN-07-1379. [DOI] [PubMed] [Google Scholar]
- 9.Lee HH, Zhu Y, Govindasamy KM, Gopalan G. Downregulation of Aurora-A overrides estrogen-mediated growth and chemoresistance in breast cancer cells. Endocr Relat Cancer. 2008;15:765–75. doi: 10.1677/ERC-07-0213. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, He L, Kruk P, Nicosia SV, Cheng JQ. Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. Int J Cancer. 2006;119:2304–12. doi: 10.1002/ijc.22154. [DOI] [PubMed] [Google Scholar]
- 11.Huang XF, Luo SK, Xu J, Li J, Xu DR, Wang LH, et al. Aurora kinase inhibitory VX-680 increases Bax/Bcl-2 ratio and induces apoptosis in Aurora-A-high acute myeloid leukemia. Blood. 2008;111:2854–65. doi: 10.1182/blood-2007-07-099325. [DOI] [PubMed] [Google Scholar]
- 12.Yao JE, Yan M, Guan Z, Pan CB, Xia LP, Li CX, et al. Aurora-A down-regulates IkappaBalpha via Akt activation and interacts with insulin-like growth factor-1 induced phosphatidylinositol 3-kinase pathway for cancer cell survival. Mol Cancer. 2009;8:95. doi: 10.1186/1476-4598-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroemer G, Jäättelä M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–97. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 14.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 15.Jin S, White E. Tumor suppression by autophagy through the management of metabolic stress. Autophagy. 2008;4:563–6. [PMC free article] [PubMed] [Google Scholar]
- 16.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine B. Unraveling the role of autophagy in cancer. Autophagy. 2006;2:65–6. doi: 10.4161/auto.2.2.2457. [DOI] [PubMed] [Google Scholar]
- 18.Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–75. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 19.Kondo Y, Kondo S. Autophagy and cancer therapy. Autophagy. 2006;2:85–90. doi: 10.4161/auto.2.2.2463. [DOI] [PubMed] [Google Scholar]
- 20.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009;16:1135–45. doi: 10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marquez RT, Xu L. Bcl-2:Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res. 2012;2:214–21. [PMC free article] [PubMed] [Google Scholar]
- 23.Germain M, Slack RS. MCL-1 regulates the balance between autophagy and apoptosis. Autophagy. 2011;7:549–51. doi: 10.4161/auto.7.5.15098. [DOI] [PubMed] [Google Scholar]
- 24.Erlich S, Mizrachy L, Segev O, Lindenboim L, Zmira O, Adi-Harel S, et al. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy. 2007;3:561–8. doi: 10.4161/auto.4713. [DOI] [PubMed] [Google Scholar]
- 25.Martin SJ. Oncogene-induced autophagy and the Goldilocks principle. Autophagy. 2011;7:922–3. doi: 10.4161/auto.7.8.15821. [DOI] [PubMed] [Google Scholar]
- 26.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–6. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–7. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 28.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ducat D, Zheng Y. Aurora kinases in spindle assembly and chromosome segregation. Exp Cell Res. 2004;301:60–7. doi: 10.1016/j.yexcr.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janes MR, Limon JJ, So L, Chen J, Lim RJ, Chavez MA, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 2010;16:205–13. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–91. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–93. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 35.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 36.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 37.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–57. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 39.Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–6. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–93. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 41.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–35. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 42.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 44.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS One. 2009;4:e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–23. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu K, Dunner KJ, Jr., McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–62. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bursch W, Ellinger A, Gerner C, Fröhwein U, Schulte-Hermann R. Programmed cell death (PCD). Apoptosis, autophagic PCD, or others? Ann N Y Acad Sci. 2000;926:1–12. doi: 10.1111/j.1749-6632.2000.tb05594.x. [DOI] [PubMed] [Google Scholar]
- 48.Greene FL. The American Joint Committee on Cancer: updating the strategies in cancer staging. Bull Am Coll Surg. 2002;87:13–5. [PubMed] [Google Scholar]
- 49.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 50.Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, et al. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–52. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- 51.Shang X, Burlingame SM, Okcu MF, Ge N, Russell HV, Egler RA, et al. Aurora A is a negative prognostic factor and a new therapeutic target in human neuroblastoma. Mol Cancer Ther. 2009;8:2461–9. doi: 10.1158/1535-7163.MCT-08-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long ZJ, Xu J, Yan M, Zhang JG, Guan Z, Xu DZ, et al. ZM 447439 inhibition of aurora kinase induces Hep2 cancer cell apoptosis in three-dimensional culture. Cell Cycle. 2008;7:1473–9. doi: 10.4161/cc.7.10.5949. [DOI] [PubMed] [Google Scholar]
- 53.Pursiheimo JP, Rantanen K, Heikkinen PT, Johansen T, Jaakkola PM. Hypoxia-activated autophagy accelerates degradation of SQSTM1/p62. Oncogene. 2009;28:334–44. doi: 10.1038/onc.2008.392. [DOI] [PubMed] [Google Scholar]
- 54.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 55.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–2. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 56.Jackson WT, Giddings TH, Jr., Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.