Abstract
The Paf1 complex was originally identified over fifteen years ago in budding yeast through its physical association with RNA polymerase II. The Paf1 complex is now known to be conserved throughout eukaryotes and is well studied for promoting RNA polymerase II transcription elongation and transcription-coupled histone modifications. Through these critical regulatory functions, the Paf1 complex participates in numerous cellular processes such as gene expression and silencing, RNA maturation, DNA repair, cell cycle progression and prevention of disease states in higher eukaryotes. In this review, we describe the historic and current research involving the eukaryotic Paf1 complex to explain the cellular roles that underlie its conservation and functional importance.
1. Introduction
Through the regulation of transcription, cells are able to mount proper responses to exogenous stimuli, initiate signaling pathways involved in development and differentiation, and proliferate in complex environments. Regulation of RNA polymerase II (pol II) transcription can occur at each of the four general steps in the transcription cycle: promoter binding by RNA pol II and initiation of transcript synthesis, promoter clearance, transcription elongation, and termination. Proteins that interact with RNA pol II can control its activity to facilitate or repress transcription at one or more of these steps. The organization of eukaryotic DNA into chromatin, the basic element of which is a nucleosome containing ~147 basepairs of DNA wrapped around an octamer of histone proteins, presents a barrier to DNA accessibility during transcription. However, alterations to nucleosomes also provide an opportunity for carefully orchestrated levels of transcriptional regulation. Given the fundamental importance of transcription and chromatin regulatory factors, intensive research in a variety of organisms has focused on identifying these proteins and elucidating their interactions, molecular activities, and gene target specificities.
This review focuses on the eukaryotic Polymerase-Associated Factor 1 (Paf1) complex (Paf1C), which is a conserved protein complex that acts globally in multiple aspects of RNA pol II transcriptional regulation. First identified and characterized in Saccharomyces cerevisiae through an interaction with RNA pol II, Paf1 complexes have now been found in many eukaryotes. The overlapping functions shared by these complexes demonstrate the functional significance of the Paf1C. Here we describe the functions of the Paf1C in promoting histone modifications and regulating transcription elongation and gene expression. We also discuss less well-understood functions of the Paf1C in RNA 3’-end formation and non-histone processes. Lastly, we address the importance of the complex in regulating development and protecting against various diseases. As further research enhances our understanding of the molecular and cellular functions of the Paf1C, we hope that the involvement of Paf1C components in disease progression in higher eukaryotes will be more fully explained.
2. Subunit composition and genetic properties of the Paf1C
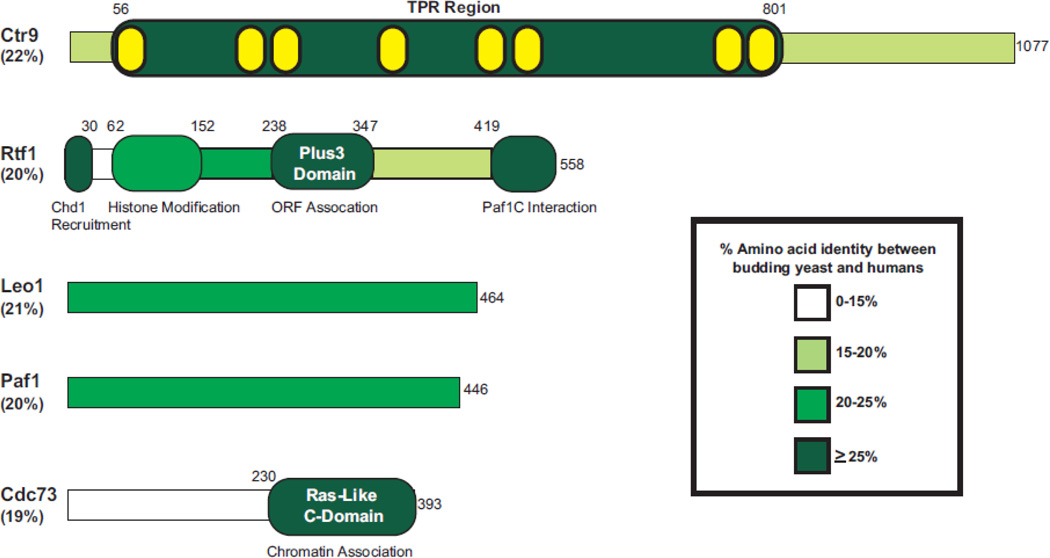
To isolate proteins associated with RNA pol II, an antibody against the conserved C-terminal repeat domain (CTD) of the largest S. cerevisiae RNA pol II subunit, Rpb1, was used for affinity purification [1]. These studies revealed a novel protein interacting with RNA pol II, which was termed Paf1 [2]. In addition, Cdc73 (Cell Division Cycle 73), a protein that had been previously shown to have connections to mating signaling pathways and cell division, was found to co-purify with Paf1 and RNA pol II in these studies [3, 4]. Cdc73 was subsequently shown to interact directly with RNA pol II in vitro [3]. Three more proteins were later identified as being part of the budding yeast Paf1C (yPaf1C): Ctr9, Leo1, and Rtf1 [5, 6]. Ctr9/Cdp1 (Cln Three Requiring 9) was genetically identified through its connections to the cell cycle, including effects on expression of the G1 cyclin genes CLN1 and CLN2 as well as microtubule formation [7–9]. Whereas the gene encoding Leo1 (Left Open Reading Frame 1) was sequenced but not characterized, Rtf1 (Restores TBP Function 1) was first identified in a yeast genetic screen for mutations that suppress the transcriptional effects of a defective TATA-binding protein and was later found to have extensive genetic interactions with transcription elongation factors [10–12]. Like yeast, the human and Drosophila Paf1 complexes have been shown to interact with RNA pol II; however, Rtf1 is less tightly associated with the rest of the Paf1C components in higher eukaryotes [13–16]. The human Paf1 complex (hPaf1C) also contains another protein, Ski8/Wdr61, which has been shown to have a role in mRNA decay as part of the Ski complex [13]. To date, structural data have been obtained for both the Ras-like C-domain of yCdc73 and the Plus-3 domain of hRtf1 [17, 18]. Extensive sequence similarity exists between Paf1C subunits in yeast and humans, suggesting conservation of structure and function [Figure 1].
Figure 1. Conservation of the yeast Paf1C subunits.
The five proteins that comprise the Paf1C in budding yeast are depicted. The overall percent amino acid identity between S. cerevisiae and H. sapiens is listed under each protein name. The amino acid identity was determined using a global pairwise alignment algorithm within EMBOSS [177]. Using published literature, regions with defined structures (Rtf1 Plus3 and Cdc73 C-domain) are indicated within each protein and areas of assigned functions are listed below. The predicted TPR motifs found within Ctr9 (depicted internally in yellow) were defined by the SMART domain server [178]. The percent amino acid identity within defined functional and/or structural domains is depicted by color (see legend). Information on domains of the human Paf1C components have been described previously [168].
Information on the interactions among members of the Paf1C is emerging. In yeast, the overexpression or deletion of individual subunits can influence the levels of other complex members. For example, the overexpression of Cdc73 or Paf1 increases the cellular levels of the other protein by enhancing protein stability [3]. Jaehning and coworkers subsequently showed that deletion of PAF1 decreases the cellular levels of Rtf1, Cdc73, and Ctr9 more than ten-fold, deletion of CDC73 decreases the levels of Rtf1, Paf1, and Leo1 at least three-fold, and deletion of CTR9 also decreases the levels of Paf1, Rtf1, and Leo1 [19]. Consistent with these findings, knockdown of hCdc73 lowers hPaf1 protein levels in human cells [20]. With respect to inter-subunit interactions, the association of Rtf1 with the rest of the Paf1C is significantly reduced in yeast cells lacking CDC73 [21]. However, in rtf1Δ or cdc73Δ cells, Paf1 still interacts with Ctr9 and Leo1, and in rtf1Δ cells, Cdc73 remains associated with Ctr9, Leo1, and Paf1 [21]. The interactions between human Paf1C components have been defined in vitro, revealing an extensive set of binary interactions between individual subunits reinforced by the function of the Paf1 component as a likely scaffold [16].
Deletion of genes encoding individual members of the yPaf1C causes mutant phenotypes that vary in their severity. The loss of Paf1 or Ctr9 is the most detrimental to cellular growth, correlating with the importance of these two subunits for overall complex integrity [22]. While none of the five Paf1C subunits are essential for S. cerevisiae viability, their functional importance is evident from the range of phenotypes caused by mutations in individual genes. These phenotypes include the Spt− (suppression of Ty) phenotype, which is indicative of defects in chromatin and transcription, sensitivity to the base analog 6-azauracil, which is frequently used as an indicator of transcription elongation defects, and sensitivity to compounds that elicit cellular stress responses, including caffeine, cycloheximide, rapamycin, hygromycin, and high temperature [5, 11, 12, 22, 23]. Loss of individual yPaf1C components can also alter the phenotypes associated with loss of other complex members [22]. For example, the deletion of RTF1 can partially rescue the caffeine and hydroxyurea sensitivity and restore proper mRNA levels of selected genes in paf1Δ cells, indicating that the reduced levels of Rtf1 in paf1Δ cells retain some activity outside the normal complex [6]. Analysis of the gene expression profiles of yeast cells lacking individual Paf1C components shows some overlap and some differences [24]. Although these observations indicate that the subunits of the Paf1C may have distinct functions, more experiments are needed to assign definitive functional classifications to individual Paf1C components. To date, specific separation-of-function alleles have only been reported for yeast RTF1, with regions identified as being important for histone modifications, interactions with other Paf1C subunits, Paf1C association with chromatin, and recruitment of transcription co-factors [23]. Unlike in S. cerevisiae, Paf1C components in higher eukaryotes are essential for viability, suggesting that the complex acquired additional functional roles over the course of evolution or that the recognized functions of the complex have essential consequences in these organisms [25, 26]. The characterization of the Paf1C in many eukaryotes has highlighted its conserved roles in transcriptional processes.
3. Connections between the Paf1C and the RNA pol II transcription elongation machinery
The initial identification of Paf1 as an RNA pol II-interacting protein implicated the Paf1C in transcription. Beyond this physical interaction, genetic studies in yeast further suggested a connection between the Paf1C and RNA pol II transcription. For example, strong synthetic growth defects were observed in yeast strains simultaneously mutated in a component of the Paf1C and other important transcriptional regulatory proteins, including subunits of the Mediator coactivator complex, transcription elongation factors, histone modifying proteins, the CTD of the Rpb1 subunit of RNA pol II, and proteins regulating CTD phosphorylation, such as the Ctk1 kinase and Fcp1 phosphatase [5, 12, 27, 28]. Subsequently, a convergence of approaches revealed a prominent role for the Paf1C in the elongation stage of the transcription cycle.
The work of multiple labs showed that the Paf1C could be purified in association with highly conserved transcription elongation factors including the Spt4-Spt5/DSIF complex and the Spt16-Pob3/FACT complex [5, 6, 29, 30]. The Spt4-Spt5/DSIF (DRB Sensitivity Inducing Factor) complex interacts directly with RNA pol II and, through regulating RNA pol II pausing and processivity, has both negative and positive effects on transcription elongation [31]. The FACT complex (Facilitates Chromatin Transcription/transactions) promotes transcription elongation through nucleosomes by functioning as a histone chaperone [32]. It has been suggested that the Paf1C mediates the optimal interaction between RNA pol II and FACT [25]. Furthermore, in HeLa cell nuclear extracts, the binding of DSIF and Paf1C with RNA pol II is cooperative [29]. These observations are supported by studies in yeast showing that while DSIF and FACT can be found at actively transcribed chromatin in the absence of Paf1C components, their association is slightly reduced compared to that in wild-type cells [19, 33]. Human Paf1C also shows interactions in vivo and in vitro with the elongation factor TFIIS, which together can bind and stimulate RNA pol II cooperatively [16]. Collectively, the physical and functional interactions with DSIF, FACT, and TFIIS suggested important roles for the Paf1C in the control of transcription elongation.
Chromatin immunoprecipitation studies, both on individual genes and on a genome-wide scale, demonstrated that the yeast Paf1C can be found at active genes from the transcriptional start site to the poly(A) site [34–36]. Although the yPaf1C was first identified through affinity purification of RNA pol II using an antibody directed against the hypophosphorylated CTD, subsequent work has shown that kinases important for RNA pol II CTD phosphorylation, including Kin28 and Bur1-2, stimulate the association of yPaf1C with coding regions [1, 37–41]. The Bur1-Bur2 cyclin-dependent kinase (CDK) complex is the ortholog of the higher eukaryotic P-TEFb kinase (Positive Transcription Elongation Factor b). The Cak1 kinase (CDK-Activating Kinase) phosphorylates Bur1 [42]. Upon its phosphorylation and activation, Bur1-Bur2 targets multiple proteins for phosphorylation, including Rpb1, Rad6, and Spt5 in its C-terminal region (CTR) [40, 43]. Experiments indicate that the Bur1-Bur2-dependent recruitment of the yPaf1C to chromatin is mediated in part by Spt5-CTR phosphorylation [37, 39, 40, 43]. Considering that P-TEFb in humans also phosphorylates the conserved Spt5-CTR, some aspects of Paf1C recruitment may be conserved [44]. Yet different mechanisms may also be utilized, as Spt5-CTR phosphorylation is not absolutely required for hPaf1C interaction with DSIF, but is necessary for stimulating elongation [29].
Within the yeast Paf1C, the Rtf1 and Cdc73 subunits play key roles in recruiting the whole complex to chromatin [17, 19, 21, 23]. The central Plus3 domain of Rtf1, also termed the ORF Association Region (OAR), contains amino acids important to Paf1C recruitment, as deletion of those residues greatly impairs chromatin association [23]. The C-terminal domain of yCdc73 is important for full levels of Paf1C recruitment to active genes, which may explain why this is the most highly conserved part of the protein [17, 41]. More work, particularly involving the identification of binding partners, is needed to define how these members of the yPaf1C ensure proper chromatin association with active open reading frames (ORFs). Interestingly, in a very recent study, the C-terminal domain of yCdc73 was shown to bind CTD and Spt5-CTR repeat peptides in a phosphorylation-specific manner, indicating that both RNA pol II and Spt5 are directly involved in Paf1C recruitment [41]. There is also some evidence from in vitro studies that the Paf1C can bind nucleic acids directly. Through the Leo1 subunit, the yeast Paf1C can bind RNA in vitro, and RNase treatment of extracts reduces the chromatin association of the Paf1C [45]. In addition, the conserved Plus3 domain of hRtf1 has been shown to exhibit DNA binding activity in vitro [18]. Finally, recent work on the hPaf1C has demonstrated a role for the hPaf1 subunit in recruiting the complex to chromatin through a specific interaction with histone H3 dimethylated on arginine 17 [46]. Cumulatively, these studies connecting the Paf1C and transcription components laid the groundwork for experiments that defined what is now the best-characterized cellular role of Paf1C in promoting histone modifications.
4. Conserved roles of the Paf1C in transcription-coupled histone modifications
The organization of eukaryotic genomes is subject to dynamic regulation. The positioning of nucleosomes is not static, as nucleosomes can be actively disassembled and reassembled by chromatin remodelers and histone chaperones. Canonical nucleosomes contain DNA wrapped around an octamer of histone proteins (two copies each of histones H2A, H2B, H3, and H4), the posttranslational modifications of which can alter DNA accessibility. The Paf1C has been shown to regulate transcription in part by promoting specific modifications of histones, including the methylation of lysine residues, which can occur in mono, di, or trimethylation states. One of the first connections between the Paf1C and histone modifications was revealed in yeast, based on the findings that rtf1Δ and paf1Δ cells showed decreased histone H3 lysine 4 (K4) trimethylation and decreased association of the Set1 histone methyltransferase responsible for this mark at the 5’-ends of genes [47, 48].
The Paf1C is now known to impact the modification of multiple histone residues associated with active transcription. For example, Paf1, Ctr9 and, to a lesser degree, Cdc73, are specifically needed for trimethylation of histone H3 K36 by the Set2 methyltransferase [Figure 2A; [27]]. The Paf1C also promotes the monoubiquitylation of histone H2B K123 by enhancing recruitment of the ubiquitin-conjugating enzyme Rad6 and the ubiquitin protein ligase Bre1 to chromatin [Figure 2B; [49–52]]. The Rtf1 subunit of the yPaf1C is thought to be primarily responsible for this modification, and amino acids 62–152 of yRtf1 are considered the histone modification domain (HMD), as these residues are necessary and sufficient for H2B K123 ubiquitylation in vivo [23, 53, 54]. Interestingly, residues within the yRtf1 HMD that are required for H2B K123 ubiquitylation are conserved across species, such as the invariant residue Glu104 [53]. H2B monoubiquitylation is a prerequisite for di- and trimethylation of histone H3 K4 and K79 by the Set1 and Dot1 methyltransferases, respectively [55–58]. The dependency of these histone modifications on the Paf1C may explain why the genetic interactions of cells lacking yPaf1C components cluster with those of cells lacking H3 K4 and H2B K123 modifying proteins [39]. The human Paf1C helps recruit hRad6 and hBre1/Rnf20 to monoubiquitylate H2B on K120, the residue that is analogous to S. cerevisiae K123 [59, 60]. An interaction was shown between the hPaf1C and hBre1 in vivo and in vitro, suggesting that the Paf1C-dependence for Bre1-Rad6 recruitment could be mediated directly by the Paf1C [59, 61]. Similar to yeast, H2B K120 monoubiquitylation facilitates hSet1/MLL1-dependent methylation of H3 K4 and hDot1-dependent methylation of H3 K79 [59, 62]. Importantly, Paf1C-dependent histone modifications in both yeast and human cells can influence the recruitment and/or activities of proteins that further impact chromatin accessibility on active genes, including those that control histone acetylation levels [63–71].
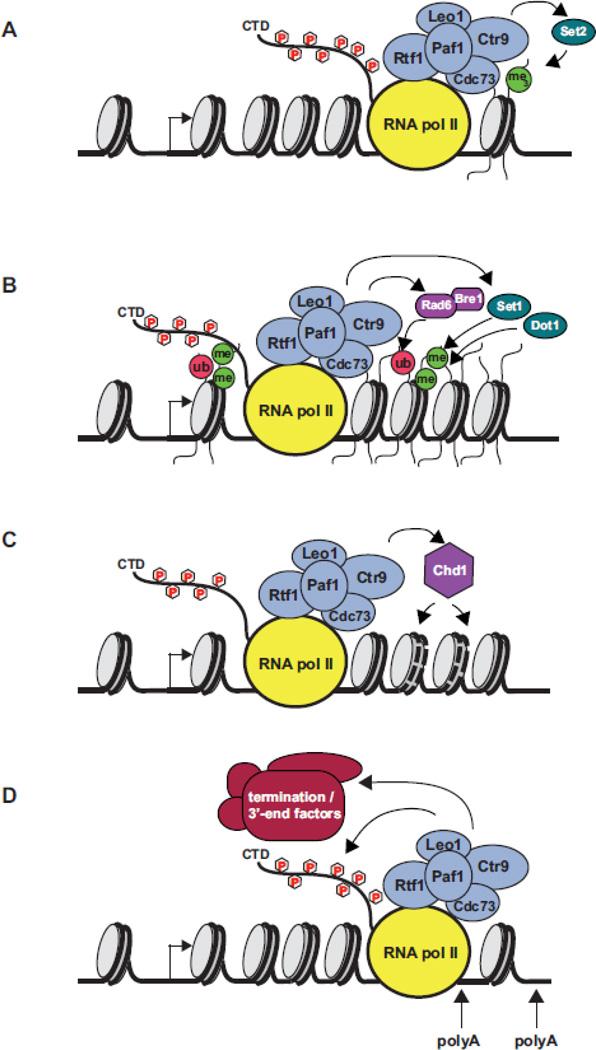
Figure 2. Molecular activities of the Paf1C.
(A) During transcription elongation, Paf1C promotes H3 K36 trimethylation by stimulating the recruitment of Set2 to ORFs. (B) The Paf1C is required for the proper recruitment and activity of Rad6-Bre1, which targets H2B K123 for monoubiquitylation in yeast. This modification facilitates di- and tri-methylation of H3 K4 and H3 K79 by the Set1/COMPASS complex and Dot1, respectively. A functional Paf1C is also required for proper recruitment of Set1 to active genes. (C) The Paf1C recruits the ATP-dependent chromatin remodeling factor Chd1 to ORFs through an interaction site near the N-terminus of Rtf1. Dotted lines on the nucleosomes represent presumed positional changes that occur as a consequence of Chd1 action. (D) Toward the 3’ ends of genes, the Paf1C is required for optimal Ser2-P levels on the RNA pol II CTD and proper recruitment of several RNA 3’-end processing and termination factors. Defects in recruiting these factors, as in paf1 mutant strains, lead to altered poly(A) site selection at some protein-coding genes and transcriptional read-through into downstream sequences at snoRNA genes (not depicted). Bent arrows indicate the start site of transcription. Factors involved in Paf1C recruitment, such as Spt5, are omitted for clarity.
As described previously, the yeast Paf1C colocalizes with RNA pol II from the transcriptional start site to the poly(A) site at all active genes examined [35, 36]. However, the recruitment of histone modifiers is not uniform throughout the whole gene, but occurs in distinct patterns along ORFs. Therefore, additional factors must affect the activity and localization of these enzymes at specific sites. This may be in part due to effects of the Paf1C on RNA pol II CTD phosphorylation. The CTD of Rpb1 consists of tandem repeats of a heptad amino acid sequence, YSPTSPS, whose serines undergo dynamic changes in phosphorylation during the transcription cycle [72, 73]. The Rpb1 CTD is in a hypophosphorylated state when RNA pol II is recruited to the promoter, becomes phosphorylated on serine 5 (Ser5-P) and serine 7 (Ser7-P) residues near the 5’ ends of genes and acquires serine 2 phosphorylation (Ser2-P) later in elongation toward the 3’ ends of genes [72]. This temporal cycle of phosphorylation represents an essential level of transcriptional regulation, which is further enhanced by RNA pol II-interacting proteins that have preferences for particular CTD phosphorylation states. Most relevant to this review, deletion of genes encoding certain yPaf1C subunits decreases the levels of Ser2-P on ORFs, an effect that could in turn impact the downstream recruitment of certain RNA pol II-interacting proteins [21]. For example, the decrease in Ser2-P levels in paf1Δ cells is likely to be partially responsible for the decreased chromatin association of Set2 and H3 K36 trimethylation levels measured in these strains [28, 47, 74–76].
The idea of the Paf1C promoting proper localization of histone modifications along a gene is supported by studies in plants. In Arabidopsis thaliana, the loss of the Paf1C was found to shift the distribution patterns of H3 K4 trimethylation and H3K36 dimethylation within ORFs, especially among highly transcribed genes [77]. Interestingly and unlike in other eukaryotes, the loss of the Paf1C in plants did not substantially impact global levels of these modifications, but instead changed histone methylation states at a specific set of FLC family genes whose expression depends on the Paf1C [77–79]. The Paf1C in plants may also be needed for proper histone H3 and nucleosome levels in highly transcribed and longer genes [77]. In support of this observation, the hPaf1C has been shown to bind to histone H3 [80]. A role for the Paf1C in affecting nucleosomes may be conserved, because in yeast the Paf1C can impact nucleosome occupancy at transcribed regions of the genome [33, 81]. The observation of altered nucleosome occupancy at an active gene in paf1Δ and ctr9Δ cells could be partially explained through the effects of H2B ubiquitylation on nucleosome stability [81, 82]. Further connections between the Paf1C and nucleosome dynamics are illustrated by the interaction with the conserved chromatin remodeling protein Chd1 [Figure 2C]. Specifically, Rtf1 is required for full Chd1 recruitment to active genes in yeast [83]. Interestingly, Chd1 was recently shown to be required for full levels of H2B K123 monoubiquitylation [84]. Together these studies support a conserved and important role for the Paf1C in promoting transcription-coupled histone modifications and impacting nucleosome dynamics.
5. Transcriptional effects of the Paf1C
Many studies suggest a central involvement of the Paf1C in transcription. Although the Paf1C interacts with RNA pol II, the absence of Paf1C subunits does not generally reduce the levels of RNA pol II associated with an active gene in vivo, though sometimes a Paf1C-dependent change in distribution of RNA pol II along a gene can be observed [19, 25, 85, 86]. Evidence supporting a direct role for the Paf1C in promoting transcription elongation came primarily from biochemical studies in yeast and humans. In vitro approaches showed hPaf1C can facilitate elongation and that the absence of yPaf1 components impaired elongation efficiency [16, 29, 60, 87]. Furthermore, Paf1C-dependent histone modifications such as H2B monoubiquitylation facilitate FACT function and transcription elongation [60]. Additional evidence that the Paf1C impacts elongation came from in vivo experiments showing elongation defects on reporter templates in the absence of individual yPaf1C components [88]. These studies defined an important role for the Paf1C in RNA pol II transcription elongation.
Although the Paf1C affects transcription elongation efficiency in vitro, not all genes are impacted by the loss of the Paf1C in yeast cells. Transcriptome analyses showed that approximately 15–20% of genes are affected, either positively or negatively, by the deletion of yPaf1 [24]. Because paf1Δ cells have altered levels of some transcription factors and phenotypes associated with cellular stress, not all of these effects on gene expression are likely to be direct. In fact the levels of occupancy of Ctr9 and Paf1 at selected genes does not always correlate with Paf1C-dependent expression changes [81]. Furthermore, while biochemical studies have demonstrated a stimulatory effect of Paf1C on chromatin templates, deletion of PAF1 leads to increased expression of several hundred yeast genes, indicating a role in gene repression [16, 24, 29, 60, 87]. For the yeast ARG1 gene, the repressive effects of the Paf1C are mediated, in part, through its histone modification functions [89, 90]. Identification of RNAs directly dependent on yPaf1 showed an overrepresentation of essential genes, which may explain why paf1Δ cells have compromised fitness [24]. Very recently, Paf1C homologues have been found to influence expression of many essential genes in the obligate parasite Trypanosoma brucei, which suggests that functions of Paf1C components may be even more broadly conserved then previously appreciated [91].
The Paf1C has also been shown to play a role in gene silencing. Certain regions of the yeast genome are transcriptionally silent and exhibit a chromatin structure similar to heterochromatin. These regions include the silent mating type cassettes, the ribosomal DNA (rDNA) locus, and telomeric regions of chromosomes [92]. As measured through a commonly used reporter assay, proper telomeric silencing is Paf1C-dependent, and this is likely due to the roles of the Paf1C in Set1-dependent H3 K4 methylation, Dot1-dependent H3 K79 methylation, and Set2-dependent H3 K36 methylation [48, 49, 93, 94]. Interestingly, telomerase RNA levels are decreased in strains lacking yPaf1C members, which leads to a shortening of telomere length [95]. Further work is needed to determine whether the Paf1C-mediated silencing of natural telomere-proximal genes mimics that of the reporter genes [96, 97]. The Paf1C is also required for H3 K4 methylation-dependent rDNA silencing [98]. The proteins comprising the yPaf1C are primarily nuclear; however, upon the loss of other complex members, Paf1C subunits show some nucleolar localization [2, 3, 99]. The nucleolus is the site of both the transcription of rDNA by RNA pol I and the subsequent assembly of ribosomes. To date, the Paf1C has been shown to associate with rDNA and impact RNA pol I transcription [100].
6. Participation of the Paf1C in RNA 3’-end formation
Beyond contributing to transcription elongation, the Paf1C also regulates transcript termination and processing [Figure 2D]. Loss of yPaf1C components results in shorter poly(A) tail lengths of mRNAs and alternative poly(A) site selection at some genes [19, 24]. This likely stems from decreased recruitment of 3’-end mRNA processing factors such as the Cft1 protein, which is part of the CPF complex (Cleavage and Polyadenylation Factors) [21]. Although the reduced recruitment of 3’-end processing factors to elongating RNA pol II might be explained by the reduced levels of CTD Ser2-P levels in paf1Δ cells, physical interactions between yPaf1C and these factors have also been reported [21]. A conserved role for the Paf1C in RNA 3’-end formation is suggested from the observations that the hPaf1C associates with the homologous CPSF complex (Cleavage and Polyadenylation Specificity Factors) and another RNA processing complex, CstF [101]. Depletion of hPaf1C components in cells results in decreased mRNA polyadenylation [102]. Furthermore, a posttranscriptional role for hCdc73 was found in histone mRNA 3’-end processing, and in accordance with this observation, the yPaf1C can affect levels of histone mRNAs [103, 104]. Finally, physical and genetic interactions link the yPaf1C to the CCR4-NOT cytoplasmic deadenylase complex and the THO-TREX complex, which play important roles in RNA processing and export [105].
In addition to regulating mRNA processing, the Paf1C is also important for the efficient 3’-end formation of nonpolyadenylated RNA pol II transcripts such as small nucleolar RNAs (snoRNAs), which are one set of non-coding RNAs (ncRNAs) [85]. After being transcribed, terminated, and processed, snoRNAs associate with proteins to form small nucleolar ribonucleoproteins (snoRNPs) that catalyze essential ribosomal RNA (rRNA) modification and cleavage reactions during rRNA processing and ribosome biogenesis [106]. To date, the Paf1C has been shown to be required for proper RNA 3’-end formation at two yeast snoRNA genes, SNR13 and SNR47 [53, 85]. In paf1Δ cells, transcripts expressed from these loci have extended 3’ ends [85]. Some Paf1C-dependent histone modifications have been connected to snoRNA 3’-end formation. For example, H2B K123 ubiquitylation is required for efficient 3’-end formation of SNR47 and SNR13 transcripts, and H3 K4 trimethylation promotes proper SNR13 termination [53, 107]. Interestingly, the gene showing the strongest decrease in expression in response to depletion of hCdc73 was a component of the Integrator complex, which mediates 3’-end processing of ncRNAs [101].
The yeast Paf1C participates within a larger regulatory pathway to promote proper snoRNA termination, which involves the two essential RNA binding proteins Nrd1 and Nab3 and the helicase Sen1 [108–110]. Specific binding sites direct Nrd1 and Nab3 to snoRNA transcripts where they interact with RNA pol II and recruit other proteins involved in processing these RNAs [111–115]. The exosome complex is recruited to ncRNA targets and cleaves the extended snoRNA transcripts to their mature length [116]. Therefore, impairing the function of Nrd1, Nab3, Sen1, the exosome, or other proteins involved in termination causes extended snoRNA transcripts that often continue into the downstream genes. Importantly, these same proteins were recently shown to be involved in regulation of another class of ncRNAs in yeast called cryptic unstable transcripts (CUTs) [117–119]. CUTs are similar to some short and unstable ncRNA transcripts found in human cells [120]. Even though the exosome quickly degrades CUTs in wild-type yeast cells, these transcripts, which occur in the sense or antisense direction, can influence gene transcription [121]. The use of genomic assays has made the pace of discovery of CUTs faster than the detailed understanding of their molecular regulation. However, one study showed that an interaction conserved from yeast to humans between the Paf1C and the MAP kinase Mpk1 can influence the recruitment of Nrd1-Nab3-Sen1, providing a possible mechanistic explanation for the genetic interactions between the Paf1C and Pkc1-Mpk1 kinase cascade in yeast [105, 122]. Understanding if the Nrd1-Nab3-Sen1 pathway regulates CUTs through mechanisms similar to those used in snoRNA termination may also uncover if and how the Paf1C participates in the regulation of CUTs.
7. Connections of the Paf1C to the cell cycle, DNA repair and other processes
Although the levels of the yeast Paf1C subunits and the mRNAs encoding them appear to be constant, the expression of hPaf1 changes during the cell cycle [123, 124]. In fact, the Paf1C has many interesting connections to cell cycle regulation, beginning with the identification of CDC73 in a yeast genetic screen for cell cycle regulators [4]. Yeast and human cells with misregulated levels of Paf1C proteins appear to accumulate in the G1 phase of the cell cycle with a delayed S phase entry, which may be due to the poor expression or activation of cyclins needed for the transition to S phase [123, 125, 126]. In yeast, Cln3 and Cdc28 activate the SBF (Swi4/6) transcription factor by promoting nuclear export of its inhibitor Whi5, which allows for synthesis of Cln1 and Cln2. Genetic connections to this process are found with yPaf1C mutants. Deletion of PAF1 or CTR9 is synthetically lethal with a deletion of CLN3, and in the absence of either SWI4 or SWI6, PAF1 is required for viability [9, 124]. In both yeast and human cells, the Paf1C has been shown to influence expression of genes involved in the cell cycle [123, 124]. Direct roles for Paf1C components in regulating c-myc and cyclin D levels, which can influence cell cycle progression, have been observed in human cells [123, 127–129]. Furthermore, connections of the Paf1C to chromosome loss and microtubule dynamics have been discovered. Yeast cells containing a CTR9 mutant allele show substantial increases in chromosome loss and multinucleated cells [8]. Cells depleted for hPaf1 show impaired mitotic spindles [123]. In addition to alterations in gene expression and histone modifications, this may be due to a direct role of the Paf1C in proper microtubule assembly, as hCdc73 interacts with actin binding proteins [130].
Interestingly, the Paf1C-dependent H2B K123 ubiquitylation mark helps promote Set1-dependent methylation of a non-histone target, the K233 residue of the kinetochore protein Dam1 [131]. The phosphorylation of Dam1 by the Aurora kinase Ipl1 destabilizes improper kinetochore-microtubule attachments [132]. It remains to be determined if methylation of K233 on Dam1 contributes to regulation of chromosome segregation during the cell cycle. More detailed analysis of microtubule assembly in synchronized cells each lacking a component of the Paf1C may help clarify how the Paf1C impacts the cell cycle and tubulin dynamics. It is also currently unclear what non-histone targets of Set1 exist in higher eukaryotes.
Histone modifications dependent on the Paf1C are well documented as being important for the repair of DNA damage in both yeast and human systems. For example, Dot1 participates in the repair of DNA double strand breaks (DSBs) and can promote recruitment of Rad9/52BP1, which is involved in DNA damage checkpoint signaling [133–135]. Rad6 and Bre1 also are involved in recruitment of downstream effectors of DSB repair like Rad51 and Brca1 [136, 137]. The repair of DNA damage by nucleotide excision repair (NER) occurs thorough two major pathways, global genome (GG-NER) repair and transcription coupled repair (TC-NER). Members of the Paf1C complex, histone modifiers and FACT have all been implicated to differing extents as participating in these NER pathways [138–140]. Additional studies will be needed to elucidate the contribution of the Paf1C to DNA damage repair pathways and determine if all of its roles require its histone modifications functions.
Additional evidence implicates the Paf1C in regulating mRNA translation. For example, the Drosophila Paf1C interacts with cytoplasmic polyadenylation element binding proteins (CPEB), which are translational regulatory proteins [20]. Also, members of the yPaf1C can influence nonsense protein suppression by the aggregated prion form of the Sup35 translation termination factor, [PSI+], without completely curing cells of this prion [141]. Future work may provide mechanistic insights into how the Paf1C impacts nonsense suppression and possibly other events in translation.
8. The involvement of the Paf1C in stem cell biology and development
Embryonic stem (ES) cells are pluripotent and capable of becoming any fetal or adult cell type through the process of differentiation. The Paf1C has roles in maintaining ES cell pluripotency and preventing expression of genes involved in lineage specification [Figure 3]. A screen in human ES cells identified hRtf1 and hCtr9 as impacting expression levels of OCT4/POU5F1 [142]. Additional characterization showed that all the other members of the hPaf1C had similar effects and could be found associated with the promoters of genes involved in ES cell biology, such as genes encoding Oct4, Nanog, and Sox2 [142]. The Oct4, Nanog, and Sox2 transcription factors are critically important for ES cell self-renewal. A physical interaction between murine Paf1C, RNA pol II, and Oct3/4 in ES cells may implicate the Paf1C in participating in the autoregulatory loop that feeds forward to enhance transcription of Oct4, Sox3 and Nanog [143]. Importantly, depletion of Paf1C components in human and murine ES cells leads to a decrease in expression of transcription factors required for pluripotency and an increase in genes that control lineage specification [142, 143]. Furthermore, it was found that histone modifications may underlie the Paf1C dependency of ES cell gene expression; upon depletion of Paf1C subunits, pluripotency gene promoters were downregulated and showed decreased H3 K4 trimethylation, which normally marks active genes, and upregulated cell fate gene promoters showed decreased levels of H3 K27 trimethylation, a repressive mark [142]. In agreement with these results, mammalian Chd1 specifically interacts with H3 trimethylated on K4 and promotes ES cell pluripotency [144]. These findings, together with the observation that differentiation is accompanied by a global decrease in Paf1C-dependent H2B ubiquitylation, demonstrate that the effects of Paf1C on histone modifications can influence the expression of key genes in ES cells and alter their pluripotency [145].
Figure 3. Roles of the human Paf1C complex in pathways of development and disease.
The Paf1C in higher eukaryotes has been found to influence signaling within Wg/Wnt, Hedgehog, and Notch pathways of development and IL-6 pathways of immunological responses. Misregulation of these pathways has been shown to lead to cancer. Additionally, the Paf1C can promote pluripotency of stem cells through effects on DNA methylation as well as increasing expression of genes dependent on the Oct4, Nanog, and Sox2 transcription factors involved in ES cell self-renewal. The hPaf1C has been shown to provide resistance to HIV and SIV infections and can trigger antiviral responses to influenza unless bound by the H2N2 flu subtype NS1 protein. Mutations in hCdc73/parafibromin can lead to hyperparathyroidism-jaw tumor (HPT-JT) syndrome. Misregulation of Paf1C components have been found in tumor cells and direct binding with MLL and the SEC implicates the Paf1C in pathways of transformation leading to leukemia.
Another way gene expression is controlled in stem cells is through DNA methylation of gene promoters, which can be misregulated in human cancer cell lines [146]. An interaction in murine ES cells between the Paf1C and the de novo DNA methyltransferases DNMT3A and B suggests that the Paf1C may also be involved in DNA methylation pathways in ES cells that influence lineage commitment [147]. Consistent with the idea that the Paf1C has pluripotency targets that need to be turned off to initiate cellular differentiation, a recent study showed that expression of hPaf1C components is downregulated during hematopoietic differentiation [148].
In light of its global roles in gene regulation, it may not be surprising that the Paf1C is important for the development of multicellular organisms [26, 149, 150]. For example, Paf1C in zebrafish is required for proper development of the heart, ears, and neural crest cells [150–152]. Furthermore, homozygous null mice lacking the gene encoding Cdc73, HRPT2, are embryonic lethal at day E6.5 [26]. Depletion of HRPT2 in adult mice results in death within approximately 20 days and homozygous null HRPT2−/− MEFs undergo apoptosis [26]. The contributions of the Paf1C to development may be mediated through effects on key developmental signaling cascades, such as the Wg/Wnt, Hedgehog, and Notch pathways [Figure 3]. The Drosophila Paf1C can bind the Hedgehog pathway Gli/Ci transcription factors and influence their effects on downstream gene expression [153]. A function for Paf1C-dependent histone modifications in transcription of Drosophila Notch target genes was further solidified by the demonstration of a role for Rtf1 in Notch signaling [149, 154]. Additionally, Drosophila hyrax (Cdc73) mediates Wg/Wnt signaling through the key transcription factor β-catenin/armadillo. Components of Paf1C were shown to directly bind β-catenin/armadillo and recruit other signaling mediators to activate gene expression [155]. Additional work in human cells suggested that this process may involve a dephosphorylation event on hCdc73 (parafibromin) [156]. Although the yeast Cdc73 protein could not rescue defects in fly development, the hCdc73/parafibromin was capable of restoring proper development [155]. Further underscoring functional conservation in higher eukaryotes, Drosophila Cdc73 could rescue Wnt signaling defects in cultured human cells [155]. The importance of these conserved signaling pathways goes beyond developmental processes, as tumor cells show perturbations in these signaling cascades. In fact, human parathyroid carcinomas, like those in patients harboring altered hCdc73/parafibromin, demonstrate aberrant Wnt/β-catenin signaling [157]. Indeed, the Paf1C has important functions in the prevention of a wide range of diseases, as discussed in greater detail below.
9. The Paf1C affects immunological responses to disease states and cancer progression
The Paf1C has general effects on immunological signaling cascades and transcription factors as well as roles in specific diseases [Figure 3]. The murine Paf1C directly recruits the STAT3 transcriptional activator to the promoters of IL-6 responsive genes, such as those encoding cyclin D and c-myc [158]. This IL-6 STAT3 pathway has been implicated in colon cancer [159]. Furthermore, H2B ubiquitylation is actively regulated at other IL-6 responsive genes, which are activated by the STAT1 transcription factor [160]. Since hPaf1C is found expressed in lymphocytic cells, it appears to be in the right place to directly impact immunological signaling responses [161].
The Paf1C has been implicated in HIV and influenza resistance in humans. Deficiency of hPaf1C renders cells more susceptible to the influenza virus [80]. hPaf1 binds directly to the H2N2 influenza subtype NS1 protein, a histone H3 mimic, which prevents the hPaf1C from activating transcription of antiviral genes [80]. The Paf1C was also isolated in a screen for resistance factors to HIV-1 replication and was shown to provide resistance to SIV and HIV-2 infections [161]. The hPaf1C was further shown to affect levels of viral transcription and integration [161]. The HIV-1 transactivator Tat can bind Paf1C, and these proteins are recruited together to viral promoters [162]. Further characterization of the role of the Paf1C in the expression of viral response genes may ultimately reveal therapeutic targets for the prevention or treatment of viral infections.
Interesting connections exist between the Paf1C and the processes of cellular transformation. The importance of MLL (Mixed Lineage Leukemia) gene translocations in leukemogenesis is well established [163, 164]. Normally MLL1 encodes a protein with similarity to ySet1 and can promote H3 K4 methylation [165, 166]. After MLL rearrangement and fusion of N-terminal amino acids with a translocation partner, the translocation product activates genes, such as the homeotic (HOX) genes, and causes cell immortalization [164]. The hPaf1C has been shown to interact with the SEC (Super Elongation Complex) that associates with all MLL chimeras and is required for HOX gene expression in leukemic cells [164]. In fact, the Paf1C has been shown to interact in vivo and in vitro with an important MLL domain in MLL proteins even after fusion [148]. Importantly this Paf1C interaction seems to stimulate the ability of MLL to activate transcription by enhancing MLL recruitment to target loci and ultimately is needed for bone marrow cell transformation by an MLL-AF9 fusion protein [148].
Much additional evidence links the hPaf1C to tumor formation. For example, hBre1/Rnf20 is thought to function as a tumor suppressor by inhibiting TFIIS and Paf1C localization at pro-oncogenic genes [167]. The misexpression of each component of the hPaf1C has been observed in different cancers, and the overexpression of hPaf1/PD2 promotes tumor formation both in vitro and in vivo [168, 169]. However, the hPaf1C subunit with the best-defined role in cancer progression is the hCdc73/parafibromin subunit, which is encoded by the hyperparathyroidism 2 gene, HRPT2, and has been classified as a tumor suppressor gene [168]. Mutations in HRPT2 are associated with hyperparathyroidism-jaw tumor (HPT-JT) syndrome, which is one of the most common disorders of the endocrine system and can frequently present with parathyroid tumors [170, 171]. Mutations that inactivate, truncate, or reduce the levels of hCdc73/parafibromin account for more than two-thirds of the patients presenting with parathyroid carcinomas [172]. Furthermore, unlike wild-type parafibromin, the protein expressed in tumors no longer binds Paf1 and RNA pol II, indicating misregulation of parafibromin in cancer cells may be in part due to its separation from the rest of the Paf1 complex [15]. hCdc73/parafibromin can suppress tumor formation by blocking cell proliferation and affecting cyclin D and c-myc expression, which gives it a role in other cancers as well, including glioblastoma and carcinomas of the liver, stomach, and breast [156, 168, 173]. Clinical research suggests that the levels of parafibromin could serve as a prognostic indicator of colon, stomach, and breast cancer [174–176]. Given that the misexpression of Paf1C subunits other than hCdc73/parafibromin was also observed in tumor cells, we believe that further research into all components of the Paf1C will better define the role of this highly conserved complex in cancer and other diseases.
10. Concluding Remarks
Since its initial discovery as an RNA pol II-interacting complex in yeast, an understanding of the structure, function, conservation, and cellular roles of the Paf1C has rapidly emerged. The finding that the Paf1C travels with RNA pol II on all actively transcribed genes examined explains the broad effects of this complex on histone modification patterns and gene expression. Moreover, the physical association of the Paf1C with RNA pol II from the beginning of an ORF to the poly(A) site provides insight into how this one complex can impact multiple steps in RNA synthesis, including transcription termination. Despite a great deal of progress in recent years, the molecular details of how the Paf1C is recruited to RNA pol II and, once there, how it facilitates diverse histone modifications, modulates CTD phosphorylation levels, and governs RNA 3’-end formation remain unclear. While the histone modification function of the Paf1C is its most recognized activity, current data indicate that the complex has functions beyond this role. Consistent with this idea, mutations that strongly impair Paf1C-dependent histone modifications do not always phenocopy absence of the entire complex. Likewise, not all functions of the Paf1C, such as its connections to DNA damage repair, are directly explained by its association with RNA pol II transcription. A challenge for the future will be to elucidate all of the molecular functions of the Paf1C, determine which of these functions are responsible for its diverse biological outcomes, and identify therapeutic strategies for overcoming mutations in the complex that underlie disease states. The importance of this challenge is evident from the large number of connections that exist between the Paf1C and human health.
Highlights.
The Paf1 complex associates with RNA polymerase II on all active genes examined
The Paf1 complex couples conserved histone modifications to transcript elongation
The Paf1 complex regulates post-transcriptional events in gene expression
In higher eukaryotes, the Paf1 complex has diverse functions in cell cycle control, development, and disease
Acknowledgements
We would like to thank Joe Martens for valuable comments on the manuscript and Andrew VanDemark and Adam Wier for assistance with Figure 1. Research in the Arndt lab is supported by a National Institute of Health grant GM52593 to K.M.A. and by Award Number F32GM093383 to B.N.T. from the National Institute of General Medical Sciences. The content of this review is solely the responsibility of the authors and does not represent the views of the funding institutions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wade PA, Werel W, Fentzke RC, Thompson NE, Leykam JF, Burgess RR, Jaehning JA, Burton ZF. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr Purif. 1996;8:85–90. doi: 10.1006/prep.1996.0077. [DOI] [PubMed] [Google Scholar]
- 2.Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed SI, Ferguson J, Jahng KY. Isolation and characterization of two genes encoding yeast mating pheromone signaling elements: CDC72 and CDC73. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):621–627. doi: 10.1101/sqb.1988.053.01.071. [DOI] [PubMed] [Google Scholar]
- 5.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Como CJ, Chang H, Arndt KT. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foreman PK, Davis RW. CDP1, a novel Saccharomyces cerevisiae gene required for proper nuclear division and chromosome segregation. Genetics. 1996;144:1387–1397. doi: 10.1093/genetics/144.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch C, Wollmann P, Dahl M, Lottspeich F. A role for Ctr9p and Paf1p in the regulation G1 cyclin expression in yeast. Nucleic Acids Res. 1999;27:2126–2134. doi: 10.1093/nar/27.10.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magdolen V, Lang P, Mages G, Hermann H, Bandlow W. The gene LEO1 on yeast chromosome XV encodes a non-essential, extremely hydrophilic protein. Biochim Biophys Acta. 1994;1218:205–209. doi: 10.1016/0167-4781(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 11.Stolinski LA, Eisenmann DM, Arndt KM. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4490–4500. doi: 10.1128/mcb.17.8.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa PJ, Arndt KM. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics. 2000;156:535–547. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, Krek W. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amrich CG, Davis CP, Rogal WP, Shirra MK, Heroux A, Gardner RG, Arndt KM, Vandemark AP. The Cdc73 subunit of the Paf1 complex contains a C-terminal Ras-like domain that promotes association of the Paf1 complex with chromatin. J Biol Chem. 2012 doi: 10.1074/jbc.M111.325647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong RN, Truffault V, Diercks T, Ab E, Daniels MA, Kaptein R, Folkers GE. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure. 2008;16:149–159. doi: 10.1016/j.str.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- 20.Zhang JH, Panicker LM, Seigneur EM, Lin L, House CD, Morgan W, Chen WC, Mehta H, Haj-Ali M, Yu ZX, Simonds WF. Cytoplasmic polyadenylation element binding protein is a conserved target of tumor suppressor HRPT2/CDC73. Cell Death Differ. 2010;17:1551–1565. doi: 10.1038/cdd.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordick K, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol Genet Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- 23.Warner MH, Roinick KL, Arndt KM. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol. 2007;27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 25.Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P, Bowl MR, Bender S, Peng J, Farber L, Chen J, Ali A, Zhang Z, Alberts AS, Thakker RV, Shilatifard A, Williams BO, Teh BT. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol Cell Biol. 2008;28:2930–2940. doi: 10.1128/MCB.00654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu Y, Simic R, Warner MH, Arndt KM, Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Formosa T. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta. 2012;1819:247–255. doi: 10.1016/j.bbagrm.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pruneski JA, Hainer SJ, Petrov KO, Martens JA. The Paf1 complex represses SER3 transcription in Saccharomyces cerevisiae by facilitating intergenic transcription-dependent nucleosome occupancy of the SER3 promoter. Eukaryot Cell. 2011;10:1283–1294. doi: 10.1128/EC.05141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 35.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3' ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 37.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol Cell Biol. 2006;26:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu H, Hu C, Gaur NA, Hinnebusch AG. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J. doi: 10.1038/emboj.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao S, Prelich G. Activation of the Bur1-Bur2 cyclin-dependent kinase complex by Cak1. Mol Cell Biol. 2002;22:6750–6758. doi: 10.1128/MCB.22.19.6750-6758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A. 2009;106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFbmediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 45.Dermody JL, Buratowski S. Leo1 subunit of the yeast paf1 complex binds RNA and contributes to complex recruitment. J Biol Chem. 2010;285:33671–33679. doi: 10.1074/jbc.M110.140764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J, Xu W. Histone H3R17me2a mark recruits human RNA polymeraseassociated factor 1 complex to activate transcription. Proc Natl Acad Sci U S A. 2012;109:5675–5680. doi: 10.1073/pnas.1114905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 48.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 49.Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- 50.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 51.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING fingerindependent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem. 2009;284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomson BN, Davis CP, Warner MH, Arndt KM. Identification of a role for histone H2B ubiquitylation in noncoding RNA 3'-end formation through mutational analysis of Rtf1 in Saccharomyces cerevisiae. Genetics. 2011;188:273–289. doi: 10.1534/genetics.111.128645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piro AS, Mayekar MK, Warner MH, Davis CP, Arndt KM. Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc Natl Acad Sci U S A. 2012;109:10837–10842. doi: 10.1073/pnas.1116994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 56.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 57.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 58.Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, Shilatifard A. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 61.Hahn MA, Dickson KA, Jackson S, Clarkson A, Gill AJ, Marsh DJ. The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum Mol Genet. 2012;21:559–568. doi: 10.1093/hmg/ddr490. [DOI] [PubMed] [Google Scholar]
- 62.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 63.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 64.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 65.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. Cotranscriptional set2 methylation of histone H3 lysine36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 66.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 67.Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The Yng1p plant homeodomain finger is a methylhistone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2012;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 72.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 74.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 75.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schaft D, Roguev A, Kotovic KM, Shevchenko A, Sarov M, Neugebauer KM, Stewart AF. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 2003;31:2475–2482. doi: 10.1093/nar/gkg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh S, Park S, van Nocker S. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 2008;4:e1000077. doi: 10.1371/journal.pgen.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalizationresponsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol. 2008;28:1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, Lee K, Garcia-Sastre A, Roeder RG, Tarakhovsky A. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marton HA, Desiderio S. The Paf1 complex promotes displacement of histones upon rapid induction of transcription by RNA polymerase II. BMC Mol Biol. 2008;9:4. doi: 10.1186/1471-2199-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee JS, Garrett AS, Yen K, Takahashi YH, Hu D, Jackson J, Seidel C, Pugh BF, Shilatifard A. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev. 2012;26:914–919. doi: 10.1101/gad.186841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sheldon KE, Mauger DM, Arndt KM. A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3' end formation. Mol Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 87.Rondon AG, Gallardo M, Garcia-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tous C, Rondon AG, Garcia-Rubio M, Gonzalez-Aguilera C, Luna R, Aguilera A. A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO J. 2011;30:1953–1964. doi: 10.1038/emboj.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crisucci EM, Arndt KM. The Paf1 complex represses ARG1 transcription in Saccharomyces cerevisiae by promoting histone modifications. Eukaryot Cell. 2011;10:712–723. doi: 10.1128/EC.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Crisucci EM, Arndt KM. Paf1 restricts Gcn4 occupancy and antisense transcription at the ARG1 promoter. Mol Cell Biol. 2012;32:1150–1163. doi: 10.1128/MCB.06262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ouna BA, Nyambega B, Manful T, Helbig C, Males M, Fadda A, Clayton C. Depletion of Trypanosome CTR9 Leads to Gene Expression Defects. PLoS One. 2012;7:e34256. doi: 10.1371/journal.pone.0034256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 93.Tompa R, Madhani HD. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics. 2007;175:585–593. doi: 10.1534/genetics.106.067751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 95.Mozdy AD, Podell ER, Cech TR. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol Cell Biol. 2008;28:4152–4161. doi: 10.1128/MCB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Takahashi YH, Schulze JM, Jackson J, Hentrich T, Seidel C, Jaspersen SL, Kobor MS, Shilatifard A. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol Cell. 2011;42:118–126. doi: 10.1016/j.molcel.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rossmann MP, Luo W, Tsaponina O, Chabes A, Stillman B. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol Cell. 2011;42:127–136. doi: 10.1016/j.molcel.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Porter SE, Penheiter KL, Jaehning JA. Separation of the Saccharomyces cerevisiae Paf1 complex from RNA polymerase II results in changes in its subnuclear localization. Eukaryot Cell. 2005;4:209–220. doi: 10.1128/EC.4.1.209-220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Sikes ML, Beyer AL, Schneider DA. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci U S A. 2009;106:2153–2158. doi: 10.1073/pnas.0812939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3' mRNA processing factors. Proc Natl Acad Sci U S A. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Farber LJ, Kort EJ, Wang P, Chen J, Teh BT. The tumor suppressor parafibromin is required for posttranscriptional processing of histone mRNA. Mol Carcinog. 2010;49:215–223. doi: 10.1002/mc.20591. [DOI] [PubMed] [Google Scholar]
- 104.Braun MA, Costa PJ, Crisucci EM, Arndt KM. Identification of Rkr1, a nuclear RING domain protein with functional connections to chromatin modification in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:2800–2811. doi: 10.1128/MCB.01947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 107.Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol Cell Biol. 2011;31:3569–3583. doi: 10.1128/MCB.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 109.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3'-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 110.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 111.Creamer TJ, Darby MM, Jamonnak N, Schaughency P, Hao H, Wheelan SJ, Corden JL. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 2011;7:e1002329. doi: 10.1371/journal.pgen.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jamonnak N, Creamer TJ, Darby MM, Schaughency P, Wheelan SJ, Corden JL. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA. 2011;17:2011–2025. doi: 10.1261/rna.2840711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt JF, Nagy PL. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 115.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1- Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3'- extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 118.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 120.Carninci P. Molecular biology: The long and short of RNAs. Nature. 2009;457:974–975. doi: 10.1038/457974b. [DOI] [PubMed] [Google Scholar]
- 121.Colin J, Libri D, Porrua O. Cryptic transcription and early termination in the control of gene expression. Genet Res Int. 2011;2011:653494. doi: 10.4061/2011/653494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim KY, Levin DE. Mpk1 MAPK association with the paf1 complex blocks sen1-mediated premature transcription termination. Cell. 2011;144:745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moniaux N, Nemos C, Deb S, Zhu B, Dornreiter I, Hollingsworth MA, Batra SK. The human RNA polymerase II-associated factor 1 (hPaf1): a new regulator of cell-cycle progression. PLoS One. 2009;4:e7077. doi: 10.1371/journal.pone.0007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Porter SE, Washburn TM, Chang M, Jaehning JA. The yeast pafl-rNA polymerase II complex is required for full expression of a subset of cell cycleregulated genes. Eukaryot Cell. 2002;1:830–842. doi: 10.1128/EC.1.5.830-842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.White MA, Riles L, Cohen BA. A systematic screen for transcriptional regulators of the yeast cell cycle. Genetics. 2009;181:435–446. doi: 10.1534/genetics.108.098145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang C, Kong D, Tan MH, Pappas DL, Jr, Wang PF, Chen J, Farber L, Zhang N, Koo HM, Weinreich M, Williams BO, Teh BT. Parafibromin inhibits cancer cell growth and causes G1 phase arrest. Biochem Biophys Res Commun. 2006;350:17–24. doi: 10.1016/j.bbrc.2006.08.169. [DOI] [PubMed] [Google Scholar]
- 127.Woodard GE, Lin L, Zhang JH, Agarwal SK, Marx SJ, Simonds WF. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2005;24:1272–1276. doi: 10.1038/sj.onc.1208274. [DOI] [PubMed] [Google Scholar]
- 128.Yang YJ, Han JW, Youn HD, Cho EJ. The tumor suppressor, parafibromin, mediates histone H3 K9 methylation for cyclin D1 repression. Nucleic Acids Res. 2010;38:382–390. doi: 10.1093/nar/gkp991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lin L, Zhang JH, Panicker LM, Simonds WF. The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc protooncogene. Proc Natl Acad Sci U S A. 2008;105:17420–17425. doi: 10.1073/pnas.0710725105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Agarwal SK, Simonds WF, Marx SJ. The parafibromin tumor suppressor protein interacts with actin-binding proteins actinin-2 and actinin-3. Mol Cancer. 2008;7:65. doi: 10.1186/1476-4598-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Latham JA, Chosed RJ, Wang S, Dent SY. Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell. 2011;146:709–719. doi: 10.1016/j.cell.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lampert F, Westermann S. A blueprint for kinetochores - new insights into the molecular mechanics of cell division. Nat Rev Mol Cell Biol. 2011;12:407–412. doi: 10.1038/nrm3133. [DOI] [PubMed] [Google Scholar]
- 133.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1- dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 136.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 137.Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci U S A. 2004;101:11380–11385. doi: 10.1073/pnas.0400078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gaillard H, Tous C, Botet J, Gonzalez-Aguilera C, Quintero MJ, Viladevall L, Garcia-Rubio ML, Rodriguez-Gil A, Marin A, Arino J, Revuelta JL, Chavez S, Aguilera A. Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genet. 2009;5:e1000364. doi: 10.1371/journal.pgen.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ding B, LeJeune D, Li S. The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. J Biol Chem. 2010;285:5317–5326. doi: 10.1074/jbc.M109.082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tatum D, Li W, Placer M, Li S. Diverse roles of RNA polymerase II-associated factor 1 complex in different subpathways of nucleotide excision repair. J Biol Chem. 2011;286:30304–30313. doi: 10.1074/jbc.M111.252981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Strawn LA, Lin CA, Tank EM, Osman MM, Simpson SA, True HL. Mutants of the Paf1 complex alter phenotypic expression of the yeast prion [PSI+] Mol Biol Cell. 2009;20:2229–2241. doi: 10.1091/mbc.E08-08-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, Hubner N, Doss MX, Sachinidis A, Hescheler J, Iacone R, Anastassiadis K, Stewart AF, Pisabarro MT, Caldarelli A, Poser I, Theis M, Buchholz F. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 143.Ponnusamy MP, Deb S, Dey P, Chakraborty S, Rachagani S, Senapati S, Batra SK. RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem Cells. 2009;27:3001–3011. doi: 10.1002/stem.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, Ramalho-Santos M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xiong L, Darwanto A, Sharma S, Herring J, Hu S, Filippova M, Filippov V, Wang Y, Chen CS, Duerksen-Hughes PJ, Sowers LC, Zhang K. Mass spectrometric studies on epigenetic interaction networks in cell differentiation. J Biol Chem. 2011;286:13657–13668. doi: 10.1074/jbc.M110.204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cantz T, Key G, Bleidissel M, Gentile L, Han DW, Brenne A, Scholer HR. Absence of OCT4 expression in somatic tumor cell lines. Stem Cells. 2008;26:692–697. doi: 10.1634/stemcells.2007-0657. [DOI] [PubMed] [Google Scholar]
- 147.Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 148.Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, Dorsett D, Eissenberg JC, Shilatifard A. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci U S A. 2006;103:11970–11974. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Akanuma T, Koshida S, Kawamura A, Kishimoto Y, Takada S. Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO Rep. 2007;8:858–863. doi: 10.1038/sj.embor.7401045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Langenbacher AD, Nguyen CT, Cavanaugh AM, Huang J, Lu F, Chen JN. The PAF1 complex differentially regulates cardiomyocyte specification. Dev Biol. 2011;353:19–28. doi: 10.1016/j.ydbio.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nguyen CT, Langenbacher A, Hsieh M, Chen JN. The PAF1 complex component Leo1 is essential for cardiac and neural crest development in zebrafish. Dev Biol. 2010;341:167–175. doi: 10.1016/j.ydbio.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mosimann C, Hausmann G, Basler K. The role of Parafibromin/Hyrax as a nuclear Gli/Ci-interacting protein in Hedgehog target gene control. Mech Dev. 2009;126:394–405. doi: 10.1016/j.mod.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 154.Bray S, Musisi H, Bienz M. Bre1 is required for Notch signaling and histone modification. Dev Cell. 2005;8:279–286. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 155.Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 156.Takahashi A, Tsutsumi R, Kikuchi I, Obuse C, Saito Y, Seidi A, Karisch R, Fernandez M, Cho T, Ohnishi N, Rozenblatt-Rosen O, Meyerson M, Neel BG, Hatakeyama M. SHP2 tyrosine phosphatase converts parafibromin/Cdc73 from a tumor suppressor to an oncogenic driver. Mol Cell. 2011;43:45–56. doi: 10.1016/j.molcel.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Juhlin CC, Haglund F, Villablanca A, Forsberg L, Sandelin K, Branstrom R, Larsson C, Hoog A. Loss of expression for the Wnt pathway components adenomatous polyposis coli and glycogen synthase kinase 3-beta in parathyroid carcinomas. Int J Oncol. 2009;34:481–492. [PubMed] [Google Scholar]
- 158.Youn MY, Yoo HS, Kim MJ, Hwang SY, Choi Y, Desiderio SV, Yoo JY. hCTR9, a component of Paf1 complex, participates in the transcription of interleukin 6-responsive genes through regulation of STAT3-DNA interactions. J Biol Chem. 2007;282:34727–34734. doi: 10.1074/jbc.M705411200. [DOI] [PubMed] [Google Scholar]
- 159.Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15:79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chipumuro E, Henriksen MA. The ubiquitin hydrolase USP22 contributes to 3'- end processing of JAK-STAT-inducible genes. FASEB J. 2012;26:842–854. doi: 10.1096/fj.11-189498. [DOI] [PubMed] [Google Scholar]
- 161.Liu L, Oliveira NM, Cheney KM, Pade C, Dreja H, Bergin AM, Borgdorff V, Beach DH, Bishop CL, Dittmar MT, McKnight A. A whole genome screen for HIV restriction factors. Retrovirology. 2011;8:94. doi: 10.1186/1742-4690-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 164.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene. 2007;26:7499–7507. doi: 10.1038/sj.onc.1210582. [DOI] [PubMed] [Google Scholar]
- 169.Moniaux N, Nemos C, Schmied BM, Chauhan SC, Deb S, Morikane K, Choudhury A, Vanlith M, Sutherlin M, Sikela JM, Hollingsworth MA, Batra SK. The human homologue of the RNA polymerase II-associated factor 1 (hPaf1), localized on the 19q13 amplicon, is associated with tumorigenesis. Oncogene. 2006;25:3247–3257. doi: 10.1038/sj.onc.1209353. [DOI] [PubMed] [Google Scholar]
- 170.Shattuck TM, Valimaki S, Obara T, Gaz RD, Clark OH, Shoback D, Wierman ME, Tojo K, Robbins CM, Carpten JD, Farnebo LO, Larsson C, Arnold A. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N Engl J Med. 2003;349:1722–1729. doi: 10.1056/NEJMoa031237. [DOI] [PubMed] [Google Scholar]
- 171.Newey PJ, Bowl MR, Thakker RV. Parafibromin--functional insights. J Intern Med. 2009;266:84–98. doi: 10.1111/j.1365-2796.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- 172.Wang PF, Tan MH, Zhang C, Morreau H, Teh BT. HRPT2, a tumor suppressor gene for hyperparathyroidism-jaw tumor syndrome. Horm Metab Res. 2005;37:380–383. doi: 10.1055/s-2005-870150. [DOI] [PubMed] [Google Scholar]
- 173.Zhao J, Yart A, Frigerio S, Perren A, Schraml P, Weisstanner C, Stallmach T, Krek W, Moch H. Sporadic human renal tumors display frequent allelic imbalances and novel mutations of the HRPT2 gene. Oncogene. 2007;26:3440–3449. doi: 10.1038/sj.onc.1210131. [DOI] [PubMed] [Google Scholar]
- 174.Selvarajan S, Sii LH, Lee A, Yip G, Bay BH, Tan MH, Teh BT, Tan PH. Parafibromin expression in breast cancer: a novel marker for prognostication? J Clin Pathol. 2008;61:64–67. doi: 10.1136/jcp.2007.048694. [DOI] [PubMed] [Google Scholar]
- 175.Zheng HC, Takahashi H, Li XH, Hara T, Masuda S, Guan YF, Takano Y. Downregulated parafibromin expression is a promising marker for pathogenesis, invasion, metastasis and prognosis of gastric carcinomas. Virchows Arch. 2008;452:147–155. doi: 10.1007/s00428-007-0551-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zheng HC, Wei ZL, Xu XY, Nie XC, Yang X, Takahashi H, Takano Y. Parafibromin expression is an independent prognostic factor for colorectal carcinomas. Hum Pathol. 2011;42:1089–1102. doi: 10.1016/j.humpath.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 177.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 178.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]